Abstract
Antibiotics are used for disease therapeutic or preventative effects in humans and animals, as well as for enhanced feed conversion efficiency in livestock. Antibiotics can also cause undesirable effects in microbial populations, including selection for antibiotic resistance, enhanced pathogen invasion, and stimulation of horizontal gene transfer. Carbadox is a veterinary antibiotic used in the US during the starter phase of swine production for improved feed efficiency and control of swine dysentery and bacterial swine enteritis. Carbadox has been shown in vitro to induce phage-encoded Shiga toxin in Shiga toxin-producing Escherichia coli (STEC) and a phage-like element transferring antibiotic resistance genes in Brachyspira hyodysenteriae, but the effect of carbadox on prophages in other bacteria is unknown. This study examined carbadox exposure on prophage induction and genetic transfer in Salmonella enterica serovar Typhimurium, a human foodborne pathogen that frequently colonizes swine without causing disease. S. Typhimurium LT2 exposed to carbadox induced prophage production, resulting in bacterial cell lysis and release of virions that were visible by electron microscopy. Carbadox induction of phage-mediated gene transfer was confirmed by monitoring the transduction of a sodCIII::neo cassette in the Fels-1 prophage from LT2 to a recipient Salmonella strain. Furthermore, carbadox frequently induced generalized transducing phages in multidrug-resistant phage type DT104 and DT120 isolates, resulting in the transfer of chromosomal and plasmid DNA that included antibiotic resistance genes. Our research indicates that exposure of Salmonella to carbadox induces prophages that can transfer virulence and antibiotic resistance genes to susceptible bacterial hosts. Carbadox-induced, phage-mediated gene transfer could serve as a contributing factor in bacterial evolution during animal production, with prophages being a reservoir for bacterial fitness genes in the environment.
Keywords: Salmonella, bacteriophage, antibiotic, carbadox, prophage, gene transfer, transduction
Introduction
Antibiotics are used to treat bacterial infections, prevent bacterial infections, or improve feed conversion efficiency in food-producing animals. However, antibiotics have broad and unintended, sometimes called collateral, effects on microorganisms and the microbial communities they inhabit. A microorganism's response to antibiotic exposure can be monitored by gene expression signatures that indicate the organism's physiological response to antibiotic-related stress (Brazas and Hancock, 2005). Sub-minimal inhibitory concentrations of antibiotics in particular have been shown to have the unintended effect of modulating gene expression in various bacteria (Davies et al., 2006). Transcription in the foodborne pathogen Salmonella enterica serovar Typhimurium is affected by sub-inhibitory concentrations of antibiotics of agricultural importance, such as quinolones (Yim et al., 2011) and tetracyclines (Brunelle et al., 2013). In both studies virulence genes were among those upregulated by these antibiotics, suggesting that low concentrations of certain antibiotics may promote rather than inhibit S. Typhimurium survival in the host.
In the US, salmonellae are the leading cause of bacterial foodborne morbidity and mortality for humans (Scallan et al., 2011). The prevalence of multidrug-resistant Salmonella isolates has increased over the last few decades, and outbreak investigations indicate that antimicrobial resistant Salmonella isolates are associated with an increased rate of hospitalization (Varma et al., 2005a). Furthermore, patients infected with antimicrobial resistant Salmonella have an increased frequency of bloodstream infections and longer lengths of hospitalization (Varma et al., 2005b). Acquisition and carriage of antibiotic resistance genes by Salmonella is therefore a critical factor in the degree of human morbidity and mortality caused by this pathogen.
Salmonella has acquired antibiotic resistance genes from the environment. Its primary habitat is within the microbial community of the intestinal tract, and this community, or gut microbiota, is a reservoir for antibiotic resistance genes (Salyers et al., 2004). Sub-inhibitory antibiotics promote resistance gene transfer among gut bacteria via transposons (Shoemaker et al., 2001; Song et al., 2009), plasmids (Feld et al., 2008), and phage-like gene transfer agents (GTAs) (Stanton et al., 2008). The agricultural antibiotic carbadox is frequently used in the US during the starter phase of swine production for performance enhancement and control of enteric diseases. Carbadox is an antibacterial agent used exclusively in animals. For growth promotion and disease prophylaxis, swine feed contains 10–25 g/ton [11–28 mg/kg or parts-per-million (ppm)] and 50 g/ton [55 mg/kg (ppm)], respectively. Carbadox, a quinoxaline-di-N-oxide, is mutagenic, causing base pair substitutions and frameshift mutations in DNA (Beutin et al., 1981). A range of carbadox concentrations from 0.5 to 8 μg/ml (ppm) has been shown to induce prophages in Shiga toxin-producing Escherichia coli (STEC) (Kohler et al., 2000) and GTAs in Brachyspira hyodysenteriae (Stanton et al., 2008). However, it is unknown what effect carbadox would have on prophages encoded by other bacterial species, including those native to S. Typhimurium strains.
Salmonella strains have multiple prophage genomes integrated into their chromosomes. For example, the genome of S. Typhimurium strain LT2 contains four functional prophages: Gifsy-1 and -2 and Fels-1 and -2 (McClelland et al., 2001; Casjens, 2011). Investigation of prophages in S. Typhimurium indicates that many of these prophages can be induced to produce infectious virions by various environmental signals including DNA damage, antibiotics such as mitomycin C, and hydrogen peroxide (Schicklmaier et al., 1998; Figueroa-Bossi and Bossi, 1999; Schmieger and Schicklmaier, 1999; Frye et al., 2005; Garcia-Russell et al., 2009). Furthermore, prophages often encode virulence genes that enhance the pathogenesis of the bacterial strain into which the prophage is integrated (Groman, 1955; O'Brien et al., 1984; Cheetham and Katz, 1995; Waldor and Mekalanos, 1996; Figueroa-Bossi and Bossi, 1999; Mirold et al., 1999; Figueroa-Bossi et al., 2001; Ho et al., 2002; Casjens and Hendrix, 2005).
Since Salmonella strains usually contain multiple functional prophages and frequently colonize the swine intestinal tract, the goal of the current study was to evaluate prophage induction and genetic transfer in S. Typhimurium following carbadox exposure. Our research demonstrates that carbadox induced prophage production, thereby generating infectious virions capable of transferring virulence and antibiotic resistance genes via a prophage genome or generalized transduction.
Methods
Bacterial strains, media, and chemicals
Bacterial strains (Table 1) were grown in LB (Lennox Laboratory Supplies, Dublin, Ireland) or E minimal medium containing 0.4% glucose (Vogel and Bonner, 1956). A 5 mg/ml carbadox (Sigma-Aldrich, St. Louis, MO, USA) stock solution was made in 0.1 N NaOH and used at a final concentration of 2.5 μg/ml unless noted otherwise. Other antibiotics were used at the following concentrations: ampicillin (100 μg/ml), kanamycin (50 μg/ml), tetracycline (20 μg/ml), chloramphenicol (30 μg/ml), and carbenicillin (50 μg/ml).
Table 1.
Bacterial strain list.
| Strain no. | Strain background | Genotype | Phenotype* | Source |
|---|---|---|---|---|
| LT2 (BSX 1) | S. enterica LT2 | Wild-type | John Foster | |
| UB-0020 | S. enterica LT2 | leuA414, Fels2−, r −, sup° | Miriam Susskind | |
| UB-1731 (BSX 97) | S. enterica LT2 TT23657 | Δ(Fels-2 Gifsy-1 Gifsy-2) | Kelly Hughes (Bunny et al., 2002) | |
| UB-1790 | S. enterica LT2 | leuA414, Fels2−, r −, sup° P22 UC-911 prophage | KnR | Padilla-Meier et al., 2012 |
| DB7004 | S. enterica LT2 | leuA414, supE | Winston et al., 1979 | |
| ATTC 14028s | S. enterica | Wild-type | Lionello Bossi (Figueroa-Bossi et al., 1997) | |
| UK1 | S. enterica UK1 | Wild-type | John Foster | |
| SL1344 | S. enterica SL1344 | Wild-type | StrR | John Foster |
| χ4232 (BSX 8) | S. enterica χ4232 | Wild-type | NalR | Tom Stabel |
| NCTC13348 | S. enterica DT104 | Wild-type | ApR, CamR, TetR, StrR, SuR, SpR | Public Health England |
| DT104-530 | S. enterica DT104 | Wild-type | ApR, CamR, TetR, StrR | This study |
| DT104-745 | S. enterica DT104 | Wild-type | ApR, CamR, TetR, StrR, KnR | This study |
| DT104b-5414 | S. enterica DT104 | Wild-type | ApR, CamR, TetR, StrR | This study |
| DT120-150 | S. enterica DT120 | Wild-type | ApR, CamR, TetR, StrR | This study |
| DT120-305 | S. enterica DT120 | Wild-type | ApR, CamR, TetR, StrR | This study |
| DT120-613 | S. enterica DT120 | Wild-type | ApR, CamR, TetR, StrR | This study |
| DT120-7055 | S. enterica DT120 | Wild-type | This study | |
| DT193-1434 | S. enterica DT193 | Wild-type | ApR, CamR, TetR, StrR, KnR, NalR | This study |
| DT208-2348 | S. enterica DT208 | Wild-type | ApR, CamR, TetR, NalR | This study |
| U302-4715 | S. enterica U302 | Wild-type | ApR, CamR, TetR, | This study |
| BSX 7 | S. enterica TT22971 (LT2) | metA22 metE551 trpD2 ilv-452 leu pto (leaky) hsdLT6 hsdSA29 hsdB strA120/pKD46 | ApR, 30°C | John Roth via Max Wu |
| BBS 119 | S. enterica LT2 | metA22 metE551 trpD2 ilv-452 leu pto (leaky) hsdLT6 hsdSA29 hsdB strA120 | BSX 7 cured of pKD46 | |
| BBS 120 | S. enterica LT2 | metA22 metE551 trpD2 ilv-452 leu pto (leaky) hsdLT6 hsdSA29 hsdB strA120/pCP20 | ApR, 30°C | BBS 119/pCP20 |
| BBS 231 | S. enterica LT2 | metA22 metE551 trpD2 ilv-452 leu pto (leaky) hsdLT6 hsdSA29 hsdB strA120 hisDCBHA::neo | KnR | BSX 7/oBBI 197/198 (hisD-A::neo) |
| BBS 233 | S. enterica χ4232 | hisDCBHA::neo | NalR, KnR | BSX 8 × HT BBS231 |
| BBS 243 | S. enterica χ4232 | ΔhisDCBHA | NalR | BBS 233 × HT BBS 120 |
| BBS 561 | S. enterica LT2 | metA22 metE551 trpD2 ilv-452 leu pto (leaky) hsdLT6 hsdSA29 hsdB strA120 sodCIII::neo | KnR | BSX 7/oBBI 300/301 neo |
| BBS 565 | S. enterica LT2 | sodCIII::neo | KnR | BSX 1 × HT BBS 561 |
| BBS 649 | S. enterica DT104-745 | ΔfloR Δtet Δpse-1 | KnR | |
| BBS 651 | S. enterica DT104-745 | ΔfloR Δtet Δpse-1/pKD46 | KnR, ApR, 30°C | BBS 649/pKD46 |
| BBS 998 | S. enterica UB-1731 | sodCIII::neo | KnR | BSX 97 × HT BBS 561 |
| BBS 1004 | S. enterica LT2 | metA22 metE551 trpD2 ilv-452 leu pto (leaky) hsdLT6 hsdSA29 hsdB strA120 Fels-1::neo | KnR | BSX 7/oBBI 302/439 neo |
| BBS 1008 | S. enterica LT2 | Δ(Fels-2 Gifsy-1 Gifsy-2) Fels-1::neo | KnR | BSX 97 × HT BBS 1004 |
| BBS 1010 | S. enterica DT104-745 | ΔfloR Δtet Δpse-1 hisDCBHA::cat | KnR, CamR | BBS 651/oBBI 197/198 cat |
| BBS 1012 | S. enterica DT104-745 | ΔfloR Δtet Δpse-1 ΔhisDCBHA | KnR | BBS 1010/pCP20 |
| UB-1703 | E. coli K-12 | λ prophage | Roger Hendrix (Hendrix and Duda, 1992) | |
| UB-1704 | E. coli K-12 | HK97 prophage | Roger Hendrix | |
| 594 | E. coli K-12 594 | StrR | Weigle, 1966 | |
| UB-1458 | S. flexneri PE577 | Wild-type | Renato Morona (Casjens et al., 2004) | |
| UB-1496 | S. flexneri PE577 | Sf6 prophage | Renato Morona |
Known antibiotic resistance phenotypes. Ap, Ampicillin; Cam, Chloramphenicol; Tet Tetracycline; Str, Streptomycin; Su, Sulfamethoxazole; Sp, Spectinomycin; Kn, Kanamycin; Nal, Nalidixic acid.
S. Typhimurium gene and prophage knockouts by recombineering
Oligonucleotide primers for PCR amplification and construction of gene and prophage knockouts are listed in Table 2. S. Typhimurium gene and prophage knockouts were constructed by recombineering (recombination-mediated genetic engineering) as previously described (Bearson and Bearson, 2008; Bearson et al., 2008). Briefly, the 5' end of a gene knockout primer (bold, Table 2) has homology to 32–44 bp of the target gene whereas the 3' end contains universal sequences (underlined) to amplify an antibiotic resistance gene and truncate potential translation of the target gene. A gene knockout primer set was used to PCR amplify either the neo or the cat gene. Gel electrophoresis was performed on the amplification product of a knockout fragment and the respective DNA fragment was gel extracted using a Freeze'n Squeeze column (Bio-Rad, Hercules, CA). Each knockout fragment was transformed (Sambrook and Russell, 2006) into an arabinose-induced S. Typhimurium strain containing the pKD46 plasmid (Datsenko and Wanner, 2000). Transformants containing the knockout were selected on LB agar medium containing kanamycin. If necessary, the gene knockout with the neo marker was moved to another strain background by transduction using a P22 phage with a high transduction frequency. Flp mediated deletion of the neo or cat gene was performed by transferring the pCP20 plasmid into the knockout strain by either transduction or transformation followed by a procedure to screen for loss of resistance to kanamycin (Cherepanov and Wackernagel, 1995).
Table 2.
Primers used in this study.
| Gene/phage target | Primer | Sequence (5'–3') | References |
|---|---|---|---|
| hisD-A | oBBI 197 | ctgatggcgctgcgcttatcaggcctacgtaatgcatagagcagtgacgtagtcgc | This study |
| oBBI 198 | cgttttgccagcattggatggcctccttaacgatagctgaatgagtgacgtgc | ||
| sodCIII | oBBI 300 | gttaaccttgtaaatgccaatggcacaggtcaaaagatcgatagctgaatgagtgacgtgc | This study |
| oBBI 301 | gtggaacagtgcctcacagagtgaattttattttataacgcatagagcagtgacgtagtcgc | ||
| Fels-1 | oBBI 302 | cattcattaaggaaggaaagagtatgactgtagaaaaatccgatagctgaatgagtgacgtgc | This study |
| oBBI 439 | cataaccacttaacatcttgttttatctaaataaaattaagcatagagcagtgacgtagtcgc | ||
| P22-like | ST104Gp1F | gacgcccgtcactgcacagtta | This study |
| ST104Gp1R | acccggcgacgcttaatctg |
Bold text indicates homology to the target gene/phage.
Underlined text indicates sequence for neo or cat amplification.
Determination of phage titers following carbadox induction of bacterial strains lysogenized with the model phages P22, λ, HK97, and SF6
Bacterial strains lysogenized with P22 (UB-1790), λ (UB-1703), HK97 (UB-1704), and Sf6 (UB-1496) were grown in LB broth at 37°C with shaking. At a density of 1 × 108 bacterial cells/ml, carbadox was added to a final concentration of 0.5 μg/ml (ppm). At the indicated times after carbadox addition, aliquots of the cultures were removed, shaken with several drops of chloroform to complete lysis, and titered on the permissive host. Strains used to titer the phage lysates were DB7004 (P22), 594 (λ and HK97), and UB-1458 (Sf6).
Carbadox induction of wild-type S. Typhimurium
S. Typhimurium strains were grown in LB 0.4% glucose at 37°C with shaking at 180 rpm. Carbadox was added to cultures at OD600 = 0.2 at a final concentration of 2.5 μg/ml (ppm). Incubation of cultures was continued to monitor for bacterial cellular lysis.
Phage transduction using carbadox-induced S. Typhimurium lysates
Supernatants from non-induced and carbadox-induced bacterial cultures were harvested by adding 100 μl of chloroform, vortexing gently, and allowing the culture to incubate for ~15 min with shaking. The cultures were centrifuged at 1000× g for 20 min and the supernatant was transferred to a fresh tube containing 200 μl of chloroform for storage. Bacterial lysates were plated to LB to ensure that viable bacterial cells were not present. An overnight culture of the transduction recipient was grown in LB or LB glucose at 37°C with shaking. The transduction was performed with equal volumes of both the S. Typhimurium recipient strain and either the non-induced or the carbadox-induced culture supernatant. The transduction was incubated at 37°C for ~1–3 h before plating on the appropriate selective medium. To determine the transduction frequency for transfer of the histidine operon, the transduction was plated onto E glucose minimal medium. Transduction frequency for antibiotic resistance gene transfer (sodCIII:: neo in Fels-1, plasmid-encoded kanamycin, and chromosomally encoded tetracycline) was determined by plating the transduction to LB containing either kanamycin or tetracycline.
Phage purification for electron microscopy
Overnight cultures were diluted 1:100 in 400 ml E glucose minimal medium and incubated at 37°C with shaking. At OD600 = 0.5, carbadox was added to a final concentration of 2.5 μg/ml, and incubation continued until lysis was achieved. Phages were purified and visualized by electron microscopy as described previously (Humphrey et al., 1997) with the following modifications. Purified phages were negatively stained by mixing phage samples with an equal volume of phosphotungstic acid (2%, pH 7.0). Samples were deposited on Formvar-coated 200-mesh carbon-reinforced copper grids (Electron Microscopy Sciences, Hatfield, PA) and viewed with a FEI Tecnai G2 BioTWIN electron microscope (80 kV; Hillsboro, OR).
Results and discussion
Carbadox induces S. Typhimurium and other enterobacteriaceae prophages to cause phage replication and cell lysis
To test whether well-characterized prophages in S. enterica and other Enterobacteriaceae species are induced by carbadox, we monitored the infectious phages produced by carbadox-treated cultures of bacterial strains that were lysogenized by the “model system” phages P22 (S. enterica), λ and HK97 (E. coli), and Sf6 (Shigella flexneri). In each case, at least partial clearing of the culture indicated a substantial fraction of the cells in the culture had lysed by about 2 h. All four prophages gave an approximately three-log increase in free phage in the culture (Figure 1), with about 10–200 progeny phage produced per bacterium that were initially present at the time of carbadox addition. Several concentrations of carbadox were tested, and 0.5 μg/ml (ppm) is shown as the minimum that gave good induction of P22. Carbadox induces the P22 Salmonella prophage as well as E. coli and S. flexneri prophages under these conditions, and this is not surprising given its apparent mechanistic similarities to the action of mitomycin C. Carbadox is a DNA damaging agent, and DNA damage induced by mitomycin C is known to induce the bacterial SOS pathway, which induces prophages. It is likely capable of induction of prophages from many if not all Enterobacteriaceae bacterial species, as well as more distantly related bacterial phyla.
Figure 1.
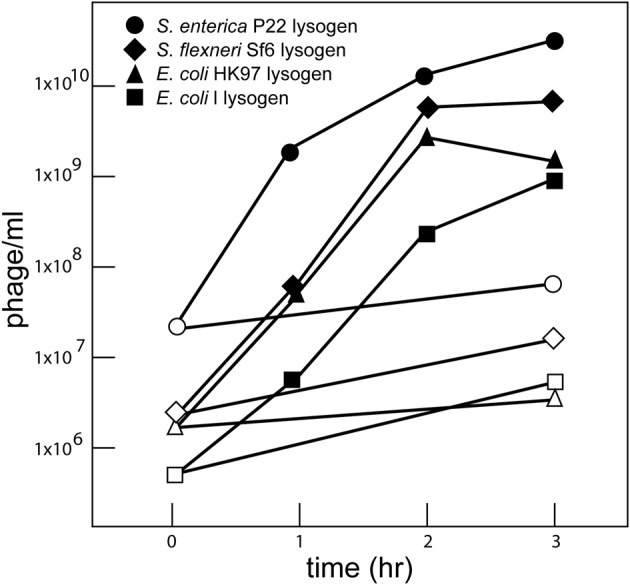
Carbadox induction of Enterobacteriaeae prophages. Bacterial strains lysogenized with P22 (UB-1790), λ (UB-1703), HK97 (UB-1704), and Sf6 (UB-1496) were grown in LB broth at 37°C with shaking. At a density of 1 × 108 bacterial cells/ml, carbadox was added to a final concentration of 0.5 μg/ml. Phage lysates were obtained by shaking with chloroform at the indicated times after carbadox addition and titered on a permissive host. Open symbols indicate cultures with no added carbadox and closed symbols indicate cultures with carbadox added. The different induced lysogens are indicated in the figure insert.
To examine prophage induction by carbadox in wild-type S. Typhimurium isolates containing their natural prophages, we initially examined S. Typhimurium LT2, a strain that is widely used in the study of Salmonella genetics in the laboratory (Lilleengen, 1948). Cultures of wild-type LT2 in early log phase growth were exposed to carbadox. The bacterial density of the culture abruptly decreased at ~2 h following exposure to 2.5 μg/ml (ppm) of carbadox (Figure 2). Mitomycin C exposure is known to result in the induction of prophage Fels-1 from strain LT2 (Garcia-Russell et al., 2009). The fact that carbadox induces wild-type phage λ (above), which is only known to be induced by the SOS function of activated RecA protein (Little, 1993), strongly supports this mechanism for carbadox-mediated induction. Thus, the decrease in LT2 cell density is almost certainly due to bacterial cell lysis resulting from prophage induction.
Figure 2.
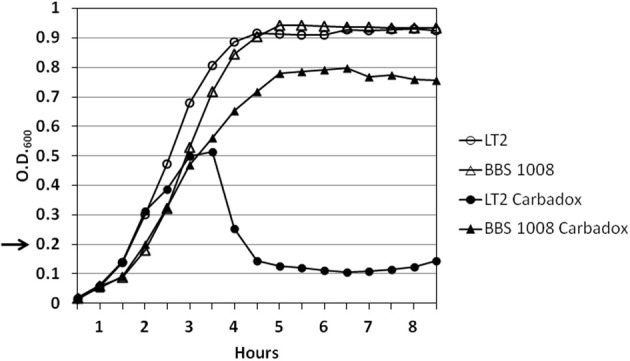
Carbadox exposure of wild-type S. Typhimurium LT2 results in bacterial cell lysis. S. Typhimurium strains (LT2 and BBS 1008) were grown in LB glucose medium at 37°C with shaking. At OD600 = 0.2 (arrow), carbadox (2.5 μg/ml) was added to cultures indicated by the closed symbols. The open symbols indicate control cultures without carbadox.
LT2 is known to carry four prophages. Amplification of the integrase gene from DNA extracted from phage heads indicated that at least one of these, the Fels-1 prophage, was induced following carbadox exposure (Stanton and Humphrey, unpublished data). The BBS 1008 strain is an LT2 derivative from which all four prophages have been deleted. Following exposure of BBS 1008 to 2.5 μg/ml of carbadox, the bacterial culture density did not decrease (Figure 2), indicating that these prophages are responsible for the bacterial cell lysis phenotype induced by carbadox exposure.
Examination of the purified phages by electron microscopy demonstrated the presence of mostly empty phage heads in the carbadox-induced culture of LT2 (data not shown). Treatment of S. Typhimurium LT2 and other strains with mitomycin C is known to induce Fels-1 plaque-forming phages with poor efficiency (Figueroa-Bossi and Bossi, 1999), so it is perhaps not surprising that whole phage particles were not seen. Nonetheless, phage heads were either greatly reduced or were not observed in the absence of carbadox for the LT2 strain and in the presence of carbadox for BBS 1008. These results confirm prophage induction in a natural wild-type S. Typhimurium isolate following carbadox exposure.
Carbadox exposure of S. Typhimurium LT2 promotes phage transduction into a susceptible bacterial host
To monitor phage transduction frequency, a neo gene was inserted by recombineering into the putative virulence factor sodCIII on the Fels-1 prophage of strain LT2 to create strain S. Typhimurium BBS 565. The carbadox-induced phage lysate from BBS 565 (LT2 sodCIII::neo) efficiently transduced the kanamycin-sensitive strain BBS 243, as demonstrated by 8 × 103 kanamycin-resistant transductants per ml of lysate. In the absence of carbadox induction, BBS 243 remained susceptible to kanamycin following transduction with the control supernatant from BBS 565. This indicates that carbadox-induced prophages can carry genetic material from a donor into a recipient bacterial strain.
Carbadox-induction of multidrug-resistant S. Typhimurium DT104 results in generalized transduction of chromosomal and plasmid DNA
Generalized transduction involves the packaging of random host DNA (genomic or plasmid) into a bacteriophage particle and the transfer of that DNA to a recipient strain. Bacteriophage P22 is a generalized transducing phage that is commonly used for genetic experiments with Salmonella (Zinder and Lederberg, 1952; Kropinski et al., 2007). Although S. Typhimurium LT2 contains multiple prophages, it does not contain a P22-like prophage that performs generalized transduction. Some multidrug-resistant S. Typhimurium isolates like phage type DT104 harbor a P22-like prophage. This prophage has been described as PDT17, ST104, and prophage 1 (Schmieger and Schicklmaier, 1999; Tanaka et al., 2004; Cooke et al., 2008). To determine whether carbadox exposure could induce generalized transduction, S. Typhimurium DT104 NCTC13348 was exposed to carbadox. The culture lysed, and the phage-containing supernatant was harvested. The BBS 243 strain, a histidine auxotroph lacking the genes hisDCBHA, could be successfully transduced to his+ with this lysate. Generalized transduction was observed by growth on minimal medium lacking histidine, demonstrating the transfer of the his operon from the his+ donor (DT104) to the his− recipient (BBS 243) (Table 3). The results show that carbadox exposure of multidrug-resistant S. Typhimurium DT104 can stimulate generalized transduction and therefore chromosomal gene transfer.
Table 3.
Average frequency of generalized transduction per 0.5 ml of lysate from numerous S. Typhimurium donor strains into BBS 243.
| Salmonella donor strain | Not induced | Carbadox induced ± s.e.m. |
|---|---|---|
| LT2 | 0 | 0 |
| UB-1731 | 0 | 0 |
| ATTC 14028s | 0 | 0 |
| UK1 | 0 | 0 |
| SL1344 | 0 | 0 |
| χ4232 | 0 | 0 |
| DT104 (NCTC13348) | 0 | 6 ± 1.7 |
| DT104-530 | 0 | 291 ± 100.8 |
| DT104b-5414 | <1 | 117 ± 52.8 |
| DT120-150 | 0 | 124 ± 19.3 |
| DT120-305 | <1 | 420 ± 94.8 |
| DT120-613 | <1 | 86 ± 16.7 |
| DT120-7055 | 0 | 0 |
| DT193-1434 | 0 | 0 |
| DT208-2348 | 0 | 0 |
| U302-4715 | 0 | 0 |
The Salmonella genomic island-1 (SGI-1) is ~43 kb and typically contains chromosomally-encoded resistance genes for multiple antibiotics including ampicillin, chloramphenicol, and tetracycline within an integron (Boyd et al., 2001; Mulvey et al., 2006). We attempted to transduce SGI-1 but were unsuccessful. Due to the size of SGI-1, transduction of this entire island by P22 into another Salmonella strain that does not already contain a segment of SGI-1 is inefficient [P22 packages about 43.4 Kbp of DNA (Kropinski et al., 2007)]. To overcome this experimental limitation, we utilized a DT104 derivative (BBS 1012) as our recipient strain. The BBS 1012 strain is a histidine auxotroph that has an internal deletion within SGI-1, resulting in sensitivity to ampicillin, chloramphenicol, and tetracycline due to the loss of multiple antibiotic resistance genes. In addition, BBS 1012 contains a natural plasmid that confers kanamycin resistance. Transduction of BBS 1012 with the carbadox-induced phage lysate from wild-type S. Typhimurium DT104-530 (kanamycin sensitive) resulted in the transfer of tetracycline resistance following selection on tetracycline-containing LB agar medium. Transduction into the BBS 1012 strain was confirmed by growth on medium containing kanamycin and the absence of growth on minimal medium without histidine. Furthermore, the initial selection of BBS 1012 on LB medium containing tetracycline resulted in 100% co-transduction of resistance to both chloramphenicol and carbenicillin. The floR and pse-1 genes encode resistance to chloramphenicol and carbenicillin, respectively, and these genes are located adjacent to tetG on SGI-1. These results indicate that exposing multidrug-resistant S. Typhimurium DT104 to carbadox can promote the transfer of numerous genes co-located within SGI-1 that encode resistance to multiple classes of antibiotics. Thus generalized transduction could participate in bacterial strain evolution by providing an assortment of antibiotic resistance genes for recombination within this important genomic region.
Although the SGI-1 integron in S. Typhimurium DT104 encodes multiple antibiotic resistance genes, some strains contain additional antibiotic resistance genes encoded on plasmids. Transduction of BBS 243 (kanamycin sensitive) with the carbadox-induced phage lysate from S. Typhimurium DT104 (745) resulted in bacterial growth on LB medium containing kanamycin, demonstrating the transfer of the plasmid encoding kanamycin resistance. Thus, carbadox exposure promoted generalized transduction of this natural plasmid as well as chromosomally-encoded antibiotic resistance genes in multidrug-resistant S. Typhimurium DT104.
Carbadox-induced gene transfer is a general phenomenon that occurs in multidrug-resistant S. Typhimurium strains DT120 and DT104
Since the prevalence of multidrug-resistant S. Typhimurium strains has increased over the last few decades, we wanted to determine whether carbadox-induced gene transfer is unique to S. Typhimurium DT104 or is a general property of multidrug-resistant S. Typhimurium strains. Phage lysates were harvested from several S. Typhimurium phage types following carbadox exposure and used to transduce the recipient strain BBS 243 (ΔhisDCBHA) with selection on E glucose minimal medium; several different phage types were investigated as these, by definition (i.e., DT), should have varying prophage content. Carbadox-induced phage lysates from several S. Typhimurium DT104 and DT120 isolates resulted in growth on minimal medium, indicating the transfer of the his operon to BBS 243 (Table 3). The results suggest that generalized transduction following carbadox induction is a common phenomenon for multidrug-resistant S. Typhimurium DT104 and DT120.
We PCR amplified the P22 gene 1 (encoding gp1/portal protein) from several of the DT104 and DT120 isolates that we have shown are capable of generalized transduction, suggesting that these multidrug-resistant isolates contain a P22-like prophage. To confirm that a phage capable of generalized transduction is required for carbadox-induced gene transfer, the P22-like prophage (prophage 1) was deleted from DT104 using recombineering. Gene transfer into BBS 243 was eliminated following transduction with a carbadox-induced phage lysate from the DT104 P22-like prophage knockout strain, indicating that the P22-like prophage is responsible for the generalized transduction from S. Typhimurium DT104. In support of this, the S. Typhimurium strains LT2, UK1, SL1344, and χ4232 (frequently investigated strains in the literature) are incapable of generalized transduction and do not contain a P22-like prophage. In contrast, genome scanning has identified integrated P22-like prophages in the genome sequences of isolates of S. enterica serovars Arizonae, Cholerae, Dublin, Hadar, Heidelberg, Houtenae, Johannesburg, Mississippi, Montevideo, Newport, Paratyphi, Rubislaw, Schwarzengrund, Tennessee, Typhimurium, Uganda, Wandsworth, and Welteverden; this suggests that P22-like prophages are common among Salmonella serovars and the potential for strain evolution due to generalized transduction is perhaps underappreciated. The capability of generalized transduction among Salmonella serovars is reinforced with the knowledge that P22 phage lysates are known to be stable for many years in the laboratory. Likewise, an ecological significance of phages in the environment is that DNA encapsulated within a phage head is protected from nucleases and therefore can survive outside of a bacterial host until encountering a recipient.
Swine environments, including swine manure, have been shown to contain abundant phage populations (McLaughlin et al., 2006; Wang et al., 2010). Bacteriophage populations present in manure could be derived principally from prophage induction of bacteria present in manure or a combination of induction from within the swine gastrointestinal tract and in manure. Prophage induction can be stimulated by various environmental signals and stresses including ultraviolet light, hydrogen peroxide, mitomycin C, and carbadox. Analysis of fecal phage metagenomes from medicated swine administered in-feed antibiotics [carbadox or ASP250 (chlortetracycline, sulfamethazine, and penicillin)] compared to non-medicated swine suggested that prophages were induced with antibiotic treatment (Allen et al., 2011). Similar work with mouse fecal phage metagenomes has shown that antibiotic treatment caused an increase in the abundance of phage-encoded antibiotic resistance genes (Modi et al., 2013). This suggests that antibiotic-induced phage-mediated transduction may contribute to antibiotic resistance gene transfer during animal production. Relatively little information is available concerning the extent of carbadox-induced prophage from bacteria, as Salmonella is only the third bacterial genus for which this response has been described (Kohler et al., 2000; Stanton et al., 2008). Additional information is needed to understand the capacity for carbadox to induce prophages during swine production since there is a potential for dissemination into the environment following manure application onto agricultural soils.
Conclusions
Prophages are a potential environmental reservoir for bacterial fitness genes and may drive the emergence of new epidemic clones (Brussow et al., 2004). Prophages integrated in the genomes of Salmonella strains can encode genes associated with virulence or antimicrobial resistance (Figueroa-Bossi and Bossi, 1999; Figueroa-Bossi et al., 2001; Moreno Switt et al., 2013). Therefore, the pathogenic potential of a particular Salmonella strain depends in part upon the prophage repertoire integrated into the bacterial genome, and acquisition of prophages could conceivably result in enhanced bacterial virulence or survival during host colonization. In this report, we demonstrate that exposure of several different S. Typhimurium isolates to the agricultural antibiotic carbadox resulted in the production of transducing particles capable of transferring the individual phage genome as well as chromosomal and plasmid DNA by generalized transduction.
Author contributions
Conceived and designed experiments: Bradley L. Bearson, Heather K. Allen, Sherwood R. Casjens, Thaddeus B. Stanton, and Brian W. Brunelle. Performed the experiments: Bradley L. Bearson, Heather K. Allen, Sherwood R. Casjens, In S. Lee, Brian W. Brunelle, and Thaddeus B. Stanton. Wrote and edited the manuscript: Bradley L. Bearson, Heather K. Allen, Sherwood R. Casjens, Brian W. Brunelle, and Thaddeus B. Stanton.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are indebted to Stephanie Jones, Kellie Winter, Samuel Humphrey, and Briony Atkinson for outstanding technical assistance, to Judi Stasko for excellence with electron microscopy, to Kassandra Wilson and Eddie Gilcrease for measurement of phage titers, and to Kelly Hughes, Roger Hendrix, and Renato Morona for the gift of bacterial strains. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. USDA is an equal opportunity provider and employer.
References
- Allen H. K., Looft T., Bayles D. O., Humphrey S., Levine U. Y., Alt D., et al. (2011). Antibiotics in feed induce prophages in swine fecal microbiomes. MBio 2, 1–9 10.1128/mBio.00260-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearson B. L., Bearson S. M. (2008). The role of the QseC quorum-sensing sensor kinase in colonization and norepinephrine-enhanced motility of Salmonella enterica serovar Typhimurium. Microb. Pathog. 44, 271–278 10.1016/j.micpath.2007.10.001 [DOI] [PubMed] [Google Scholar]
- Bearson B. L., Bearson S. M., Uthe J. J., Dowd S. E., Houghton J. O., Lee I., et al. (2008). Iron regulated genes of Salmonella enterica serovar Typhimurium in response to norepinephrine and the requirement of fepDGC for norepinephrine-enhanced growth. Microbes Infect. 10, 807–816 10.1016/j.micinf.2008.04.011 [DOI] [PubMed] [Google Scholar]
- Beutin L., Preller E., Kowalski B. (1981). Mutagenicity of quindoxin, its metabolites, and two substituted quinoxaline-di-N-oxides. Antimicrob. Agents Chemother. 20, 336–343 10.1128/AAC.20.3.336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd D., Peters G. A., Cloeckaert A., Boumedine K. S., Chaslus-Dancla E., Imberechts H., et al. (2001). Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183, 5725–5732 10.1128/JB.183.19.5725-5732.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazas M. D., Hancock R. E. (2005). Using microarray gene signatures to elucidate mechanisms of antibiotic action and resistance. Drug Discov. Today 10, 1245–1252 10.1016/S1359-6446(05)03566-X [DOI] [PubMed] [Google Scholar]
- Brunelle B. W., Bearson S. M., Bearson B. L. (2013). Tetracycline accelerates the temporally-regulated invasion response in specific isolates of multidrug-resistant Salmonella enterica serovar Typhimurium. BMC Microbiol. 13:202 10.1186/1471-2180-13-202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussow H., Canchaya C., Hardt W. D. (2004). Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68, 560–602, table of contents. 10.1128/MMBR.68.3.560-602.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunny K., Liu J., Roth J. (2002). Phenotypes of lexA mutations in Salmonella enterica: evidence for a lethal lexA null phenotype due to the Fels-2 prophage. J. Bacteriol. 184, 6235–6249 10.1128/JB.184.22.6235-6249.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S. (2011). A plethora of putative phages and prophages, in The Lure of Bacterial Genetics: A Tribute to John Roth, eds Maloy S., Hughes K., Casadesus J. (Washington, DC: ASM Press; ), 291–306 [Google Scholar]
- Casjens S., Hendrix R. W. (2005). Bacteriophages and the bacterial genome, in The Bacterial Chromosome, ed Higgins N. P. (Washington, DC: ASM Press; ), 39–52 [Google Scholar]
- Casjens S., Winn-Stapley D. A., Gilcrease E. B., Morona R., Kuhlewein C., Chua J. E., et al. (2004). The chromosome of Shigella flexneri bacteriophage Sf6: complete nucleotide sequence, genetic mosaicism, and DNA packaging. J. Mol. Biol. 339, 379–394 10.1016/j.jmb.2004.03.068 [DOI] [PubMed] [Google Scholar]
- Cheetham B. F., Katz M. E. (1995). A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol. Microbiol. 18, 201–208 10.1111/j.1365-2958.1995.mmi_18020201.x [DOI] [PubMed] [Google Scholar]
- Cherepanov P. P., Wackernagel W. (1995). Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14 10.1016/0378-1119(95)00193-A [DOI] [PubMed] [Google Scholar]
- Cooke F. J., Brown D. J., Fookes M., Pickard D., Ivens A., Wain J., et al. (2008). Characterization of the genomes of a diverse collection of Salmonella enterica serovar Typhimurium definitive phage type 104. J. Bacteriol. 190, 8155–8162 10.1128/JB.00636-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A., Wanner B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J., Spiegelman G. B., Yim G. (2006). The world of subinhibitory antibiotic concentrations. Curr. Opin. Microbiol. 9, 445–453 10.1016/j.mib.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Feld L., Schjorring S., Hammer K., Licht T. R., Danielsen M., Krogfelt K., et al. (2008). Selective pressure affects transfer and establishment of a Lactobacillus plantarum resistance plasmid in the gastrointestinal environment. J. Antimicrob. Chemother. 61, 845–852 10.1093/jac/dkn033 [DOI] [PubMed] [Google Scholar]
- Figueroa-Bossi N., Bossi L. (1999). Inducible prophages contribute to Salmonella virulence in mice. Mol. Microbiol. 33, 167–176 10.1046/j.1365-2958.1999.01461.x [DOI] [PubMed] [Google Scholar]
- Figueroa-Bossi N., Coissac E., Netter P., Bossi L. (1997). Unsuspected prophage-like elements in Salmonella typhimurium. Mol. Microbiol. 25, 161–173 10.1046/j.1365-2958.1997.4451807.x [DOI] [PubMed] [Google Scholar]
- Figueroa-Bossi N., Uzzau S., Maloriol D., Bossi L. (2001). Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol. Microbiol. 39, 260–271 10.1046/j.1365-2958.2001.02234.x [DOI] [PubMed] [Google Scholar]
- Frye J. G., Porwollik S., Blackmer F., Cheng P., McClelland M. (2005). Host gene expression changes and DNA amplification during temperate phage induction. J. Bacteriol. 187, 1485–1492 10.1128/JB.187.4.1485-1492.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Russell N., Elrod B., Dominguez K. (2009). Stress-induced prophage DNA replication in Salmonella enterica serovar Typhimurium. Infect. Genet. Evol. 9, 889–895 10.1016/j.meegid.2009.05.017 [DOI] [PubMed] [Google Scholar]
- Groman N. B. (1955). Evidence for the active role of bacteriophage in the conversion of nontoxigenic Corynebacterium diphtheriae to toxin production. J. Bacteriol. 69, 9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix R. W., Duda R. L. (1992). Bacteriophage lambda PaPa: not the mother of all lambda phages. Science 258, 1145–1148 10.1126/science.1439823 [DOI] [PubMed] [Google Scholar]
- Ho T. D., Figueroa-Bossi N., Wang M., Uzzau S., Bossi L., Slauch J. M. (2002). Identification of GtgE, a novel virulence factor encoded on the Gifsy-2 bacteriophage of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184, 5234–5239 10.1128/JB.184.19.5234-5239.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey S. B., Stanton T. B., Jensen N. S., Zuerner R. L. (1997). Purification and characterization of VSH-1, a generalized transducing bacteriophage of Serpulina hyodysenteriae. J. Bacteriol. 179, 323–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler B., Karch H., Schmidt H. (2000). Antibacterials that are used as growth promoters in animal husbandry can affect the release of Shiga-toxin-2-converting bacteriophages and Shiga toxin 2 from Escherichia coli strains. Microbiology 146, 1085–1090 [DOI] [PubMed] [Google Scholar]
- Kropinski A. M., Sulakvelidze A., Konczy P., Poppe C. (2007). Salmonella phages and prophages–genomics and practical aspects. Methods Mol. Biol. 394, 133–175 10.1007/978-1-59745-512-1_9 [DOI] [PubMed] [Google Scholar]
- Lilleengen K. (1948). Typing Salmonella typhimurium by means of bacteriophage. Acta Pathol. Microbiol. Scand. Suppl. 77, 11–25 [Google Scholar]
- Little J. W. (1993). LexA cleavage and other self-processing reactions. J. Bacteriol. 175, 4943–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Sanderson K. E., Spieth J., Clifton S. W., Latreille P., Courtney L., et al. (2001). Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413, 852–856 10.1038/35101614 [DOI] [PubMed] [Google Scholar]
- McLaughlin M. R., Balaa M. F., Sims J., King R. (2006). Isolation of Salmonella bacteriophages from swine effluent lagoons. J. Environ. Qual. 35, 522–528 10.2134/jeq2005.0080 [DOI] [PubMed] [Google Scholar]
- Mirold S., Rabsch W., Rohde M., Stender S., Tschape H., Russmann H., et al. (1999). Isolation of a temperate bacteriophage encoding the type III effector protein SopE from an epidemic Salmonella typhimurium strain. Proc. Natl. Acad. Sci. U.S.A. 96, 9845–9850 10.1073/pnas.96.17.9845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi S. R., Lee H. H., Spina C. S., Collins J. J. (2013). Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 499, 219–222 10.1038/nature12212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno Switt A. I., Orsi R. H., den Bakker H. C., Vongkamjan K., Altier C., Wiedmann M. (2013). Genomic characterization provides new insight into Salmonella phage diversity. BMC Genomics 14:481 10.1186/1471-2164-14-481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey M. R., Boyd D. A., Olson A. B., Doublet B., Cloeckaert A. (2006). The genetics of Salmonella genomic island 1. Microbes Infect. 8, 1915–1922 10.1016/j.micinf.2005.12.028 [DOI] [PubMed] [Google Scholar]
- O'Brien A. D., Newland J. W., Miller S. F., Holmes R. K., Smith H. W., Formal S. B. (1984). Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226, 694–696 10.1126/science.6387911 [DOI] [PubMed] [Google Scholar]
- Padilla-Meier G. P., Gilcrease E. B., Weigele P. R., Cortines J. R., Siegel M., Leavitt J. C., et al. (2012). Unraveling the role of the C-terminal helix turn helix of the coat-binding domain of bacteriophage P22 scaffolding protein. J. Biol. Chem. 287, 33766–33780 10.1074/jbc.M112.393132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers A. A., Gupta A., Wang Y. (2004). Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 12, 412–416 10.1016/j.tim.2004.07.004 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W. (2006). Transformation of E. coli by Electroporation. CSH Protoc. 2006 10.1101/pdb.prot3933 [DOI] [PubMed] [Google Scholar]
- Scallan E., Hoekstra R. M., Angulo F. J., Tauxe R. V., Widdowson M. A., Roy S. L., et al. (2011). Foodborne illness acquired in the United States–major pathogens. Emerg. Infect. Dis. 17, 7–15 10.3201/eid1701.091101p1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schicklmaier P., Moser E., Wieland T., Rabsch W., Schmieger H. (1998). A comparative study on the frequency of prophages among natural isolates of Salmonella and Escherichia coli with emphasis on generalized transducers. Antonie Van Leeuwenhoek 73, 49–54 10.1023/A:1000748505550 [DOI] [PubMed] [Google Scholar]
- Schmieger H., Schicklmaier P. (1999). Transduction of multiple drug resistance of Salmonella enterica serovar typhimurium DT104. FEMS Microbiol. Lett. 170, 251–256 10.1111/j.1574-6968.1999.tb13381.x [DOI] [PubMed] [Google Scholar]
- Shoemaker N. B., Vlamakis H., Hayes K., Salyers A. A. (2001). Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl. Environ. Microbiol. 67, 561–568 10.1128/AEM.67.2.561-568.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B., Wang G. R., Shoemaker N. B., Salyers A. A. (2009). An unexpected effect of tetracycline concentration: growth phase-associated excision of the Bacteroides mobilizable transposon NBU1. J. Bacteriol. 191, 1078–1082 10.1128/JB.00637-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton T. B., Humphrey S. B., Sharma V. K., Zuerner R. L. (2008). Collateral effects of antibiotics: carbadox and metronidazole induce VSH-1 and facilitate gene transfer among Brachyspira hyodysenteriae strains. Appl. Environ. Microbiol. 74, 2950–2956 10.1128/AEM.00189-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Nishimori K., Makino S., Nishimori T., Kanno T., Ishihara R., et al. (2004). Molecular characterization of a prophage of Salmonella enterica serotype Typhimurium DT104. J. Clin. Microbiol. 42, 1807–1812 10.1128/JCM.42.4.1807-1812.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma J. K., Greene K. D., Ovitt J., Barrett T. J., Medalla F., Angulo F. J. (2005a). Hospitalization and antimicrobial resistance in Salmonella outbreaks, 1984–2002. Emerg. Infect. Dis. 11, 943–946 10.3201/eid1106.041231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma J. K., Molbak K., Barrett T. J., Beebe J. L., Jones T. F., Rabatsky-Ehr T., et al. (2005b). Antimicrobial-resistant nontyphoidal Salmonella is associated with excess bloodstream infections and hospitalizations. J. Infect. Dis. 191, 554–561 10.1086/427263 [DOI] [PubMed] [Google Scholar]
- Vogel H. J., Bonner D. M. (1956). Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218, 97–106 [PubMed] [Google Scholar]
- Waldor M. K., Mekalanos J. J. (1996). Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272, 1910–1914 10.1126/science.272.5270.1910 [DOI] [PubMed] [Google Scholar]
- Wang S., Zhao W., Raza A., Friendship R., Johnson R., Kostrzynska M., et al. (2010). Prevalence of Salmonella infecting bacteriophages associated with Ontario pig farms and the holding area of a high capacity pork processing facility. J. Sci. Food Agric. 90, 2318–2325 10.1002/jsfa.4090 [DOI] [PubMed] [Google Scholar]
- Weigle J. (1966). Assembly of phage lambda in vitro. Proc. Natl. Acad. Sci. U.S.A. 55, 1462–1466 10.1073/pnas.55.6.1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Botstein D., Miller J. H. (1979). Characterization of amber and ochre suppressors in Salmonella typhimurium. J. Bacteriol. 137, 433–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim G., McClure J., Surette M. G., Davies J. E. (2011). Modulation of Salmonella gene expression by subinhibitory concentrations of quinolones. J. Antibiot. 64, 73–78 10.1038/ja.2010.137 [DOI] [PubMed] [Google Scholar]
- Zinder N. D., Lederberg J. (1952). Genetic exchange in Salmonella. J. Bacteriol. 64, 679–699 [DOI] [PMC free article] [PubMed] [Google Scholar]


