Abstract
Objectives
This work aims at uncovering challenges in biomedical knowledge representation research by providing an understanding of what was historically called "medical concept representation" and used as the name for a working group of the International Medical Informatics Association.
Methods
Bibliometrics, text mining, and a social media survey compare the research done in this area between two periods, before and after 2000.
Results
Both the opinion of socially active groups of researchers and the interpretation of bibliometric data since 1988 suggest that the focus of research has moved from "medical concept representation" to "medical ontologies".
Conclusions
It remains debatable whether the observed change amounts to a paradigm shift or whether it simply reflects changes in naming, following the natural evolution of ontology research and engineering activities in the 1990s. The availability of powerful tools to handle ontologies devoted to certain areas of biomedicine has not resulted in a large-scale breakthrough beyond advances in basic research.
Keywords: Data Mining, Terminology, Semantics, Vocabulary, Publishing
I. Introduction
The study of the meaning of language expressions has a long history in health informatics, both regarding narratives (e.g., text in clinical reports and from the biomedical literature) and structured information (e.g., terms from standard vocabularies used for clinical research, health statistics, quality assessment and billing). It motivated the activities of the International Medical Informatics Association (IMIA)'s Working Group on Medical Concept Representation (MCR WG) [1], which was an influential body in the late 1980s and the 1990s, publishing regular overviews [2].
The evolution of ontologies for biomedical research, the proliferation of clinical vocabularies, advances in human language technologies with increasingly large amounts of training data have changed the health information science landscape profoundly. New scientific communities have arisen like the Semantic Web community, and social media are changing communication between researchers. In this context the MCR WG, now renamed to "Language and Meaning in Biomedicine (LaMB)", will have to find a new ecological niche. In order to better define the future activities of this working group, the authors have investigated the evolution of the field of biomedical language and representation of meaning over the years, and will discuss some persistent research areas to be addressed in the future.
II. Methods
The analysis of literature over time can provide insight in how a research field develops [3]. We have used bibliographics, on-line text mining tools and a social media survey tool, in order to investigate how the research area, known as "Medical Knowledge Representation" has evolved since the 1990s.
The phrase "medical concept representation" (not to be mixed with "concept representation" as a category used in the science of psychology) was key in that period-a reason to name the working group accordingly. Therefore, we placed this phrase in the centre of our investigation, divided into the following steps:
Time line analysis of the occurrence of the phrase "medical concept representation" using the Scopus term analyser [4], extraction of the contextual environment using Ultimate Research Assistant [5] and visualization of the results using a tag cloud [6];
Using the tool Publish or Perish [7] to identify the authors of the most influential papers, using seven sources, viz. Web of Science, Scopus, Embase, PubMed, Google Scholar, Cochrane Library, British Library on-line catalogue. The question was to have an idea of the persistence of the influential authors from the first period to the second one. The Boolean search expression "concept representation" AND ("medical" OR "medicine") AND ("knowledge" OR "information") was submitted to all of them, with variations according to their proprietary syntax. For identifying the top ten papers, the results of the seven lists were consolidated into a common table. For this, available citation ranks were taken, otherwise the source's own ranking mechanism was used. In the following, the top ten papers were the source for extracting the top thirty authors, which were ranked in a second step. For this, the following heuristics was used: The nth author in the list was assigned a score of 11 - n, the eleventh and following authors was given a zero value. The scoring was weighted, favouring multiple appearances of authors in different sources: a final score was calculated as a net score (0.8 + 0.2 × occurrence).
In the post-2000 analysis, due to the significant drop of the usage of the exact phrase "medical concept representation" the resulting paper population would have been too small for applying the same procedure as described for the first period. Therefore, instead of summing up the citation data only for papers matching the query, here the citation data for all papers per author were used. This same method, however, could not be used for same analysis backwards to the previous period, due to limitations of the tool used [7].
The hypothesis of a paradigm shift was studied, comparing relevant papers published during the years from 1988 to 1999 with those appearing between 2000 and 2012, focusing the same subject area. The reason for starting with 1988 was the availability of bibliographic databases, being almost accordant with the period of our interest, viz. the activities of the IMIA WG on Medical Knowledge Representation. Author lists were compared and all the titles of the two full paper sets were text mined using Textalyser [8].
The second, more recent set was cross-checked against a third set from the same period, obtained by an online survey targeted to the specifically interested audience. For this survey (open from August to October 2012) the primary source was the LinkedIn group of the MCR WG, having at that time over fifty members of widely various backgrounds. Secondary sources were additional LinkedIn Groups in broader domain. Participants were asked to quote and to share the papers they found to be most influential in their work or research. We used Datagle [9] and a Google document to collect survey data.
III. Results
1. Looking Back: 'Medical Concept Representation' before the Turn of the Millennium
Scopus has revealed that the exact phrase "medical concept representation" was used mostly in the nineties (Figure 1). Scopus data were available for 1993-2008. The targeted semantic search revealed a wide conceptual domain related to this phrase, as shown in Figure 2.
Figure 1.
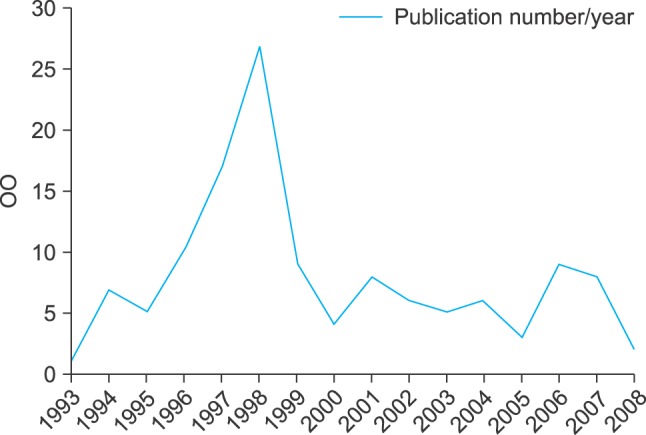
Scopus time line analytics results for the exact phrase "medical concept representation".
Figure 2.
Wordle tag cloud generated from result of catchphrase search using Ultimate Research Assistant [5].
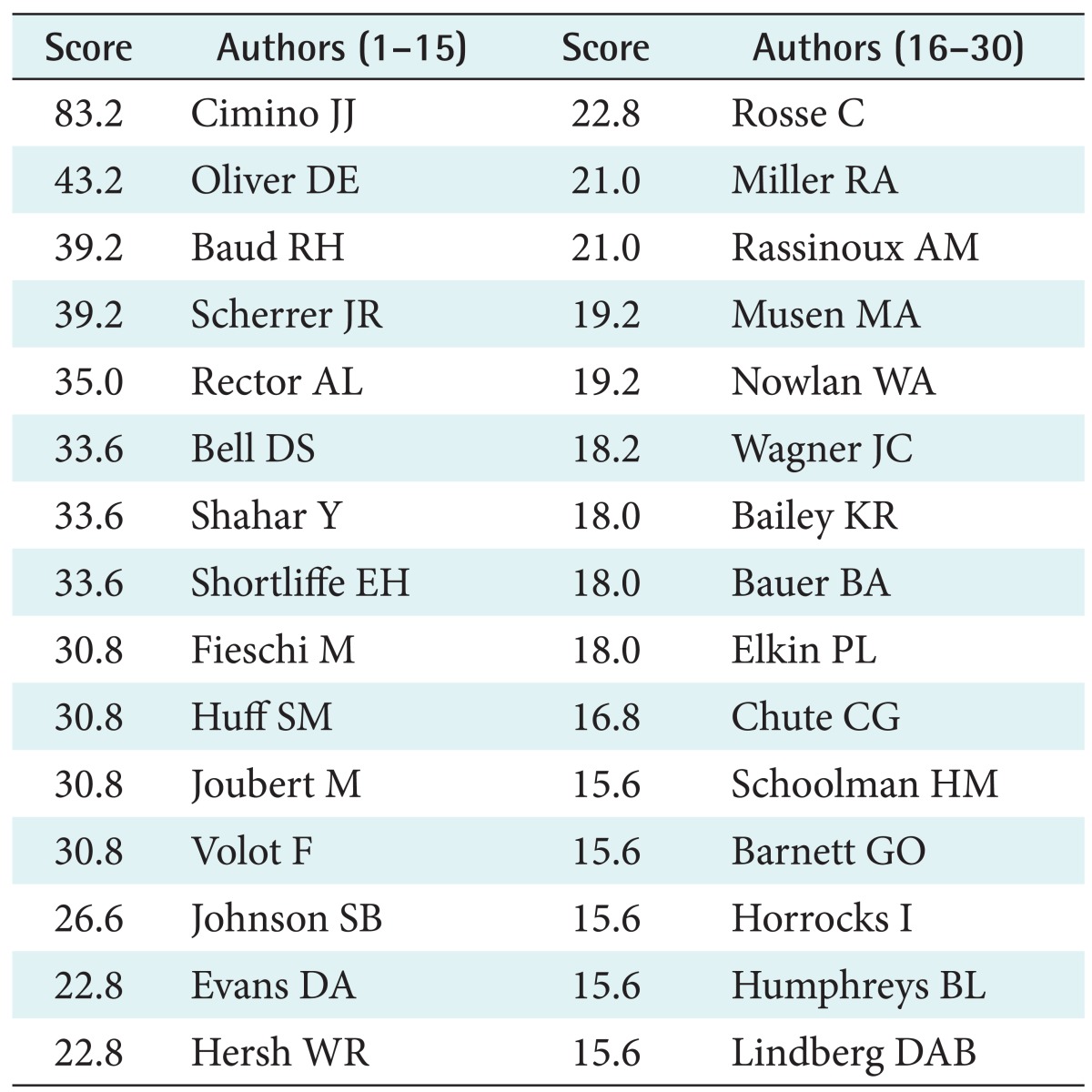
The top thirty authors of the ten most influential papers 1988-1999 were identified (the starting date of the study was justified by the availability of electronic bibliographic databases and the comparability of the investigated periods before and after 2000). The tool Publish or Perish [7] showed the average number of authors to be 2.45. The results of the extraction of the first three author names per paper are shown in Table 1. Our querying strategy was found effective for excluding papers regarded irrelevant for our purpose, e.g., in the domain of concept representation in psychology.
Table 1.
The thirty most influential authors of the period 1988-1999 that used the phrase 'medical concept representation'
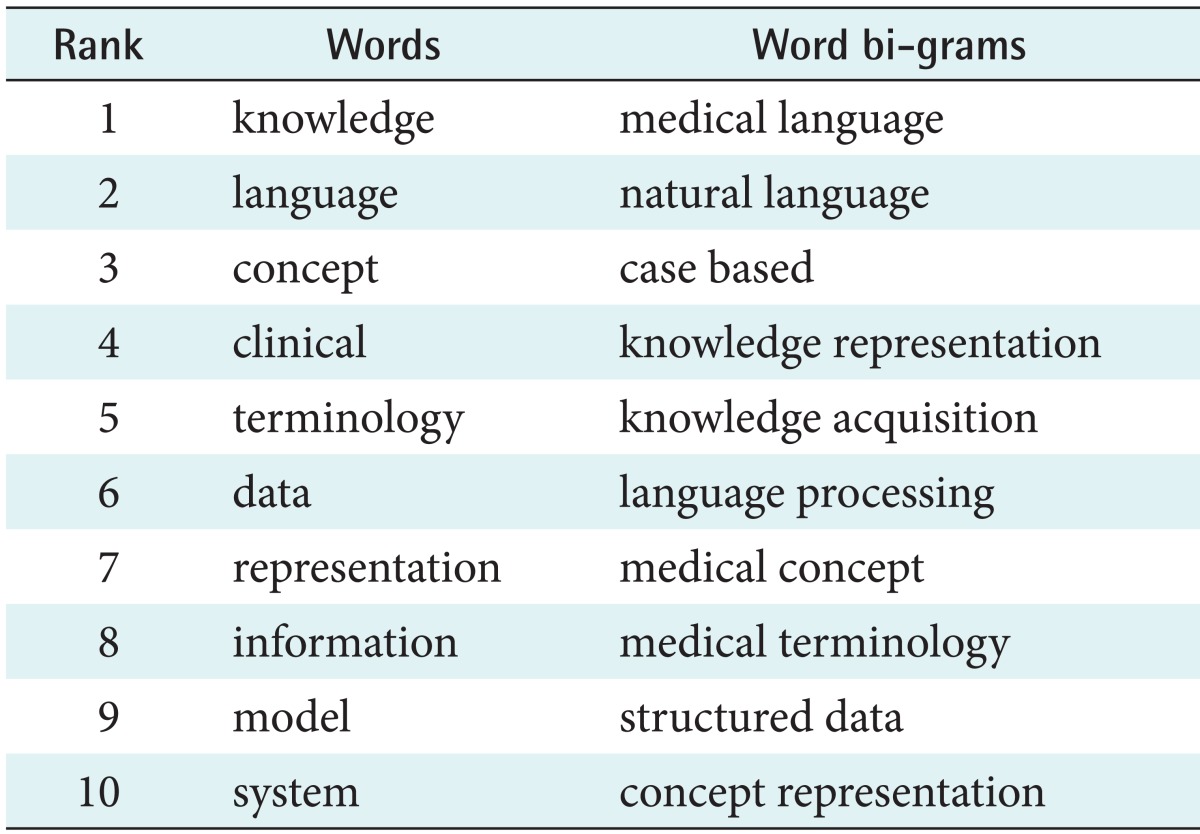
A frequency analysis of the title words of the papers in the same period shows the most frequently used uni- and bi-grams (single noun phrases and meaningful two-word phrases) in Table 2. Note that 'ontology' was not among the most frequently used terms at that period.
Table 2.
List of most frequently used uni- and bi-grams of the period 1988-1999
2. "Medical Concept Representation" Since the Year 2000
Table 3 presents the list of the top thirty authors of most cited publications, using the same Boolean expression applied to the period of 2000-2012. However, as the methodology was different for the reasons explained above, the comparison should be interpreted with reservation. Nevertheless it is striking that the two lists only overlap in three authors (in bold). In addition, the word frequency analysis of the period 2000-2012 shows a clearly distinct result (Table 4).
Table 3.
Set of most cited authors, between 2000 and 2012, covering the whole domain of all authors publishing on medical concept representation
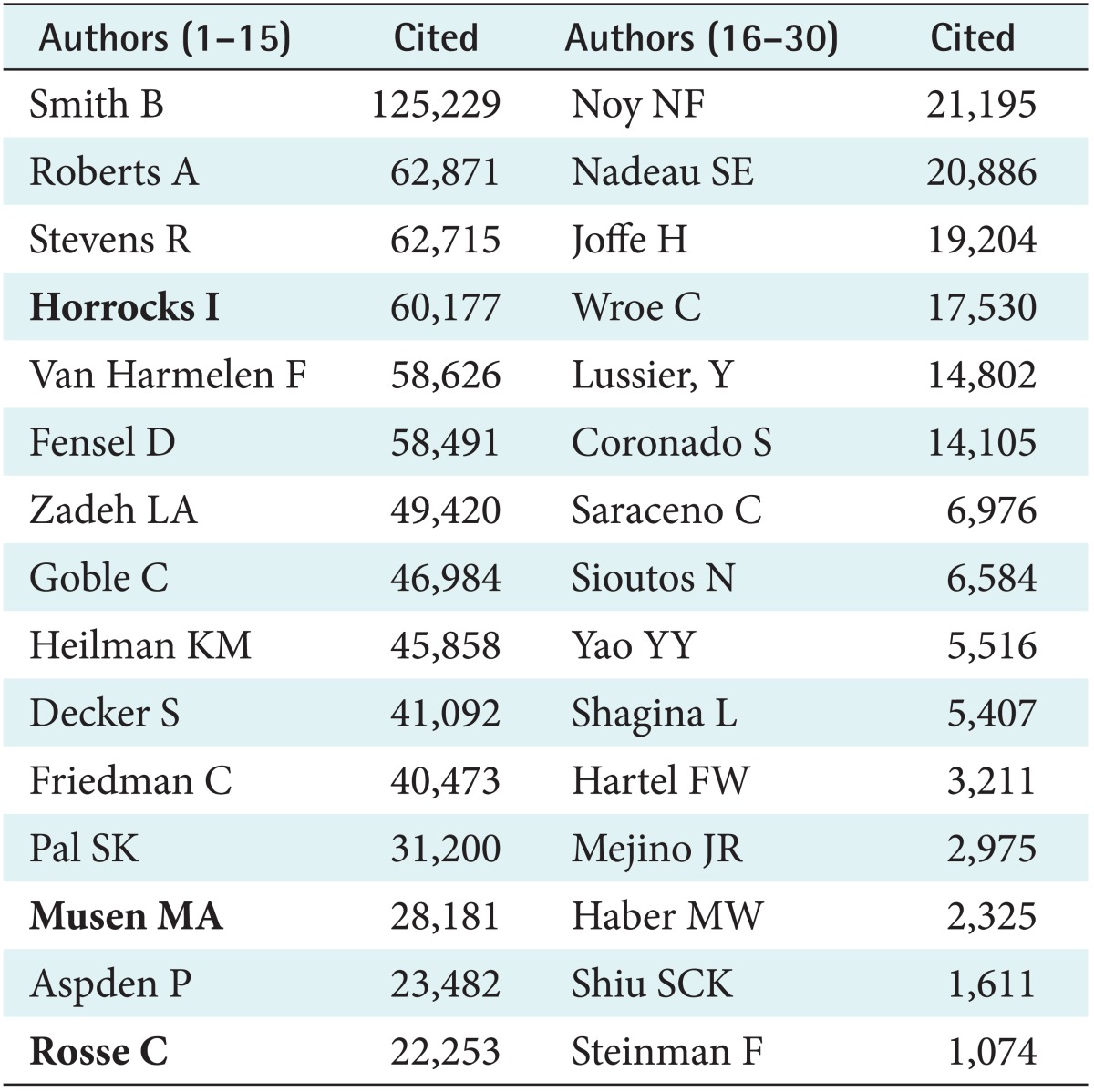
Names that are in the 1988-1999 ranking are in bold face.
Table 4.
List of most frequently used uni- and bi-grams of the period of the period 2000-2012 in the domain of medical concept representation
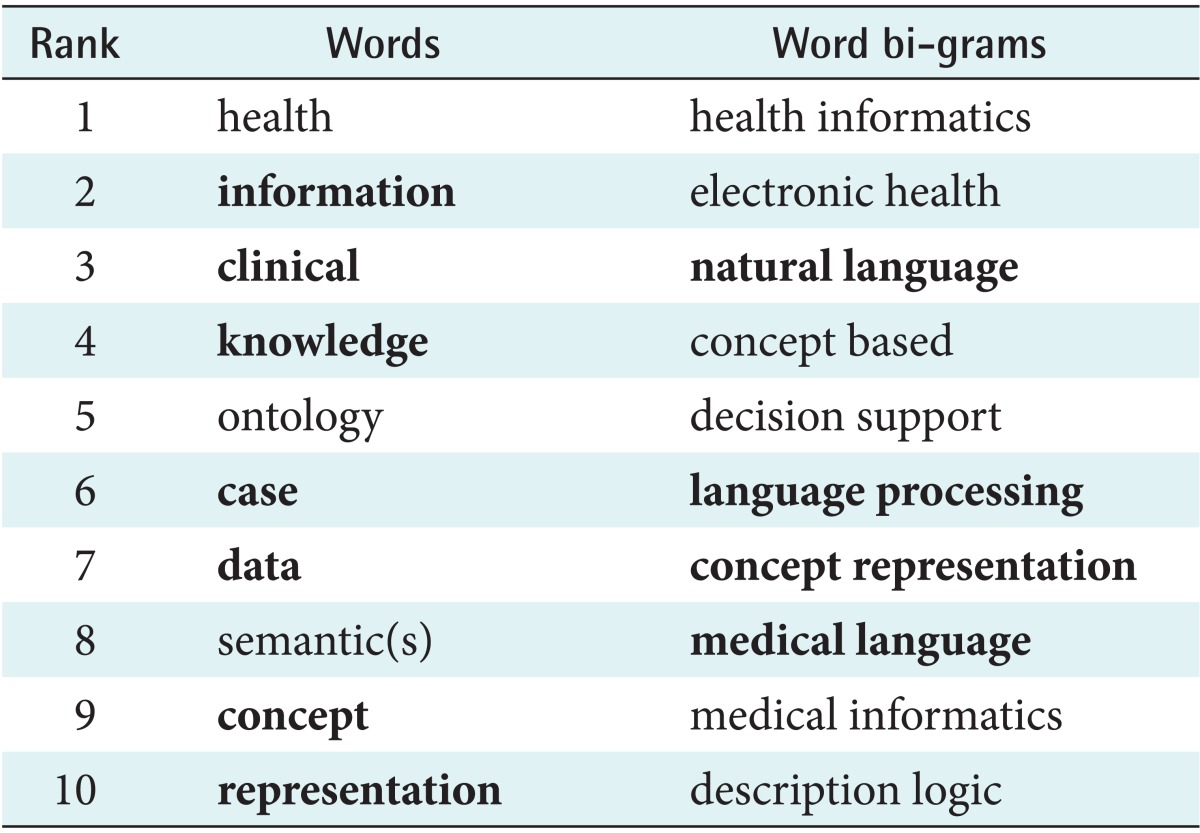
Bold face highlights the terms that also occur in the top-ten list from the 1988-1999 period (Table 2).
3. Mapping the Conceptual Context of Most Influential Papers Based on Text Mining of Titles
Figure 3 shows how the terms in the titles were changing. "Old" terms that are no longer found among the "new" top ten are depicted in white. New terms appearing in the 2000-2012 list are shown in red. The top ten terms also suggest that the subject matter of "concept representation" was broadened (from focusing on "medical" to areas as "health" and "clinical"). In addition, the words "semantics" and "ontology" suggest that new ideas have influenced the concept representation domain. The fact that "language", "model" and "terminology" disappeared may suggest that some more differentiated areas branched off the previously common roots.
Figure 3.
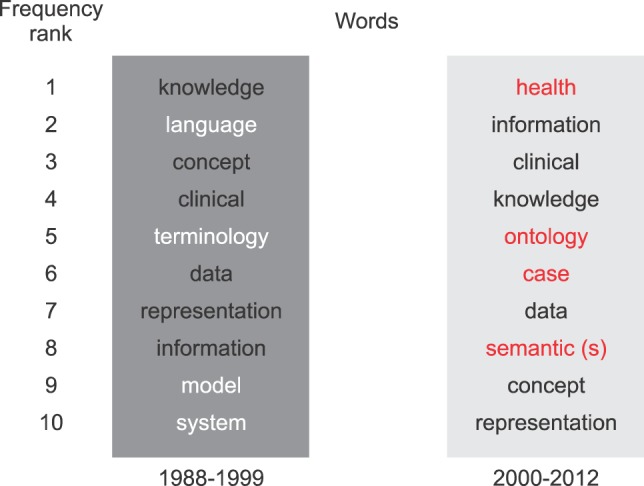
Changes in the most frequent title words of papers on medical concept representation.
4. Results of the Survey Taken Show the Opinion of Socially Active Researchers Interested in the Domain
The survey had 42 respondents. Not surprisingly, the central role of ontologies is clearly reflected in the list of the twenty most influential papers (Table 5). Recurring resources include the Open Biological and Biomedical Ontologies (OBO) Foundry [10], the Gene Ontology [11], Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT) [12], and the Unified Medical Language System (UMLS) [13].
Table 5.
Titles of the twenty most influential papers as listed by LinkedIn MCR WG members
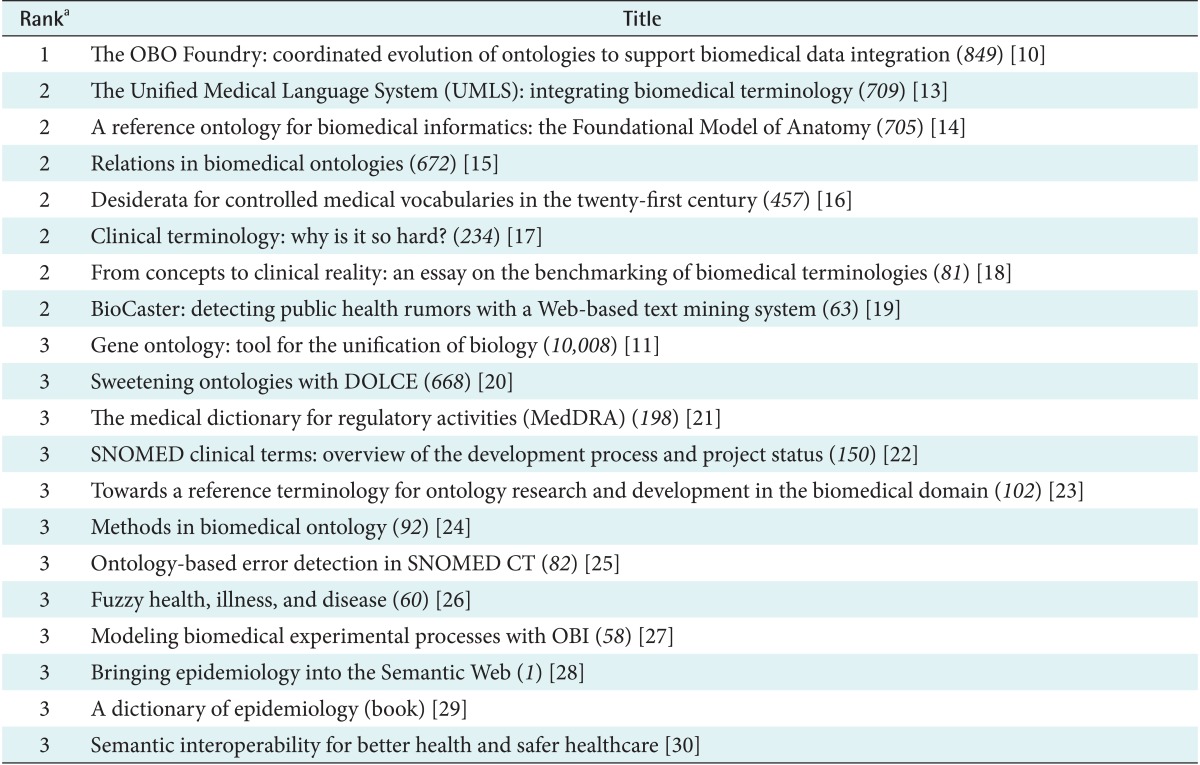
MCR WG: Working Group on Medical Concept Representation, OBO: Open Biological and Biomedical Ontologies, SNOMED CT: Systematized Nomenclature of Medicine Clinical Terms, OBI: Ontology for Biomedical Investigations.
aFrequency ranking is based on incidence in survey lists. The first ranked paper was mentioned the most times in various lists. Papers with rank '2' shared the second highest number of occurrence and so on. Papers with same rank are in order of their citation frequency, shown above in italics.
IV. Discussion
1. Methodology Issues Regarding the Literature Study
Although the methodology applied in this paper does not aim at establishing a new scientometric index or a generalizable tool, it clearly demonstrated that on-line searchable library databases, bibliometric services, and simple text mining tools enable the creation of study-focused tool sets as used in this study without investing much effort and resources. Using multiple, large bibliographic source databases helped to alleviate the possible bias in such studies that are limited to one particular source or aspect of the field.
2. Current Trends
The tools we used in this study were aimed at exploring the specific area of medical concept representation with the focus on testing the complementary question as to whether the observed changes amount to a significant paradigm shift.
Our results show that researchers active in this area for several decades have pursued the main goal of being able to make health-related information machine readable and processable. This has been a major driver of the development of clinical information systems in general. The use of formal languages, such as description logics, has been a step in this direction. In 1990s, "medical concept representation" was seen as a solution by proposing just one general method: practical conceptualization of information in medical research and practice. However, these efforts were hindered by theoretical issues, difficulties of modelling a domain, and the explosion of knowledge in general [31].
Building on this background, our investigation has taken the pulse of a group of researchers interested in what we could refer to, generally speaking, as the study of meaning of structured and unstructured representations. First of all the use of the term "concept" has decreased, which we attribute to the following factors:
Propagation of the paradigm of ontological realism, the proponents of which have been arguing against the usage of this word in the context of ontologies, contending that the representation of concepts as "entities of thought" is inappropriate for the representation of a scientific domain and obfuscates the difference between the entities and names given to them [32];
The preference of "class" over "concept" in the Semantic Web and description logics community, especially regarding the influential OWL family of representation languages [33];
The obvious polysemy of the word itself [34].
In addition, the popularity of the word "ontology" shows a new tendency in which artefacts that represent types of domain entities are more clearly distinguished by some researchers from artefacts that describe language items. The importance of ontology-based artefacts can be seen by the central place the OBO Foundry and SNOMED CT occupy in publications and importance judgments. However, the boundaries between ontologies and knowledge representation artefacts are less clear, although relatively crisp criteria can be formulated. In practice, "ontology" is used by many to refer to a wide array of resources across the semantic spectrum, encompassing terminologies, thesauri, classifications and formal ontologies [35].
At the same time important areas as medical language processing and medical terminologies, but also metadata, semantic annotation and folksonomies have gained importance, so that they are no longer subsumed under "concept representation".
The analysis of influential authors faced methodological difficulties, as the selection criterion-namely the phrase "concept representation" turned out to be a moving target. The comparability of the two lists of authors is therefore limited. Nevertheless, it is noteworthy that only three authors appeared on both lists. Note that this comparison is additionally biased by the following: it is very likely that there are relevant authors in the second period that were not retrieved, simply because they did not use the-already outmoded-phrase "concept representation", at all. There are authors of the papers in Table 5 that are not among the top 20 (Table 4), simply because they avoid that phrase. If they would have been included, the overlap were probably even lower.
V. Conclusion
There are several indications that the turn of the new millennium coincided with a change in the focus of research in medical domain representation and semantics. The millennium marked the emergence of the establishment of applied ontology [36] and the Semantic Web [37] as new disciplines. The central role of the term "concept" has been gradually abandoned. Whether this really amounts to a paradigm shift, or a simple change in terminological preferences, may be argued. Undoubtedly, the ontology research and engineering efforts, which started around 1990, yielded important results, including the development of description logics [38], tools like Protégé [39], as well as the groundbreaking GALEN project [40].
The following directions for the future have emerged from our analysis:
The capture of medical information and knowledge leverages (standards) ontologies;
Open reference resources for content are developed collaboratively, shared, and reused;
Web enabled standards help achieve transparent results;
"Big data" opens new ways for knowledge acquisition;
However, a large part of clinical information continues being recorded as free text, which keeps the need of processing medical language on the research agenda.
All these topics justify, more than ever, collaborative research and development efforts, for which the IMIA WG Language and Meaning in Biomedicine (LaMB) [41] can be an effective catalyst.
Acknowledgments
This work was supported in part by the Intramural Research Program of the NIH, National Library of Medicine. We also thank participants of the IMIA LaMB Working Group for their participation in the survey.
Footnotes
No potential conflict of interest relevant to this article was reported.
References
- 1.International Medical Informatics Association [Internet] Geneva, Switzerland: International Medical Informatics Association; c2013. [cited at 2013 Nov 15]. Available from: http://www.imia-medinfo.org/new2/ [Google Scholar]
- 2.Cimino JJ, Smith B. Introduction: international medical informatics association working group 6 and the 2005 Rome conference. J Biomed Inform. 2006;39(3):249–251. [Google Scholar]
- 3.Schuemie MJ, Talmon JL, Moorman PW, Kors JA. Mapping the domain of medical informatics. Methods Inf Med. 2009;48(1):76–83. doi: 10.3414/me0576. [DOI] [PubMed] [Google Scholar]
- 4.Scopus [Internet] Amsterdam, The Netherlands: Elsevier; c2013. [cited at 2013 Nov 15]. Available from: http://www.scopus.com. [Google Scholar]
- 5.Ultimate Research Assistant [Internet] Herndon (VA): Andy Hoskinson, LLC; [cited at 2013 Nov 15]. Available from: http://ultimate-research-assistant.com. [Google Scholar]
- 6.Wordle [Internet] [unknown]: Jonathan Feinberg; c2013. [cited at 2013 Nov 15]. Available from: http://www.wordle.net/ [Google Scholar]
- 7.Harzing AW. Publish or Perish [Internet] [unknown]: Harzing.com; c2013. [cited at 2013 Nov 15]. Available from: http://www.harzing.com/pop.htm. [Google Scholar]
- 8.Textalyzer [Internet] [unknown]: textalyzer.net; c2004. [cited at 2013 Nov 15]. Available from: http://textalyser.net/ [Google Scholar]
- 9.Datagle [Internet] [unknown]: Datagle LLC; [cited at 2013 Nov 15]. Available from: http://www.datagle.com/ [Google Scholar]
- 10.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007;25(11):1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.International Health Terminology Standards Development Organisation. SNOMED CT [Internet] Copenhagen, Denmark: International Health Terminology Standards Development Organisation; c2013. [cited at 2013 Nov 15]. Available from: http://www.ihtsdo.org/snomed-ct. [Google Scholar]
- 13.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32(Database issue):D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosse C, Mejino JL., Jr A reference ontology for biomedical informatics: the Foundational Model of Anatomy. J Biomed Inform. 2003;36(6):478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Smith B, Ceusters W, Klagges B, Kohler J, Kumar A, Lomax J, et al. Relations in biomedical ontologies. Genome Biol. 2005;6(5):R46. doi: 10.1186/gb-2005-6-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med. 1998;37(4-5):394–403. [PMC free article] [PubMed] [Google Scholar]
- 17.Rector AL. Clinical terminology: why is it so hard? Methods Inf Med. 1999;38(4-5):239–252. [PubMed] [Google Scholar]
- 18.Smith B. From concepts to clinical reality: an essay on the benchmarking of biomedical terminologies. J Biomed Inform. 2006;39(3):288–298. doi: 10.1016/j.jbi.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 19.Collier N, Doan S, Kawazoe A, Goodwin RM, Conway M, Tateno Y, et al. BioCaster: detecting public health rumors with a Web-based text mining system. Bioinformatics. 2008;24(24):2940–2941. doi: 10.1093/bioinformatics/btn534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gangemi A, Guarino N, Masolo C, Oltramari A, Schneider L. Knowledge engineering and knowledge management: ontologies and the Semantic Web. Heidelberg, Germany: Springer; 2002. Sweetening ontologies with DOLCE; pp. 166–181. [Google Scholar]
- 21.Brown EG, Wood L, Wood S. The medical dictionary for regulatory activities (MedDRA) Drug Saf. 1999;20(2):109–117. doi: 10.2165/00002018-199920020-00002. [DOI] [PubMed] [Google Scholar]
- 22.Stearns MQ, Price C, Spackman KA, Wang AY. SNOMED clinical terms: overview of the development process and project status. Proc AMIA Symp. 2001:662–666. [PMC free article] [PubMed] [Google Scholar]
- 23.Smith B, Kusnierczyk W, Schober D, Ceusters W. Towards a reference terminology for ontology research and development in the biomedical domain; Proceedings of KR-MED; 2006 Nov 8; Baltimore, MD. pp. 56–67. [Google Scholar]
- 24.Yu AC. Methods in biomedical ontology. J Biomed Inform. 2006;39(3):252–266. doi: 10.1016/j.jbi.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 25.Ceusters W, Smith B, Kumar A, Dhaen C. Ontology-based error detection in SNOMED-CT. Stud Health Technol Inform. 2004;107(Pt 1):482–486. [PubMed] [Google Scholar]
- 26.Sadegh-Zadeh K. Fuzzy health, illness, and disease. J Med Philos. 2000;25(5):605–638. doi: 10.1076/0360-5310(200010)25:5;1-W;FT605. [DOI] [PubMed] [Google Scholar]
- 27.Brinkman RR, Courtot M, Derom D, Fostel JM, He Y, Lord P, et al. Modeling biomedical experimental processes with OBI. J Biomed Semantics. 2010;1(Suppl 1):S7. doi: 10.1186/2041-1480-1-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferreira JD, Pesquita C, Couto FM, Silva MJ. Bringing epidemiology into the Semantic Web; Proceedings of the 3rd International Conference on Biomedical Ontology; 2012 Jul 21-25; Graz, Austria. [Google Scholar]
- 29.Porta M. A dictionary of epidemiology. 5th ed. New York (NY): Oxford University Press; 2008. [Google Scholar]
- 30.European Commission. Semantic interoperability for better health and safer healthcare: research and development roadmap for Europe. Luxembourg: European Commission; 2009. [Google Scholar]
- 31.Gillam M, Feied C, Handler J, Moody E, Shneiderman B, Plaisant C, et al. The healthcare singularity and the age of semantic medicine. In: Hey T, Tansley S, Tolle K, editors. The fourth paradigm: data-intensive scientific discovery. Redmond (WA): Microsoft Research; 2009. pp. 57–64. [Google Scholar]
- 32.Smith B. Beyond concepts: ontology as reality representation; Proceedings of the 3rd International Conference on Formal Ontology in Information Systems; 2004 Nov 4-6; Torino, Italy. pp. 73–84. [Google Scholar]
- 33.W3C OWL Working Group. OWL 2 Web ontology language document overview (second edition) [Internet] [unknown]: W3C; c2012. [cited at 2013 Nov 15]. Available from: http://www.w3.org/TR/owl2-overview/ [Google Scholar]
- 34.Klein GO, Smith B. Concept systems and ontologies: recommendations for basic terminology. Trans Jpn Soc Artif Intell. 2010;25(3):433–441. doi: 10.1527/tjsai.25.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schulz S, Jansen L. Formal ontologies in biomedical knowledge representation. Yearb Med Inform. 2013;8(1):132–146. [PubMed] [Google Scholar]
- 36.Smith B. Applied ontology: a new discipline is born. Philos Today. 1998;12(29):5–6. [Google Scholar]
- 37.Berners-Lee T, Hendler J, Lassila O. The Semantic Web. Sci Am. 2001;284(5):28–37. [Google Scholar]
- 38.Baader F, Calvanese D, McGuinness DL, Nardi D, Patel-Schneider P. The description logic handbook: theory, implementation, and applications. 2nd ed. Cambridge, UK: Cambridge University Press; 2007. [Google Scholar]
- 39.Gennari JH, Musen MA, Fergerson RW, Grosso WE, Crubezy M, Eriksson H, et al. The evolution of Protégé: an environment for knowledge-based systems development. Int J Hum Comput Stud. 2003;58(1):89–123. [Google Scholar]
- 40.Rector AL, Glowinski AJ, Nowlan WA, Rossi-Mori A. Medical-concept models and medical records: an approach based on GALEN and PEN&PAD. J Am Med Inform Assoc. 1995;2(1):19–35. doi: 10.1136/jamia.1995.95202545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.IMIA LaMB Working Group [Internet] Mountain View (CA): LinkedIn; c2013. [cited at 2013 Nov 15]. Available from: http://www.linkedin.com/groups/IMIA-Medical-Concept-Representation-Working-3680642/about. [Google Scholar]



