Abstract
This review gives an overview on the occurrence of sulfatases in Prokaryota, Eukaryota and Archaea. The mechanism of enzymes acting with retention or inversion of configuration during sulfate ester hydrolysis is discussed taking two complementary examples. Methods for the discovery of novel alkyl sulfatases are described by way of sequence-based search and enzyme induction. A comprehensive list of organisms with their respective substrate scope regarding prim- and sec-alkyl sulfate esters allows to assess the capabilities and limitations of various biocatalysts employed as whole cell systems or as purified enzymes with respect to their activities and enantioselectivities. Methods for immobilization and selectivity enhancement by addition of metal ions or organic (co)solvents are summarised.
Keywords: Alkyl sulfatase, Inversion, Retention, Mechanism, Biocatalysis
Introduction — inverting hydrolases
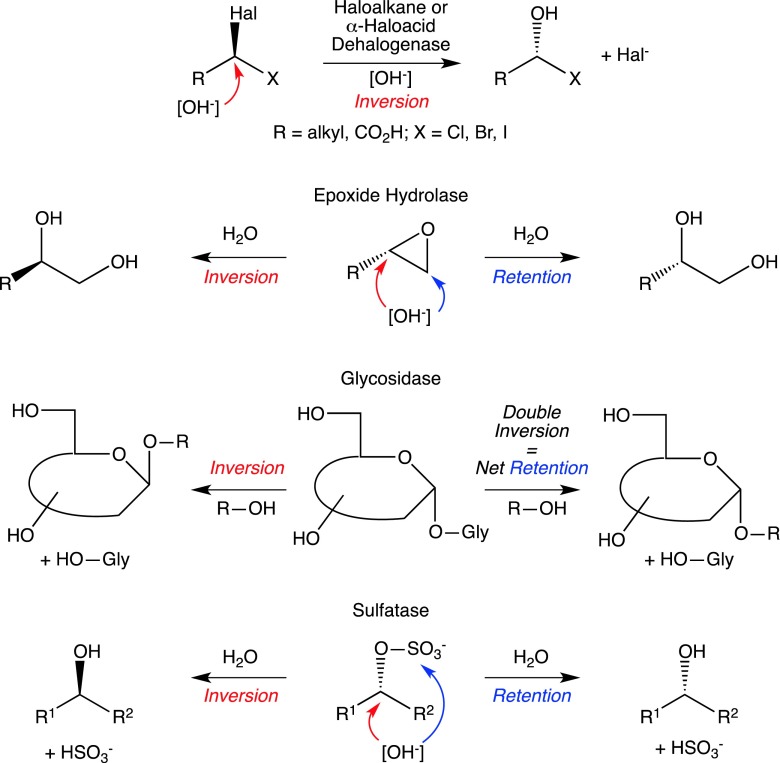
Stereochemical changes, such as inversion of configuration involving a chiral carbon center require mechanistically sophisticated pathways (But and Toy 2007). In nature, several types of hydrolytic enzymes are able to facilitate such reactions by operating either via inversion or retention of configuration, commonly via an SN2-type mechanism (Schober and Faber 2013): haloalkane and haloacid dehalogenases (Verschueren et al. 1993), epoxide hydrolases (Kotik et al. 2012), glycosidases (Lairson and Withers 2004) and sulfatases (Pogorevc and Faber 2002). The enzymes comprising the latter group are covered in this review with respect to their occurrence across all kingdoms of life, their substrate scope and the discovery of novel sulfatase enzymes.
Dehalogenases (EC 3.8.1.5) offer a wide variety of biotechnological applications and hence these enzymes were studied in great detail over the last decades. They are employed for the bioremediation of soils including the decontamination of chemical warfare agents (Fetzner 1998). For biocatalytic applications, they were exploited for the kinetic resolution of racemic sec-haloorganic compounds, such as haloalkanes and α-halocarboxylic acids to obtain valuable chiral building blocks for organic synthesis (Koudelakova et al. 2013; Janssen 2007; van Leeuwen et al. 2012). The majority of haloalkane and α-haloacid dehalogenases operate via an SN2 mechanism leading to inversion of configuration at the chiral carbon atom bearing the halide (Scheme 1). An aspartate residue within the active site acts as nucleophile, by replacing the halide to form a transient 'alkyl-enzyme intermediate'. The latter is hydrolytically cleaved by H2O, which is activated by a histidine residue thereby releasing the inverted product alcohol and liberating the Asp residue, which closes the catalytic cycle (Verschueren et al. 1993).
Scheme 1.
Stereochemical consequences of catalysis by retaining and inverting hydrolases, i.e., dehalogenases, epoxide hydrolases, glycosidases and sulfatases
The enzymatic ring opening of oxiranes is achieved by epoxide hydrolases (EC 3.3.2.3), which serves as detoxification strategy to yield more innocuous diols (Kotik et al. 2012). Mechanistically, hydrolysis can occur either under retention or inversion of configuration (Scheme 1). If the water molecule attacks a substituted carbon atom, the stereochemical outcome is inversion of configuration. Retention is achieved by attack at an unsubstituted carbon yielding a retained diol. From detailed studies on epoxide hydrolase from Aspergillus niger, it was shown that nucleophilic attack is caused by an aspartate residue, which forms a 'glycol-monoester enzyme-intermediate'. Attack by a histidine-activated water molecule releases the diol (Widersten et al. 2010).
Another example for stereochemical alteration of a substrate is the enzyme class of glycosidases (EC 3.2.1.x) cleaving the glycosidic bonds. Depending on their mechanistic pathway, both inversion and retention of configuration are possible (Scheme 1). The former reaction is facilitated by a glutamate residue in the active site activating the nucleophile (ROH), which attacks the anomeric carbon atom through an SN2-type mechanism with inversion of configuration going in hand with release of the leaving group. Alternatively, glycoside cleavage may proceed via double inversion, leading to net retention. In this case, two glutamic acid residues are involved in catalysis. The first acts as acid via protonation of the anomeric oxygen forming a glycosyl oxonium species. In the second step, the other glutamate residue acts as base and deprotonates the nucleophile (ROH), which in turn attacks the enzyme-bound glycosyl species and releases the retained product (O'Hagan and Schmidberger 2010; Lairson and Withers 2004).
Sulfatases
Due to the large number and broad diversity of sources, where sulfatases were obtained, they constitute a very heterogenic group of enzymes (Gadler and Faber 2007a; Hanson et al. 2004). So far, three distinct classes have been identified according to their substrate spectrum and mechanistic aspects (Kertesz 2000).
Aryl sulfatases
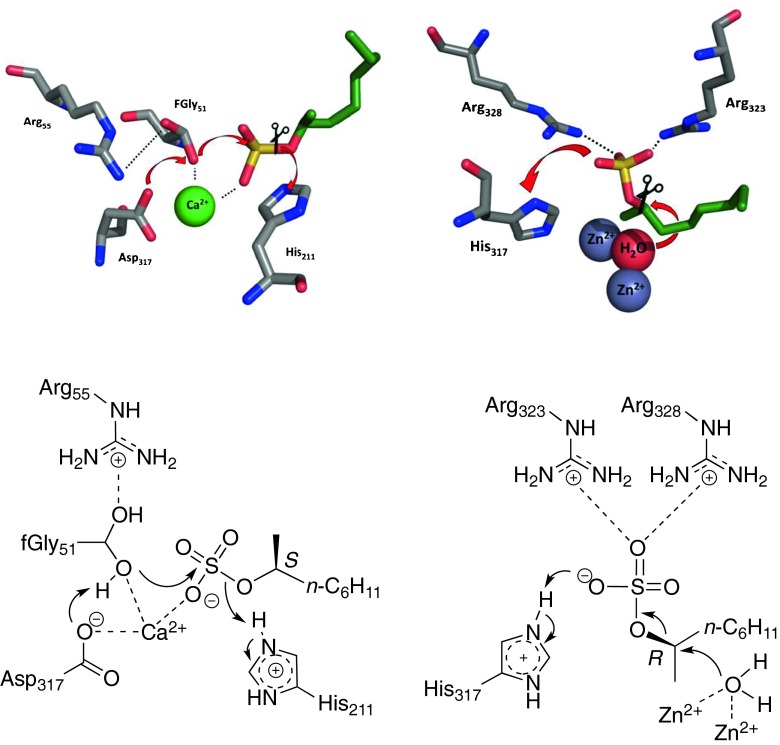
The name of the class is derived from the aromatic sulfate ester p-nitrophenyl sulfate, which is used as standard substrate for activity detection. However, many of them also act on sulfated carbohydrates, such as chondroitin sulfate and dermatan sulfate (Anson et al. 1992; Tomatsu et al. 1991). Due to their significance in humans, they have been studied in great detail with respect to their structure and mechanism and they clearly represent the best studied group of sulfatases. Fifteen of these enzymes have been identified in humans, which are linked to a lysosomal storage disease known as MSD (multiple sulfatase deficiency) (Hopwood and Ballabio 1999). Their function is to control the sulfation state of messenger molecules (Ahmed and James 1999). Several aryl sulfatases have been crystallised and structurally elucidated, including the human aryl sulfatase B (NP_000037) and the prokaryotic aryl sulfatase from P. aeruginosa (PA0183) (Fig. 1). The most notable aspect of aryl sulfatases is the highly conserved consensus motif C/S-X-P-X-A-X4-T-G (Kertesz 2000) which applies to all of the identified enzymes within this class, except Rhodococcus ruber sulfatase (G352_02444) (Gadler and Faber 2007b). The lead residue in this motif is either a cysteine or serine, which is post-translationally modified into a catalytically active aldehyde (hydrate) known as Cα-formylglycine. This residue is unique to this class of enzymes and has not yet been observed in other active sites, except for phosphatases (Jonas et al. 2008). So far, cysteine is only found in eukaryotic sources, while bacterial hosts contain either a cysteine or a serine in the 'precursor' enzyme (Benjdia et al. 2010). Depending on the catalytic residue, cysteine-type sulfatases are typically found in the cytosol, whereas serine-type sulfatases are located in the periplasm (Cloves et al. 1977; Marquordt et al. 2003; Murooka et al. 1990). Due to the non-chiral nature of aryl sulfates, the stereochemical consequences of aryl sulfatase catalysis were not investigated.
Fig. 1.
Catalytic residues and their mode of action for the retaining sulfatase PAS from Pseudomonas aeruginosa (PDB 1HDH, top left) and the inverting sulfatase Pisa1 from P. sp. DSM 6611 (PDB 4AXH, top right). Preferred enantiomers of the substrate 2-octyl sulfate (green) were docked into the active site using Schrödinger Maestro (Schrödinger Maestro Software Suite 2013) The flow of electrons implying nucleophilic attack is indicated by red arrows, and the S–O/C–O bonds being broken are marked by scissor symbols. The schematic mechanism is given below. Some amino acid residues were omitted for clarity. Pictures were generated using Pymol (Pymol Software 2013)
Fe2+-dependent sulfatases
The second class of sulfatases belong to the Fe2+-dependent dioxygenases, which catalyse an oxidative cleavage of the sulfate ester moiety to form the corresponding aldehyde and inorganic sulfate (Müller et al. 2004). In contrast to the other sulfatases, this reaction requires α-ketoglutarate as a redox-cosubstrate. The most prominent representative of this group is the alkyl sulfatase Atsk from Pseudomonas putida (PP4_02270) (Kahnert and Kertesz 2000). For biocatalytic applications, this class of sulfatases is less valuable, since a stereocenter is destroyed during the course of the reaction.
Alkyl sulfatases
Inspired by the observation that the bacterium Pseudomonas sp. C12B (NCIB 11753) is able to grow on the common surfactant sodium dodecyl sulfate (SDS) in the 1960s (Payne et al. 1965; Williams and Payne 1964), sulfatase research turned into a hot topic due to potential applications in bioremediation. Initially, only little attention was paid to the stereochemical implications of sulfate ester hydrolysis, but investigations on sec-alkyl sulfate substrates revealed that there were two modes of operation involved in sulfate ester cleavage (Matcham et al. 1977). The retaining pathway cleaves the S–O ester bond, releasing the product alcohol without stereochemical alteration. In contrast, the inverting mechanism involves attack of an activated water molecule on the chiral carbon atom bearing the sulfate ester, thereby breaking the C–O bond and releasing the inverted alcohol (Fig. 1). While the retaining pathway was elucidated already in 2001 (Boltes et al. 2001), the inverting mechanism was elucidated only recently with the sec-alkyl sulfatase Pisa1 (FR850678) (Knaus et al. 2012). The catalytic mechanism of this β-lactamase type enzyme involves a binuclear Zn2+ cluster activating a water molecule which launches a nucleophilic attack on the chiral carbon atom. A histidine residue protonates the sulfate moiety, transforming it into a good leaving group (Knaus et al. 2012). The synthetic potential of this enzyme was demonstrated in a sulfatase-assisted total synthesis of the anti-leukemic agent (R)-Lasiodiplodin methyl ether (Fuchs et al. 2013). Further enzymes of this class include the prim-alkyl sulfatase SdsA1 (PA0740) (Hagelueken et al. 2006) and the sec-alkyl sulfatase SdsAP (HQ189533) (Long et al. 2011), both of which originate from Pseudomonas sp. and show high sequence similarity to Pisa1. To date, only limited structural information is available for β-lactamase-type alkyl sulfatases, i.e., Pisa1 and SdsA1.
Search for sulfatase activity
Discovery of novel sulfatases
Considering the vast amount of possible microbial sources for sulfatases, guidelines facilitating the search for novel sulfatase activities are desirable, which can be delineated from successful case stories in the literature.
The strongest indication for aryl sulfatase activity is probably the existence of sulfatase maturation enzymes, required for the post-translational modification of a cysteine or serine into the catalytically active Cα-formylglycine aldehyde, or the corresponding catalytically active hydrate (Fig. 1). Given the uniqueness of this residue, it constitutes a strong marker for aryl sulfatase activity (Scheme 2).
Scheme 2.
Partial sequence alignment of SUMF1 gene derived proteins. PJDR2, Paenibacillus sp. JDR-2 (YP_003009726); CM, Cupriavidus metallidurans (YP_586663); AC, Acinetobacter calcoaceticus (YP_004994666); RS, putative FGE-protein from Ralstonia solanacearum RFBP2957 (YP_003747422); human (NP_877437); mouse (NP_666049); sea urchin (XP_782973). Sequence alignment was done with clustal omega (Sievers et al. 2011). Numbers in brackets indicate the aligned amino acid residues. Letters highlighted in bold display the conserved sequence across Eukaryota and Prokaryota
Two systems are known to promote the maturation of Cys and/or Ser into the Cα-formylglycine residue. The sulfatase maturation factor 1 (SUMF1) gene codes for the so called formylglycine-generating enzyme (FGE) (Dierks et al. 2003), which acts on Cys. The second system, termed anaerobic sulfatase maturating enzymes (anSME), was initially believed to be specific for serine-type sulfatases (Hanson et al. 2004). Recent studies, however, have shown that cysteine-type enzymes are also accepted via this S-adenosyl-l-methionine mediated pathway (Berteau et al. 2006). The genetic origin can be found within homologues of the Klebsiella pneumoniae-derived AtsA gene (AF262989_2) and has been investigated with the sulfatase from Clostridium perfringens ATCC 13124 (Q0TTH1) (Benjdia et al. 2007). However, due to limited sequence information, this approach is not always applicable.
Metabolic demand for sulfur
As an essential element for growth, sulfur uptake occurs through various pathways, depending on the organism. Usually inorganic sulfate is available and is transformed through a cascade of reactions to yield the high-energy intermediate 3′-phosphoadenosine-5′-phosphosulfate (PAPS). Ultimately, sulfur is incorporated into the essential amino acids cysteine and methionine. In case inorganic sulfate is unavailable, organisms are forced to express other sulfur metabolizing enzymes, such as alkyl sulfatases, which are able to hydrolytically cleave inorganic sulfate from organic sulfate esters. The dependence of organisms on sulfate for growth opens a window for opportunities to induce sulfatases when sulfate esters are offered as the sole sulfur source.
Another and more general approach lies in the identification of a rich and robust (in)organic sulfate metabolism as a pointer for potential sulfatase activity, following the reasoning that the activity on (in)organic sulfate might be connected to alkyl sulfate esters and hence sulfatase activity. Sulfur metabolising organisms are typically found within the kingdom of Archaea due to their evolutionary background (Huber and Stetter 1998). This approach proved to be highly successful and led to the discovery of several highly selective sulfatase activities, such as from Sulfolobus acidocaldarius DSM 639 (Wallner et al. 2005b).
The most common indicator for sulfatase activity is the ability of organisms to grow on detergent-contaminated soil or wastewater and several bacteria have been isolated from such sources (Dodgson et al. 1954; Payne et al. 1965; Williams and Payne 1964). However, it has been shown that the reverse argument, i.e., the inability to degrade SDS should exclude sulfatase activity, cannot be drawn. A broad screening of bacteria and fungi by Gadler and Faber in 2007 for potential sulfatase activity on 2-octyl sulfate resulted in several highly active strains from Pseudomonas sp., together with many inactive candidates (Gadler and Faber 2007b). The most recent example is the sec-alkyl sulfatase Pisa 1 isolated from Pseudomonas sp. DSM 6611. Although this enzyme is inactive on SDS, it has a remarkably broad activity on a wide variety of prim- and sec-sulfate esters (Schober et al. 2012).
Growth conditions
Forcing an organism to express the desired sulfatase activity is usually achieved by limiting the available sulfur to organically bound sources, such as short chain alkyl sulfates or (more common) the surfactant SDS. Since uptake of the preferred sulfur source (inorganic sulfate) (Kertesz and Wietek 2001) proceeds through different metabolic pathways, in comparison to organically bound sulfate (Cook et al. 1998; Lie et al. 1998; Marzluf 1994), supplementing the growth medium with sulfate esters drives the metabolism towards the desired direction. Importantly though, the chosen growth conditions should only be selective, but not growth limiting (Kertesz 2000). Under such constraints, organisms express sulfate starvation-induced (SSI) proteins. They usually include low sulfur containing enzymes, where cysteine and methionine in non-essential positions are substituted by other amino acids. P. putida strain KT2440, for example, is able to reduce the total soluble cellular thiol content 5-fold under sulfur limiting conditions (Scott et al. 2007).
Due to the fact that the expression of aryl sulfatases in several bacteria is strongly upregulated when sulfate is unavailable (Dodgson et al. 1982; Kertesz et al. 1993), it has been suggested that sulfatase enzymes are expressed at a lower level than other proteins (Kertesz 2000), which makes selective growth constraints all the more important to successfully obtain sulfate ester hydrolysing cells. Consequently, when the growth medium of P. aeruginosa IFO 3901 was supplemented with intermediates of the primary sulfur assimilation pathway, such as sufate, sulfite or cysteine, no enzyme expression was observed (Harada 1964). Conversely, supplementing the growth medium of Coryneform sp. B1a with phenyl sulfate allowed the organism to accept several primary alkyl sulfates as substrates (Matts et al. 1994).
It is noteworthy that some alkyl sulfatases are only expressed at certain stages of growth or in presence of different sulfur sources, which may complicate the screening and might lead to 'false negatives' in search for sulfatases.
Prime examples for this case are the Pseudomonas strains C12B (NCIB 11753) and AE-A. The C12B strain is probably the most thoroughly examined organism regarding sulfatase activity and harbours alkyl sulfatases P1 and P2. While P1 is constitutive and expressed onwards from the late exponential phase, the P2 enzyme is inducible and expressed only transiently during the exponential phase (Cloves et al. 1977, 1980; Ellis et al. 2002). A zymographic analysis of Pseudomonas strain AE-A showed up to three different alkyl sulfatases, depending on the nature of the supplemented sulfur source. In nutrient broth, only one sulfatase could be identified. With addition of SDS to the medium, a second alkyl sulfatase was observed, which migrated further towards the anode than the first enzyme. Finally, with the branched 2-butyl-1-octyl sulfate as supplement, a third alkyl sulfatase could be identified.
Occurrence of microbial alkyl sulfatases
Several higher eukaryotes have been shown to harbour sulfatases, a few have even been put to use in technical applications, such as soil sulfate analysis using a sulfatase from Helix pomatia (AF109924) (Burgundy edible snail) (Whalen and Warman 1996). Others, like human sulfatases, are connected to the lysosomal storage disorder and are an important subject of biomedical studies (Hanson et al. 2004). These enzymes, however, originate from higher eukaryotic sources, which are usually not applicable to biotransformations. Besides the difficulties in obtaining substantial amounts of biomass in a reproducible way, post-translational modification, such as glycosylation or acetylation are additional but unavoidable challenges in the expression of active proteins. In contrast, prokaryotes, lower eukaryotes and several Archaea species are advantageous in terms of fast growth rates, facile enzyme cloning and heterologous expression and usually omit the need for cellular post-translational enzyme modification, which speeds up the screening for sulfatase-harbouring organisms. The following section gives an overview of microorganisms with proven sulfatase activities together with the respective substrate scope (Table 1).
Table 1.
Classification of sulfate esters employed as test substrates for the screening of whole microbial cells and/or purified enzymes for sulfatase activity
| Substrate | Type |
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | ||||||
| R 1 | R 2 | R 1 | R 2 | R 1 | R 2 | R 1 | R 2 | R 1 | R 2 | |
| 1 | H | n-C3H7 | CH3 | n-C3H7 | CH=CH2 | n-C4H9 | CH3 | C≡C–C2H5 | CH3 | 3,5-(CF3)2C6H3 |
| 2 | H | n-C4H9 | CH3 | n-C4H9 | CH=CH2 | n-C5H11 | CH3 | CH2–C≡C–CH3 | CH3 | p-F-C6H4-CH2 |
| 3 | H | n-C5H11 | CH3 | n-C5H11 | CH=CH2 | n-C6H13 | CH3 | C≡C–Ph | CH3 | p-Cl-C6H4-CH2 |
| 4 | H | n-C6H13 | CH3 | n-C6H13 | CH=CH2 | n-C7H15 | C2H5 | C≡C–CH3 | ||
| 5 | H | n-C7H15 | CH3 | n-C7H15 | CH3 | (CH2)2CH=C(CH3)2 | C2H5 | C≡C–C2H5 | ||
| 6 | H | n-C8H17 | CH3 | n-C8H17 | CH3 | CH2–CH=CH2 | C≡CH | n-C3H7 | ||
| 7 | H | n-C9H19 | CH3 | n-C9H19 | CH3 | (CH2)2CH=CH2 | C≡CH | CH2–CH(CH2)3 | ||
| 8 | H | n-C11H23 | CH3 | n-C10H21 | C≡CH | n-C4H9 | ||||
| 9 | H | n-C12H25 | CH3 | n-C12H25 | C≡CH | n-C5H11 | ||||
| 10 | H | n-C13H27 | C2H5 | C2H5 | ||||||
| 11 | H | n-C14H29 | C2H5 | n-C4H9 | ||||||
| 12 | SLES | C2H5 | n-C5H11 | |||||||
| 13 | 2-BOS | n-C3H7 | n-C3H7 | |||||||
| 14 | n-C3H7 | n-C4H9 | ||||||||
| 15 | n-C3H7 | n-C6H13 | ||||||||
| 16 | n-C4H9 | n-C4H9 | ||||||||
| 17 | CH3 | c-C6H11 | ||||||||
| 18 | CH3 | Ph | ||||||||
| 19 | CH3 | CH2-Ph | ||||||||
| 20 | CH3 | (CH2)2-Ph | ||||||||
SLES sodium lauryl ether sulfate (Na+ n-C12H25–O–(CH2)2–O–SO3 −), 2-BOS 2-butyl-1-octyl sulfate
Whole cell preparations
Whole cell preparations enable fast screening procedures and less work-up compared to the use of purified enzymes, which has to be paid for by reduced average activities. A comprehensive overview of microbial strains harbouring sulfatase activities is given in Table 2.
Table 2.
Substrate scope, activities and enantioselectivities for sulfatase-activities of whole cell preparations
| Organism | Substrate type | Conversion (%)a | E b | Refs. |
|---|---|---|---|---|
| Prokaryota | ||||
| Proteobacteria | ||||
| Pseudomonas sp. C12B (NCIMB 11753) | A 4,5,6,8,9,10,11,12 | n.d. | n.a. | (Gadler and Faber 2007b; Thomas and White 1991; Dodgson et al. 1982) |
| B 3,4,5,12,14 | <5–10 | 4–13 | ||
| C 5 | 5–10 | 1 | ||
| Pseudomonas sp. DSM 6978 | B 3,4,5,12,14 | <5–10 | 4– > 200 | (Gadler and Faber 2007b) |
| Pseudomonas sp. DSM 6611 | B 3,4,5,9,14,18,20 C 5 | 5–21 | 6– > 200 | (Gadler and Faber 2007b) |
| Pseudomonas sp. S7 | A 9 | n.d. | n.a. | (Yeldho et al. 2011) |
| Pseudomonas sp. AE-A | A 9 | n.d. | n.a. | (Ellis et al. 2002) |
| Pseudomonas sp. AE-D | A 13 | n.d. | n.a. | (Ellis et al. 2002) |
| Pseudomonas ATCC 19151 | A 9 | n.d. | n.a. | (Jovcic et al. 2010) |
| Klebsiella oxytoca | A 9 | n.d. | n.a. | (Shukor et al. 2009) |
| Comamonas sp. DSM 115091 | B 4 | 5–10 | 16 | (Gadler and Faber 2007b) |
| Citrobacter braakii | A 9,12 | n.d. | n.a. | (Dhouib et al. 2003) |
| Rhizobiaceae sp. FCC 175 | B 4 | >10 | 2 | (Gadler and Faber 2007b) |
| Cupriavidus necator DSM 5536 | B 4 | <5 | 4 | (Gadler and Faber 2007b) |
| Paracoccus sp. DSM 6392 | B 3,4,12,14 | <5–10 | 1–3 | (Gadler et al. 2009) |
| C 5 | 5–10 | 2– > 200d | ||
| Ralstonia eutropha SPT0002 FCC120 | B 4 | >10 | 1 | (Gadler and Faber 2007b) |
| Ralstonia eutropha sp. DSM 6428 | B 4 | 5–10 | 21 | (Gadler and Faber 2007b) |
| Xanothbacter autotrophicus DSM 431 | B 4 | >10 | 2 | (Gadler and Faber 2007b) |
| Xanothbacter autotrophicus DSM 6696 | B 4 | <5 | 2 | (Gadler and Faber 2007b) |
| Xanothbacter flavus DSM 338 | B 4 | <5 | 2 | (Gadler and Faber 2007b) |
| Xanothbacter flavus autotrophicus DSM 3874 | B 4 | <5 | 7 | (Gadler and Faber 2007b) |
| Achromobacter sp. FCC 175 | B 4 | <5 | 2 | (Gadler and Faber 2007b) |
| Actinobacteria | ||||
| Rhodococcus ruber DSM 44541e | A 5,8 | <5 | n.a. | (Gadler and Faber 2007a) |
| B 3,4,5,6,7,12,14,15,19 C 2 | 4–68 | 1–21 | (Pogorevc and Faber 2002) | |
| Gulosibacter molinativorax DSM 13485 | B 4 | >10 | 5 | (Gadler and Faber 2007b) |
| Nocardia nova DSM 43843 | B 4 | >10 | (Gadler and Faber 2007b) | |
| Planctomycetes | ||||
| Rhodopirellula baltica DSM 10527 | A 5 | 26 | n.a. | (Wallner et al. 2005a) |
| B 3,4,5,12,14 C 2,5 | <5–18 | 2– > 200 | ||
| Cyanobacteria | ||||
| Synechococcus sp. PCC 7942 | B 3,4,5,12,14 | <5–24 | 1–3 | (Gadler et al. 2009) |
| C 5 | 5–10 | 4– > 200d | ||
| Synechococcus sp. RCC 556 | B 3,4,5,12,14 | <5–10 | n.d./1 | (Gadler et al. 2009) |
| Firmicutes | ||||
| Bacillus cereus | A 9 | n.d. | n.a. | (Singh et al. 1998) |
| Bacillus sphaericus FCC 098 | B 4 | <5 | 1 | (Gadler and Faber 2007b) |
| Strain combination | ||||
| Acinetobacter calcoaceticus + Pantoea agglomerans | A 9 | n.d. | n.a. | (Abboud et al. 2007) |
| Archaea | ||||
| Crenarchaeota | ||||
| Sulfolobus acidocaldarius DSM 639 | B 3,4,5,12,14,19 | 10–43 | 5– > 200 | (Gadler and Faber 2007a; Wallner et al. 2004) |
| C 5 | 5–10 | n.d. | ||
| E 2,3 | 5–10 | 2 | ||
| Sulfolobus solfataricus DSM 1617 | B 4,19 | 20–56 | 2–35 | (Wallner et al. 2005b) |
| Sulfolobus shibatae DSM 5389 | B 4,19 | 20–43 | 2–48 | (Wallner et al. 2005b) |
| Sulfolobus metallicus DSM 6482 | B 4 | 5–10 | 1 | (Wallner et al. 2005b) |
| Sulfolobus hakoniensis DSM 7519 | B 4 | 5–10 | 1 | (Wallner et al. 2005b) |
| Acidianus brierley DSM 1651 | B 4 | 5–10 | 1 | (Wallner et al. 2005b) |
| Acidianus infernus DSM 3191 | B 4 | 25 | 1 | (Wallner et al. 2005b) |
| Acidianus ambivalens DSM 3772 | B 4 | 13 | 1 | (Wallner et al. 2005b) |
| Metallosphaera sedula DSM 5348 | B 4 | 5–10 | 1 | (Wallner et al. 2005b) |
| Sulfurisphaera ohwakuensis DSM 12421 | B 4 | 11 | 1 | (Wallner et al. 2005b) |
n.a. not applicable, n.d. not determined
aIn kinetic resolutions showing high enantioselectivity, the maximum conversion is 50 %
bEnantioselectivity is expressed as the ratio of the reaction rate of enantiomers ('enantiomeric ratio', E) (Straathof and Jongejan 1997)
cUnpublished results
dImproved E values in presence of organic cosolvents
eCrude enzyme preparation
Prokaryotic strains
Prokaryotes encompass the largest group among all potential sulfatase harbouring organisms. In addition to their abundance, they can be easily accessed under simple growth conditions. Several strains possessing the ability to degrade sulfate-based detergents have recently been identified, which indicates the existence of sulfatases. The enantioselectivities — expressed as 'enantiomeric ratio' (E) (Straathof and Jongejan 1997) — range from virtually nil (E ~ 1) to perfect (E > 200) depending on the strains.
The largest phylum is occupied by proteobactera. In particular, Pseudomonas strains have been positively identified for their alkyl sulfatase activity. Newly discovered strains include Citrobacter braakii and a previously unknown Pseudomonas strain MTC 10311. Even a combination of strains — Acinetobacter calcoaecticus and Pantoea agglomerans — was successfully employed. C. braakii (strain CTM 50600) is able to degrade sodium lauryl ether sulfate over the course of 6 h starting from a concentration of 0.6 g l−1 and the bacterium also exhibited growth up to an OD600 of 1.0 with SDS as a sole carbon source (Dhouib et al. 2003). These properties indicate the occurrence of sulfatases which enable the cells to use also the carbon moiety of the detergent for growth. A BLAST search with the (limited) available sequence information from C. braakii strain CTM 50600 on the basis of the protein sequences of the sulfatases from P. aeruginosa and Pisa1 did not reveal any positive hits.
The aforementioned Pseudomonas species have shown to harbour sulfatases which were thoroughly characterised and which exhibited a broad applicability for biocatalytic processes (Gadler and Faber 2007a,b; Schober et al. 2012). Another promising candidate is the strain P. aeruginosa MTC 10311. This bacterium was recently isolated from detergent contaminated soil and could degrade 1.4 g of SDS within 48 h. It also showed activity at 40 °C with a depletion capacity of 90 % compared to the initial amount of surfactant at this temperature (Ambily and Jisha 2012).
Interestingly, some strains only show degrading properties when they are embedded within a consortium of other bacteria, such as Acinetobacter calcoaceticus and Pantoea agglomerans. Neither A. calcoaceticus nor P. agglomerans alone was able to degrade SDS, whereas the combination of both bacteria resulted in 60 % degradation within 50 h and complete degradation within 130 h (Abboud et al. 2007).
Eukaryotic strains
In comparison to prokaryotic sources for sulfatases, activities in eukaryotes are more scarce. Lower eukaryotic organisms, such as the green algae Volvox carteri is a potential hosts for alkyl sulfatase activity (Hallmann and Sumper 1994; Selmer et al. 1996). Higher eukaryotes, such as the Burgundy edible snail Helix pomatia contains sulfatases, whose genes have been identified and cloned (Wittstock et al. 2000), and which were found to be active on aryl sulfates (Whalen and Warman 1996; Yegles et al. 1997). Hence, they were also tested with several alkyl sulfates to investigate latent activity towards this substrate class. Moderate activity was obtained towards prim-alkyl sulfates but unfortunately no conversion was observed when sec-alkyl sulfatases were employed (Schober 2013).
A sulfatase from V. carteri f. nagariensis (strain HK10) could be purified to homogeneity and was assayed with aryl sulfates, such as p-nitrophenyl sulfate, 4-nitrocatechol sulfate and 5-bromo-4-chloro-3-indolyl sulfate and activity was detected for all of those compounds. In contrast, the alkyl sulfate SDS was not converted (Hallmann and Sumper 1994).
Sulfatases from Archaea
Owing to the importance of sulfur metabolism during the first billion of years of life, the third domain — Archaea — complement a particularly rich source of sulfatases. These hyperthermophilic organisms prefer a strongly acidic environment (pH 2–3) and temperatures ranging between 55 °C and 95 °C, which is well above the usual 37 °C (Wallner et al. 2005b). On the premise of harbouring a robust sulfur metabolism, several hyperthermophilic strains were examined in search for novel alkyl sulfatase activities by Wallner et. al. in 2005 (Wallner et al. 2005b). The occurrence of sulfatase activity was strongly influenced by the growth conditions: While cells could be obtained under availability of oxygen, no growth was observed in the absence of O2. Optimisation in terms of the carbon source revealed that sucrose was most beneficial. Among the tested species, Sulfolobus acidocaldarius DSM 639 turned out to be most promising. This strain was able to convert both linear and branched sec-alkyl sulfate substrates with low to good conversion and (occasionally) also excellent enantioselectivity (E > 200) (Wallner et al. 2004).
Purified sulfatases
Compared to the data available for whole cell preparations, only very few purified sulfatase enzymes have been characterised with respect to their substrate tolerance towards alkyl sulfates (Table 3). Since whole cells displaying low enantioselectivity most likely contain multiple sulfatases with different (or even opposite) enantiopreference, which makes protein isolation tedious and complicated, the prime target for the isolation of novel sulfatases were strains showing excellent stereoselectivites, because they are (most likely) expected to contain only a single sulfatase, which is easier to identify on the protein level.
Table 3.
Substrate scope, activities and enantioselectivities for purified sulfatases
| Kingdom | Substrate type | Conversiona (%) | E b | References |
|---|---|---|---|---|
| Prokaryota | ||||
| Pisa1 | A 5 | n.d. | n.a. | (Schober et al. 2011, 2012, 2013) |
| B 1,2,4,6,11,12,14,18 | 5–57 | 10– > 200 | ||
| C 1,2,3,4,5,6,7 | 5–50 | 17– > 200c | ||
| D 1,2,3,4,5,6,7,8,9 | 5–57 | 8– > 200 | ||
| E 1 | 49 | >200 | ||
| PAS | D 3,4,5,7,8,9 | 46–65 | 2– > 200 | (Schober et al. 2013) |
| E 1 | 30 | >200 | ||
| SdsA1 | A 3,5,7,9 | n.d. | n.a. | (Knaus et al. 2012; Hagelueken et al. 2006) |
| B 4d | n.d. | n.d. | ||
| SdsAP | A 9 | n.d. | n.a. | (Long et al. 2011) |
| Comamonas terrigena CS1 | B 4 | n.d. | n.d. | (Gadler and Faber 2007a) |
| Comamonas terrigena CS2 | B 3,4,5,6,8,9 | n.d. | n.d. | (Gadler and Faber 2007a) |
| Rhodococcus ruber S2 | B 4 | n.d. | 21– > 200e | (Gadler and Faber 2007a) |
| Pseudomonas S1 | B 3,4,6 | n.d. | n.d. | (Gadler and Faber 2007a) |
| Pseudomonas S2 | B 4 | n.d. | n.d. | (Gadler and Faber 2007a) |
| Pseudomonas S3 | B 4,10,13,16 | n.d. | n.d. | (Gadler and Faber 2007a) |
| Coryneform sp. B1a | A 1,2,3,4,5 | n.d. | n.a. | (Matts et al. 1994) |
| Aerobacter aerogenes f | A 6 | n.d. | n.a. | (Schober 2013) |
| Eukaryota | ||||
| Helix pomatia f | A 6 | n.d. | n.a. | (Schober 2013) |
| D 9 | n.d. | n.d. | ||
n.a. not applicable, n.d. not determined
aIn kinetic resolutions showing high enantioselectivity, the maximum conversion is 50 %
bEnantioselectivity is expressed as the ratio of the reaction rate of enantiomers ('enantiomeric ratio', E) (Straathof and Jongejan 1997)
cImproved E values in presence of organic cosolvents
dOnly the (R)-enantiomer of B4 was tested
eIn the presence of Fe3+
fCommercially available
Considering the source of purified enzymes, the majority of isolated sulfatases so far were derived from prokaryotic sources, with few exceptions, such as Helix pomatia aryl sulfatase (Thies 1979). To date, no enzyme could be purified from Archaea, despite their promising activity towards alkyl sulfates (Wallner et al. 2004, 2005b).
With Pisa1, PAS, SdsA1, Pseudomonas S1-3 (NCIB 11753) and SdsAP, the majority of purified sulfatases were derived from the phylum of Proteobacteria, which is not suprising, given the fact that these strains ranked among those exhibiting the highest E values and activities observed in whole cell screenings. The Comamonas terrigena enzymes S1 and S2 also accepted several sec-alkyl sulfates; however, no quantitative data are available regarding their enantioselectivity (Barrett et al. 1980). The enzyme from Coryneform sp. B1a accepted several primary alkyl sulfate esters with chain lengths ranging from three to seven carbon atoms. Aerobacter aerogenes ATCC 9621 and Helix pomatia sulfatases are commercially available and have been tested in our laboratory for their respective substrate scopes; however, they only showed a very limited substrate spectrum in comparison to other sulfatases, since they were only active on prim-sulfate esters but did not accept sec-sulfates (Schober et al. 2013).
Selectivity enhancement and immobilisation
When using whole cell systems or partially purified enzyme preparations, insufficient enantioselectivites are often encountered, which most likely are due to the existence of competing multiple sulfatases possessing different (or even opposite) stereoselectivities. In order to circumvent this drawback, several methods for the selectivity enhancement of alkyl sulfatase reactions were developed.
Cosolvents
The most convenient technique is the use of organic cosolvents, which have been applied to purified enzymes and whole cell biocatalysts alike. For the purified enzyme Pisa1 from Pseudomonas sp. DSM 6611, a thorough study for a range of common polar and apolar organic solvents was conducted. Among them, DMSO was found to suppress background hydrolysis of sulfate esters bearing activated allylic and benzylic functional groups. The latter increased E values significantly. Enhanced enantioselectivities were also observed for whole cell preparations of cyanobacteria, such as Synechococcus elongatus PCC 7942 and Paracoccus denitrificans DSM 6392 in combination with lower alcohols, such as MeOH, EtOH or t-ButOH. E values could be improved up to >200 for both strains, albeit at the expense of decreased conversion (Gadler et al. 2009).
Metal ions
Several aryl and alkyl sulfatases depend on metal ions required for catalysis. Aryl sulfatases, for example the enzyme PAS from P. aeruginosa, relies on Ca2+ for proper orientation and binding of the negatively charged substrate (Boltes et al. 2001), whereas the alkyl sulfatase Pisa 1 from Pseudomonas sp. DSM 6611 needs two Zn2+ ions for water activation to provide a good nucleophile [OH−] for the hydrolytic cleavage. Supplementation with other metal ions, such as Fe2+ and Fe3+, were found to have a dramatic effect on the enatioselectivity of the Rhodococcus ruber DSM 44541 RS2 enzyme. In presence of 5 mM of FeCl3, the selectivity could be improved from an E value of 3.6 to >200 (Pogorevc et al. 2002).
Immobilisation
Reusability and recyclability are important features of biocatalysts to make them industrially more appealing. At present, only few sulfatase preparations have been subjected to immobilisation. First successful experiments were conducted with immobilised cells of Pseudomonas C12B on polyacrylamide beads, which resulted in similar activities towards primary alkyl sulfate esters compared to the free enzyme. Total degradation of sulfate esters was observed within 48 h and an encouraging residual activity of 13 % was retained after 13 days of use (Thomas and White 1991). Further whole cell experiments with Pseudomonas C12B and Comamonas terrigena N3H were performed on various supports to evaluate their applicability as biofilms in wastewater treatment plants (Roig et al. 1998a, b). While C. terrigena N3H did not exhibit activity towards SDS, Pseudomonas C12B, which is a well known SDS-degrader (Dodgson et al. 1982), was also able hydrolyse the surfactant in immobilized form (Roig et al. 1998a; Thomas and White 1990).
In 2002 Pogorevc et al. were able to bind a crude preparation of the Rhodococcus ruber DSM 44541 RS2 enzyme onto a DEAE- and Ecteola-cellulose carrier resulting in ~100 % and 22 % residual activity, respectively (Pogorevc et al. 2002). Further studies in this direction would certainly lead to more widespread biotechnological applications of sulfatases, given the fact that several new sulfatases have emerged since the initial immobilization studies were conducted.
Although the phylogenetic relationship of organisms harbouring alkyl sulfatases is very heterogenic, the majority of enzymes identified and characterised on a molecular level derive from prokaryotic sources. So far, a broad range of more than fifty different prim- and sec-alkyl sulfate esters tested as substrates were successfully converted to the corresponding alcohol with inversion or retention of configuration, depending on the source and nature of the enzyme. Although there are several crystal structures available, mostly of the class of aryl sulfatases, sequence and structural information are still scarce with regard to alkyl sulfatases. Given the rapid advances in molecular biology, the number of alkyl sulfatases is expected to increase significantly in the near future, which will broaden the applicability of these useful hydrolytic enzymes in the design of enantioconvergent processes for the generation of single stereoisomeric sec-alcohols from the corresponding rac-sec-sulfate esters (Schober and Faber 2013; Schober et al. 2013).
Acknowledgments
Funding by the Austrian Science Fund (FWF) within the DK Molecular Enzymology (project W9) is gratefully acknowledged.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Abboud MM, Khleifat KM, Batarseh M, Tarawneh KA, Al-Mustafa A, Al-Madadhah M. Different optimization conditions required for enhancing the biodegradation of linear alkylbenzosulfonate and sodium dodecyl sulfate surfactants by novel consortium of Acinetobacter calcoaceticus and Pantoea agglomerans. Enzyme Microb Technol. 2007;41:432–439. doi: 10.1016/j.enzmictec.2007.03.011. [DOI] [Google Scholar]
- Ahmed S, James K. Derivation of a possible transition-state for the reaction catalysed by the enzyme estrone sulfatase (ES) Bioorg Med Chem Lett. 1999;9:1645–1650. doi: 10.1016/S0960-894X(99)00245-0. [DOI] [PubMed] [Google Scholar]
- Ambily PS, Jisha MS. Biodegradation of anionic surfactant, sodium dodecyl sulphate by Pseudomonas aeruginosa MTCC 10311. J Environ Biol. 2012;33:717–720. [PubMed] [Google Scholar]
- Anson DS, Taylor JA, Bielicki J, Harper SG, Peters C, Gibson GJ, Hopwood JJ. Correction of human mucopolysaccharidosis type-VI fibroblasts with recombinant N-acetylgalactosamine-4-sulphatase. Biochem J. 1992;284:789–794. doi: 10.1042/bj2840789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett CH, Dodgson KS, White GF. Further studies on the substrate specificity and inhibition of the stereospecific CS2 secondary alkylsulphohydrolase of Comamonas terrigena. Biochem J. 1980;191:467–473. doi: 10.1042/bj1910467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjdia A, Leprince J, Guillot A, Vaudry H, Rabot S, Berteau O. Anaerobic sulfatase-maturating enzymes: radical SAM enzymes able to catalyze in vitro sulfatase post-translational modification. J Am Chem Soc. 2007;129:3462–3463. doi: 10.1021/ja067175e. [DOI] [PubMed] [Google Scholar]
- Benjdia A, Subramanian S, Leprince J, Vaudry H, Johnson MK, Berteau O. Anaerobic sulfatase-maturating enzyme — a mechanistic link with glycyl radical-activating enzymes? FEBS J. 2010;277:1906–1920. doi: 10.1111/j.1742-4658.2010.07613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berteau O, Guillot A, Benjdia A, Rabot S. A new type of bacterial sulfatase reveals a novel maturation pathway in prokaryotes. J Biol Chem. 2006;281:22464–22470. doi: 10.1074/jbc.M602504200. [DOI] [PubMed] [Google Scholar]
- Boltes I, Czapinska H, Kahnert A, von Bulow R, Dierks T, Schmidt B, von Figura K, Kertesz MA, Uson I. 1.3 Å Structure of arylsulfatase from Pseudomonas aeruginosa establishes the catalytic mechanism of sulfate ester cleavage in the sulfatase family. Structure. 2001;9:483–491. doi: 10.1016/S0969-2126(01)00609-8. [DOI] [PubMed] [Google Scholar]
- But TYS, Toy PH. The Mitsunobu reaction: origin, mechanism, improvements, and applications. Chem Asian J. 2007;2:1340–1355. doi: 10.1002/asia.200700182. [DOI] [PubMed] [Google Scholar]
- Cloves JM, Dodgson KS, Games DE, Shaw DJ, White GF. The mechanism of action of primary alkylsulphohydrolase and arylsulphohydrolase from a detergent-degrading micro-organism. Biochem J. 1977;167:843–846. doi: 10.1042/bj1670843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloves JM, Dodgson KS, White GF, Fitzgerald JW. Specificity of P2 primary alkylsulphohydrolase induction in the detergent-degrading bacterium Pseudomonas C12B. Effects of alkanesulphonates, alkyl sulphates and other related compounds. Biochem J. 1980;185:13–21. doi: 10.1042/bj1850013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook AM, Laue H, Junker F. Microbial desulfonation. FEMS Microbiol Rev. 1998;22:399–419. doi: 10.1111/j.1574-6976.1998.tb00378.x. [DOI] [PubMed] [Google Scholar]
- Dhouib A, Hamad N, Hassairi I, Sayadi S. Degradation of anionic surfactants by Citrobacter braakii. Process Biochem. 2003;38:1245–1250. doi: 10.1016/S0032-9592(02)00322-9. [DOI] [Google Scholar]
- Dierks T, Schmidt B, Borissenko LV, Peng JH, Preusser A, Mariappan M, von Figura K. Multiple sulfatase deficiency is caused by mutations in the gene encoding the human C-alpha-formylglycine generating enzyme. Cell. 2003;113:435–444. doi: 10.1016/S0092-8674(03)00347-7. [DOI] [PubMed] [Google Scholar]
- Dodgson KS, Melville TH, Spencer B, Williams K. Studies on sulphatases. 8. The arylsulphatase of a strain of Alcaligenes metalcaligenes isolated from inter-tidal mud. Biochem J. 1954;58:182–187. doi: 10.1042/bj0580182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodgson KS, White GF, Fitzgerald JW. Sulfatases of microbial origin. Boca Raton: CRC Press; 1982. [Google Scholar]
- Ellis AJ, Hales SG, Ur-Rehman NGA, White GF. Novel alkylsulfatases required for biodegradation of the branched primary alkyl sulfate surfactant 2-butyloctyl sulfate. Appl Environ Microbiol. 2002;68:31–36. doi: 10.1128/AEM.68.1.31-36.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetzner S. Bacterial dehalogenation. Appl Microbiol Biotechnol. 1998;50:633–657. doi: 10.1007/s002530051346. [DOI] [PubMed] [Google Scholar]
- Fuchs M, Toesch M, Schober M, Wuensch C, Faber K (2013) Chemoenzymatic asymmetric total synthesis of (R)-lasiodiplodin methyl ether through a sulfatase-based deracemization process. Eur J Org Chem 356–361
- Gadler P, Faber K. New enzymes for biotransformations: microbial alkyl sulfatases displaying stereo- and enantioselectivity. Trends Biotechnol. 2007;25:83–88. doi: 10.1016/j.tibtech.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Gadler P, Faber K (2007b) Highly enantioselective biohydrolysis of sec-alkyl sulfate esters with inversion of configuration catalysed by Pseudomonas spp. Eur J Org Chem 5527–5530
- Gadler P, Reiter TC, Hoelsch K, Weuster-Botz D, Faber K. Enantiocomplementary inverting sec-alkylsulfatase activity in cyano- and thio-bacteria Synechococcus and Paracoccus spp.: selectivity enhancement by medium engineering. Tetrahedron Asymmetry. 2009;20:115–118. doi: 10.1016/j.tetasy.2009.01.007. [DOI] [Google Scholar]
- Hagelueken G, Adams TM, Wiehlmann L, Widow U, Kolmar H, Tümmler B, Heinz DW, Schubert W-D. The crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa, defines a third class of sulfatases. Proc Natl Acad Sci U S A. 2006;103:7631–7636. doi: 10.1073/pnas.0510501103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmann A, Sumper M. An inducible arylsulfatase of Volvox carteri with properties suitable for a reporter-gene system — purification, characterization and molecular cloning. Eur J Biochem. 1994;221:143–150. doi: 10.1111/j.1432-1033.1994.tb18723.x. [DOI] [PubMed] [Google Scholar]
- Hanson SR, Best MD, Wong C-H. Sulfatases: structure, mechanism, biological activity, inhibition, and synthetic utility. Angew Chem Int Ed. 2004;43:5736–5763. doi: 10.1002/anie.200300632. [DOI] [PubMed] [Google Scholar]
- Harada T. Formation of sulphatases by Pseudomonas aeruginosa. Biochim Biophys Acta. 1964;81:193–196. [Google Scholar]
- Hopwood JJ, Ballabio A. The metabolic and molecular bases of inherited disease. 8. New York: McGraw-Hill; 1999. [Google Scholar]
- Huber H, Stetter KO. Hyperthermophiles and their possible potential in biotechnology. J Biotechnol. 1998;64:39–52. doi: 10.1016/S0168-1656(98)00102-3. [DOI] [Google Scholar]
- Janssen DB (2007) Biocatalysis by dehalogenating enzymes. In: Laskin AI, Sariaslani S, Gadd GM (eds) Adv Appl Microbiol 61:233–252 [DOI] [PubMed]
- Jonas S, van Loo B, Hyvönen M, Hollfelder F. A new member of the alkaline phosphatase superfamily with a formylglycine nucleophile: structural and kinetic characterisation of a phosphonate monoester hydrolase/phosphodiesterase from Rhizobium leguminosarum. J Mol Biol. 2008;384:120–136. doi: 10.1016/j.jmb.2008.08.072. [DOI] [PubMed] [Google Scholar]
- Jovcic B, Venturi V, Davison J, Topisirovic L, Kojic M. Regulation of the SdsA alkyl sulfatase of Pseudomonas sp. ATCC19151 and its involvement in degradation of anionic surfactants. J Appl Microbiol. 2010;109:1076–1083. doi: 10.1111/j.1365-2672.2010.04738.x. [DOI] [PubMed] [Google Scholar]
- Kahnert A, Kertesz MA. Characterization of a sulfur-regulated oxygenative alkylsulfatase from Pseudomonas putida S-313. J Biol Chem. 2000;275:31661–31667. doi: 10.1074/jbc.M005820200. [DOI] [PubMed] [Google Scholar]
- Kertesz MA. Riding the sulfur cycle – metabolism of sulfonates and sulfate esters in Gram-negative bacteria. FEMS Microbiol Rev. 2000;24:135–175. doi: 10.1016/S0168-6445(99)00033-9. [DOI] [PubMed] [Google Scholar]
- Kertesz MA, Wietek C. Desulfurization and desulfonation: applications of sulfur-controlled gene expression in bacteria. Appl Microbiol Biotechnol. 2001;57:460–466. doi: 10.1007/s002530100800. [DOI] [PubMed] [Google Scholar]
- Kertesz MA, Leisinger T, Cook AM. Proteins induced by sulfate limitation in Escherichia coli, Pseudomonas putida, or Staphylococcus aureus. J Bacteriol. 1993;175:1187–1190. doi: 10.1128/jb.175.4.1187-1190.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaus T, Schober M, Kepplinger B, Faccinelli M, Pitzer J, Faber K, Macheroux P, Wagner U. Structure and mechanism of an inverting alkylsulfatase from Pseudomonas sp. DSM6611 specific for secondary alkyl sulfates. FEBS J. 2012;279:4374–4384. doi: 10.1111/febs.12027. [DOI] [PubMed] [Google Scholar]
- Kotik M, Archelas A, Wohlgemuth R. Epoxide hydrolases and their application in organic synthesis. Curr Org Chem. 2012;16:451–482. doi: 10.2174/138527212799499840. [DOI] [Google Scholar]
- Koudelakova T, Bidmanova S, Dvorak P, Pavelka A, Chaloupkova R, Prokop Z, Damborsky J. Haloalkane dehalogenases: biotechnological applications. Biotechnol J. 2013;8:32–45. doi: 10.1002/biot.201100486. [DOI] [PubMed] [Google Scholar]
- Lairson LL, Withers SG (2004) Mechanistic analogies amongst carbohydrate modifying enzymes. Chem Commun 2243–2248 [DOI] [PubMed]
- Lie TJ, Leadbetter JR, Leadbetter ER. Metabolism of sulfonic acids and other organosulfur compounds by sulfate-reducing bacteria. Geomicrobiol J. 1998;15:135–149. doi: 10.1080/01490459809378070. [DOI] [Google Scholar]
- Long M, Ruan L, Li F, Yu Z, Xu X. Heterologous expression and characterization of a recombinant thermostable alkylsulfatase (sdsAP) Extremophiles. 2011;15:293–301. doi: 10.1007/s00792-011-0357-4. [DOI] [PubMed] [Google Scholar]
- Marquordt C, Fang QH, Will E, Peng JH, von Figura K, Dierks T. Posttranslational modification of serine to formylglycine in bacterial sulfatases — recognition of the modification motif by the iron–sulfur protein AtsB. J Biol Chem. 2003;278:2212–2218. doi: 10.1074/jbc.M209435200. [DOI] [PubMed] [Google Scholar]
- Marzluf GA. Genetics and molecular genetics of sulfur assimilation in the fungi. Adv Genet. 1994;31:187–206. doi: 10.1016/S0065-2660(08)60398-3. [DOI] [PubMed] [Google Scholar]
- Matcham GWJ, Bartholomew B, Dodgson KS, Fitzgerald JW, Payne WJ. Stereospecificity and complexity of microbial sulphohydrolases involved in the biodegradation of secondary alkylsulphate detergents. FEMS Microbiol Lett. 1977;1:197–199. doi: 10.1111/j.1574-6968.1977.tb00613.x. [DOI] [Google Scholar]
- Matts PJ, White GF, Payne WJ. Purification and characterization of the short-chain alkylsulphatase of coryneform Bla. Biochem J. 1994;304:937–943. doi: 10.1042/bj3040937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller I, Kahnert A, Pape T, Sheldrick GM, Meyer-Klaucke W, Dierks T, Kertesz M, Usón I. Crystal structure of the alkylsulfatase AtsK: insights into the catalytic mechanism of the Fe(II) α-ketoglutarate-dependent dioxygenase superfamily. Biochemistry. 2004;43:3075–3088. doi: 10.1021/bi035752v. [DOI] [PubMed] [Google Scholar]
- Murooka Y, Ishibashi K, Yasumoto M, Sasaki M, Sugino H, Azakami H, Yamashita M. A sulfur- and tyramine-regulated Klebsiella aerogenes operon containing the arylsulfatase (atsA) gene and the atsB gene. J Bacteriol. 1990;172:2131–2140. doi: 10.1128/jb.172.4.2131-2140.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan D, Schmidberger JW. Enzymes that catalyse SN2 reaction mechanisms. Nat Prod Rep. 2010;27:900–918. doi: 10.1039/b919371p. [DOI] [PubMed] [Google Scholar]
- Payne WJ, Williams JP, Mayberry WR. Primary alcohol sulfatase in a Pseudomonas species. Appl Microbiol. 1965;13:698–701. doi: 10.1128/am.13.5.698-701.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogorevc M, Faber K. Enantioselective stereoinversion of sec-alkyl sulfates by an alkylsulfatase from Rhodococcus ruber DSM 44541. Tetrahedron Asymmetry. 2002;13:1435–1441. doi: 10.1016/S0957-4166(02)00362-2. [DOI] [PubMed] [Google Scholar]
- Pogorevc M, Strauss UT, Riermeier T, Faber K. Selectivity-enhancement in enantioselective hydrolysis of sec-alkyl sulfates by an alkylsulfatase from Rhodococcus ruber DSM 44541. Tetrahedron Asymmetry. 2002;13:1443–1447. doi: 10.1016/S0957-4166(02)00363-4. [DOI] [Google Scholar]
- PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC.
- Roig MG, Pedraz MA, Sanchez JM. Sorption isotherms and kinetics in the primary biodegradation of anionic surfactants by immobilized bacteria: I. Pseudomonas C12B. J Mol Catal B: Enzym. 1998;4:253–270. doi: 10.1016/S1381-1177(98)00005-8. [DOI] [Google Scholar]
- Roig MG, Pedraz MA, Sanchez JM, Huska J, Tóth D. Sorption isotherms and kinetics in the primary biodegradation of anionic surfactants by immobilized bacteria: II. Comamonas terrigena N3H. J Mol Catal B: Enzym. 1998;4:271–281. doi: 10.1016/S1381-1177(98)00006-X. [DOI] [Google Scholar]
- Schober M (2013) Characterization and biocatalytic application of sulfatases. PhD thesis, Karl-Franzens University, Graz
- Schober M, Faber K. Inverting hydrolases and their use in enantioconvergent biotransformations. Trends Biotechnol. 2013;31:468–478. doi: 10.1016/j.tibtech.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober M, Gadler P, Knaus T, Kayer H, Birner-Gruenberger R, Guelly C, Macheroux P, Wagner U, Faber K. A stereoselective inverting sec-alkylsulfatase for the deracemization of sec-alcohols. Org Lett. 2011;13:4296–4299. doi: 10.1021/ol201635y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober M, Knaus T, Toesch M, Macheroux P, Wagner U, Faber K. The substrate spectrum of the inverting sec-alkylsulfatase Pisa1. Adv Synth Catal. 2012;354:1737–1742. doi: 10.1002/adsc.201100864. [DOI] [Google Scholar]
- Schober M, Toesch M, Knaus T, Strohmeier GA, van Loo B, Fuchs M, Hollfelder F, Macheroux P, Faber K. One-pot deracemization of sec-alcohols: enantioconvergent enzymatic hydrolysis of alkyl sulfates using stereocomplementary sulfatases. Angew Chem Int Ed. 2013;52:3277–3279. doi: 10.1002/anie.201209946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger Release 2013-1: Maestro, version 9.4, Schrödinger, LLC, New York, NY, 2013.
- Scott C, Hilton ME, Coppin CW, Russell RJ, Oakeshott JG, Sutherland TD. A global response to sulfur starvation in Pseudomonas putida and its relationship to the expression of low-sulfur-content proteins. FEMS Microbiol Lett. 2007;267:184–193. doi: 10.1111/j.1574-6968.2006.00575.x. [DOI] [PubMed] [Google Scholar]
- Selmer T, Hallmann A, Schmidt B, Sumper M, vonFigura K. The evolutionary conservation of a novel protein modification, the conversion of cysteine to serine semialdehyde in arylsulfatase from Volvox carteri. Eur J Biochem. 1996;238:341–345. doi: 10.1111/j.1432-1033.1996.0341z.x. [DOI] [PubMed] [Google Scholar]
- Shukor MY, Husin WS, Rahman MFA, Shamaan NA, Syed MA. Isolation and characterization of an SDS-degrading Klebsiella oxytoca. J Environ Biol. 2009;30:129–134. [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539–547. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Kumar A, Kumar A. Bacillus cereus capable of degrading SDS shows growth with a variety of detergents. World J Microb Biot. 1998;14:777–779. doi: 10.1023/A:1008883915003. [DOI] [Google Scholar]
- Straathof AJJ, Jongejan JA. The enantiomeric ratio: origin, determination and prediction. Enzyme Microb Technol. 1997;21:559–571. doi: 10.1016/S0141-0229(97)00066-5. [DOI] [Google Scholar]
- Thies W. Detection and utilization of a glucosinolate sulfohydrolase in the edible snail, Helix pomatia. Naturwissenschaften. 1979;66:364–365. doi: 10.1007/BF00368477. [DOI] [Google Scholar]
- Thomas ORT, White GF. Immobilization of the surfactant-degrading bacterium Pseudomonas C12B in polyacrylamide gel: II. Optimizing SDS-degrading activity and stability. Enzyme Microb Technol. 1990;12:969–975. doi: 10.1016/0141-0229(90)90119-B. [DOI] [PubMed] [Google Scholar]
- Thomas ORT, White GF. Immobilization of the surfactant-degrading bacterium Pseudomonas C12B in polyacrylamide gel: III. Biodegradation specificity for raw surfactants and industrial wastes. Enzyme Microb Technol. 1991;13:338–343. doi: 10.1016/0141-0229(91)90154-3. [DOI] [PubMed] [Google Scholar]
- Tomatsu S, Fukuda S, Masue M, Sukegawa K, Fukao T, Yamagishi A, Hori T, Iwata H, Ogawa T, Nakashima Y, Hanyu Y, Hashimoto T, Titani K, Oyama R, Suzuki M, Yagi K, Hayashi Y, Orii T. Morquio disease: isolation, characterization and expression of full-length cDNA for human N-acetylgalactosamine-6-sulfate sulfatase. Biochem Biophys Res Commun. 1991;181:677–683. doi: 10.1016/0006-291X(91)91244-7. [DOI] [PubMed] [Google Scholar]
- van Leeuwen JGE, Wijma HJ, Floor RJ, van der Laan J-M, Janssen DB. Directed evolution strategies for enantiocomplementary haloalkane dehalogenases: from chemical waste to enantiopure building blocks. Chem Bio Chem. 2012;13:137–148. doi: 10.1002/cbic.201100579. [DOI] [PubMed] [Google Scholar]
- Verschueren KHG, Seljee F, Rozeboom HJ, Kalk KH, Dijkstra BW. Crystallographic analysis of the catalytic mechanism of haloalkane dehalogenase. Nature. 1993;363:693–698. doi: 10.1038/363693a0. [DOI] [PubMed] [Google Scholar]
- Wallner SR, Nestl BM, Faber K. Highly enantioselective sec-alkyl sulfatase activity of Sulfolobus acidocaldarius DSM 639. Org Lett. 2004;6:5009–5010. doi: 10.1021/ol0477778. [DOI] [PubMed] [Google Scholar]
- Wallner SR, Bauer M, Würdemann C, Wecker P, Glöckner FO, Faber K. Highly enantioselective sec-alkyl sulfatase activity of the marine planctomycete Rhodopirellula baltica shows retention of configuration. Angew Chem Int Ed. 2005;44:6381–6384. doi: 10.1002/anie.200501955. [DOI] [PubMed] [Google Scholar]
- Wallner SR, Nestl BM, Faber K. Highly enantioselective stereo-inverting sec-alkylsulfatase activity of hyperthermophilic Archaea. Org Biomol Chem. 2005;3:2652–2656. doi: 10.1039/b504883d. [DOI] [PubMed] [Google Scholar]
- Whalen JK, Warman PR. Examination of ester sulfates in Podzolic and Regosolic soils using an immobilized arylsulfatase reactor. Biol Fert Soils. 1996;23:64–69. doi: 10.1007/BF00335820. [DOI] [Google Scholar]
- Widersten M, Gurell A, Lindberg D. Structure–function relationships of epoxide hydrolases and their potential use in biocatalysis. Biochim Biophys Acta. 2010;1800:316–326. doi: 10.1016/j.bbagen.2009.11.014. [DOI] [PubMed] [Google Scholar]
- Williams J, Payne WJ. Enzymes induced in a bacterium by growth on sodium dodecyl sulfate. Appl Microbiol. 1964;12:360–362. doi: 10.1128/am.12.4.360-362.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittstock U, Fischer M, Svendsen I, Halkier BA. Cloning and characterization of two cDNAs encoding sulfatases in the Roman snail, Helix pomatia. IUBMB Life. 2000;49:71–76. doi: 10.1080/713803591. [DOI] [PubMed] [Google Scholar]
- Yegles M, Mersch F, Wennig R. Detection of benzodiazepines and other psychotropic drugs in human hair by GC/MS. Forensic Sci Int. 1997;84:211–218. doi: 10.1016/S0379-0738(96)02064-6. [DOI] [PubMed] [Google Scholar]
- Yeldho D, Rebello S, Jisha MS. Plasmid-mediated biodegradation of the anionic surfactant sodium dodecyl sulphate by Pseudomonas aeruginosa S7. Bull Environ Contam Toxicol. 2011;86:110–113. doi: 10.1007/s00128-010-0162-2. [DOI] [PubMed] [Google Scholar]