Abstract
The study objectives were: to mine the complete exome to identify putative rare single nucleotide variants (SNVs) associated with irritable bowel syndrome (IBS)-diarrhea (IBS-D) phenotype, to assess genes that regulate bile acids in IBS-D, and to explore univariate associations of SNVs with symptom phenotype and quantitative traits in an independent IBS cohort. Using principal components analysis, we identified two groups of IBS-D (n = 16) with increased fecal bile acids: rapid colonic transit or high bile acids synthesis. DNA was sequenced in depth, analyzing SNVs in bile acid genes (ASBT, FXR, OSTα/β, FGF19, FGFR4, KLB, SHP, CYP7A1, LRH-1, and FABP6). Exome findings were compared with those of 50 similar ethnicity controls. We assessed univariate associations of each SNV with quantitative traits and a principal components analysis and associations between SNVs in KLB and FGFR4 and symptom phenotype in 405 IBS, 228 controls and colonic transit in 70 IBS-D, 71 IBS-constipation. Mining the complete exome did not reveal significant associations with IBS-D over controls. There were 54 SNVs in 10 of 11 bile acid-regulating genes, with no SNVs in FGF19; 15 nonsynonymous SNVs were identified in similar proportions of IBS-D and controls. Variations in KLB (rs1015450, downstream) and FGFR4 [rs434434 (intronic), rs1966265, and rs351855 (nonsynonymous)] were associated with colonic transit (rs1966265; P = 0.043), fecal bile acids (rs1015450; P = 0.064), and principal components analysis groups (all 3 FGFR4 SNVs; P < 0.05). In the 633-person cohort, FGFR4 rs434434 was associated with symptom phenotype (P = 0.027) and rs1966265 with 24-h colonic transit (P = 0.066). Thus exome sequencing identified additional variants in KLB and FGFR4 associated with bile acids or colonic transit in IBS-D.
Keywords: klotho B, susceptibility, risk factor, symptom
there may be a genetic predisposition to the development of irritable bowel syndrome (IBS), based on studies demonstrating associations of genetic variation in patients with IBS with symptom phenotype and intermediate phenotypes or quantitative traits (predominantly colonic transit, reviewed in Ref. 7) as well as studies of mRNA expression in rectal and colonic mucosa in patients with IBS (1). Significant associations (6) are reported between IBS phenotypes and genetic variations in mucosal immunity or susceptibility to inflammation (e.g., TNFSF15, TLR 9), barrier function (e.g., PRDM1), protective biochemical processes against luminal microorganisms (e.g., mucosal expression of oxidases), bile acid homeostasis [e.g., Klotho β (KLB) rs17618244 and fibroblast growth factor receptor 4 (FGFR4) rs1966265 and rs351855], as well as expression of neurotransmitters and cytokines, such as cannabinoid (e.g., FAAH, and CNR1) and adrenergic (e.g., α2C) receptors, and serotonin transporter (SLC6A4).
Candidate genes controlling KLB and FGFR4 proteins are associated with colonic transit in diarrhea-predominant IBS (IBS-D), the response of colonic transit to the bile acid (BA) sequestrant colesevelam in IBS-D, and the colonic transit and stool responses to chenodeoxycholic acid in patients with constipation-predominant IBS (IBS-C) (28, 42, 43). About 25% of patients with IBS-D have evidence of increased BA synthesis or excretion (41). Two other genes involved in BA function are liver receptor homolog-1 (LRH1), which is involved in bile acid homeostasis, and fatty acid binding protein 6 (FABP6), which is required for efficient absorption and transport of bile acids. We have previously reported on associations of the bile acid receptor gene, TGR5 (GpBAR1) single nucleotide polymorphism (SNP) rs11554825 [minor allele frequency (MAF) 41%], with small bowel and colonic transit (11). We included TGR5 in this study in assessing association with IBS-D but excluded it from the analysis of genetics of BA uptake, synthesis, and homeostasis.
Whereas genomewide association studies (GWAS) have identified genetic markers associated with common diseases, (22), there have been no GWAS pertinent to the common gastrointestinal disorder IBS. For meaningful GWAS, it was recommended that there should be accurate phenotype assessment and building of pathway/network models to link inherited and acquired variation together in a unified framework (22).
We have previously documented colonic transit by scintigraphy as a biomarker of colonic function in IBS (9) and used it extensively in genotype-intermediate phenotype studies in the last decade. BA malabsorption in patients with functional diarrhea and IBS-D is identified by measurement of BA synthesis, BA retention, or fecal BA excretion (10, 13, 15, 31, 36–38, 40, 41).
The aims of the present study were, first, to mine the exome DNA in genes reported to be associated with dysfunctions in IBS-D in published literature, and second, to mine the complete exome to identify putative rare single nucleotide variants (SNVs) associated with irritable bowel syndrome-diarrhea (IBS-D) phenotype. For these first two aims, DNA from 16 IBS-D patients was extracted and analyzed by exome-capture and next-generation sequencing. The data analysis was stratified to initially focus on a candidate list of genes previously associated with IBS or its treatment (selected from a review of the literature) and, as a second step, to discover rare, putatively functional variants with a phenotype association in the remaining exome.
In a third aim, we assessed the complete exome of genes that regulate BA absorption, feedback regulation, and synthesis in IBS-D in two groups of patients with increased fecal BAs (32), selected on the basis of a principal components analysis: one group with rapid colonic transit and “passive” increase in fecal bile acid excretion; and a second group of patients who have increased synthesis of bile acids, as might occur as a result of reduced FGF-19 feedback regulation, in accordance with the data published by Walters et al. (37) and confirmed in a prior study from our laboratory (28). The analysis was then extended to evaluate the univariate association of identified genetic mutations with the established phenotypes and principal component subgroups. Significant findings from this analysis were validated in the fourth aim in a separate cohort of participants previously characterized in our laboratory in whom we explored univariate associations of SNVs of interest, identified by the exome analysis of BA-related genes, with symptoms phenotype (n = 633 IBS or healthy controls) and colonic transit (n = 141 IBS).
METHODS
Study design, participants, and ethical review for exome sequencing study.
Our study design was based on the evaluation of detailed genetic associations in 16 patients with IBS-D. We chose to focus on IBS-D patients rather than the whole spectrum of phenotypes and the diverse underlying mechanisms in IBS patients across the spectrum, some having constipation or alternating bowel function. It is estimated that ∼25% of IBS-D is associated with increased BA synthesis or excretion. These patients had participated in a prior study that included a total of 22 patients with IBS-D who had undergone studies of bile acid homeostasis [altered BA feedback regulation (serum FGF-19), hepatic synthesis (serum C4; 7α-hydroxy-4-cholesten-3-one), and fecal BA excretion (fecal BA)] and colonic transit (41). Thus, to explore the mechanisms causing alterations in BA synthesis or excretion, we first conducted a principal components analysis to identify two cohorts, each consisting of eight patients, in whom the increased fecal BA excretion was associated with either accelerated colonic transit or increased hepatic synthesis (serum C4).
Principal components analysis.
We have recently shown that there are independent contributions of colonic transit and fecal bile acids to stool form and number (32); in addition, fecal BA may be increased as a result of increased hepatic BA synthesis. However, the previously reported significant correlation between serum C4 and fecal BA excretion [rs = 0.606, P < 0.001 (41)] showed that increased synthesis accounts for <40% in the variance of fecal BA. Another cause of increased fecal BA excretion is accelerated intestinal transit (30) or colonic transit at 48 h [rs = 0.41, P < 0.01 (32)]. Therefore, it is possible for patients with IBS-D to have elevated fecal bile acids as a result of at least two mechanisms: accelerated transit or increased bile acid synthesis. To explore the potential genetic mechanisms involved in IBS-D that may operate by altering bile acid control mechanisms, it was therefore important to identify patient cohorts that reflected a phenotype of accelerated transit or increased bile acid synthesis, even though both groups had increased fecal bile acids.
To achieve this goal, we used a principal component analysis that identifies linear combinations of the pathophysiological variables (29), which are weighted (“loading factor”) sums of the variables to be considered together when more than one mechanism is conceivably responsible for the phenotype, specifically IBS-D. Each linear combination is chosen to explain the most between-persons variation subject to the constraint that they are not correlated with other combinations. Thus the first principal component score (or PC1) was a weighted linear combination of the four variables in transit and bile acid homeostasis (serum FGF-19, serum C4, stool BA) that accounted for the maximum between-persons variation. The weight (loading) for a specific variable is the coefficient multiplier used for that variable in the given principal component score. The second linear combination (PC2) explained the maximum possible remaining variation and was not correlated with PC1. A third principal component was identified after the study. Ultimately, a total of three principal component scores were assessed (including two a priori selected principal component scores). These three principal components accounted for ∼92% of the total between subjects variation in the four variables.
Subgroup 1 had fast colonic transit [24-h geometric center (GC24)], high serum FGF-19, moderate fecal BA, and low serum C4. This cohort was selected to reflect low likelihood of the phenotype being secondary to excess BA synthesis.
Subgroup 2 was characterized by moderate colonic GC24, moderate levels of FGF-19, and high fecal BA and serum C4, which collectively reflect greater likelihood of BA excess synthesis or malabsorption. The characteristics of these two groups appear in Table 1.
Table 1.
PC analysis identifying 2 subgroups of IBS-D
| IBS-D Subgroup PC 1, n = 8 | IBS-D Subgroup PC 2, n = 8 | |
|---|---|---|
| Age | 41.5 (28, 50.5) | 28.3 (25, 40) |
| Sex | 6 women/2 men | 8 women/0 men |
| BMI, kg/m2 | 30.1 (25.4,34.8) | 31.5 (26.3, 33.3) |
| Colonic GC at 24 h | 4.57 (3.10, 4.79) | 2.41 (1.62, 3.54) |
| Serum FGF-19, pg/ml | 179.5 (88.4, 276.3) | 77.6 (62.4, 127.4) |
| Serum C4, ng/ml | 21.1 (12.9, 29.0) | 68.0 (33.1, 75.2) |
| Fecal BA, μM/24 h | 655 (334,1,236) | 889 (611, 1,236) |
| Overall phenotype | Fast colonic transit | Increased BA synthesis diarrhea |
Values are median and intraquartile range (IQR). PC, principal component; IBS-D, irritable bowel syndrome (IBS)-diarrhea; BMI, body mass index; GC, geometric center (range 1–5); FGF-19, ileal hormone fibroblast growth factor 19, feedback regulator of hepatic synthesis; C4, 7α-hydroxy-4-cholesten-3-one, surrogate for bile acid synthesis rate; BA, bile acid. Normal values in 22 healthy volunteers (Ref. 41): serum FGF-19: 88.5 (IQR 79.3–131.1) pg/ml; serum C4: 16.9 (IQR 6.1–28.0) ng/ml; fecal BA: 363.4 (IQR 193.7–762.1) μM/24 h. Normal colonic transit values (Ref. 18) at 24 h from 220 healthy adults: median 2.3 (5th, 95th percentile 1.3, 4.4); data are provided as 5–95th percentile to identify acceleration of colonic transit by median value of PC1 subgroup. Values for serum C4 in a study of 111 healthy volunteers (Ref. 10) were median 14.3 (IQR 9.5–29.2).
Intermediate phenotypes (or quantitative traits) were characterized for the 16 patients identified, DNA availability confirmed, and research authorization for use of DNA and medical records obtained from the Mayo Clinic Institutional Review Board.
Exome and in-depth DNA sequencing method and bioinformatics analysis.
Patient DNA was captured by using Agilent SureSelectXT All Exome V4 and sequenced by multiplexing four samples per lane on an Illumina HiSeq 2000. The resulting data were analyzed by using an internally developed next-generation sequencing workflow consisting of three steps: alignment, single nucleotide and small insertion/deletion variant calling, and annotation. FASTQ files were aligned to the hg19 reference genome by using Novoalign (VN:V2.07.13; www.novocraft.com) with the following options: –hdrhd off -v 120 -c 4 -i PE 425,80 -x 5 -r Random. Realignment and recalibration was performed by use of GATK (VN:1.6–7-g2be5704) (21) Best Practices version 3. Germline variations were called with GATK's UnifiedGenotyper using default parameters. Variant quality score recalibration is also done with the following command line optimizations: for SNVs, -an QD -an HaplotypeScore -an MQRankSum -an ReadPosRankSum -an FS -an MQ -an DP -nt 2 –maxGaussians 4 –percentBadVariants 0.05; and for insertions or deletions of bases in the DNA (INDELs), -an QD -an FS -an HaplotypeScore -an ReadPosRankSum –maxGaussians 4 -nt 2 –percentBadVariants 0.12 -std 10.0. VQSR “PASS” filter variants were selected for further analysis. VCF files were subsequently annotated by using Mayo Clinic's BioR annotation repository, which includes variant population frequencies (HapMap, 1k Genomes, ESP6500), frequency of occurrence in 50 internal-to-Mayo control samples, and function predictions of variant effects with SNP-Eff (effects of SNPs) (12) and SIFT (Sorting Intolerant from Tolerant) (26). “High-impact” variants identified by SNP-Eff were selected for further analysis and consisted of coding variants that affect canonical splice site acceptors and donors, disrupt start sites, cause frame shifts, and disrupt or induce stop codons.
The 50 control samples were used to identify and exclude variants with discontinuity between the reported population-wide frequency and the observed internal-to-Mayo frequency; these variants have a high likelihood of being false positive arising from local analytical pipeline bias. These samples were captured with Agilent SureSelect All Exome V4 + UTR and sequenced to an average depth of 120 million reads per sample on an Illumina Hiseq2000. The samples were obtained from Mayo Clinic's BioBank and consisted of deceased individuals with confirmed cause of death.
Targeted analysis of exome data in genes associated with bile acid and IBS.
The candidate list was extracted from information found in the literature and includes genes associated with IBS or its treatment. The list included genes with function related to drug metabolism, receptors of neurotransmitters or hormones, an alternative bile acid receptor (TGR5 or GPBAR1), and cytokines and inflammatory mediators (Table 2). All variants identified within these genes were reported for further consideration.
Table 2.
SNVs or INDELs in candidate gene list associated with IBS-D
| Gene Symbol | Chr | Position | Base Change | rs ID | Controls | IBS Subjects | HapMap CEU MAF |
|---|---|---|---|---|---|---|---|
| NR0B2 | chr1 | 27239920 | C>G | rs6659176 | 9/50 | 3/16 | 0.092 |
| IL23R | chr1 | 67705958 | G>A | rs11209026 | 4/50 | 4/16 | 0.067 |
| BDNF | chr11 | 27720937 | C>T | rs66866077 | 3/50 | 1/16 | n/a |
| C11orf30 | chr11 | 76257232 | C>T | 0/50 | 1/16 | n/a | |
| GNB3 | chr12 | 6954864 | G>A | rs5442 | 3/50 | 1/16 | 0.1 |
| TPH1 | chr11 | 18047154 | C>T | rs145855109 | 1/50 | 1/16 | n/a |
| TPH2 | chr12 | 72425419 | G>A | rs147025898 | 0/50 | 1/16 | n/a |
| HTR2A | chr13 | 47409034 | G>A | rs6314 | 12/50 | 4/16 | 0.075 |
| HTR2A | chr13 | 47469968 | G>T | rs1805055 | 1/50 | 1/16 | n/a |
| HTR3D | chr3 | 183754278 | C>G | rs73183412 | 4/50 | 1/16 | n/a |
| HTR3D | chr3 | 183756391 | C>G | rs75040538 | 2/50 | 1/16 | n/a |
| HTR3E | chr3 | 183818221 | G>− | rs141100381 | 9/50 | 3/16 | n/a |
| SLC6A4 | chr17 | 28548810 | C>G | rs6355 | 2/50 | 2/16 | 0.025 |
| SLC10A2 | chr13 | 103701672 | A>G | rs71640248 | 0/50 | 1/16 | n/a |
| SLC10A2 | chr13 | 103710635 | C>T | rs60380298 | 4/50 | 1/16 | n/a |
| SLC10A2 | chr13 | 103710656 | A>G | 0/50 | 1/16 | n/a | |
| NOD2 | chr16 | 50733785 | G>A | rs146054564 | 0/50 | 1/16 | n/a |
| NOD2 | chr16 | 50745596 | G>A | rs148683734 | 0/50 | 1/16 | n/a |
| NOD2 | chr16 | 50745926 | C>T | rs2066844 | 3/50 | 2/16 | n/a |
| NOD2 | chr16 | 50750842 | A>G | rs104895447 | 0/50 | 1/16 | n/a |
| NOD2 | chr16 | 50756540 | G>C | rs2066845 | 0/50 | 1/16 | n/a |
| NOD2 | chr16 | 50757276 | G>A | rs5743291 | 10/50 | 3/16 | 0.108 |
| NOD2 | chr16 | 50763778 | −>C | rs20668847 | 0/50 | 1/16 | n/a |
| TGFB1 | chr19 | 41858876 | C>G | rs1800471 | 12/50 | 2/16 | n/a |
| CARD8 | chr19 | 48715196 | T>C | rs34632751 | 2/50 | 1/16 | n/a |
| CARD8 | chr19 | 48735017 | −>TT | 3/50 | 1/16 | n/a | |
| IL1R2 | chr2 | 102626104 | T>G | 0/50 | 1/16 | n/a | |
| IL1R1 | chr2 | 102781649 | C>G | rs2228139 | 7/50 | 2/16 | 0.075 |
| COMT | chr22 | 19951897 | G>C | rs4646315 | 21/50 | 5/16 | n/a |
| CYP2D6 | chr22 | 42522601 | T>C | 0/50 | 1/16 | n/a | |
| CYP2D6 | chr22 | 42523528 | C>T | rs1058172 | 6/50 | 2/16 | n/a |
| CYP2D6 | chr22 | 42523805 | C>T | rs28371725 | 6/50 | 3/16 | n/a |
| CYP2D6 | chr22 | 42525811 | T>C | rs28371704 | 20/50 | 5/16 | n/a |
| CYP2D6 | chr22 | 42525821 | G>T | rs28371703 | 20/50 | 5/16 | n/a |
| MGLL | chr3 | 127411064 | C>T | rs138910107 | 2/50 | 1/16 | n/a |
| GPBAR1 | chr2 | 219128261 | A>G | rs144467445 | 0/50 | 1/16 | n/a |
| KLB | chr4 | 39448586 | C>T | rs35372803 | 2/50 | 2/16 | n/a |
| PRDM1 | chr6 | 106536253 | G>A | rs2185379 | 5/50 | 1/16 | 0.034 |
| PRDM1 | chr6 | 106553096 | G>A | rs143040512 | 1/50 | 1/16 | n/a |
| OPRM1 | chr6 | 154360508 | G>T | rs6912029 | 6/50 | 1/16 | 0.05 |
| NPSR1 | chr7 | 34867124 | G>T | rs34705969 | 6/50 | 1/16 | n/a |
| NPSR1 | chr7 | 34917702 | G>A | rs28480169 | 5/50 | 3/16 | n/a |
SNV, single nucleotide variant; INDEL, insertion/deletion of bases in the DNA; Chr, chromosome; MAF, minor allele frequency; CEU, Utah residents with ancestry from northern and western Europe; rs ID, reference single-nucleotide polymorphism (SNP) identification tag assigned by NCBI to a cluster of SNPs that map to an identical location.
Association of whole exome and IBS-D phenotype.
The complete exome was analyzed to identify rare variants with potentially damaging effect on the encoded protein that might be associated with the IBS-D phenotype. Results were filtered to select for nonsynonymous variants and exclude all variants with a reported MAF >2% in any population dataset from HapMap, 1k Genomes, as well as the ESP4500. The results were subsequently annotated with the SIFT prediction for functional impact.
Association of genetic variations in bile acid genes.
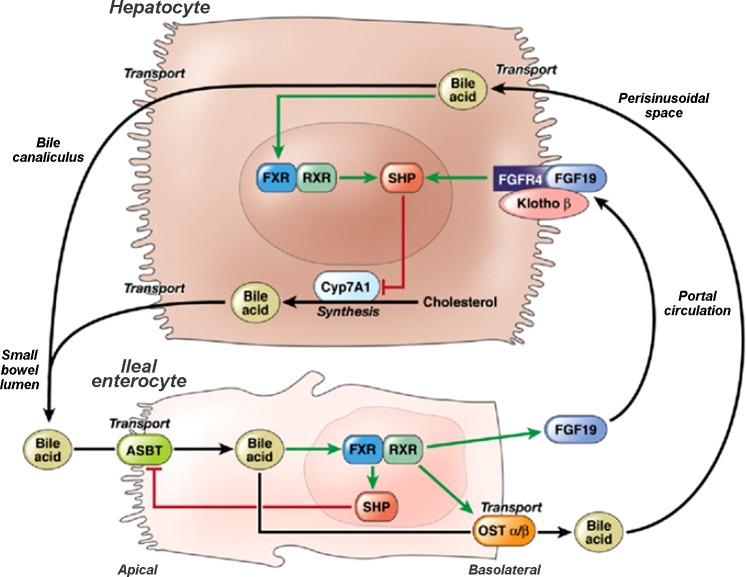
To investigate possible molecular differences in IBS-D symptom phenotype subsets, a list of 11 genes associated with bile acid homeostasis was created from the established pathway for bile acid synthesis (Fig. 1). This list included ASBT (SLC10A2), FXR (NR1H4), OSTα/β (SLC51A/B), FGF19, FGFR4, KLB, SHP (NROB2), CYP7A1, as well as LRH1 (also known as NR5A2 or nuclear receptor subfamily 5, group A, member 2), which is involved in bile acid homeostasis and steroidogenesis, and FABP6, which is required for efficient absorption and transport of bile acids in the distal portion of the murine small intestine (27), but a prior study concluded that most cases of primary BA malabsorption are unlikely to be caused by genetic variation in FABP6 (3). All variants identified within these genes were reported for further consideration.
Fig. 1.
Molecular mechanisms in homeostatic control of bile acid synthesis. Fibroblast growth factor 19 (FGF19) plays an important role in bile acid homeostasis by binding to FGF receptor 4 (FGFR4) on the hepatocyte cell membrane, triggering intracellular signaling in a klotho B (KLB)-dependent manner to downregulate cytochrome P 450 7A1 (CYP7A1) expression and, thereby, suppress bile acid synthesis. ASBT, apical sodium-coupled bile acid transporter; FXR, farnesoid X receptor; OSTα/β, organic solute transporter α/β; RXR, retinoid X receptor; SHP, short heterodimer partner. Reproduced from Wong et al. (42) with permission from Elsevier.
Association of SNVs of interest in KLB and FGFR4 in a larger cohort of IBS and controls.
After identifying three SNVs that had different prevalence in the IBS-D patients than the controls or were univariately associated with one or more of the principal components, we validated these findings by interrogating our database of 633 people (405 IBS patients and 228 controls), as described elsewhere (8, 42). There were 152 patients with IBS-C, 170 with IBS-D, and 83 with IBS-Alternators (IBS-A). Of these IBS patients with IBS-D or -C, 141 had undergone measurement of scintigraphic colonic transit: 70 IBS-D, 71 IBS-C. This ancillary study was also approved by Mayo Clinic Institutional Review Board for patients who had provided consent for use of their stored DNA and medical record in research conducted at Mayo Clinic.
We included intron FGFR4 rs434434 because we noted that the proportion of IBS-D patients with this allele (15/16) far exceeded the reported frequency of this allele in HapMap (CEU) of 0.21. Genotyping of KLB (rs1015450) and FGFR4 (rs434434, rs1966265, and rs351855) was performed with TaqMan SNP Genotyping Assays (Life Technologies, Grand Island, NY) according to the manufacturer's instructions using 10–20 ng of genomic DNA for each sample. Following polymerase chain reaction (PCR) amplification, end reactions were read on the 7300 Real-Time PCR System by use of Sequence Detection Software.
Statistical analysis.
We assessed the univariate associations of each SNV with colonic transit, fecal BA, serum FGF-19, serum C4, and two subgroups based on the first two principal components from a principal components analysis selected a priori. The associations with a third principal component obtained in the principal components analysis were also assessed post hoc. In the separate set of data from previous studies, the association of symptom phenotype (IBS subtype vs. health) with four selected SNVs was assessed by the χ2 test (general genetic model) and, for colonic transit quantitative trait (in IBS-C and IBS-D), by the Wilcoxon rank sum test (dominant genetic model).
RESULTS
Organization of results section.
The results are organized in the following sections: first, we present demographics of 16 participants whose DNA was sequenced in depth, and a high-level review of the exome-sequencing data; second, we address the first aim (to mine the complete exome to identify putative SNVs associated with IBS-D phenotype) through an in-depth analysis of SNVs previously reported in association with IBS and its treatment, followed by a mining of the remaining exome for rare, putatively functional variants associated with IBS-D; third, we analyze the association of SNVs identified by in-depth DNA sequencing with principal component subgroups based on quantitative traits of colonic transit, fecal BA excretion, and principal component groups; fourth, we assess the symptom phenotypes and colonic transit in association with SNVs in FGFR4 and KLB of each IBS group (IBS-C, IBS-D, and IBS-A) based on a cohort of 405 IBS patients and 228 healthy controls. Finally, we compared the prevalence of all the 54 SNVs in 10 of the 11 BA homeostasis genes in patients with IBS-D and 50 controls.
Demographics of participants.
The age, gender distribution, body mass index, quantitative transit, and bile acid parameters of patients in the two subgroups designated according to the principal components analysis are shown in Table 1.
Exome-sequencing data.
The 16 IBS-D samples were sequenced with 100 base paired-end reads, multiplexed four to a lane, on an Illumina HiSeq2000. An average of 104 million reads per sample were generated, with 97.9% mapping to the reference genome, resulting in greater than 90% of the exome-capture region covered by at least 30 reads. There were 40,957 SNVs identified on average, per sample. An average of 145 variants computationally classified as “High Impact,” potentially detrimental to the encoded protein, were identified per sample, including five novel (as defined by the absence of an associated dbSNP135 entry) variants.
Associations of literature-reported genetic variations in IBS with the present cohort.
Forty-five of 50 genes identified from the literature as associated with IBS or its treatment carried at least one SNV in our 16 IBS-D samples. In total, there were 189 position-unique SNVs identified, of which 74 were nonsynonymous changes. We used a permissive MAF filter (20%) to exclude extremely common mutations from further consideration. The frequency filter was based on a level twice the reported global community-based prevalence of IBS of 11.2%, 95% CI 9.8–12.8% (20). The 39 nonsynonymous mutations that passed the MAF filter are summarized in Table 2 and show the frequency of IBS-D subjects and control samples carrying each mutation. There was no difference in the proportion of IBS-D subjects and controls with these 39 nonsynonymous mutations, as shown in Table 2.
There were also 21 position-unique INDELs identified in the target genes, of which 3 caused frame shift mutations (Table 2). A G deletion in HTR3E (HTR3E 5-hydroxytryptamine receptor 3E) was found in three samples, a C insertion in NOD2 (nucleotide-binding oligomerization domain containing 2) was found in a single sample, and a TT insertion in CARD8 was found in another single sample. All three frame shift INDELs appeared to be heterozygous mutations. The HTR3E and CARD8 (caspase recruitment domain family, member 8) INDELs were identified at comparable frequencies in the 50 control samples, whereas the NOD2 insertion was not identified in any control sample.
Rare functional variants in association with IBS-D.
In addition to the candidate gene query, we also surveyed the remaining exome for possibly variants of interest. Given epidemiological data suggesting a pooled prevalence of IBS-D among women with IBS of 31% [95% CI 22–41% (19)], the most conservative estimate of IBS-D would be 22% of 9.8%, that is, ∼2%. Results were filtered to identify putatively functional, rare variants associated with the IBS-D phenotype, by selecting variants with an associated minor-allele-frequency <5%, found in <20% of the control samples, and annotated as High Impact by SNP Effect Predictor. High-impact classified mutations include changes impacting splice sites (acceptor and donor motifs), loss of a start site, and gain or loss of stop sites. A total of 210 variants meeting these criteria were found across the 16 samples. One hundred and ninety of these variants were unique private mutations (found in 1/16 samples analyzed). Of the remaining, 16 variants were found in 2/16 samples and 3 in 3/16 samples. Three of the variants found in 3/16 samples were enriched in the IBS-D samples compared with the control samples. All variants found in more than one sample are listed in Appendix Table A1.
Additionally, there were 302 INDELs identified in the exome data that were classified by SNP Effect Prediction as High Impact and identified in <20% of the control samples. Of these, 203 INDELs were found in only 1 sample, 55 in two samples, 21 in three samples, 9 in four samples, 6 in five samples, 3 in six samples, 2 in seven samples, and 1 in each of 8, 11, and 15 samples (Appendix Table A1).
We identified 19 SNVs that were absent in the 50 controls and were identified in at least 2 of the 16 patients: ASB15, ATP13A5, ATP2C2, C15orf48, C6orf203, CCDC138, CTBP2, CYB5R2, EIF5B, FADS6 (two SNVs), KRTAP10–7, MICA, MMP1, OR2T35, OR2T4, XAF1, ZNF626, and ZNF717. The full names of the genes and the potential specific mechanisms that might be related to IBS, based on literature reports on the genes, are listed in Appendix Table A2.
Interestingly, a 57-base deletion in keratin-associated protein 4–1 (KRTAP4–1) was identified in 15 samples and only 3 of the control samples. However, this deletion leads to the loss of an intron region between exons 1 and 2 but does not impact the coding sequence.
Principal component identification.
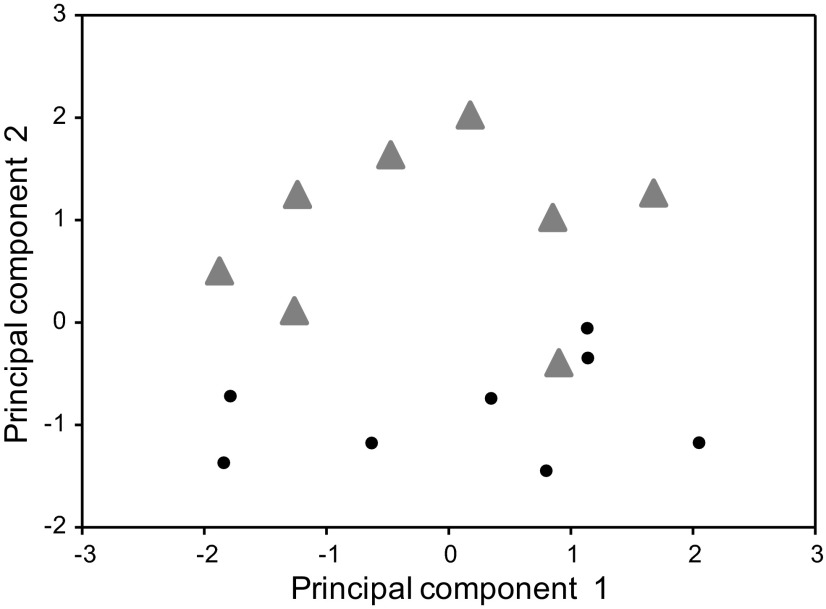
As shown in Fig. 2, the four selected quantitative parameters were able to identify two preselected groups, each consisting of eight patients: those with fast colonic transit/moderate BA excretion, and those with high BA excretion. Figure 2 clearly shows the separation of the two groups of patients; one patient was included in principal components analysis subgroup 1 based on very rapid colonic transit with normal serum C4. The analysis of the entire 16 patients also identified a third principal component (designated in Table 3 as PC3). This component was positively associated with FGF-19 (r = 0.58, P = 0.02) and inversely correlated with colonic transit GC24 (r = −0.53, P = 0.03).
Fig. 2.
Separation of principal component groups of patients whose DNA was in-depth sequenced. Note clear separation of the 2 groups with the exception of 1 patient who was included in subgroup 1 on the basis of very rapid colonic transit with normal serum C4 (7α-hydroxy-4-cholesten-3-one).
Table 3.
Associations of SNVs identified by in-depth DNA sequencing with quantitative traits of colonic transit and fecal BA excretion and principal component groups
| SNV | Effect | Prevalence in Controls | Prevalence in IBS | GC24* | Stool BA* | PC3* | PC Group† |
|---|---|---|---|---|---|---|---|
| rs1015450 (KLB) | Downstream | 11/50 | 6/16 | ns | 0.064 | ns | ns |
| rs434434 (FGFR4) | Intron | 49/50 | 15/16 | ns | ns | 0.061 | 0.031 |
| rs1966265 (FGFR4) | Nonsynonymous | 26/50 | 8/16 | 0.043 | ns | 0.026 | 0.077 |
| rs351855 (FGFR4) | Nonsynonymous | 26/50 | 6/16 | 0.056 | ns | 0.024 | 0.085 |
Associations are based on PC1 and PC2. The third principal component assessed posthoc is a quantitative score. PC groups are summarized as: PC1 fast colonic transit, PC2 increased bile acid synthesis diarrhea, and PC3 decreased bile acid synthesis (due to increased feedback regulation by FGF-19) with relatively slower colonic transit.
Kruskal-Wallis test and †Fisher's exact test: P values for test of association with SNV.
Principal component 3 was positively associated with (rank) FGF19 (r = 0. 44, P = 0.08) and negatively associated with (rank) GC24 (r = −0.59, P = 0.022).
Gene variants associated with colonic transit, fecal BA, and principal components.
Genetic variations in KLB and FGFR4 (but not FGF-19, ASBT, FXR, SHP, or CYP7A1) were associated with colonic transit, fecal BA, and two subgroups based on a principal components analysis (Table 3). It is noteworthy that, among these SNVs, one appeared to be more frequently present [KLB (rs1015450)] and one somewhat less prevalent (FGFR4 rs351855) in the 16 IBS-D patients than in the 50 controls. In addition FGFR4 rs434434 was identified in 15/16 IBS-D patients, and this frequency was much higher than the 0.21 frequency reported in HapMap CEU; however, it was also found in 49/50 control samples. The KLB variant (rs1015450) was associated with fecal BA excretion, FGFR4 rs1966265 was associated with increased colonic transit or the principal components, and FGFR4 rs434434 was associated with principal components subgroup. On the other hand, FGFR4 rs351855 was associated with both colonic transit (GC24) and with the third principal component (a quantitative score related to decreased BA synthesis and relatively slower colonic transit).
The symptom and colonic transit phenotypes in association with SNVs of interest in FGFR4 and KLB in a larger cohort of IBS-D and IBS-C are detailed in Tables 4 and 5 and discussed more extensively below.
Table 4.
Symptom phenotypes in association with SNVs in FGFR4 and KLB with demographics of each group
| IBS-C, n = 152 | IBS-D, n = 170 | IBS-A, n = 83 | Healthy, n = 228 | P Value* | |
|---|---|---|---|---|---|
| Mean (SE) age, yr | 45(1.2) | 45(1.2) | 41(1.5) | 37(0.9) | |
| Sex (% female) | 95 | 82 | 92 | 71 | |
| FGFR4 | |||||
| rs434434 | 0.027 | ||||
| AA (n = 16, 2.5%) | 31.2 | 50.0 | 12.5 | 6.2 | |
| GA (n = 222, 35.1%) | 26.6 | 28.8 | 9.0 | 35.6 | |
| GG (n = 395, 62.4%) | 22.3 | 24.8 | 15.5 | 37.5 | |
| rs1966265 | 0.404 | ||||
| AA (n = 32, 5.1%) | 28.1 | 28.1 | 15.6 | 28.1 | |
| GA (n = 244, 38.5%) | 26.2 | 23.8 | 13.9 | 36.1 | |
| GG (n = 357, 56.4%) | 22.1 | 28.8 | 12.3 | 36.7 | |
| rs351855 † | 0.715 | ||||
| AA (n = 52, 8.2%) | 21.1 | 34.6 | 13.5 | 30.8 | |
| GA (n = 282, 44.6%) | 23.8 | 22.7 | 12.8 | 40.8 | |
| GG (n = 298, 47.1%) | 24.8 | 29.5 | 13.1 | 32.6 | |
| KLB | |||||
| rs1015450 | 0.301 | ||||
| CC (n = 11, 1.7%) | 9.1 | 54.6 | 0 | 36.4 | |
| CT (n = 156, 24.6%) | 23.7 | 26.9 | 14.7 | 34.6 | |
| TT (n = 466, 73.6%) | 24.5 | 26.2 | 12.9 | 36.5 | |
| rs17618244† | |||||
| AA (n = 21, 3.3%) | 28.6 | 33.3 | 19.0 | 19.0 | 0.62 |
| AG (n = 198, 31.3%) | 26.3 | 25.2 | 11.1 | 37.4 | |
| GG (n = 413, 65.3%) | 22.8 | 27.4 | 13.6 | 36.3 | |
Data show percentages of symptom phenotypes within specified genotype for each of the 5 SNVs. IBS-C, IBS-constipation; IBS-A, IBS with alternating bowel function.
χ2 test.
One subject did not have this genotype measured due to technical reasons.
Table 5.
Association of rs434434 and rs17618244 and colonic transit in IBS-C and IBS-D
| No. IBS-C | IBS-C GC24 | IBS-C GC48 | No. IBS-D | IBS-D GC24 | IBS-D GC48 | |
|---|---|---|---|---|---|---|
| rs434434 AA | 5 | 1.92 ± 0.46 | 3.40 ± 1.30 | 3 | 3.69 ± 1.14 | 5.00 ± 0 |
| GA | 23 | 2.12 ± 1.03 | 2.96 ± 1.01 | 31 | 3.02 ± 1.17 | 4.16 ± 1.05 |
| GG | 32 | 1.92 ± 0.66 | 2.95 ± 1.00 | 35 | 3.32 ± 1.26 | 4.37 ± 0.94 |
| rs17618244 AA | 1 | 1.77 | 2.86 | 1 | 3.49 | 4.91 |
| AG | 24 | 1.84 ± 0.68 | 2.92 ± 1.04 | 23 | 2.56 ± 1.16 | 3.79 ± 1.24 |
| GG | 35 | 2.11 ± 0.89 | 3.04 ± 1.03 | 44 | 3.52 ± 1.13 | 4.56 ± 0.71 |
Values are means ± SD. Association of rs17618244 with both GC24 (P = 0.005) and GC48 (P = 0.034) in the combined IBS-C and IBS-D subtypes (based on the Kruskal-Wallis test). No significant associations were detected for rs434434.
Mutation data for the 11 genes in the bile acid pathways.
We found a total of 55 single-nucleotide variations in 10 of the 11 genes involved in BA homeostasis [including LRH-1 and FABP6 (Table 6)], with no SNVs identified in the 11th gene, FGF19. The variations were annotated with the frequency observed in 50 control samples of similar ethnicity. There were 15/55 variations that were observed in less than 20% of the control samples. Sixteen SNVs were nonsynonymous, including coding variations in SHP(NROB2) (rs6659176), KLB (rs17618244, rs35372803, rs4975017), ASBT (SLC10A2) (rs71640248, rs188096, rs60380298, and a rare variant at chr13:103710656 A→G), FGFR4 (rs1966265, rs376618, rs351855, rs442856), OSTα (rs939885, rs7642243), and FABP6 (rs116237330 and rs1130435). There were no significant differences in proportions of these SNVs in IBS-D relative to normal controls; however, in addition to the significant associations with colonic transit (nonsynonymous FGFR4 rs1966265) or principal component (intron FGFR4 rs434434) discussed above in Table 3, three nonsynonymous SNVs exhibited possible association with high vs. medium FGF-19 status. These were KLB rs17618244 [which was previously associated with accelerated transit in IBS overall and IBS-D (5 high BA excretion and 1 medium BA excretion)], KLB rs4975017 (2 high BA excretion and 5 medium BA excretion); and NROB2 (SHP) rs6659176 (0 high BA excretion and 3 medium BA excretion).
Table 6.
Characterization of 55 SNVs in 10 of the 11 BA homeostasis genes detected in 16 patients with IBS-D
| Gene Symbol | Chr | Position | Base Change | rs ID | Controls | IBS-D | BA Output High/Medium | Region | SNP Type | SIFT Prediction |
|---|---|---|---|---|---|---|---|---|---|---|
| NR0B2 | 1 | 27238150 | A>G | rs7504 | 46/50 | 1/16 | 0/1 | NonGENIC | NA | Not scored |
| 27238235 | G>C | rs41303639 | 6/50 | 1/16 | 0/1 | NonGENIC | NA | Not scored | ||
| 27239920 | C>G | rs6659176 | 9/50 | 3/16 | 0/3 | EXON CDS | Nonsyn | DAMAGING | ||
| NR5A2 | 1 | 199998281 | G<T | rs112399233 | 12/50 | 2/16 | 0/2 | NonGENIC | NA | Not scored |
| 199998304 | G<A | rs60535681 | 8/50 | 1/16 | 1/0 | NonGENIC | NA | Not scored | ||
| 200014560 | C<T | rs41300849 | 12/50 | 2/16 | 0/2 | NonGENIC | NA | Not scored | ||
| 200017586 | C<G | rs2821368 | 14/50 | 4/16 | 2/2 | EXON CDS | Syn | NA | ||
| 200080463 | C<T | rs3828110 | 31/50 | 5/16 | 3/2 | NonGENIC | NA | Not scored | ||
| 200090120 | G<A | rs3762398 | 25/50 | 5/16 | 3/2 | NonGENIC | NA | Not scored | ||
| 200143281 | C<T | rs1060060 | 28/50 | 8/16 | 3/5 | EXON CDS | Syn | NA | ||
| 200143442 | T<C | rs1060061 | 36/50 | 12/16 | 5/7 | 3′ UTR | Not scored | |||
| NR1H4 | 12 | 100887101 | G>T | rs56163822 | 0/50 | 1/16 | 1/0 | EXON.3 | Not scored | |
| 100930213 | C>G | rs17030285 | 14/50 | 4/16 | 3/1 | NonGENIC | NA | Not scored | ||
| SLC10A2 | 13 | 103698363 | A>G | rs2301157 | 42/50 | 14/16 | 8/6 | 3′ UTR | Not scored | |
| 103698630 | C>T | rs279940 | 50/50 | 16/16 | 8/8 | NonGENIC | NA | Not scored | ||
| 103701672 | A>G | rs71640248 | 0/50 | 1/16 | 0/1 | EXON CDS | Nonsyn | DAMAGING | ||
| 103703579 | T>C | rs8000956 | 42/50 | 14/16 | 8/6 | NonGENIC | NA | Not scored | ||
| 103705044 | A>C | rs188096 | 50/50 | 16/16 | 8/8 | EXON CDS | Nonsyn | TOLERATED | ||
| 103705098 | T>A | rs66842575 | 4/50 | 1/16 | 0/1 | NonGENIC | NA | Not scored | ||
| 103705132 | G>T | rs67736127 | 4/50 | 1/16 | 0/1 | NonGENIC | NA | Not scored | ||
| 103710635 | C>T | rs60380298 | 4/50 | 1/16 | 0/1 | EXON CDS | Nonsyn | TOLERATED | ||
| 103710656 | A>G | 0/50 | 1/16 | 1/0 | EXON CDS | Nonsyn | DAMAGING | |||
| 103718534 | A>G | rs138387807 | 0/50 | 1/16 | 0/1 | EXON CDS | Syn | NA | ||
| OSTβ | 15 | 65345962 | G<T | rs1046482 | 47/50 | 1/16 | 0/1 | 3′ UTR | Not scored | |
| OSTα | 3 | 195941216 | C<G | rs7642243 | 42/50 | 13/16 | 7/6 | EXON CDS | Nonsyn | TOLERATED |
| 195943481 | C<T | rs56030157 | 21/50 | 1/16 | 0/1 | 5′ UTR | Not scored | |||
| 195950622 | G<A | rs113298137 | 8/50 | 1/16 | 1/0 | NonGENIC | NA | Not scored | ||
| 195955762 | G<A | rs939885 | 31/50 | 12/16 | 7/5 | EXON CDS | Nonsyn | TOLERATED | ||
| 195956827 | T<C | rs17852687 | 33/50 | 9/16 | 5/4 | EXON CDS | Syn | NA | ||
| 195956970 | C<T | rs67261052 | 18/50 | 6/16 | 4/2 | NonGENIC | NA | Not scored | ||
| 195960225 | A<G | rs9343 | 34/50 | 9/16 | 5/4 | 3′ UTR | Not scored | |||
| KLB | 4 | 39448529 | G>A | rs17618244 | 12/50 | 6/16 | 5/1 | EXON CDS | Nonsyn | TOLERATED |
| 39448542 | C>G | rs7685429 | 47/50 | 15/16 | 8/7 | EXON CDS | Syn | NA | ||
| 39448586 | C>T | rs35372803 | 2/50 | 2/16 | 0/2 | EXON CDS | Nonsyn | TOLERATED | ||
| 39448848 | G>A | 0/50 | 1/16 | 0/1 | EXON CDS | Syn | NA | |||
| 39450229 | C>A | rs4975017 | 32/50 | 7/16 | 2/5 | EXON CDS | Nonsyn | TOLERATED | ||
| 39456635 | T>C | rs10025155 | 13/50 | 6/16 | 3/3 | NonGENIC | NA | Not scored | ||
| 39457857 | T>C | rs1015450 | 11/50 | 6/16 | 3/3 | NonGENIC | NA | Not scored | ||
| FABP6 | 5 | 159655498 | G<A | rs72812227 | 5/50 | 3/16 | 2/1 | NonGENIC | NA | Not scored |
| 159659262 | G<A | rs116237330 | 6/50 | 1/16 | 1/0 | EXON CDS | Nonsyn | TOLERATED | ||
| 159659273 | C<T | rs1130435 | 30/50 | 10/16 | 7/3 | EXON CDS | Nonsyn | DAMAGING | ||
| 159665726 | C<T | rs10071871 | 50/50 | 15/16 | 8/7 | 3′ UTR | Not scored | |||
| 159665733 | C<T | rs2277953 | 49/50 | 15/16 | 8/7 | NonGENIC | NA | Not scored | ||
| FGFR4 | 5 | 176516631 | G>A | rs1966265 | 26/50 | 8/16 | 5/3 | EXON CDS | Nonsyn | TOLERATED |
| 176516953 | A>G | rs434434 | 49/50 | 15/16 | 7/8 | NonGENIC | NA | Not scored | ||
| 176517170 | G>A | rs387598 | 48/50 | 14/16 | 6/8 | EXON CDS | Syn | NA | ||
| 176517292 | A>G | rs442856 | 49/50 | 15/16 | 7/8 | EXON CDS | Nonsyn | DAMAGING | ||
| 176517326 | T>C | rs422421 | 49/50 | 15/16 | 7/8 | EXON CDS | Syns | NA | ||
| 176517461 | T>G | rs446382 | 48/50 | 14/16 | 7/7 | EXON CDS | Syn | NA | ||
| 176517797 | C>T | rs376618 | 48/50 | 15/16 | 7/8 | EXON CDS | Nonsyn | TOLERATED | ||
| 176518784 | C>T | rs452885 | 48/50 | 15/16 | 7/8 | EXON CDS | Syn | NA | ||
| 176520243 | G>A | rs351855 | 26/50 | 6/16 | 3/3 | EXON CDS | Nonsyn | TOLERATED | ||
| 176523562 | C>A | rs31777 | 49/50 | 14/16 | 6/8 | NonGENIC | NA | Not scored | ||
| 176523597 | A>G | rs31776 | 48/50 | 14/16 | 7/7 | NonGENIC | NA | Not scored | ||
| CYP7A1 | 8 | 59411042 | G>A | rs2162459 | 43/50 | 13/16 | 6/7 | NonGENIC | NA | Not scored |
There were no SNVs detected in the FGF-19 gene. Syn, synonymous; nonsyn, nonsynonymous; UTR, untranslated region; NA, not applicable; CDS, coding sequence.
Other than FGFR4 rs434434 (intron) association with principal component group (Table 2), the remaining 39 SNVs had unclear significance: 22 nongenic, 10 synonymous, 2 in the 5′ untranslated region (UTR), and 5 in the 3′ UTR. There were also 5 INDELs identified in these BA-associated genes. All five INDELs were located in noncoding regions and none of the INDELs showed enrichment in the two cohorts of patients with high BA vs. those with faster colonic transit and moderate BA excretion.
Associations of SNVs in FGFR4 and KLB with symptom phenotype or colonic transit in larger cohorts of IBS and health.
Given the univariate associations of four SNVs in KLB (rs1015450) and FGFR4 (rs434434, rs1966265, rs351855) with colonic transit, BA quantitative traits, or principal components group (Table 3), we analyzed the associations of these genetic variants with IBS symptom phenotype (633 total participants) and colonic transit in 141 patients with IBS. The percentages of patients in each of the symptom phenotypes in association with the four SNVs are shown in Table 4.
FGFR4 rs434434 is intronic and is likely in linkage disequilibrium (r2 = 0.781) with rs376618. In our prior study, we detected that rs376618 was not in Hardy-Weinberg equilibrium (P = 0.048) (8), and, in the present study, it was present in similar proportions in IBS-D (15/16) and controls (49/50).
However, rs434434 was associated with symptom phenotype in the 633 participants (P = 0.027), but not with colonic transit at 24 h (P = 0.78) and 48 h (P = 0.89), based on the 141 patients with colonic transit measured in IBS-C or IBS-D (Table 5).
FGFR4 rs351855 was previously identified as a characteristic involved significantly with KLB rs17618244, which was associated with colonic transit at 24 h in the IBS-D subgroup (8). In the present study, rs351855 was not significantly associated with symptom phenotype (P = 0.30), and there was not an association with colonic transit at 24 h (P = 0.81) and at 48 h (P = 0.76).
FGFR4 rs1966265 was previously identified as a significant characteristic involved with KLB rs17618244, which was associated with colonic transit (GC24) in the IBS-D subgroup (8). In the present study, rs1966265 was not significantly associated with symptom phenotype (P = 0.70), but there was a modest association with colonic transit at 24 h (P = 0.066), less evident at 48 h (P = 0.256).
KLB (rs1015450) was not significantly associated with symptom phenotype (P = 0.40) or colonic transit at 24 h (P = 0.85) and at 48 h (P = 0.98).
KLB (rs17618244) was not significantly associated with symptom phenotype (P = 0.67), but, as in our prior study (43), there was significant association with colonic transit. Specifically, we identified association of rs17618244 with colonic transit (Table 5) at both 24 h (P = 0.005) and 48 h (P = 0.034) in the combined IBS-C and IBS-D subtypes (based on Kruskal-Wallis test).
DISCUSSION
There are several observations pertaining to the main aims of the present study.
Mining genetic mechanisms putatively involved in the control of dysfunctions in IBS-D.
Our analysis explored variants in the main putative mechanisms involved in the control of dysfunctions in IBS-D, specifically receptors of neurotransmitters or hormones (e.g., 5-HT and 5-HT3 receptors), and cytokines or inflammatory factors (e.g., IL-6, TNF-α). The nonsynonymous SNVs in the 45 genes (Table 2) were also similarly distributed in the IBS-D and 50 controls, and these included the SNVs in GPBAR1, the gene controlling the synthesis of TGR5 receptor protein.
Specifically, this analysis identified two variant-containing genes controlling 5-HT3 receptors and IBS-D, although the prevalence in controls and IBS-D in the present study was not different. A G deletion in HTR3E was found in 3 of the 16 samples from IBS-D patients. This observation is consistent with a study of genetic variation of 5-HT3 receptor genes in IBS patients from the United Kingdom and Germany (16), which showed that the novel HTR3E 3′-UTR variant c.*76G>A (rs62625044) was associated with D-IBS in female subjects. Using a reporter assay, Kapeller et al. (16) showed that gene variant affected the binding of miRNA-510 to the HTR3E 3′-UTR and caused elevated protein expression in two different cell lines.
Similarly, a SNV in the HTR3D gene was identified in 2 of 16 patients. It is relevant to note that 5-HT3D receptors are expressed in both myenteric and submucosal neurons (17) and therefore conceivably alter secretory and motor functions in the intestines or conceivably the response to treatment with 5-HT3 antagonists such as alosetron, which is efficacious for treatment of IBS-D (2).
Mining the complete exome.
Our study mining the complete exome identified a total of 118 candidate mutations of interest.
One candidate mutation of interest following a review of the literature was an SNV in ZNF77 (zinc finger protein 77) in 2 of 16 patients was also found (compared to 2 of 50 controls). This mutation, causing a glutamine to stop codon change, has been shown to be significantly associated with fibromyalgia syndrome (14); a systematic review reported that 49% of fibromyalgia patients suffer IBS (39). Overall, these data suggest a potential role for the disruption of this zinc finger protein in IBS.
There were four SNVs that may have biological effects that may be relevant to peripheral pathophysiological mechanisms associated with IBS (6). A SNV in ATP2C2 [the gene encoding secretory pathway calcium-transporting ATPase, type2C, member2, which is a Golgi-localized pump with high affinity for Ca2+ ions (34)] could affect motor, sensory, or secretory mechanisms. A second variant was located in XAF1 (X-linked inhibitor of apoptosis-associated factor 1) and annotated to induce a premature stop codon. A third SNV in MHC class I polypeptide-related sequence A (MICA) gene could impact immune processes that are relevant in the microscopic immune activation in some patients with IBS. A fourth SNV in MMP1 (matrix metallopeptidase 1) that encodes for an interstitial collagenase could conceivably impact colonic compliance or tone which may be relevant to IBS.
In addition to the direct associations reported through the literature above, potentially relevant functional associations are described for two additional genes harboring SNVs in 2 of 16 patients, each. ATP2C2 [the gene encoding secretory pathway calcium-transporting ATPase, type2C, member2, which is a Golgi-localized pump with high affinity for Ca2+ ions (34)] was annotated as being in a splice site acceptor motif. The second variant was located in XAF1 and annotated to induce a premature stop codon. These are potentially relevant because the former influences intracellular calcium transport, which is involved in function of the enterocytes, muscle, and nerve function, whereas the latter influences apoptosis or autophagic cell death. However, there are no hitherto reported effects or associations with IBS. It is interesting to note that rats fed the secretory bile salts chenodeoxycholate and deoxycholate showed colonic mucosa histology with a significantly reduced apoptotic index (4). The relatively low prevalence (2/16 patients) suggests relevance in a small subgroup of IBS-D.
Case-control analysis of whole exome in two cohorts based on principal components.
Our third approach in this study followed a case-control design, enhanced by a principal components analysis to compare two subgroups of patients with IBS-D, based on two pathophysiological mechanisms: accelerated transit [which was documented in 46% patients with IBS-D (9)] with moderate fecal BA excretion and a separate group with increased bile acid synthesis [by serum C4 in 9% patients with IBS-D (41)]. Although whole exome sequencing in complex disorders is typically applied to family-based studies with affected and nonaffected individuals, this approach is impractical in IBS in which data from familial aggregation and twin studies are conflicting (5) and their pathophysiological mechanisms of IBS-D are diverse (6). Therefore, we perceive that genetic association studies in IBS are strengthened by robust identification of the quantitative phenotype of included participants.
In this analysis, we identified additional genetic variants in KLB and FGFR4 in association with fecal BA excretion or colonic transit in patients with IBS-D; SNVs in 8 of 9 other BA-related genes were not associated with these quantitative traits in IBS-D patients. These findings generally confirm our prior, candidate gene-based study that KLB and FGFR4 are associated with IBS-D phenotype (42). Walters et al. (37) had provided evidence that deficiency of FGF-19 was associated with increased serum C4, suggesting that feedback regulation by FGF-19 influenced the rate of synthesis of BA in the hepatocytes and the development of IBS-D as a symptom phenotype. No measurements of ileal mucosal FGF-19 have been reported in patients with IBS-D. Our data suggest that changes in FGF-19 in IBS-D may not be associated with variation in FGF19 gene.
The FGF-19 deficiency might conceivably be due to genetic variations in the ileal enterocyte proteins that control its synthesis, such as SHP and FXR. We found several SNVs in these genes; however, we were not able to show an association with SNVs in the FGF-19 gene. Similarly, in the ASBT gene for the ileal BA transporter (SLC10A2), we identified 10 SNVs, seven of which were common polymorphisms (MAF > 1%), but none were associated with FGF-19 deficiency. The previous literature had shown no mutations in the ASBT gene for SLC10A2 in individuals or families of patients with idiopathic bile acid malabsorption (24, 25).
Our study approach does not exclude the potential contribution of disorders at the levels of transcription or posttranscriptional modification to the development of the IBS-D phenotype or alterations in quantitative traits (transit and fecal BA excretion). In addition, future studies will also need to be directed at the expression of these proteins in tissues such as ileal mucosa or liver.
Association of KLB and FGFR4 with quantitative traits in the current and independent IBS cohorts.
Our exome-based analysis identified SNVs in the genes for KLB and FGFR4 in association with the subgroups of patients and with fecal BA excretion or colonic transit. These data are summarized in Table 3. It is noteworthy that the newly identified KLB variant was associated with fecal BA, in view of the role played by the KLB-FGFR4 receptor proteins on hepatocyte BA synthesis.
Moreover, when the four SNVs identified by the association with principal components in 16 IBS-D patients were examined in a larger cohort of 633 patients or healthy controls, FGFR4 rs434434 was associated with symptom phenotype (Table 3), and FGFR4 rs1966265 was possibly associated with colonic transit at 24 h in 141 IBS patients.
We are encouraged that the associations observed are consistent with our prior studies (42), demonstrating associations of genes for KLB and FGFR4 with IBS phenotype and colonic transit. The most illustrative example is that rs17618244 (Arg728Gln) in KLB is associated with colonic transit at 24 h, and the amino acid change is associated with reduced stability of the KLB protein; dysfunction of the KLB-FGFR4 combined protein complex results in failure of FGF-19 feedback regulation of hepatocyte BA synthesis. Thus, with the KLB genotype variant, greater BA synthesis and excretion would be expected. The interaction of KLB and FGFR4 is complex. For example, there is evidence that KLB may promote FGFR4's activation by specifically binding to dominant negative glycosylated forms of FGFR4 in the endoplasmic reticulum, preferentially mediating their proteasomal degradation to permit FGF-19-mediated signaling through active forms of FGFR4 (33).
Significance of current exome DNA studies relative to published literature.
In the present study, KLB rs17618244 was not associated with the principal component subgroups. Although we had previously shown that the SNV was functionally relevant and associated with colonic transit in IBS-D (42), we confirmed that there was no significant association with IBS symptom phenotype (Tables 4 and 5) in the current cohort of 633 participants, all of whom were included in the previously reported cohort (42). In addition, as in a prior study (43), we identified a significant association of rs17618244 and colonic transit at 24 and 48 h.
Variations in KLB and FGFR4 also modified responses to BA-directed therapy (chenodeoxycholic acid and colesevelam, respectively) in IBS-C and IBS-D. The exome in-depth analysis identified other genetic variants, although these are not necessarily functional. In fact, Table 6 shows that the rs434434 SNV is intronic, suggesting that it may not be directly causing a functional change in the protein FGFR4.
On the other hand, two SNVs are nonsynonymous and potentially functional.
First, FGFR4 rs1966265(Val10Ile) was univariately associated with stool level of BA (41) in a prior study of 26 healthy volunteers, 26 patients with IBS-C and 26 with IBS-D; rs1966265 modulated the association of KLB rs1768244 (Arg728Gln) with colonic transit in IBS-D (42) and, in the present study, there was a possible association of the SNV rs1966265 on its own and colonic transit at 24 h in IBS-D patients (P = 0.043 in the exome sequencing study), but no association with colonic transit in the larger cohort of IBS-D and IBS-C. Second, FGFR4 rs351855 (Gly388Arg) modulates KLB rs1768244's association with colonic transit in IBS-D (42) and was possibly associated with colonic transit in IBS-D in the exome sequencing study (P = 0.056), but not with symptom phenotype or colonic transit in the larger cohorts.
Perspective on application of deep sequencing of DNA in IBS-D.
There is much enthusiasm and hope that next generation, including exome sequencing, will provide diagnostic information through the identification of de novo mutations (23, 35). However, complex disorders or phenotypes with multiple potential mechanisms will present additional challenges.
We perceive that the approach we have chosen has potential advantages. First, we identify subgroups based on principal components analysis to narrow the spectrum of phenotypes that are explored by the nonselective genome sequencing approach. Second, we focus on the putative genes in a pathway that alters a specific pathophysiological mechanism in the disorder, thereby providing a means to interrogate the role of the pathway and its associated genetic variations without inflating the false detection rate for any SNV. Third, we interrogate a larger cohort of patients using symptom phenotype and quantitative trait (colonic transit) to assess in a larger cohort of patients the potential functional relevance of the SNVs identified in the focused group, relative to ethnically similar controls. Overall, the data from the present in-depth DNA analysis enhance our understanding of mechanisms that are associated with colonic transit, as well as with subgroups of IBS-D patients identified by principal components analysis. Given the multifactorial nature of IBS, and especially IBS-D (6), it is unlikely that a specific genetic predisposition will account for more than a minor component of the variance of the disorder. Further studies of the interaction of stress, parental influence, the environment, and epigenetic modification of the FGFR4 gene may further elucidate the potential role of genetic predisposition in IBS.
Summary and conclusion.
In summary, in-depth DNA sequencing identified additional genetic coding and noncoding variants in KLB and FGFR4 that are univariately associated with fecal BA excretion or colonic transit in IBS-D. Deficiency of FGF-19 in patients with IBS-D is not associated with variation in FGF19 gene. These hypothesis-generating data provide additional weight to the rationale to identify and treat alterations of BA kinetics and colonic transit in patients with IBS; indeed, rational treatment may involve modulation of the BA pathway itself with agents such as BA, BA transport inhibitors in IBS-C (e.g., elobixibat), or BA sequestrants (e.g., cholestyramine and colesevelam) in IBS-D.
GRANTS
This study was supported by NIH grant R01-DK092179 to M. Camilleri; facilities provided in the Endoscopy and Physiology Core of the Mayo Clinic Clinical Research Unit were supported by NIH CTSA grant UL1-TR000135; and the exome sequencing and bioinformatics were supported by a Mayo Clinic grant from the Center of Individualized Medicine.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
M.C. conception and design of research; M.C., E.W.K., M.G., and A.R.Z. interpreted results of experiments; M.C. prepared figures; M.C. drafted manuscript; M.C., E.W.K., A.S., P.C., Y.L., M.G., and A.R.Z. edited and revised manuscript; M.C., E.W.K., A.S., P.C., Y.L., M.G., and A.R.Z. approved final version of manuscript; E.W.K., Y.L., and A.R.Z. analyzed data; A.S. and P.C. performed experiments.
ACKNOWLEDGMENTS
We thank Cindy Stanislav for secretarial support.
Appendix
All other mutations identified in mining the complete genome in more than one sample during the EXOME DNA analysis are listed in Appendix Table A1. Note that for the 118 mutations listed here, the HapMap CEU MAF is not available except for ANXA9 rs 267731, whose MAF is 0.017.
Appendix Table A1.
Genes identified in IBS but not in controls and their potential effects in IBS
| Gene Symbol | Chr | Position | Base Change | rs ID | Controls | IBS Subjects |
|---|---|---|---|---|---|---|
| A2M | 12 | 9246176 | CTATGG>C | rs3832852 | 9/50 | 5/16 |
| ABO | 9 | 136131058 | GG>G | rs8176750 | 3/50 | 2/16 |
| ACOT4 | 14 | 74060511 | T>TTCAA | 5/50 | 2/16 | |
| ACOT4 | 14 | 74060513 | GCTTA>G | 6/50 | 2/16 | |
| ACTR3C | 7 | 149983566 | G>A | rs181975624 | 7/50 | 2/16 |
| AGAP6 | 10 | 51768674 | CAA>C | rs141217862 | 6/50 | 5/16 |
| AHCTF1 | 1 | 247007232 | T>A | rs148600466 | 1/50 | 2/16 |
| ANXA9 | 1 | 150955582 | A>G | rs267731 | 2/50 | 2/16 |
| ASB15 | 7 | 123267185 | CG>C | rs138215101 | 0/50 | 2/16 |
| ATP13A5 | 3 | 193081961 | TC>T | rs148224926 | 0/50 | 2/16 |
| ATP2C2 | 16 | 84495318 | A>C | rs4782970 | 0/50 | 2/16 |
| BEST3 | 12 | 70088219 | GT>G | 4/50 | 2/16 | |
| C10orf68 | 10 | 33136818 | TAA>T | 1/50 | 6/16 | |
| C15orf48 | 15 | 45724278 | G>GA | 0/50 | 2/16 | |
| C16orf55 | 16 | 89735891 | C>CTG | rs147716338 | 1/50 | 2/16 |
| C19orf26 | 19 | 1236080 | A>G | rs113298601 | 2/50 | 3/16 |
| C1QTNF8 | 16 | 1143496 | C>A | rs150337362 | 1/50 | 2/16 |
| C20orf132 | 20 | 35807790 | G>GCTTATAGACAGGGCCCCGCGGCCGGCACT | rs11467214 | 8/50 | 4/16 |
| C20orf96 | 20 | 271225 | T>TTTA | rs3835237 | 3/50 | 2/16 |
| C20orf96 | 20 | 271225 | T>TTA | rs3835237 | 3/50 | 2/16 |
| C6orf203 | 6 | 107351312 | G>A | rs117058816 | 0/50 | 2/16 |
| C7orf33 | 7 | 148288020 | G>T | rs62624492 | 6/50 | 3/16 |
| C9orf152 | 9 | 112969744 | AT>A | 4/50 | 2/16 | |
| CAPN11 | 6 | 44140054 | T>TGGCTGCC | 5/50 | 2/16 | |
| CCDC138 | 2 | 109404485 | T>TT | rs35693674 | 0/50 | 3/16 |
| CCDC39 | 3 | 180334077 | T>TA | 3/50 | 2/16 | |
| CCNL2 | 1 | 1334052 | CTAGAG>C | rs3831366 | 6/50 | 2/16 |
| CDC27 | 17 | 45234406 | CA>C | rs112848754 | 7/50 | 2/16 |
| CDCP2 | 1 | 54605326 | G>GGG | rs3841798 | 2/50 | 3/16 |
| CDSN | 6 | 31083800 | TCTT>T | rs56899166 | 8/50 | 11/16 |
| CHMP4C | 8 | 82665476 | TGTGA>T | rs137960856 | 6/50 | 3/16 |
| CNKSR1 | 1 | 26510310 | AC>A | 6/50 | 2/16 | |
| COPZ2 | 17 | 46115079 | C>CT | 1/50 | 2/16 | |
| COX7A1 | 19 | 36642568 | C>CC | rs3214131 | 4/50 | 3/16 |
| CPNE1 | 20 | 34215234 | C>CA | 5/50 | 4/16 | |
| CRIPAK | 4 | 1388379 | G>GCA | 4/50 | 3/16 | |
| CTBP2 | 10 | 126678122 | GT>G | rs140913543 | 0/50 | 2/16 |
| CXorf59 | X | 36162684 | C>CTG | 4/50 | 2/16 | |
| CYB5R2 | 11 | 7686606 | T>TTGTT | rs56663591 | 0/50 | 2/16 |
| CYP3A43 | 7 | 99434077 | TA>T | rs139781484 | 2/50 | 2/16 |
| DCDC2B | 1 | 32674742 | T>TG | 2/50 | 2/16 | |
| DDIT4L | 4 | 101108875 | GT>G | rs148719835 | 4/50 | 2/16 |
| DEFB128 | 20 | 168625 | G>GT | rs11396059 | 5/50 | 2/16 |
| DHDH | 19 | 49447748 | GGA>G | rs3835153 | 2/50 | 3/16 |
| DNAH14 | 1 | 225380561 | CAAAG>C | rs144339803 | 4/50 | 3/16 |
| DNAJA4 | 15 | 78558584 | CAT>C | rs142025971 | 2/50 | 3/16 |
| EBLN2 | 3 | 73111481 | C>CA | rs3832186 | 8/50 | 4/16 |
| EFCAB2 | 1 | 245133628 | C>CCCTCC | rs78699431 | 6/50 | 6/16 |
| EIF5B | 2 | 99988116 | CAGA>C | 0/50 | 2/16 | |
| FADS6 | 17 | 72889676 | G>GGGCTCCGTAGGTTCCATGGGCT | 0/50 | 2/16 | |
| FADS6 | 17 | 72889676 | G>GGGCTCCGTAGGTTCCATGGGCTCCGT | 0/50 | 2/16 | |
| FANK1 | 10 | 127685987 | A>G | rs140634850 | 2/50 | 2/16 |
| FIG4 | 6 | 110053837 | T>TT | rs11429845 | 3/50 | 7/16 |
| FMO2 | 1 | 171165802 | TG>T | rs142989335 | 6/50 | 2/16 |
| GEN1 | 2 | 17962993 | CAAGTT>C | rs149936944 | 7/50 | 2/16 |
| GLT8D1 | 3 | 52729205 | C>CTTAC | rs60069075 | 1/50 | 2/16 |
| HELB | 12 | 66704275 | CA>C | 1/50 | 2/16 | |
| KCNMB3 | 3 | 178960766 | CT>C | rs148909269 | 8/50 | 4/16 |
| KIAA1751 | 1 | 1887112 | CC>C | rs34659723 | 4/50 | 4/16 |
| KIAA1841 | 2 | 61361325 | TG>T | rs142269591 | 7/50 | 2/16 |
| KRTAP15 | 17 | 39183011 | AG>A | 5/50 | 2/16 | |
| KRTAP10-7 | 21 | 46020655 | CCTGCTGCGCCCCCAG>C | rs36208679 | 0/50 | 4/16 |
| KRTAP4-1 | 17 | 39340795 | ACGGCAGCAGCTGGACATACCACAGCTGGGGTGGCAGGTGGTCTGACAGCAGAGTGGG>A | 3/50 | 15/16 | |
| KRTAP4-8 | 17 | 39254247 | A>T | rs137943557 | 5/50 | 2/16 |
| MICA | 6 | 31380002 | GG>G | 0/50 | 2/16 | |
| MICA | 6 | 31380161 | G>GCT | rs113353287 | 6/50 | 3/16 |
| MICA | 6 | 31380161 | G>GCTGCTGCTGCT | rs113353287 | 7/50 | 2/16 |
| MMP1 | 11 | 102667446 | CA>C | rs17879749 | 0/50 | 3/16 |
| MOGAT1 | 2 | 223559986 | AG>A | 1/50 | 2/16 | |
| NOTCH2 | 1 | 120612002 | CGG>C | 4/50 | 2/16 | |
| NOTCH2 | 1 | 120612004 | GGG>G | 1/50 | 2/16 | |
| NUDT17 | 1 | 145586678 | AC>A | 2/50 | 2/16 | |
| OR13C2 | 9 | 107367665 | GCG>G | 1/50 | 4/16 | |
| OR2C1 | 16 | 3406756 | GT>G | rs142397376 | 5/50 | 5/16 |
| OR2D3 | 11 | 6942588 | TC>T | rs150992591 | 2/50 | 2/16 |
| OR2T35 | 1 | 248801944 | TCAGCACG>T | rs143010547 | 0/50 | 3/16 |
| OR2T4 | 1 | 248525330 | A>ACG | 0/50 | 2/16 | |
| OR4L1 | 14 | 20528448 | TCATAGATTTGCTCACTGAC>T | rs112192573 | 7/50 | 5/16 |
| OR52N1 | 11 | 5809806 | GA>G | rs142442713 | 5/50 | 2/16 |
| OR5M10 | 11 | 56344843 | CCATTGAAG>C | rs148438199 | 6/50 | 2/16 |
| OR5P2 | 11 | 7818383 | G>GATATGGTTACCAGGTAGATGC | 3/50 | 2/16 | |
| PBOV1 | 6 | 138539183 | A>AG | rs3841283 | 5/50 | 3/16 |
| PCDHGA10 | 5 | 140795208 | T>TA | rs113784532 | 6/50 | 2/16 |
| PIK3C2G | 12 | 18800961 | G>C | rs61757718 | 4/50 | 2/16 |
| POLDIP2 | 17 | 26684395 | G>GG | 8/50 | 7/16 | |
| PRKRA | 2 | 179315734 | GGC>G | rs141354030 | 9/50 | 2/16 |
| PRSS55 | 8 | 10411512 | TG>T | rs142551217 | 3/50 | 2/16 |
| PSORS1C1 | 6 | 31106500 | TC>T | rs111966729 | 3/50 | 3/16 |
| PSORS1C2 | 6 | 31105857 | AG>A | rs144826968 | 5/50 | 4/16 |
| PTPRQ | 12 | 80943505 | CTGAAACAGGTAACTAACG>C | rs141686707 | 7/50 | 3/16 |
| RECQL4 | 8 | 145738768 | GG>G | rs11342077 | 2/50 | 8/16 |
| RNF133 | 7 | 122338970 | C>T | rs71574716 | 4/50 | 2/16 |
| RYK | 3 | 133969438 | G>GG | 3/50 | 2/16 | |
| SARM1 | 17 | 26699207 | AA>A | rs11340312 | 9/50 | 2/16 |
| SIGLEC12 | 19 | 52004795 | G>GT | 4/50 | 2/16 | |
| SLC25A5 | X | 118603706 | A>AG | rs113356560 | 1/50 | 3/16 |
| SLC39A9 | 14 | 69928413 | G>A | rs111495442 | 1/50 | 2/16 |
| SLC5A4 | 22 | 32643447 | AC>A | 1/50 | 2/16 | |
| SLC5A4 | 22 | 32643460 | C>A | rs62239058 | 2/50 | 3/16 |
| SLC7A13 | 8 | 87226634 | TCCGACATC>T | rs56993779 | 8/50 | 3/16 |
| SPANXN2 | X | 142803691 | AT>A | 1/50 | 2/16 | |
| SPATA4 | 4 | 177106009 | TTCTC>T | rs28381989 | 9/50 | 5/15 |
| SSTR1 | 14 | 38679763 | C>CGCTCTGAGCCCGGGCCACGCAGGG | 1/50 | 3/16 | |
| TDG | 12 | 104373728 | G>GA | 5/50 | 6/16 | |
| TEX13A | X | 104464277 | AA>A | rs56000396 | 8/50 | 3/16 |
| TEX13A | X | 104464284 | CC>C | rs56118606 | 8/50 | 3/16 |
| UGT2A1 | 4 | 70512787 | A>T | rs111696697 | 4/50 | 2/16 |
| WNK1 | 12 | 974311 | C>CC | rs35706572 | 8/50 | 5/16 |
| WWTR1 | 3 | 149238595 | C>CTTAA | rs112399999 | 7/50 | 4/16 |
| XAF1 | 17 | 6663899 | G>T | rs146752602 | 0/50 | 2/16 |
| ZNF101 | 19 | 19790876 | CAT>C | rs34704748 | 4/50 | 3/16 |
| ZNF193 | 6 | 28198122 | C>T | rs76542212 | 7/50 | 2/16 |
| ZNF492 | 19 | 22817276 | GGT>G | rs148713444 | 1/50 | 2/16 |
| ZNF626 | 19 | 20807178 | A>AA | rs71174721 | 0/50 | 2/16 |
| ZNF717 | 3 | 75786555 | C>CTG | rs141276988 | 3/50 | 3/16 |
| ZNF717 | 3 | 75790815 | T>TT | 0/50 | 2/16 | |
| ZNF77 | 19 | 2936535 | G>A | rs35699176 | 2/50 | 2/16 |
| ZNF780B | 19 | 40554705 | T>C | rs112293642 | 1/50 | 2/16 |
Appendix Table A2 lists functions and potential significance of gene variants among 118 SNVs that were identified in mining the complete genome in IBS patients, but not in 50 controls.
Appendix Table A2.
Genes that might be related to IBS
| SNV | Full Name of Gene or Protein Encoded | Potential Specific Mechanism Related to IBS, Based on Literature Reports on the Genes |
|---|---|---|
| ASB15 | Ankyrin repeat and suppressor of cytokine signaling (SOCS) box-containing protein (ASB) 15 | None (predominantly expressed in skeletal muscle) |
| ATP13A5 | ATPase type 13A5 | None |
| ATP2C2 | ATPase, type2C, member2 | Secretory pathway for Ca2+ ions, e.g., motor, sensory, secretory |
| C15orf48 | Chr 15 open reading frame 48 | None (differentiation of skin cancers) |
| C6orf203 | Chr 6 open reading frame 203 | None (protein polymerization) |
| CCDC138 | Coiled-coil domain containing 138 | None (ubiquitylation) |
| CTBP2 | COOH-terminal binding protein 2 | None (transcriptional activator, regulator of the cytoskeleton and transcriptional corepressor) |
| CYB5R2 | Cytochrome b5 reductase 2 | None (protein-protein interaction) |
| EIF5B | Eukaryotic translation initiation factor 5B | None (translation initiation) |
| FADS6 | Fatty acid desaturase 6 | None (fatty acid desaturation in metabolic disease) |
| KRTAP10-7 | Keratin-associated protein 10 | None |
| MICA | MHC class I polypeptide-related sequence A (MICA) gene | Controls immune process (balancing natural killer cells, γδ T cells, αβ CD8 T cells, CD4 T cells) |
| MMP1 | Matrix metallopeptidase 1 (interstitial collagenase) | Conceivable impact on colonic compliance, tone |
| OR2T35 | Olfactory receptor, family 2, subfamily T, member 35 | None |
| OR2T4 | Olfactory receptor, family 2, subfamily T, member 4 | None |
| XAF1 | X-linked inhibitor of apoptosis-associated factor 1 | Apoptosis, e.g., epithelial integrity, barrier function |
| ZNF626 | Zinc finger protein 626 | None |
| ZNF717 | Zinc finger protein 717 | None |
REFERENCES
- 1.Aerssens J, Camilleri M, Talloen W, Thielemans L, Göhlmann HW, Van Den Wyngaert I, Thielemans T, De Hoogt R, Andrews CN, Bharucha AE, Carlson PJ, Busciglio I, Burton DD, Smyrk T, Urrutia R, Coulie B. Alterations in mucosal immunity identified in the colon of patients with irritable bowel syndrome. Clin Gastroenterol Hepatol 6: 194–205, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andresen V, Montori VM, Keller J, West C, Layer P, Camilleri M. Effects of 5-hydroxytryptamine (serotonin) type 3 antagonists on symptom relief and constipation in non-constipated irritable bowel syndrome: a systematic review and meta-analysis of randomized controlled trials. Clin Gastroenterol Hepatol 6: 545–555, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balesaria S, Pell RJ, Abbott LJ, Tasleem A, Chavele KM, Barley NF, Khair U, Simon A, Moriarty KJ, Brydon WG, Walters JR. Exploring possible mechanisms for primary bile acid malabsorption: evidence for different regulation of ileal bile acid transporter transcripts in chronic diarrhoea. Eur J Gastroenterol Hepatol 20: 413–422, 2008 [DOI] [PubMed] [Google Scholar]
- 4.Barone M, Berloco P, Ladisa R, Ierardi E, Caruso ML, Valentini AM, Notarnicola M, Di LA, Francavilla A. Demonstration of a direct stimulatory effect of bile salts on rat colonic epithelial cell proliferation. Scand J Gastroenterol 37: 88–94, 2002 [DOI] [PubMed] [Google Scholar]
- 5.Camilleri M. Genetics and irritable bowel syndrome: from genomics to intermediate phenotype and pharmacogenetics. Dig Dis Sci 54: 2318–2324, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camilleri M. Peripheral mechanisms in irritable bowel syndrome. N Engl J Med 367: 1626–1635, 2012 [DOI] [PubMed] [Google Scholar]
- 7.Camilleri M, Katzka DA. Irritable bowel syndrome: methods, mechanisms, and pathophysiology. Genetic epidemiology and pharmacogenetics in irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol 302: G1075–G1084, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camilleri M, Kolar GJ, Vazquez-Roque MI, Carlson P, Burton DD, Zinsmeister AR. Cannabinoid receptor 1 gene and irritable bowel syndrome: phenotype and quantitative traits. Am J Physiol Gastrointest Liver Physiol 304: G553–G560, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camilleri M, McKinzie S, Busciglio I, Low PA, Sweetser S, Burton D, Baxter K, Ryks M, Zinsmeister AR. Prospective study of motor, sensory, psychologic, and autonomic functions in patients with irritable bowel syndrome. Clin Gastroenterol Hepatol 6: 772–781, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camilleri M, Nadeau A, Tremaine WJ, Lamsam J, Burton D, Odunsi S, Sweetser S, Singh R. Measurement of serum 7α-hydroxy-4-cholesten-3-one (or 7αC4), a surrogate test for bile acid malabsorption in health, ileal disease and irritable bowel syndrome using liquid chromatography-tandem mass spectrometry. Neurogastroenterol Motil 21: 734-e43, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camilleri M, Vazquez-Roque MI, Carlson P, Burton D, Wong BS, Zinsmeister AR. Association of bile acid receptor TGR5 variation and transit in health and lower functional gastrointestinal disorders. Neurogastroenterol Motil 23: 995–999, e458, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cingolani P, Platts A, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eusufzai S. Bile acid malabsorption in patients with chronic diarrhoea. Scand J Gastroenterol 28: 865–868, 1993 [DOI] [PubMed] [Google Scholar]
- 14.Feng J, Zhang Z, Wu X, Mao A, Chang F, Deng X, Gao H, Ouyang C, Dery KJ, Le K, Longmate J, Marek C, St Amand RP, Krontiris TG, Shively JE. Discovery of potential new gene variants and inflammatory cytokine associations with fibromyalgia syndrome by whole exome sequencing. PLoS One 8: e65033, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez-Banares F, Esteve M, Salas A, Alsina M, Farré C, González C, Buxeda M, Forné M, Rosinach M, Espinós JC, Maria Viver J. Systematic evaluation of the causes of chronic watery diarrhea with functional characteristics. Am J Gastroenterol 102: 2520–2528, 2007 [DOI] [PubMed] [Google Scholar]
- 16.Kapeller J, Houghton LA, Mönnikes H, Walstab J, Möller D, Bönisch H, Burwinkel B, Autschbach F, Funke B, Lasitschka F, Gassler N, Fischer C, Whorwell PJ, Atkinson W, Fell C, Büchner KJ, Schmidtmann M, van der Voort I, Wisser AS, Berg T, Rappold G, Niesler B. First evidence for an association of a functional variant in the microRNA-510 target site of the serotonin receptor-type 3E gene with diarrhea predominant irritable bowel syndrome. Hum Mol Genet 17: 2967–2977, 2008 [DOI] [PubMed] [Google Scholar]
- 17.Kapeller J, Möller D, Lasitschka F, Autschbach F, Hovius R, Rappold G, Brüss M, Gershon MD, Niesler B. Serotonin receptor diversity in the human colon: expression of serotonin type 3 receptor subunits 5-HT3C, 5-HT3D, and 5-HT3E. J Comp Neurol 519: 420–432, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolar GJ, Camilleri M, Burton D, Nadeau A, Zinsmeister AR. Prevalence of colonic motor or evacuation disorders in patients presenting with chronic nausea and vomiting evaluated by a single gastroenterologist in a tertiary referral practice. Neurogastroenterol Motil. 2013. October 10. 10.1111/nmo.12242 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lovell RM, Ford AC. Effect of gender on prevalence of irritable bowel syndrome in the community: systematic review and meta-analysis. Am J Gastroenterol 107: 991–1000, 2012 [DOI] [PubMed] [Google Scholar]
- 20.Lovell RM, Ford AC. Global prevalence of and risk factors for irritable bowel syndrome: a meta-analysis. Clin Gastroenterol Hepatol 10: 712–721, 2012 [DOI] [PubMed] [Google Scholar]
- 21.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mechanic LE, Chen HS, Amos CI, Chatterjee N, Cox NJ, Divi RL, Fan R, Harris EL, Jacobs K, Kraft P, Leal SM, McAllister K, Moore JH, Paltoo DN, Province MA, Ramos EM, Ritchie MD, Roeder K, Schaid DJ, Stephens M, Thomas DC, Weinberg CR, Witte JS, Zhang S, Zöllner S, Feuer EJ, Gillanders EM. Next generation analytic tools for large scale genetic epidemiology studies of complex diseases. Genet Epidemiol 36: 22–35, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mefford HC. Diagnostic exome sequencing—are we there yet? N Engl J Med 367: 1951–1953, 2012 [DOI] [PubMed] [Google Scholar]
- 24.Montagnani M, Abrahamsson A, Gälman C, Eggertsen G, Marschall HU, Ravaioli E, Einarsson C, Dawson PA. Analysis of ileal sodium/bile acid cotransporter and related nuclear receptor genes in a family with multiple cases of idiopathic bile acid malabsorption. World J Gastroenterol 12: 7710–7714, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montagnani M, Love MW, Rössel P, Dawson PA, Qvist P. Absence of dysfunctional ileal sodium-bile acid cotransporter gene mutations in patients with adult onset idiopathic bile acid malabsorption. Scand J Gastroenterol 36: 1077–1080, 2001 [DOI] [PubMed] [Google Scholar]
- 26.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res 11: 863–874, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Praslickova D, Torchia EC, Sugiyama MG, Magrane EJ, Zwicker BL, Kolodzieyski L, Agellon LB. The ileal lipid binding protein is required for efficient absorption and transport of bile acids in the distal portion of the murine small intestine. PLoS One 7: e50810, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rao AS, Wong BS, Camilleri M, Odunsi-Shiyanbade ST, McKinzie S, Ryks M, Burton D, Carlson P, Lamsam J, Singh R, Zinsmeister AR. Chenodeoxycholate in females with irritable bowel syndrome-constipation: a pharmacodynamic and pharmacogenetic analysis. Gastroenterology 139: 1549–1558, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ratuapli SK, Bharucha AE, Noelting J, Harvey DM, Zinsmeister AR. Phenotypic identification and classification of functional defecatory disorders using high-resolution anorectal manometry. Gastroenterology 144: 314–322.e2, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sciarretta G, Fagioli G, Furno A, Vicini G, Cecchetti L, Grigolo B, Verri A, Malaguti P. 75Se HCAT test in the detection of bile acid malabsorption in functional diarrhoea and its correlation with small bowel transit. Gut 28: 970–975, 1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sciarretta G, Furno A, Morrone B, Malaguti P. Absence of histopathological changes of ileum and colon in functional chronic diarrhea associated with bile acid malabsorption, assessed by SeHCAT test: a prospective study. Am J Gastroenterol 89: 1058–1061, 1994 [PubMed] [Google Scholar]
- 32.Shin A, Camilleri M, Vijayvargiya P, Busciglio I, Burton D, Ryks M, Rhoten D, Lueke A, Saenger A, Girtman A, Zinsmeister AR. Bowel functions, fecal excretion of unconjugated primary and secondary bile acids, and colonic transit in irritable bowel syndrome. Clin Gastroenterol Hepatol 11: 1270–1275.e1, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Triantis V, Saeland E, Bijl N, Oude-Elferink RP, Jansen PL. Glycosylation of fibroblast growth factor receptor 4 is a key regulator of fibroblast growth factor 19-mediated down-regulation of cytochrome P450 7A1. Hepatology 52: 656–666, 2010 [DOI] [PubMed] [Google Scholar]
- 34.Vanoevelen J, Dode L, Van Baelen K, Fairclough RJ, Missiaen L, Raeymaekers L, Wuytack F. The secretory pathway Ca2+/Mn2+-ATPase 2 is a Golgi-localized pump with high affinity for Ca2+ ions. J Biol Chem 280: 22800–22808, 2005 [DOI] [PubMed] [Google Scholar]
- 35.Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nat Rev Genet 13: 565–575, 2012 [DOI] [PubMed] [Google Scholar]
- 36.Vijayvargiya P, Camilleri M, Shin A, Saenger A. Methods for diagnosis of bile acid malabsorption in clinical practice. Clin Gastroenterol Hepatol 11: 1232–1239, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walters JR, Tasleem AM, Omer OS, Brydon WG, Dew T, le Roux CW. A new mechanism for bile acid diarrhea: defective feedback inhibition of bile acid biosynthesis. Clin Gastroenterol Hepatol 7: 1189–1194, 2009 [DOI] [PubMed] [Google Scholar]
- 38.Wedlake L, A'Hern R, Russell D, Thomas K, Walters JR, Andreyev HJ. Systematic review: the prevalence of idiopathic bile acid malabsorption as diagnosed by SeHCAT scanning in patients with diarrhoea-predominant irritable bowel syndrome. Aliment Pharmacol Ther 30: 707–717, 2009 [DOI] [PubMed] [Google Scholar]
- 39.Whitehead WE, Palsson O, Jones KR. Systematic review of the comorbidity of irritable bowel syndrome with other disorders: what are the causes and implications? Gastroenterology 122: 1140–1156, 2002 [DOI] [PubMed] [Google Scholar]
- 40.Wildt S, Norby Rasmussen S, Lysgard Madsen J, Rumessen JJ. Bile acid malabsorption in patients with chronic diarrhoea: clinical value of SeHCAT test. Scand J Gastroenterol 38: 826–830, 2003 [DOI] [PubMed] [Google Scholar]
- 41.Wong BS, Camilleri M, Carlson P, McKinzie S, Busciglio I, Bondar O, Dyer RB, Lamsam J, Zinsmeister AR. Increased bile acid biosynthesis is associated with irritable bowel syndrome with diarrhea. Clin Gastroenterol Hepatol 10: 1009–1015, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong BS, Camilleri M, Carlson PJ, Guicciardi ME, Burton D, McKinzie S, Rao AS, Zinsmeister AR, Gores GJ. A Klothoβ variant mediates protein stability and associates with colon transit in irritable bowel syndrome with diarrhea. Gastroenterology 140: 1934–1942, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong BS, Camilleri M, Carlson PJ, Odunsi-Shiyanbade S, McKinzie S, Busciglio I, Burton D, Zinsmeister AR. Pharmacogenetics of the effects of colesevelam on colonic transit in irritable bowel syndrome with diarrhea. Dig Dis Sci 57: 1222–1226, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]