Abstract
The epigenomic era has revealed a well-connected network of molecular processes that shape the chromatin landscape. These processes comprise abnormal methylomes, transcriptosomes, genome-wide histone post-transcriptional modifications patterns, histone variants, and noncoding RNAs. The mapping of these processes in large scale by chromatin immunoprecipitation sequencing and other methodologies in both cancer and normal cells reveals novel therapeutic opportunities for anticancer intervention. The goal of this minireview is to summarize pharmacological strategies to modify the epigenetic landscape of cancer cells. These approaches include the use of novel small molecule inhibitors of epigenetic processes specifically deregulated in cancer cells and the design of engineered proteins able to stably reprogram the epigenetic code in cancer cells in a way that is similar to normal cells.
Introduction
The concept of “epigenetics” was pioneered by Conrad H. Waddington in 1942 as “the branch of biology which studies the causal interactions between genes and their products, which bring the phenotype into being” (Goldberg et al., 2007; Waddington, 2012). “Epi” comes from the Greek word “over,” and thereby epigenetics is the study of the molecular, cellular, and environmental aspects of heredity, which are not explained by simple changes in the underlying DNA sequence. Epigenetic processes influence the chromatin structure, not the gene sequence per se, but the tridimensional folding and package of the DNA and associated proteins, referred to as chromatin (Strahl and Allis, 2000; Kouzarides, 2007). Heterochromatin, comprising pericentromeric regions, satellite DNA, and retroviral long terminal repeats are condensed and silenced areas of the genome (Zakrzewski et al., 2011). In contrast, euchromatin comprises the loci being actively transcribed (Gilbert et al., 2004). There are two important characteristics of the epigenetic state of cells. First, unlike somatic mutations and other genetic alterations, which irreversibly alter the genes, epigenetic processes are reversible and thus provide environmental plasticity and cell adaptation to new microenvironments. Second, the chromatin structure is inherited, and in absence of perturbations, the chromatin state has an intrinsic memory and it is transmitted over cell generations (Lange and Schneider, 2010; Margueron and Reinberg, 2010).
In the last decade it has become clear that epigenetic disruptions play a critical role in tumor initiation and progression (Berdasco and Esteller, 2010). Recent research in cancer has focused on the prognosis value of epigenetic signatures in cancer, with the advent of genome-wide large-scale profiling of tumor specimens. The emerging view of cancer epigenetics is that epimutations might affect early events of cancer, which could prime subsequent events in the progression of the disease (Carmona and Esteller, 2011; Rodriguez-Paredes and Esteller, 2011; Sandoval and Esteller, 2012). Epigenetic aberrations in cancer include both abnormal silencing (for example, silencing of tumor/metastasis suppressors) and reactivation of silenced regions (which leads to oncogenic activation). In many cases, epigenetic aberrations can collaborate with genetic changes, as for example in the case of silencing of a second allele of a tumor suppressor gene, such as BRCA (Liu et al., 2008a; Lim et al., 2009) and p16 (Arima et al., 2012). Numerous examples involve mutations in chromatin modifiers themselves, which have a profound impact in neoplastic progression. In addition to epigenetic modifiers and chromatin remodelers, multiple molecular constituents impact chromatin structure, such as transcription factors (TFs), signal transduction proteins, histone variants, and noncoding RNA, outlining the complexity and molecular crosstalk that sculpts the epigenome (Baylin and Jones, 2011).
In light of the recognition of the importance of epigenetic disruptions in cancer cells, strategies to block or revert these changes are of growing interest in cancer therapeutics. This review will focus on novel pharmacological approaches to reprogram and remodel the epigenome of cancer cells into a pattern that is similar to normal somatic cells. Ideally, this reprogramming would entail hereditary changes in chromatin resulting in long-lasting phenotypic alterations of tumor cells, namely phenotypic memory. In the first part of this minireview we will summarize the language of chemical modifications in chromatin and the main chromatin modifiers (“writers”) whose expression/and or activity is disrupted in cancer cells relative to normal tissue. Next, we will discuss novel state-of-the art approaches to revert or “rewrite” those aberrant epigenetic processes in cancer cells using both small molecule inhibitors and targeted engineered proteins.
The “Language” of Chromatin Modifications
The genome-wide epigenetic status (“epigenetic landscape”) of cells is dictated by comprehensive sets of chemical modifications that occur at both the DNA and in the histone tails. Methylation of the DNA (DNAme) occurs in position 5 in cytosine residues (Fig. 1). In mammals, the vast majority (98%) of DNAme occurs in CpG (cytosine-phosphate-guanine) dinucleotides in somatic cells. In embryonic stem (ES) cells, however, about one-quarter of all DNAme occurs in non-CpG context (Lister et al., 2009). In contrast to DNAme, histones are subjected to several chemical modifications in their N-terminal tails (Strahl and Allis, 2000), which are rather unstructured and largely exposed in the context of the three-dimensional structure of the nucleosomes (Dutnall and Ramakrishnan, 1997; Luger et al., 1997) (Fig. 2). There is large chemical diversity and a great number of histone modifications, including methylation, acetylation, phosphorylation, ubiquitination, sumoylation, ADP-ribosylation, deamination, and protein isomerization. Some histone modifications, such as H3K9me, H3K27me, and H4K20me, are typically associated with repressive chromatin. In contrast, other modifications, such as H3K4me, H3K9Ac, H3K36me, and H3K79 are connected with actively expressed genes (Kouzarides, 2007; Henikoff and Shilatifard, 2011; Turner, 2012).
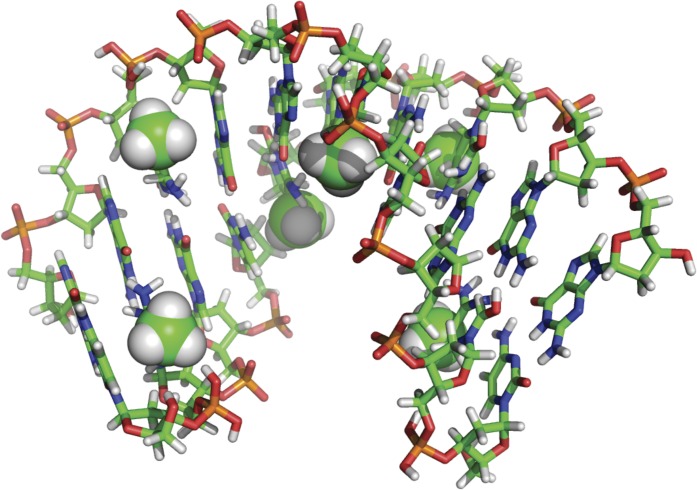
Fig. 1.
Structure of a double-stranded DNA duplex containing meCpG (383D).
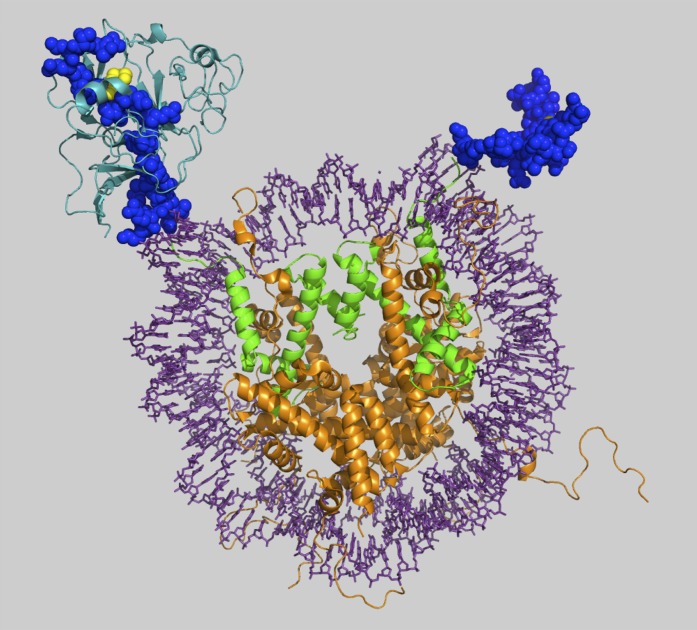
Fig. 2.
Structure of a nucleosome showing the core histones H3 in green and the histone tail protruding out of the nucleosome outlining the H3K9me2 mark in yellow (1KX5) and superimposed with the H3K9 lysine methyltransferase (2RFI).
Why is chemical language of histones so degenerate? In principle, chemical modifications can modulate chromatin structure by primarily altering the physical properties of the DNA and or/associated proteins. For example, among all the histone modifications, phosphorylation adds negative charges to the histones, whereas acetylation merely reduces the number of positive charges in chromatin. Because the DNA helix is negatively charged, chemical modifications could act to unravel the DNA from the histones, promoting chromatin relaxation. Second, chemical modifications act as binding platforms or epitopes for specific endogenous proteins (“binders” or “readers”). For example, DNAme marks are recognized by methyl-binding proteins, such as methyl-binding protein-1, histone methylation is recognized by chromo-like domains, acetylation by bromodomains, and phosphorylation by 14-3-3 proteins. “Readers” are selective proteins able to recognize specific histones and even different states of methylation, for example heterochromatin protein 1 (HP1) recognizes H3K9me3, polycomb 2 proteins recognize the H3K27me3 (Kouzarides, 2007). In the cell, endogenous readers form specific complexes with chromatin modifiers possessing catalytic activity (“modifiers” or “writers”). The complexity of epigenetic regulation is further exemplified by the discovery of multiple enzymes able to erase specific epigenetic marks. These include histone deacetylases, lysine demethylases (Klose et al., 2006; Arrowsmith et al., 2012), and the more recently discovered DNA-demethylases, which are involved in excision-repair DNA pathways (Rai et al., 2008; Bhutani et al., 2010, 2011) erasing DNA methylation patterns during development, differentiation, and reprogramming.
It has long been proposed that the chemical modifications in chromatin act in a combinatorial manner to define an epigenetic code, which coordinately shapes chromatin structure (Strahl and Allis, 2000). However, the large number of histone modifications and the possible combinations of all these primary modifications (many of them in the same histone tail) clearly add several layers of complexity and outline the plasticity of the epigenome. Among all the epigenetic modifications, DNAme and H3K9me are regarded as stable and inherited repressive marks (Margueron and Reinberg, 2010; Han and Brunet, 2012). Notably, DNAme is a key modification that has traditionally been thought to “irreversibly canalize” or “force cells” to engage into specific cell fates (Hochedlinger and Plath, 2009). In the following sections, the relationships and crosstalks between epigenetic modifications, particularly DNAme and histone modifications, will be outlined. Finally, we will briefly describe some of the key mechanisms associated with DNA demethylation in mammalian cells.
Interconnections between DNA Methylation and Histone Methylation
The interactions between “readers” and “writers” provide regulatory opportunities for the physical transmission of epigenetic marks along the chromosome (spreading) for the transmission of epigenetic marks from a mother cell to a daughter cell (inheritance of epigenetic information) and define crosstalks of multiple epigenetic constituents (Cheng and Blumenthal, 2010; Hashimoto et al., 2010). The interactions between readers and writers can be direct or indirect. Direct interactions include physical associations between epigenetic modulators. For example, the lysine methyltransferase G9a, which deposits H3K9me2 and H3K9me1 in euchromatin (Wu et al., 2010), interacts with DNA methyltransferases through ankyrin domains (Feldman et al., 2006). In self-renewal gene promoters, such as Oct4, this recruitment promotes an irreversible epigenetic switch, which forces stem cells to differentiate (Feldman et al., 2006; Tachibana et al., 2008). During DNA replication, the protein UHRF1 recognizes hemimethylated DNA and forms a complex with several proteins, including DNA methyltransferase 1 (DNMT1) (which maintains DNAme patterns during replication) and lysine methyltransferase G9a. These physical interactions link DNAme with H3K9me in the replication fork, making sure that both DNAme and H3K9me are spatiotemporally coupled processes faithfully transmitted during replication (Bostick et al., 2007; Sharif et al., 2007; Kim et al., 2009; Hashimoto et al., 2010). Another crosstalk between DNAme and H3K9me during DNA replication is mediated by methyl-CpG-binding-proteins, such as the methyl-CpG binding protein 1 (MBD1). In heterochromatin, methylated DNA is bound by MBD1, which recruits the H3K9 methyltransferase SET domain, bifurcated 1 (SETDB1) (Schultz et al., 2002), an enzyme that deposits H3K9me3 (Ichimura et al., 2005; Uchimura et al., 2006; Cheng and Blumenthal, 2010; Hashimoto et al., 2010). The latter mark is recognized by HP1 (Bannister et al., 2001; Lachner et al., 2001), which in turn recruits histone lysine methyltransferases, such as SUV39H1/2 (Krouwels et al., 2005). In addition, HP1 directly binds to the PhD-like domain of DNMT3A in vitro, which suggests that H3K9me could in turn influence DNAme (Fuks et al., 2003). The above suggest that DNAme and H3K9me are mutually reinforcing silencing marks, which might serve to synchronize these two processes during DNA replication, ensuring the inheritance of the epigenetic information (Fig. 3). The crosstalk between DNAme and H3K9me has also been described in plants (Johnson et al., 2007).
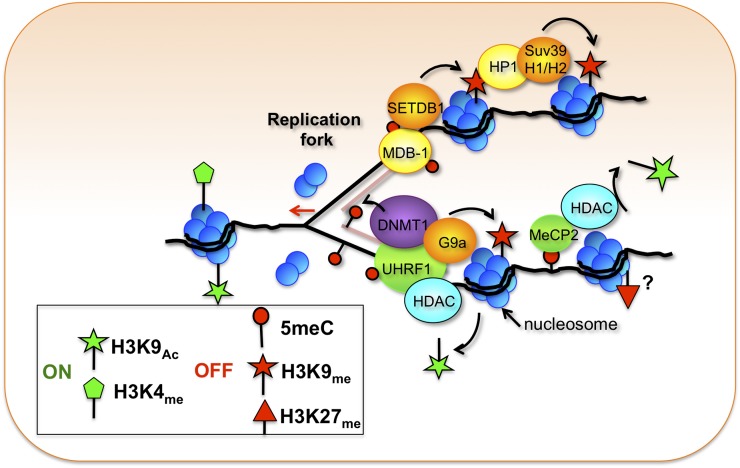
Fig. 3.
A model outlining the crosstalk of some epigenetic modifications during replication and after the replication fork. HDAC, histone deacetylase; Suv39, suppression of variegation 39 homolog 1 or 2 (Drosophila); UHRF, Ubiquitin-like with PHD and Ring Finger domains 1.
A link between DNAme and histone deacetylation has been long demonstrated through the discovery of the methyl-CpG-binding protein 2 (Scarsdale et al., 2011). Methyl-CpG-binding protein 2 carries a transcriptional repressor domain, which recruits the corepressor histone deacetylase (Singal et al., 2002). Reciprocally, histone acetylation is a mark associated with promoter activation and gene transcription. H3/H4 lysine acetylation is recognized by bromodomain-containing proteins, often physically linked to plant homeodomain (PHD) fingers and catalytic domains of histone acetyltransferases, such as pCAF (Clements et al., 1999; Nagy and Tora, 2007) and p300 (Liu et al., 2008b; Wang et al., 2008a). Bromodomains in the subunits of chromatin-remodeling enzymes, such as switch/sucrose nonfermentable family, interact with acetylated histones (Reinke and Horz, 2003; Kundu et al., 2007; Gao et al., 2008; Kizilyaprak et al., 2010), which anchor remodeling activities in the nucleosomes and serve to promote nucleosome eviction (Suganuma and Workman, 2011). In summary, direct physical interactions between readers and writers, and readers and erasers, could contribute to a quick spread of a given mark(s) along the genome, leading to effective chromosomal condensation and gene silencing or chromatin relaxation and gene activation.
The interaction or crosstalk between epigenetic processes could be also indirect. A residue of a given histone can be a substrate of multiple modifications. For example, the incorporation of H3K9me by histone lysine methyltransferases could indirectly block the acetylation of the same histone residues by histone acetyltransferases. Another indirect example of epigenetic crosstalks is the incorporation of DNAme, which might negatively affect H3K4me. Methylated DNA is particularly refractory to binding of some DNA-binding proteins, including the zinc finger protein Cfp1, which preferentially binds unmethylated DNA. Cfp1 interacts and recruits H3K4 methyltransferase SETD1, and thereby induction of DNAme might indirectly trigger a decrease on H3K9me (Xu et al., 2011), potentially explaining the dramatic reverse correlation between these two marks (Thomson et al., 2010). Reciprocally, high levels of Cfp1 in the genome could protect these regions from DNA methylation and gene silencing.
The relationship between DNAme and the silencing mark H3K27me3 is currently less well understood. Multiple groups had proposed that in cancer cells H3K27me3 could mark genes subsequently silenced by DNAme, although many developmentally regulated genes are marked H3K27me3 in the absence of DNAme (Ohm et al., 2007; Schlesinger et al., 2007; Widschwendter et al., 2007). The existence of a mutually reinforcing mechanism linking DNAme and H3K27me3 is a topic of intensive research (Hon et al., 2012).
Context-Dependence of the Epigenetic Modifications
The advent of genome-wide approaches for the mapping of chromatin modifications, such as chromatin immunoprecipitation sequencing, has established the relationships and context dependence of epigenetic modifications, both in embryonic, induced pluripotent, and somatic cells (Chari et al., 2010; Zhou et al., 2011). For example, in promoter regions, DNAme strongly correlates with some histone modifications, specifically with high levels of H3K9me and low levels of H3K4me. In fact, DNAme is a better predictor of H3K9me and H3K4me than the underlying genome context (Mikkelsen et al., 2007; Meissner et al., 2008). Unlike promoter regions, DNAme inside the gene bodies is not correlated with silencing. Widespread low-density of methylation has been proposed to serve as a mechanism to avoid inappropriate transcription initiation from cryptic promoters (Bird, 1993; Tran et al., 2005).
Genome-wide maps of histone modifications in stem and lineage-committed progenitor cells have revealed that H3K4me3 marks the genes that are actively expressed. The “bivalent state” (H3K4me3 and H3K27me3) labels developmental genes poorly expressed that are poised for either expression or repression (Bernstein et al., 2006). In addition, the H3K27me3 mark is retained in genes that are stably repressed. H3K9me3 and H4K20me3 are characteristic of satellite, telomeric, and long-terminal repeats. Finally, H3K36me3 marks coding and noncoding transcripts (Mikkelsen et al., 2007; Meissner et al., 2008).
Mechanisms of DNA Demethylation
The discovery of active and passive mechanisms to erase DNAme has opened the door to a new paradigm: DNAme and demethylation may not be “one-way” streets as initially thought but highly dynamic and regulated processes (Bhutani et al., 2011). The current view of DNA demethylation involves the integration of multiple interconnected pathways. DNA demethylation can be the result of passive mechanisms, for example, by dilution during replication or inactivation of DNMTs. In addition to passive demethylation, several enzymatic activities have been recently associated with active demethylation and DNA repair. Much excitement has surfaced since the discovery of 5-hydroxymethylcytosine in the mammalian genome in neuronal and ES cells (Kriaucionis and Heintz, 2009; Tahiliani et al., 2009) and its role as an intermediate and key player in DNA demethylation during DNA repair. The ten-eleven translocation (TET) protein family members catalyze the active modification of 5meC to give 5-hydroxymethylcytosine. TET proteins are 2-oxoglutarate and Fe(II)-dependent dioxygenases that catalyze the conversion of 5-methylcytosine in 5hC. TET1 was first identified in acute myeloid leukemia as a fusion protein with the mixed lineage leukemia factor and was designated as leukemia associated protein (Ono et al., 2002). TET1 contains a CXXC zinc finger DNA-binding domain that binds preferentially CpG-rich sequences near the transcription start site of the targeted genes (Williams et al., 2011; Wu et al., 2011). TET1 and TET2 are expressed in mouse ES cells and are necessary for cell lineage specification (Ficz et al., 2011; Koh et al., 2011). TET3 mediates the conversion of 5-methylcytosine to 5hC in the paternal pronucleus after fertilization (Gu et al., 2011; Iqbal et al., 2011).
Recently, the activation-induced cytidine deaminase (AID) has been recognized to participate in DNA demethylation. AID has a pivotal role in generating antibody diversity in lymphocytes. Unlike TET proteins, AID lacks of a DNA binding activity. IAD catalyzes deamination of cytosine residues to uracil. This step is followed by an excision repair or mismatch repair pathway, which is error prone. In addition to this function to generate the antibody repertoire, the AID enzyme has been implicated in demethylation in zebrafish (Rai et al., 2008) and in mammals. Hence, mice lacking AID exhibit global DNA hypermethylation in the primordial germ cells (Popp et al., 2010). Active DNA demethylation is believed to be enzymatically coupled with the replacement of the modified cytosine via DNA repair. DNA glycosylases, such as those belonging to the base excision repair pathway have been involved in the removal of modified cytosine in plants (Gehring et al., 2009). In mammals, these enzymes could act in concert with TET and AID enzymes to promote demethylation. In this model, 5-methylcytosine and 5-hydroxymethylcytosine are first deaminated by AID to thymine and 5-hydroxymethyluracil, respectively, followed by glycoylase-mediated thymine and 5-hydroxymethyluracil excision repair (Cortellino et al., 2011).
Epigenetic Alterations in Cancer
Tumor cells show multidimensional aberrations affecting many levels of regulation, including genetic changes, but also profound modifications in the epigenetic landscape. The “cancer methylome” is therefore distinct from normal, nontransformed tissue. Characteristic hallmarks of cancer include a global genome-wide hypomethylation and also hypermethylation of CpG islands (which is typically associated with gene repression) (Berdasco and Esteller, 2010; Portela and Esteller, 2010; Rodriguez-Paredes and Esteller, 2011; Sandoval and Esteller, 2012). However, a substantial fraction of DNA methylation occurs in the cancer methylome away from the transcription start site (~2 kb or more), comprising microRNAs and mirtrons (Dudziec et al., 2011)—these dinucleotides are named CpG shores (Irizarry et al., 2009; Sandoval et al., 2011).
Global DNA hypomethylation and local hypermethylation has recently been confirmed by whole genome shotgun sequencing of bisulfite-treated DNA (methyl-sequencing) of a low-passage breast cancer cell HCC1954 relative to normal epithelial cells (HUMEC cell line). This study revealed that the majority of hypomethylated regions in cancer cells correlated with partially methylated domains in normal cells. Surprisingly, in the tumor cells, these regions exhibited mono-allelic DNA methylation, one allele displaying DNAme and the other allele possessing the repressive histone marks H3K9me3 or H3K27me3. Thus, hypomethylated regions in cancer cells were correlated with silenced gene expression and repressed chromatin (Hon et al., 2012). In an independent study, differentially methylated regions have been mapped in colon cancer relative to normal tissue. These regions were associated with high gene expression variability. Whole genome bisulfite sequencing has also confirmed hypomethylation in discrete blocks in about half of the cancer genome. These data suggest a model of neoplastic progression involving loss of epigenetic stability and tumor heterogeneity (Hansen et al., 2011). In another recent study in colon cancers versus matched normal tissue, focal CpG island hypermethylation was also detected within long-range (>100 base pairs) hypomethylation regions. In this case, the widespread changes of DNA methylation in cancer was correlated with repressive chromatin and by the three-dimensional organization of the chromatin inside the nucleus (Berman et al., 2012)
In mammalian cells, DNA methylation outside promoter regions frequently occurs in long-terminal repeats of viral origin and other repetitive regions named satellite regions. The methylation of these sequences in healthy cells could serve to protect the genomes by preventing the expression of exogenous promoters. Hypomethylation of preferentially repetitive sequence classes including short interspersed elements, long interspersed elements (e.g., LINE-1 elements), satellite, subtelomeric, and Alu repeats, has been associated with tumor progression in several cancer models, such as colon (Rodriguez et al., 2008; Sunami et al., 2011), squamous cell carcinomas of the lung (Pfeifer and Rauch, 2009), head, neck (Szpakowski et al., 2009), and mammary tumors (Costa et al., 2006). Hypomethylation of repetitive regions could prime genomic instability and also reactivate potential oncogenes during transformation, tumor progression, and metastasis. Importantly, the use of global demethylating agents for the treatment of cancers (e.g., 5-Aza-2′dC) in in vitro models has indicated that these inhibitors could prime tumor progression by re-expression of silenced oncogenes, such as Sox2 in chondrosarcoma cells (Hamm et al., 2009) and c-Met in colon carcinoma cell lines (Weber et al., 2010).
The genome-wide mapping of DNAme changes in tumor versus normal cells could help investigators gain valuable prognosis information in cancer patients and help not only to identify single gene hits important for tumor progression, but also to define prognosis signatures. Esteller et al. (2000) demonstrated that hypermethylation of CpG islands in the promoter of the gene O-6-methylguanine-DNA methyltransferase is a predictor of good response of glioma cancer patients to alkylating agents (Esteller et al., 2000). Since then, genomic approaches have identified groups of biomarkers associated with poor prognosis and responsiveness to therapeutic drugs, such as in pancreatic (Gupta et al., 2012) and colon cancers (Esteller et al., 2001). In glioblastoma multiforme, a genome-wide study by Martinez and colleague demonstrated that genes hypermethylated in glioblastoma multiforme were highly enriched for targets of the polycomb repressive complex 2 in embryonic stem cells, and thus, these tumors exhibited a methylome with stem cell-like features (Martinez et al., 2009; Martinez and Esteller, 2010).
During the process of immortalization, tumorigenesis, and tumor progression precancerous cells might acquire a “hypermethylator phenotype” affecting several tumor suppressor gene promoters (Sato et al., 2010). Methylation of p16 is a key gateway control for cells to overcome senescence barriers, and together with overexpression of telomerase these events might be crucial for the onset of the tumorigenic process (Gallardo et al., 2004; Beltran et al., 2011a). Another example in breast and ovarian cancers is the methylation of the BRCA1 tumor suppressor (Jacinto and Esteller, 2007; Stefansson et al., 2011), a gene associated with DNA repair, thereby promoting further genomic instability (Jacinto and Esteller, 2007). Mutations of BRCA genes have been associated with expansion of stem/progenitor cells and basal-like cancer (Liu et al., 2008a; Rakha et al., 2008). For these genes, methylation and genetic alterations can contribute to inactivating both copies of the tumor suppressor. Some tumor suppressors, namely class II tumor suppressor genes, are solely inactivated by epigenetic mechanisms. One classic example is the tumor suppressor mammary serine protease inhibitor (maspin), in which the degree of promoter methylation clearly correlates with disease stage and metastatic behavior in multiple cancer models, such as breast, lung, and prostate cancer (Zou et al., 1994; Futscher et al., 2002, 2004). In the maspin promoter, DNAme clearly correlates with H3K9me2 (Wozniak et al., 2007), which could be explained by the tight epigenetic crosstalk established between DNA methyltransferases and the H3K9 methyltransferase G9a.
The era of cancer epigenomics has recently identified the extent of the histone modifications in the cancer genome relative to normal tissue, leading to aberrant gene activation or repression (Rodriguez-Paredes and Esteller, 2011; Sandoval and Esteller, 2012). In non-small lung cancer, high dimethyl-H3K4 and low acetyl-H3K9 has been associated with favorable prognosis (Song et al., 2012). As discussed earlier, aberrant H3K4me and H3K27me have also been detected in prostate cancer cells (Ke et al., 2009) and human colorectal cancer (Enroth et al., 2011).
What is the mechanistic basis of aberrant epigenetic regulation in cancer cells? In many cases, mutations or disruption in gene expression of the chromatin modifiers themselves have been identified. For example, the polycomb group of methyltransferases, such as enhancer of zeste homolog 2 (drosophila), are often overexpressed in basal-like breast cancers, particularly in African-American women (Chase and Cross, 2011; Pang et al., 2012). The histone methyltransferase G9a (Chen et al., 2010) and other lysine methyltransferases have also been associated with tumorigenesis and targeted by screening and structure-based design for the identification of specific inhibitors (Unoki, 2011). Deregulation of TF gene expression might cause profound effects in the epigenetic landscape of cells. During reprogramming of somatic cells toward pluripotency, the exogenous delivery of Oct4, Sox2, Klf4, and c-Myc TFs is sufficient to change the epigenetic architecture of the genome in a way that is similar to embryonic ES cells (Takahashi and Yamanaka, 2006; Takahashi et al., 2007). Moreover, intensive research revealed that induced pluripotent stem cells are not identical to ES cells, retaining epigenetic features (epigenetic memory) from the cell of origin (Takahashi and Yamanaka, 2006; Takahashi et al., 2007; Kim et al., 2010; Polo et al., 2010; Lister et al., 2011; Ziller et al., 2011). The “pluripotency” TFs activate a cellular network involving many other self-renewal TFs and chromatin remodelers. An imbalance in normal gene expression due to changes in expression or activity of specific TFs could lead to dysregulation of the epigenome of cells, resulting in cancer predisposition. In the mammary gland, our laboratory has demonstrated that overexpression of the TF Oct4 leads to aberrant epithelial to mesenchymal transition and to the generation of clones possessing tumor initiation capabilities (Beltran et al., 2011a).
In addition to coding regions, many noncoding RNAs have been associated with tumor progression (Veeck and Esteller, 2010). An explosion of studies has demonstrated global changes in expression of micro-RNA signatures between normal and tumor cells in multiple tumor types, including a recent study in lung cancer (Guan et al., 2012). Interestingly, some microRNAs are epigenetically silenced and act as tumor suppressors, whereas many others act as oncogenes (Cho, 2012). The significance of the complex noncoding-RNA circuitry in cancer development is not well understood; microRNAs could fine tune the threshold levels of critical targets involved in development and perhaps in cancer, illustrating the complexity of the regulatory network controlling cancer progression.
Strategies to Revert the Epigenetic Landscape of Cancer Cells Using Small Molecules and Engineered Proteins
The epigenetic deregulation of tumor cells comprising the aberrant silencing of tumor suppressors and the hypomethylation of potential oncogenes creates the basis of novel therapeutic strategies to revert their chromatin state. The endogenous silencing of tumor suppressors can be reverted with epigenetic inhibitors, such as DNA methyltransferase, histone methyltransferase, and histone deacetylase inhibitors. An active area of research aims at developing chemical approaches to inhibit specific “writers” or “erasers,” some of which will be reviewed in the following sections. Moreover, it is believed that these agents’ lack of locus selectivity may result in potential off targets and might cause adverse effects in cancer patients. Another approach is the design of artificial proteins or artificial transcription factors (ATFs) able to recognize specific sequences in the genome, directing a chromatin modification or combinations of chromatin modifications, in specific loci.
Selective Inhibitors of Bromodomains and Methyl-Lysine Readers
The acetylation and methylation of lysine residues play a central role in chromatin function, substantially through creation of binding sites for the readers of these marks (Taverna et al., 2007; Wang et al., 2008b). Although acetylation of lysine eliminates the residue’s positive charge (akin to the significant change in chemical properties and charge upon phosphorylation of serine, threonine, and tyrosine), methylation is a more subtle post-translational modification, simply shifting the chemical properties of lysine toward a more diffuse, polarizable positive charge as methylation proceeds from monomethyl (Kme1) to trimethyl (Kme3) (Zacharias and Dougherty, 2002; Hughes et al., 2007). Recently, small molecule probes of bromodomains, the readers of acetylated lysine, have been reported, and their utilization has revealed bromodomain regulation of critical gene transcription events such as the regulation of the c-myc oncogene (Filippakopoulos et al., 2010; Dawson et al., 2011; Delmore et al., 2011). Although the current repertoire of acetyl lysine readers in the human genome is limited primarily to the structurally homologous 57 bromodomains and possibly a subset of PHDs (Zeng et al., 2010), there are more than 200 methyl-lysine (Kme) reader domains described within several protein families: PHD; the so-called "royal family" made up of Tudor, Agenet, chromo, (Pro-Trp-Trp-Pro), and MBT (malignant brain tumor) domains; the WD40 repeat proteins consisting of WDR5, and EED (Adams-Cioaba and Min, 2009; Margueron et al., 2009). This large Kme reader family seems destined to expand further as research in epigenetics continues (Kuo et al., 2012; Nakamura et al., 2012). A unifying feature of all of these domains is the presence of an “aromatic cage” that comprises the Kme binding pocket (Li et al., 2007b; Santiago et al., 2011). Computational analysis suggests that this target class has significant drugability (Santiago et al., 2011), and progress toward chemical probes for Kme readers is summarized here (Frye et al., 2010; Herold et al., 2011a).
In contrast to many protein target classes, such as G protein-coupled receptors, nuclear receptors, and protein kinases, where ligand discovery sometimes proceeded determination of high-resolution protein structures by decades, Kme binding sites were relatively well explored structurally prior to any reports of potent, small molecule ligands (Ruthenburg et al., 2007; Adams-Cioaba and Min, 2009). As a consequence, approaches toward ligands have been based primarily upon computational and structural techniques (Campagna-Slater and Schapira, 2010; Campagna-Slater et al., 2010; Kireev et al., 2010). In the first report of experimentally confirmed Kme binding domain small molecule ligands (MBT), Kireev et al. (2010) used both a two-dimensional and three-dimensional virtual screening approach to select ligands from a computationally curated and procurable set of 5.8 million small molecules. Of 51 compounds tested in a histone-peptide competitive dose response proximity assay (Wigle et al., 2010a), 19 compounds produced a measurable IC50 in the 5–100 μM range. All active compounds from this screen contained a mono- or dialkylamine that presumably satisfies the mono- and dimethyl-lysine histone peptide preference of the MBT domains (Kireev et al., 2010). Subsequently, Herold et al. (2011b) described the structure- and pharmacophore-based design of UNC669 (Fig. 4) that demonstrates a 5 μM dissociation constant (Kd) versus L3MBTL1 (Herold et al., 2011b). The conformationally rigid piperidino-pyrrolidine Kme mimic in UNC669 results in a ligand with fivefold greater affinity for L3MBTL1 than the H4K20me1 peptide—a presumed endogenous binding partner. UNC669 demonstrated moderate to high selectivity as a ligand for L3MBTL1 versus a small panel of other Kme binders with the next highest affinity being for L3MBTL3 (Herold et al., 2011b). A subsequent structure-activity relationship study of the UNC669 template revealed that pyrrolidine is an optimal size and shape for binding in the aromatic cage of L3MBTL1 that recognizes Kme and that an increase in ring size to piperidine was not tolerated (Herold et al., 2012).
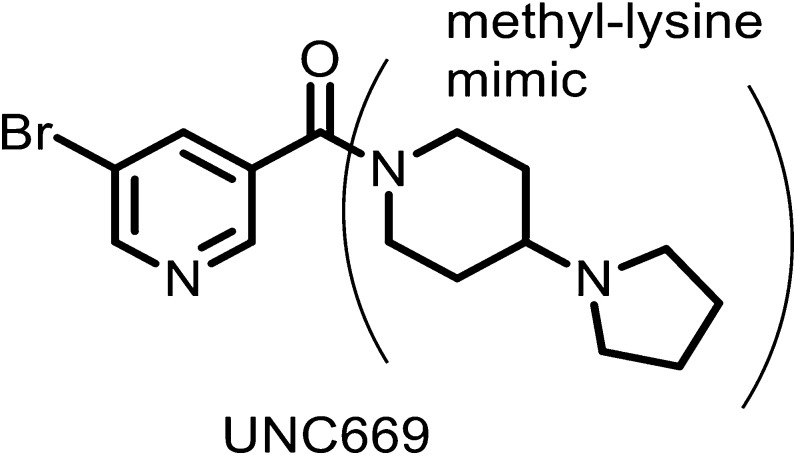
Fig. 4.
Structure of UNC669. See PDB 3P8H for structure of UNC669 bound to L3MBTL1.
Although these first reports of antagonists of Kme binding domains have described ligands of modest potency, the functional conservation of the pi-cation Kme recognition motif across this large and structurally diverse domain family bodes well for improvements in affinity as a further structure-activity relationship is developed (Herold et al., 2011a).
Selective Inhibitors of Protein Lysine Methyltransferases
Protein lysine methylation is increasingly recognized as an important type of post-translational modification. This modification has been heavily studied in the context of epigenetic regulation of gene expression through histone lysine methylation (Strahl and Allis, 2000; Jenuwein and Allis, 2001; Martin and Zhang, 2005; Bernstein et al., 2007; Kouzarides, 2007; Gelato and Fischle, 2008), but numerous nonhistone substrates suggest that the impact of lysine methylation is not limited to chromatin biology (Huang et al., 2006, 2007, 2010; Rathert et al., 2008). Protein lysine methyltransferases (PKMTs, also known as histone methyltransferases) catalyze mono-, di-, and/or trimethylation of lysine residues of histone and nonhistone proteins (Kouzarides, 2007; Copeland et al., 2009). Since the first PKMT was characterized over a decade ago (Rea et al., 2000), more than 50 PKMTs have been identified (Kouzarides, 2007; Copeland et al., 2009; Wu et al., 2010). Growing evidence suggests that PKMTs play critical roles in various human diseases including cancer (Bedford and Richard, 2005; Martin and Zhang, 2005; Fog et al., 2007; Cole, 2008; Esteller, 2008; Copeland et al., 2009; Spannhoff et al., 2009a, 2009b; Bissinger et al., 2010; Frye et al., 2010; Yost et al., 2011; He et al., 2012 ). Selective inhibitors of PKMTs are valuable tools for dissecting biologic functions of these proteins and assessing their potential as therapeutic targets. In this part of the review, we briefly discuss the selective PKMT inhibitors discovered to date (Fig. 5).
Fig. 5.
Selective Inhibitors of PKMTs. DOT1L, DOT1-like; SMYD2, SET and MYND domain containing 2.
The discovery of BIX01294, the first selective PKMT inhibitor by Jenuwein and colleagues (Kubicek et al., 2007) was a major milestone in the PKMT inhibitor field. BIX01294 has good in vitro potency against G9a and G9a-like protein (GLP) and is selective for G9a and GLP over SUV39H1 and SETDB1 and SETD7 (Kubicek et al., 2007; Chang et al., 2009). BIX01294 is also active in cells. However, it has a poor separation of functional potency and cell toxicity (Chang et al., 2010; Vedadi et al., 2011; Yuan et al., 2012). Optimization of this quinazoline scaffold via structure-based design and synthesis by Jin and coworkers led to the discovery of UNC0638 (Vedadi et al., 2011), a chemical probe of G9a and GLP and several other potent and selective inhibitors of G9a and GLP (Liu et al., 2009, 2010, 2011). UNC0638 has balanced high in vitro potency versus G9a/GLP and desirable physicochemical properties and is >100-fold selective for G9a and GLP over a broad range of epigenetic and nonepigenetic targets (Vedadi et al., 2011). Importantly, UNC0638 has robust on-target activities in cells and an excellent separation between functional potency and cell toxicity (Vedadi et al., 2011). Cheng et al. (2010) also discovered a potent G9a and GLP inhibitor E72 based on the quinazoline template via structure-based design and synthesis. E72 has high in vitro potency versus G9a and GLP and is inactive against SUV39H2 (Chang et al., 2010). E72 is also active in cells and shows reduced cell toxicity compared with BIX01294 (Chang et al., 2010). X-ray cocrystal structures of these G9a/GLP inhibitors in complex with GLP or G9a reveal that these quinazoline-based inhibitors occupy the substrate binding groove and do not occupy the cofactor binding site (Chang et al., 2009, 2010; Liu et al., 2009; Vedadi et al., 2011). These findings were confirmed by mechanism of action studies, which reveal that UNC0638 and its analog are competitive with the peptide substrate and noncompetitive with the cofactor (Wigle et al., 2010b; Vedadi et al., 2011). More recently, Schreiber and coworkers discovered a cofactor-competitive G9a inhibitor BRD9539 (Yuan et al., 2012), which is structurally distinct from quinazoline-based G9a/GLP inhibitors. BRD9539 has moderate in vitro potency versus G9a and EZH2 and is selective over SUV39H1, NSD2, DNMT1, 16 other chromatin-modifying enzymes, and 100 kinases (Yuan et al., 2012). BRD4770, the methyl ester of BRD9539, is active in cells with a cellular potency similar to the in vitro potency of BRD9539 (Yuan et al., 2012).
The discovery of EPZ004777, the first highly selective, cofactor-competitive PKMT inhibitor by Pollock and coworkers (Daigle et al., 2011) was also a significant advancement in the PKMT inhibitor discovery field. EPZ004777, a nucleoside-based inhibitor, is highly potent versus DOT1L (Ki = 0.3 nM) and >1,000-fold selective for DOT1L over other PKMTs and PRMTs (protein arginine methyltransferases) tested (Daigle et al., 2011). In addition, EPZ004777 selectively kills off cells bearing mixed lineage leukemia translocation and continuous infusion of EPZ004777 via a mini-pump increases animal survival in a mouse mixed lineage leukemia xenograft model (Daigle et al., 2011). Song and coworkers also discovered potent, selective, cofactor-competitive, nucleoside-based inhibitors of DOT1L and obtained an X-ray cocrystal structure of DOT1L in complex with one of their inhibitors (Yao et al., 2011). Compound 1, the most potent and selective inhibitor in their series, was proposed to be capable of covalently modifying the enzyme (Yao et al., 2011).
In 2011, Ferguson et al. (2011) reported the discovery of a potent SMYD2 inhibitor AZ505 (IC50 = 120 nM), which is selective for SMYD2 over a panel of PKMTs including SMYD3, DOT1L, EZH2, GLP, G9a, and SETD7. The X-ray cocrystal structure of AZ505 in complex with SMYD2 reveals that AZ505 occupies the peptide binding groove, which was confirmed by the findings from mechanism of action studies: AZ505 is competitive with the peptide substrate and uncompetitive with the cofactor (Ferguson et al., 2011). Most recently, an X-ray cocrystal structure of compound 2 in complex with SETD7 has been reported in the protein data bank (4E47). Although the in vitro potency of this SETD7 inhibitor has not been reported, the X-ray cocrystal structure reveals that this inhibitor occupies the substrate-binding groove and does not occupy the cofactor binding site.
GSK-A was recently reported as the first selective, cofactor-competitive inhibitor of EZH2 by Diaz et al. (2012). GSK-A (Ki = 700 nM), which was identified via high throughput screening, is selective for EZH2 over a panel of PKMTs, PRMTs, and DNMTs. It is competitive with the cofactor and noncompetitive with either the peptide or nucleosome substrate. GSK-A is cell permeable and reduces cellular levels of H3K27me3 with an IC50 of 8 μM. Most recently, Knutson et al. (2012) discovered EPZ005687, a highly potent, selective, and cofactor-competitive inhibitor of EZH2, via high throughput screening and subsequent optimization. EPZ005687 (Ki = 24 nM) is 50-fold selective for EZH2 over EZH1, >500-fold selective over 15 other PKMTs and PRMTs, and at least 60-fold selective over a broad panel of G protein-coupled receptors and ion channels. Similar to GSK-A, EPZ005687 is competitive with the cofactor and noncompetitive with the nucleosome substrate. It inhibits wild-type EZH2 and 6 Y641 or A677 EZH2 mutants with similar in vitro potencies and reduces H3K27 methylation in various lymphoma cells with high cellular potencies. Importantly, EPZ005687 selectively induces apoptotic cell killing in heterozygous Y641F or A677G mutant cells and has little effect on the proliferation of wild-type lymphoma cells.
Artificial Transcription Factors as Locus-specific Epigenetic Modulators
Locus-specific targeting of epigenetic domains in chromatin requires minimally two modules: a DNA-recognition domain, which has sequence selectivity, and an effector domain, which is engineered to modify chromatin (Beltran and Blancafort, 2010). Most engineered proteins were constructed using zinc finger (ZF) scaffolds, and more recently, transcription activator-like effectors (TALEs), which are bacterial proteins of the genus Xanthomonas that are injected into infected plants (Mussolino and Cathomen, 2012). The DNA-binding specificity of a TALE is a tandem array of ~15.5–19.5 single repeats, each repeat containing ~34 conserved residues. The binding specificity is dictated by positions 12 and 13 of the repeats, known as repeat variable residues. Thereby, the TALE code is a simple 1:1 interaction, one repeat unit to one nucleotide (Moscou and Bogdanove, 2009; Deng et al., 2012;). Recently, several laboratories have designed artificial TALEs for gene regulation and genome editing using nuclease technology (Bogdanove and Voytas, 2011; Bultmann et al., 2012). The on-targets and potential off-targets of these emerging technologies are still under investigation.
Sequence specific Cys2-His2 ZFs have been engineered by several approaches, including “helix grafting” (directed mutations in the α-helical recognition sequence), and by in vitro as well as in vivo selection experiments in both bacteria and yeast (Beltran et al., 2006)). These selection experiments have overcome the context dependence of the assembly of the ZF units, generating proteins with potentially improved specificity for the target DNA. Each ZF module recognizes primarily three base pairs of DNA, and most studies have engineered polydactyl proteins consisting of three or more ZF proteins. Thereby, 6ZF proteins will recognize an 18-base pair recognition site in the DNA with potentially unique specificity in the genome (Tan et al., 2003), although potential off-targets might also occur in the cell due to the binding of the engineered protein to closely related binding sites. ZF proteins have been linked to several activator or repressor domains, promoting specific activation or repression of the targeted gene by recruiting specific coactivator or corepressors (Sera, 2009; Stolzenburg et al., 2010). However, such a first generation of effector domains does not possess intrinsic catalytic activity to modify either the histones or the DNA. Consequently, these effectors are expected to have a transient effect in the target cell and are not capable of inducing epigenetic memory (transmission of epigenetic modifications over the cell progeny). In contrast, the linkage of ZFs with epigenetic modifiers possessing enzymatic activity is expected to directly modify the histones or the DNA. Thus, some of these effectors are potentially able to promote hereditable changes in chromatin (Rivenbark et al., 2012).
Among all epigenetic marks, DNAme and H3K9me are recognized as being hereditable marks, or transmitted over the cell progeny. Several studies have demonstrated that linkage of a catalytic domain of bacterial methyltransferases with designer ZFs leads to targeted DNAme in plasmid-based assays or integrated reporters (Carvin et al., 2003; Minczuk et al., 2006; Li et al., 2007a; Smith and Ford, 2007; Smith et al., 2008). To avoid untargeted methylation, programmable DNA methyltransferases have been constructed, in which the expression of two independent ZF proteins can complement or restore the activity of a functional DNA methyltransferase enzyme (Nomura and Barbas, 2007; Meister et al., 2010). Interestingly, triple-helix forming oligonucleotides have been coupled with a DNA methyltransferase M.SssI to promote targeted methylation of the EpCAM oncogenic promoter (van der Gun et al., 2010). Our laboratory recently reported the first endogenous targeted DNA methyltransferase activity in a cancer genome, by tethering 6ZF domains with the catalytic domain of human Dnmt3a, an enzyme that catalyzes the de novo DNAme (Fig. 6). When linked with a 6ZF promoting homing to a tumor suppressor gene promoter, such as Maspin, the ZFs-Dnmt3a fusions promoted targeted DNAme, which was accompanied by an increased tumorigenic ability of the breast cancer cells in vitro. In contrast, in the context of an oncogene, such as Sox2, 6ZF linked to Dnmt3a promoted cancer cell growth arrest. Importantly, these studies suggested that the silencing initiated by the ZFs-Dnmt3a fusions was maintained over more than 50 cell generations. In fact, DNAme patterns were faithfully transmitted over cell generations even when after the ZF protein was no longer expressed in the tumor cells (Rivenbark et al., 2012). Overall, these studies suggested that ZFs-Dnmt3a constructs were able to stably reprogram the epigenetic state of the targeted promoters. In addition to DNA methyltransferases, ZF proteins have been linked to histone methyltransferase domains, such as SUV39H1 and G9a, targeting H3K9me marks in the context of the vascular endothelial growth factor A gene. These chimeras were sufficient to target silencing of the endogenous gene (Snowden et al., 2002).
Fig. 6.
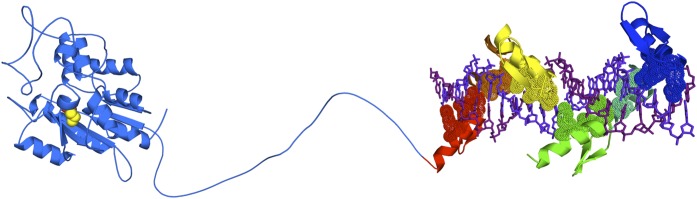
Schematic structure of the catalytic domain of Dnmt3a (2QRV) linked to a 6ZF-DNA complex.
Reactivation of epigenetically silenced genes has also been achieved with ZF proteins. Gene activation, however, appears to be fundamentally more challenging than targeted silencing due to the decreased accessibility of the targeted region. Reactivation of silenced and proapoptotic genes has been reported for maspin (Beltran and Blancafort, 2011), Bax (Falke et al., 2003), and p53-dependent pathways using ZF library screenings (Blancafort et al., 2008). Highly methylated promoters, for example tumor suppressors, are associated with condensed chromatin, which could impede TFs to find their targets in chromatin. In the case of the maspin tumor suppressor, for example, the degree or extent of transactivation by ZF proteins is highly dependent on the targeted region of the promoter and in the DNA methylation level of the cell line examined (Beltran et al., 2011b). We have characterized an activator ATF able to reactivate silenced maspin in breast and lung cancer lines having high degree of promoter methylation. When delivered in these cell lines, the ATF suppressed breast cancer growth and totally abolished metastatic colonization (Lund et al., 2004; Beltran, 2011; Beltran et al., 2011b). Moreover, the effect of the ATF in reactivating endogenous maspin was highly synergistic with small molecule inhibitors of DNA methyltransferases and histone deacetylases (Beltran et al., 2008). Similarly, in the context of an epigenetically silenced oncogene, such as Oct4, TALE activators designed against the promoter were able to reactivate Oct4 only in presence of epigenetic inhibitors, such as 5-Aza-dC and trichostatin A (Bultmann et al., 2012). These experiments indicated that compact chromatin represented a partial blockade for ZFs and TALE technologies and generally TFs to reach high levels of regulation of silenced genes. Future engineering of both ZFs and TALEs might benefit from linkage of the DNA binding domains with chromatin remodelers, which might help to unravel the silenced chromatin. The discovery of novel DNA demethylases able to erase the 5meC of DNA represents a powerful new engineering avenue.
ATFs Versus Small Molecule Inhibitors
The “pros” and “cons” for the use of ATFs and small molecules for anticancer treatment are summarized in Table 1. The design and optimization of DNA-binding domains in ATF backbones opens the possibility to regulate single genes in the cancer genome. Nevertheless, the spectrum of targets regulated by the ATF highly depends on the choice and primary selectivity of the DNA-binding domains used for the construction of the proteins. Investigators can now take advantage of genome-wide approaches to map the ATF-binding events in the cell and to study the specificity of regulation by chromatin immunoprecipitation sequencing and RNA-sequencing (or gene expression microarrays), respectively. Nevertheless, work done in our laboratories and others demonstrate that the genomic context of the host cell type, for example, target site selectivity and promoter context, are key factors defining the specificity of action of ZF proteins (Beltran and Blancafort, 2010; Beltran, 2011). As is often observed with siRNA technology, several ATF proteins need to be constructed to effectively regulate the desired gene (Beltran et al., 2007).
TABLE 1.
Comparison between artificial transcription factors and small molecule epigenetic inhibitors for anticancer intervention
| Features | Artificial Transcription Factors | Epigenetic Molecules |
|---|---|---|
| Genomic/functional (e.g., cell type) specificity | Design of sequence-specific DNA-binding domains can provide high selectivity for the target | Broad spectrum of cellular targets, some specificity depending on the compound |
| Selective targeting for cancer cells by regulation of genes differentially expressed in cancer cells | Potentially high toxicity due to undesirable targeting of normal cells | |
| Transcriptional/regulatory potency | Coupling the DNA-binding domain with transactivators/repressors can result in potent regulation | Can result in transcriptional regulation of targeted promoters |
| Combinatorial delivery of effector functions, diversity of epigenetic domains | Combinatorial delivery of small molecules results in potent synergisms | |
| Delivery in cells/tumors/tissues | Viral/nonviral (targeted nanoparticles) | Intravenous delivery, targeted nanoparticle delivery |
| Probability of acquired resistance mechanisms | Potentially diminished by using epigenetic effector domains (e.g., methyltransferase enzymes) | Drug resistance/disease recurrence is often observed during treatment |
| Cost of synthesis/delivery | Potentially very high cost | Large spectrum of small molecules available for treatment or in clinical trials |
Genomic and functional studies have defined critical target genes deregulated in cancer cells relative to normal tissue, providing investigators with ad hoc information regarding which genes are the most important to correct and in which subtypes of cancers. For example, investigators can selectively downregulate specific oncogenes overexpressed in cancer tissues without altering the behavior of normal cells. In contrast, the inhibition of endogenous epigenetic enzymes using small molecules potentially yields toxicity or undesired pharmacology due to the lack of locus selectivity of these chemical-based approaches. Because both the biologic consequence of chromatin regulator inhibition and the potential off-target profile of small molecules are not predictable, experimental data, not debate, will reveal the magnitude of this challenge. By analogy to the selectivity issues often debated for protein kinase inhibitors, we anticipate clinically useful levels of efficacy and selectivity to be obtainable in the epigenetics field with progress driven by thoughtful experimentation. The potency of transcriptional regulation using ATFs is highly enhanced by using transactivator or repressor domains. In our hands, the degree of upregulation of tumor suppressors genes is considerably stronger using ATFs compared with small molecule inhibitors. This result could be explained by the effective recruitment of RNA polymerase II complex and associated factors by ATFs, which results in enhanced transactivation and strong anticancer effects (Lara et al., 2012). A linkage of multiple epigenetic effectors (e.g., methyltransferase and histone deacetylase functions) in the same ATF could improve the effect of the engineered protein. Similarly, small molecules targeting different epigenetic processes have been shown to be highly synergistic in upregulating endogenous genes.
Certainly the strongest known limitation of the ATF technology is effective delivery into the target tissue. Targeted delivery for gene therapy has been historically addressed with the use of tissue-specific viral vectors (e.g., adeno- and adeno-associated vectors). In addition, targeted nanoparticles encapsulating a more stable and modified ATF mRNA have been used in animal models of metastatic ovarian cancer (Lara et al., 2012). These nanoparticles were able to accumulate with high doses in the tumor site and were able to inhibit tumor growth when administered intravenously. In addition to delivering the ATF in the tumor compartment, another fundamental problem of the ATFs is the high cost associated with the bulk production of ATF-containing nanoparticles required for treatment. Moreover, the ultimate hope of these ATF approaches is the generation of more stable epigenetic treatments for aggressive cancers, which might increase the longevity of the therapeutic approach, while providing a window of opportunity to combat incurable and highly drug-resistant tumors. Chimeric nanoparticles containing multiple ATFs and other anticancer agents, could be envisioned in the future to enhance therapeutic outcome.
Summary
In conclusion, the epigenomics era has provided a multilayer view of fundamental processes disrupted in cancer cells, which include changes in DNA and histone-methylation in many different loci. The development of small molecule inhibitors for specific methyltransferases and methyl-readers (bromodomains) has provided novel strategies to target the epigenetic processes that are disrupted in malignant cells. Sequence-specific, engineered proteins attached to chromatin remodeling enzymes have the pivotal advantage to target specific locus associated with malignant progression. Combinatorial treatments of cancer cells with several distinct pharmacological approaches, for example small molecules and artificial chromatin remodeling enzymes, represent attractive approaches to stably reprogram the epigenetic landscape of cancer cells.
Acknowledgments
The authors thank Dr. Foteini Hassiotou for critically reading the manuscript, and Drs. Brenda Temple and Adriana S. Beltran for providing assistance in formatting the figures of this manuscript.
Abbreviations
- AID
activation-induced cytidine deaminase
- ATF
artificial transcription factor
- CpG
cytosine-phosphate-guanine
- DNAme
DNA methylation
- DNMT
DNA methyltransferase
- ES
embryonic stem cells
- GLP
G9a-like protein
- HP1
heterochromatin protein 1
- MBT
malignant brain tumor domains
- PHDs
plant homeodomains
- PKMTs
protein lysine methyltransferases
- PRMTs
protein arginine methyltransferases
- SETD7
SET domain containing (lysine methyltransferase) 7
- SETDB1
SET domain, bifurcated 1
- SUV39H1
suppressor of variegation 3-9 homolog 1 (Drosophila)
- TALEs
transcription activator-like effectors
- TET proteins
ten-eleven translocation family of proteins
- TF
transcription factor
- ZF
zinc finger
Authorship Contributions
Wrote or contributed to the writing of the manuscript: Blancafort, Jin, Frye.
Footnotes
References
- Adams-Cioaba MA, Min J. (2009) Structure and function of histone methylation binding proteins. Biochem Cell Biol 87:93–105 [DOI] [PubMed] [Google Scholar]
- Arima Y, Hayashi N, Hayashi H, Sasaki M, Kai K, Sugihara E, Abe E, Yoshida A, Mikami S, Nakamura S, et al. (2012) Loss of p16 expression is associated with the stem cell characteristics of surface markers and therapeutic resistance in estrogen receptor-negative breast cancer. Int J Cancer 130:2568–2579 [DOI] [PubMed] [Google Scholar]
- Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. (2012) Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov 11:384–400 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120–124 [DOI] [PubMed] [Google Scholar]
- Baylin S.B., Jones P.A. (2011). A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer 11:726–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Richard S. (2005) Arginine methylation an emerging regulator of protein function. Mol Cell 18:263–272 [DOI] [PubMed] [Google Scholar]
- Beltran AS, Blancafort P. (2011). Reactivation of MASPIN in non-small cell lung carcinoma (NSCLC) cells by artificial transcription factors (ATFs). Epigenetics 6:224–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran A, Liu Y, Parikh S, Temple B, Blancafort P. (2006) Interrogating genomes with combinatorial artificial transcription factor libraries: asking zinc finger questions. Assay Drug Dev Technol 4:317–331 [DOI] [PubMed] [Google Scholar]
- Beltran A, Parikh S, Liu Y, Cuevas BD, Johnson GL, Futscher BW, Blancafort P. (2007) Re-activation of a dormant tumor suppressor gene maspin by designed transcription factors. Oncogene 26:2791–2798 [DOI] [PubMed] [Google Scholar]
- Beltran AS, Blancafort P. (2010) Remodeling genomes with artificial transcription factors (ATFs). Methods Mol Biol 649:163–182 [DOI] [PubMed] [Google Scholar]
- Beltran AS, Rivenbark AG, Richardson BT, Yuan X, Quian H, Hunt JP, Zimmerman E, Graves LM, Blancafort P. (2011a) Generation of tumor-initiating cells by exogenous delivery of OCT4 transcription factor. Breast Cancer Res 13:R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran AS, Russo A, Lara H, Fan C, Lizardi PM, Blancafort P. (2011b) Suppression of breast tumor growth and metastasis by an engineered transcription factor. PLoS ONE 6:e24595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran AS, Sun X, Lizardi PM, Blancafort P. (2008) Reprogramming epigenetic silencing: artificial transcription factors synergize with chromatin remodeling drugs to reactivate the tumor suppressor mammary serine protease inhibitor. Mol Cancer Ther 7:1080–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdasco M, Esteller M. (2010) Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev Cell 19:698–711 [DOI] [PubMed] [Google Scholar]
- Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CP, van Dijk CM, Tollenaar RA, et al. (2012) Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet 44:40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. (2007) The mammalian epigenome. Cell 128:669–681 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125:315–326 [DOI] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. (2010) Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature 463:1042–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani N, Burns DM, Blau HM. (2011) DNA demethylation dynamics. Cell 146:866–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AP. (1993) Functions for DNA methylation in vertebrates. Cold Spring Harb Symp Quant Biol 58:281–285 [DOI] [PubMed] [Google Scholar]
- Bissinger E-M, Heinke R, Sippl W, Jung M. (2010) Targeting epigenetic modifiers: Inhibitors of histone methyltransferases. MedChemComm 1:114–124 [Google Scholar]
- Blancafort P, Tschan MP, Bergquist S, Guthy D, Brachat A, Sheeter DA, Torbett BE, Erdmann D, Barbas CF., 3rd (2008) Modulation of drug resistance by artificial transcription factors. Mol Cancer Ther 7:688–697 [DOI] [PubMed] [Google Scholar]
- Bogdanove AJ, Voytas DF. (2011) TAL effectors: customizable proteins for DNA targeting. Science 333:1843–1846 [DOI] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Estève PO, Clark A, Pradhan S, Jacobsen SE. (2007) UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317:1760–1764 [DOI] [PubMed] [Google Scholar]
- Bultmann S, Morbitzer R, Schmidt CS, Thanisch K, Spada F, Elsaesser J, Lahaye T, Leonhardt H. (2012) Targeted transcriptional activation of silent oct4 pluripotency gene by combining designer TALEs and inhibition of epigenetic modifiers. Nucleic Acids Res 40:5368–5377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagna-Slater V, Arrowsmith AG, Zhao Y, Schapira M. (2010) Pharmacophore screening of the protein data bank for specific binding site chemistry. J Chem Inf Model 50:358–367 [DOI] [PubMed] [Google Scholar]
- Campagna-Slater V, Schapira M. (2010) Finding inspiration in the protein data bank to chemically antagonize readers of the histone code. Molecular Informatics 29:322–331 [DOI] [PubMed] [Google Scholar]
- Carmona FJ, Esteller M. (2011) DNA methylation in early neoplasia. Cancer Biomark 9:101–111 [DOI] [PubMed] [Google Scholar]
- Carvin CD, Parr RD, Kladde MP. (2003) Site-selective in vivo targeting of cytosine-5 DNA methylation by zinc-finger proteins. Nucleic Acids Res 31:6493–6501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Ganesh T, Horton JR, Spannhoff A, Liu J, Sun A, Zhang X, Bedford MT, Shinkai Y, Snyder JP, et al. (2010) Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases. J Mol Biol 400:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Zhang X, Horton JR, Upadhyay AK, Spannhoff A, Liu J, Snyder JP, Bedford MT, Cheng X. (2009) Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294. Nat Struct Mol Biol 16:312–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari R, Thu KL, Wilson IM, Lockwood WW, Lonergan KM, Coe BP, Malloff CA, Gazdar AF, Lam S, Garnis C, et al. (2010) Integrating the multiple dimensions of genomic and epigenomic landscapes of cancer. Cancer Metastasis Rev 29:73–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase A., Cross N.C. (2011). Aberrations of EZH2 in cancer. Clin Cancer Res 17:2613–2618 [DOI] [PubMed] [Google Scholar]
- Chen MW, Hua KT, Kao HJ, Chi CC, Wei LH, Johansson G, Shiah SG, Chen PS, Jeng YM, Cheng TY, et al. (2010) H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Res 70:7830–7840 [DOI] [PubMed] [Google Scholar]
- Cheng X, Blumenthal RM. (2010) Coordinated chromatin control: structural and functional linkage of DNA and histone methylation. Biochemistry 49:2999–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho WC. (2012) MicroRNAs as therapeutic targets and their potential applications in cancer therapy. Expert Opin Ther Targets 16:747–759 [DOI] [PubMed] [Google Scholar]
- Clements A, Rojas JR, Trievel RC, Wang L, Berger SL, Marmorstein R. (1999) Crystal structure of the histone acetyltransferase domain of the human PCAF transcriptional regulator bound to coenzyme A. EMBO J 18:3521–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole PA. (2008) Chemical probes for histone-modifying enzymes. Nat Chem Biol 4:590–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland RA, Solomon ME, Richon VM. (2009) Protein methyltransferases as a target class for drug discovery. Nat Rev Drug Discov 8:724–732 [DOI] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. (2011) Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146:67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa FF, Paixão VA, Cavalher FP, Ribeiro KB, Cunha IW, Rinck JA, Jr, O’Hare M, Mackay A, Soares FA, Brentani RR, et al. (2006) SATR-1 hypomethylation is a common and early event in breast cancer. Cancer Genet Cytogenet 165:135–143 [DOI] [PubMed] [Google Scholar]
- Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, Johnston LD, Scott MP, Smith JJ, Xiao Y, et al. (2011) Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 20:53–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan W-I, Robson SC, Chung CW, Hopf C, Savitski MM, et al. (2011) Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478:529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. (2011) BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146:904–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng D, Yan C, Pan X, Mahfouz M, Wang J, Zhu JK, Shi Y, Yan N. (2012) Structural basis for sequence-specific recognition of DNA by TAL effectors. Science 335:720–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz E., Machutta C.A., Chen S., Jiang Y., Nixon C., Hofmann G., Key D., Sweitzer S., Patel M., Wu Z., et al. (2012). Development and validation of reagents and assays for EZH2 peptide and nucleosome high-throughput screens. J Biomol Screen 17:1279–1292 [DOI] [PubMed] [Google Scholar]
- Dudziec E., Miah S., Choudhry H.M., Owen H.C., Blizard S., Glover M., Hamdy F.C., Catto J.W. (2011). Hypermethylation of CpG islands and shores around specific microRNAs and mirtrons is associated with the phenotype and presence of bladder cancer. Clin Cancer Res 17:1287–1296 [DOI] [PubMed]
- Dutnall RN, Ramakrishnan V. (1997) Twists and turns of the nucleosome: tails without ends. Structure 5:1255–1259 [DOI] [PubMed] [Google Scholar]
- Enroth S, Rada-Iglesisas A, Andersson R, Wallerman O, Wanders A, Påhlman L, Komorowski J, Wadelius C. (2011) Cancer associated epigenetic transitions identified by genome-wide histone methylation binding profiles in human colorectal cancer samples and paired normal mucosa. BMC Cancer 11:450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. (2008) Epigenetics in cancer. N Engl J Med 358:1148–1159 [DOI] [PubMed] [Google Scholar]
- Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG. (2000) Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med 343:1350–1354 [DOI] [PubMed] [Google Scholar]
- Esteller M, Risques RA, Toyota M, Capella G, Moreno V, Peinado MA, Baylin SB, Herman JG. (2001) Promoter hypermethylation of the DNA repair gene O(6)-methylguanine-DNA methyltransferase is associated with the presence of G:C to A:T transition mutations in p53 in human colorectal tumorigenesis. Cancer Res 61:4689–4692 [PubMed] [Google Scholar]
- Falke D, Fisher M, Ye D, Juliano RL. (2003) Design of artificial transcription factors to selectively regulate the pro-apoptotic bax gene. Nucleic Acids Res 31:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, Cedar H, Bergman Y. (2006) G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol 8:188–194 [DOI] [PubMed] [Google Scholar]
- Ferguson AD, Larsen NA, Howard T, Pollard H, Green I, Grande C, Cheung T, Garcia-Arenas R, Cowen S, Wu J, et al. (2011) Structural basis of substrate methylation and inhibition of SMYD2. Structure 19:1262–1273 [DOI] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. (2011) Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473:398–402 [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. (2010) Selective inhibition of BET bromodomains. Nature 468:1067–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fog CK, Jensen KT, Lund AH. (2007) Chromatin-modifying proteins in cancer. APMIS 115:1060–1089 [DOI] [PubMed] [Google Scholar]
- Frye SV, Heightman T, Jin J. (2010) Targeting methyl lysine. Annu Rep Med Chem 45:329–343 [Google Scholar]
- Fuks F, Hurd PJ, Deplus R, Kouzarides T. (2003) The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res 31:2305–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futscher BW, O’Meara MM, Kim CJ, Rennels MA, Lu D, Gruman LM, Seftor RE, Hendrix MJ, Domann FE. (2004) Aberrant methylation of the maspin promoter is an early event in human breast cancer. Neoplasia 6:380–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futscher BW, Oshiro MM, Wozniak RJ, Holtan N, Hanigan CL, Duan H, Domann FE. (2002) Role for DNA methylation in the control of cell type specific maspin expression. Nat Genet 31:175–179 [DOI] [PubMed] [Google Scholar]
- Gallardo F, Esteller M, Pujol RM, Costa C, Estrach T, Servitje O. (2004) Methylation status of the p15, p16 and MGMT promoter genes in primary cutaneous T-cell lymphomas. Haematologica 89:1401–1403 [PubMed] [Google Scholar]
- Gao X, Tate P, Hu P, Tjian R, Skarnes WC, Wang Z. (2008) ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci USA 105:6656–6661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Reik W, Henikoff S. (2009) DNA demethylation by DNA repair. Trends Genet 25:82–90 [DOI] [PubMed] [Google Scholar]
- Gelato KA, Fischle W. (2008) Role of histone modifications in defining chromatin structure and function. Biol Chem 389:353–363 [DOI] [PubMed] [Google Scholar]
- Gilbert N, Boyle S, Fiegler H, Woodfine K, Carter NP, Bickmore WA. (2004) Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell 118:555–566 [DOI] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. (2007) Epigenetics: a landscape takes shape. Cell 128:635–638 [DOI] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. (2011) The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477:606–610 [DOI] [PubMed] [Google Scholar]
- Guan P, Yin Z, Li X, Wu W, Zhou B. (2012) Meta-analysis of human lung cancer microRNA expression profiling studies comparing cancer tissues with normal tissues. J Exp Clin Cancer Res 31:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Pramanik D., Mukherjee R., Campbell N.R., Elumalai S., de Wilde R.F., Hong S.M., Goggins M.G., De Jesus-Acosta A., Laheru D., et al. (2012). Molecular determinants of retinoic acid sensitivity in pancreatic cancer. Clin Cancer Res 18:280–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm CA, Xie H, Costa FF, Vanin EF, Seftor EA, Sredni ST, Bischof J, Wang D, Bonaldo MF, Hendrix MJ, et al. (2009) Global demethylation of rat chondrosarcoma cells after treatment with 5-aza-2′-deoxycytidine results in increased tumorigenicity. PLoS ONE 4:e8340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Brunet A. (2012) Histone methylation makes its mark on longevity. Trends Cell Biol 22:42–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, et al. (2011) Increased methylation variation in epigenetic domains across cancer types. Nat Genet 43:768–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Vertino PM, Cheng X. (2010) Molecular coupling of DNA methylation and histone methylation. Epigenomics 2:657–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Korboukh I, Jin J, Huang J. (2012) Targeting protein lysine methylation and demethylation in cancers. Acta Biochim Biophys Sin (Shanghai) 44:70–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Shilatifard A. (2011) Histone modification: cause or cog? Trends Genet 27:389–396 [DOI] [PubMed] [Google Scholar]
- Herold JM, Ingerman LA, Gao C, Frye SV. (2011a) Drug discovery toward antagonists of methyl-lysine binding proteins. Curr Chem Genomics 5:51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold JM, James LI, Korboukh VK, Gao C, Coil KE, Bua DJ, Norris JL, Kireev DB, Brown PJ, Jin J, et al. (2012) Structure-activity relationships of methyl-lysine reader antagonists. Med Chem Comm 3:45–51 [Google Scholar]
- Herold JM, Wigle TJ, Norris JL, Lam R, Korboukh VK, Gao C, Ingerman LA, Kireev DB, Senisterra G, Vedadi M, et al. , (2011b) Small-molecule ligands of methyl-lysine binding proteins. J Med Chem 54:2504–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K, Plath K. (2009) Epigenetic reprogramming and induced pluripotency. Development 136:509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, Pelizzola M, Valsesia A, Ye Z, Kuan S, Edsall LE, et al. (2012) Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res 22:246–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Dorsey J, Chuikov S, Pérez-Burgos L, Zhang X, Jenuwein T, Reinberg D, Berger SL. (2010) G9a and Glp methylate lysine 373 in the tumor suppressor p53. J Biol Chem 285:9636–9641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL. (2006) Repression of p53 activity by Smyd2-mediated methylation. Nature 444:629–632 [DOI] [PubMed] [Google Scholar]
- Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, et al. (2007) p53 is regulated by the lysine demethylase LSD1. Nature 449:105–108 [DOI] [PubMed] [Google Scholar]
- Hughes RM, Wiggins KR, Khorasanizadeh S, Waters ML. (2007) Recognition of trimethyllysine by a chromodomain is not driven by the hydrophobic effect. Proc Natl Acad Sci USA 104:11184–11188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura T, Watanabe S, Sakamoto Y, Aoto T, Fujita N, Nakao M. (2005) Transcriptional repression and heterochromatin formation by MBD1 and MCAF/AM family proteins. J Biol Chem 280:13928–13935 [DOI] [PubMed] [Google Scholar]
- Iqbal K, Jin SG, Pfeifer GP, Szabó PE. (2011) Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci USA 108:3642–3647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. (2009) The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 41:178–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto FV, Esteller M. (2007) Mutator pathways unleashed by epigenetic silencing in human cancer. Mutagenesis 22:247–253 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. (2001) Translating the histone code. Science 293:1074–1080 [DOI] [PubMed] [Google Scholar]
- Johnson L.M., Bostick M., Zhang X., Kraft E., Henderson I., Callis J., Jacobsen S.E. (2007). The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol 17: 379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke XS, Qu Y, Rostad K, Li WC, Lin B, Halvorsen OJ, Haukaas SA, Jonassen I, Petersen K, Goldfinger N, et al. (2009) Genome-wide profiling of histone h3 lysine 4 and lysine 27 trimethylation reveals an epigenetic signature in prostate carcinogenesis. PLoS ONE 4:e4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Estève PO, Jacobsen SE, Pradhan S. (2009) UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res 37:493–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, et al. (2010) Epigenetic memory in induced pluripotent stem cells. Nature 467:285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireev D, Wigle TJ, Norris-Drouin J, Herold JM, Janzen WP, Frye SV. (2010) Identification of non-peptide malignant brain tumor (MBT) repeat antagonists by virtual screening of commercially available compounds. J Med Chem 53:7625–7631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizilyaprak C, Spehner D, Devys D, Schultz P. (2010) In vivo chromatin organization of mouse rod photoreceptors correlates with histone modifications. PLoS ONE 5:e11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y. (2006) JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 7:715–727 [DOI] [PubMed] [Google Scholar]
- Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, Sacks JD, Raimondi A, Majer CR, Song J, et al. (2012) A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol 8:890–896 [DOI] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. (2011) Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8:200–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. (2007) Chromatin modifications and their function. Cell 128:693–705 [DOI] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. (2009) The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324:929–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouwels IM, Wiesmeijer K, Abraham TE, Molenaar C, Verwoerd NP, Tanke HJ, Dirks RW. (2005) A glue for heterochromatin maintenance: stable SUV39H1 binding to heterochromatin is reinforced by the SET domain. J Cell Biol 170:537–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek S, O’Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, Rea S, Mechtler K, Kowalski JA, Homon CA, et al. (2007) Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell 25:473–481 [DOI] [PubMed] [Google Scholar]
- Kundu S, Horn PJ, Peterson CL. (2007) SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster. Genes Dev 21:997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo AJ, Song J, Cheung P, Ishibe-Murakami S, Yamazoe S, Chen JK, Patel DJ, Gozani O. (2012) The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature 484:115–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116–120 [DOI] [PubMed] [Google Scholar]
- Lange UC, Schneider R. (2010) What an epigenome remembers. Bioessays 32:659–668 [DOI] [PubMed] [Google Scholar]
- Lara H, Wang Y, Beltran AS, Juárez-Moreno K, Yuan X, Kato S, Leisewitz AV, Cuello Fredes M, Licea AF, Connolly DC, et al. (2012) Targeting serous epithelial ovarian cancer with designer zinc finger transcription factors. J Biol Chem 287:29873–29886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Papworth M, Minczuk M, Rohde C, Zhang Y, Ragozin S, Jeltsch A. (2007a) Chimeric DNA methyltransferases target DNA methylation to specific DNA sequences and repress expression of target genes. Nucleic Acids Res 35:100–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Fischle W, Wang W, Duncan EM, Liang L, Murakami-Ishibe S, Allis CD, Patel DJ. (2007b) Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol Cell 28:677–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, et al. kConFab (2009) Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med 15:907–913 [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. (2009) Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462:315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O’Malley R, Castanon R, Klugman S, et al. (2011) Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471:68–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Barsyte-Lovejoy D, Allali-Hassani A, He Y, Herold JM, Chen X, Yates CM, Frye SV, Brown PJ, Huang J, et al. (2011) Optimization of cellular activity of G9a inhibitors 7-aminoalkoxy-quinazolines. J Med Chem 54:6139–6150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Chen X, Allali-Hassani A, Quinn AM, Wasney GA, Dong A, Barsyte D, Kozieradzki I, Senisterra G, Chau I, et al. (2009) Discovery of a 2,4-diamino-7-aminoalkoxyquinazoline as a potent and selective inhibitor of histone lysine methyltransferase G9a. J Med Chem 52:7950–7953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Chen X, Allali-Hassani A, Quinn AM, Wigle TJ, Wasney GA, Dong A, Senisterra G, Chau I, Siarheyeva A, et al. (2010) Protein lysine methyltransferase G9a inhibitors: design, synthesis, and structure activity relationships of 2,4-diamino-7-aminoalkoxy-quinazolines. J Med Chem 53:5844–5857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Ginestier C, Charafe-Jauffret E, Foco H, Kleer CG, Merajver SD, Dontu G, Wicha MS. (2008a) BRCA1 regulates human mammary stem/progenitor cell fate. Proc Natl Acad Sci USA 105:1680–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang L, Zhao K, Thompson PR, Hwang Y, Marmorstein R, Cole PA. (2008b) The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature 451:846–850 [DOI] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251–260 [DOI] [PubMed] [Google Scholar]
- Lund CV, Blancafort P, Popkov M, Barbas CF., 3rd (2004) Promoter-targeted phage display selections with preassembled synthetic zinc finger libraries for endogenous gene regulation. J Mol Biol 340:599–613 [DOI] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. (2009) Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461:762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. (2010) Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet 11:285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Zhang Y. (2005) The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 6:838–849 [DOI] [PubMed] [Google Scholar]
- Martinez R, Esteller M. (2010) The DNA methylome of glioblastoma multiforme. Neurobiol Dis 39:40–46 [DOI] [PubMed] [Google Scholar]
- Martinez R, Martin-Subero JI, Rohde V, Kirsch M, Alaminos M, Fernandez AF, Ropero S, Schackert G, Esteller M. (2009) A microarray-based DNA methylation study of glioblastoma multiforme. Epigenetics 4:255–264 [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. (2008) Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454:766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister GE, Chandrasegaran S, Ostermeier M. (2010) Heterodimeric DNA methyltransferases as a platform for creating designer zinc finger methyltransferases for targeted DNA methylation in cells. Nucleic Acids Res 38:1749–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448:553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minczuk M, Papworth MA, Kolasinska P, Murphy MP, Klug A. (2006) Sequence-specific modification of mitochondrial DNA using a chimeric zinc finger methylase. Proc Natl Acad Sci USA 103:19689–19694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. (2009) A simple cipher governs DNA recognition by TAL effectors. Science 326:1501. [DOI] [PubMed] [Google Scholar]
- Mussolino C, Cathomen T. (2012) TALE nucleases: tailored genome engineering made easy. Curr Opin Biotechnol 23:644–650 [DOI] [PubMed] [Google Scholar]
- Nagy Z, Tora L. (2007) Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene 26:5341–5357 [DOI] [PubMed] [Google Scholar]
- Nakamura T., Liu Y.-J., Nakashima H., Umehara H., Inoue K., Matoba S., Tachibana M., Ogura A., Shinkai Y., Nakano T. (2012). PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature 486:415–419 [DOI] [PubMed] [Google Scholar]
- Nomura W, Barbas CF., 3rd (2007) In vivo site-specific DNA methylation with a designed sequence-enabled DNA methylase. J Am Chem Soc 129:8676–8677 [DOI] [PubMed] [Google Scholar]
- Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. (2007) A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet 39:237–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y. (2002) LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23). Cancer Res 62:4075–4080 [PubMed] [Google Scholar]
- Pang J, Toy KA, Griffith KA, Awuah B, Quayson S, Newman LA, Kleer CG. (2012) Invasive breast carcinomas in Ghana: high frequency of high grade, basal-like histology and high EZH2 expression. Breast Cancer Res Treat 135:59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer GP, Rauch TA. (2009) DNA methylation patterns in lung carcinomas. Semin Cancer Biol 19:181–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, et al. (2010) Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol 28:848–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. (2010) Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 463:1101–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portela A, Esteller M. (2010) Epigenetic modifications and human disease. Nat Biotechnol 28:1057–1068 [DOI] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. (2008) DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell 135:1201–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakha EA, Reis-Filho JS, Ellis IO. (2008) Basal-like breast cancer: a critical review. J Clin Oncol 26:2568–2581 [DOI] [PubMed] [Google Scholar]
- Rathert P, Dhayalan A, Murakami M, Zhang X, Tamas R, Jurkowska R, Komatsu Y, Shinkai Y, Cheng X, Jeltsch A. (2008) Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol 4:344–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593–599 [DOI] [PubMed] [Google Scholar]
- Reinke H, Hörz W. (2003) Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol Cell 11:1599–1607 [DOI] [PubMed] [Google Scholar]
- Rivenbark AG, Stolzenburg S, Beltran AS, Yuan X, Rots MG, Strahl BD, Blancafort P. (2012) Epigenetic reprogramming of cancer cells via targeted DNA methylation. Epigenetics 7:350–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J, Vives L, Jordà M, Morales C, Muñoz M, Vendrell E, Peinado MA. (2008) Genome-wide tracking of unmethylated DNA Alu repeats in normal and cancer cells. Nucleic Acids Res 36:770–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Paredes M, Esteller M. (2011) Cancer epigenetics reaches mainstream oncology. Nat Med 17:330–339 [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. (2007) Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol 8:983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval J, Esteller M. (2012) Cancer epigenomics: beyond genomics. Curr Opin Genet Dev 22:50–55 [DOI] [PubMed] [Google Scholar]
- Sandoval J, Heyn H, Moran S, Serra-Musach J, Pujana MA, Bibikova M, Esteller M. (2011) Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics 6:692–702 [DOI] [PubMed] [Google Scholar]
- Santiago C, Nguyen K, Schapira M. (2011) Druggability of methyl-lysine binding sites. J Comput Aided Mol Des 25:1171–1178 [DOI] [PubMed] [Google Scholar]
- Sato H, Oka T, Shinnou Y, Kondo T, Washio K, Takano M, Takata K, Morito T, Huang X, Tamura M, et al. (2010) Multi-step aberrant CpG island hyper-methylation is associated with the progression of adult T-cell leukemia/lymphoma. Am J Pathol 176:402–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarsdale JN, Webb HD, Ginder GD, Williams DC., Jr (2011) Solution structure and dynamic analysis of chicken MBD2 methyl binding domain bound to a target-methylated DNA sequence. Nucleic Acids Res 39:6741–6752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. (2007) Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 39:232–236 [DOI] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd (2002) SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 16:919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sera T. (2009) Zinc-finger-based artificial transcription factors and their applications. Adv Drug Deliv Rev 61:513–526 [DOI] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. (2007) The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450:908–912 [DOI] [PubMed] [Google Scholar]
- Singal R, Wang SZ, Sargent T, Zhu SZ, Ginder GD. (2002) Methylation of promoter proximal-transcribed sequences of an embryonic globin gene inhibits transcription in primary erythroid cells and promotes formation of a cell type-specific methyl cytosine binding complex. J Biol Chem 277:1897–1905 [DOI] [PubMed] [Google Scholar]
- Smith AE, Ford KG. (2007) Specific targeting of cytosine methylation to DNA sequences in vivo. Nucleic Acids Res 35:740–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AE, Hurd PJ, Bannister AJ, Kouzarides T, Ford KG. (2008) Heritable gene repression through the action of a directed DNA methyltransferase at a chromosomal locus. J Biol Chem 283:9878–9885 [DOI] [PubMed] [Google Scholar]
- Snowden AW, Gregory PD, Case CC, Pabo CO. (2002) Gene-specific targeting of H3K9 methylation is sufficient for initiating repression in vivo. Curr Biol 12:2159–2166 [DOI] [PubMed] [Google Scholar]
- Song JS, Kim YS, Kim DK, Park SI, Jang SJ. (2012) Global histone modification pattern associated with recurrence and disease-free survival in non-small cell lung cancer patients. Pathol Int 62:182–190 [DOI] [PubMed] [Google Scholar]
- Spannhoff A, Hauser AT, Heinke R, Sippl W, Jung M. (2009a) The emerging therapeutic potential of histone methyltransferase and demethylase inhibitors. ChemMedChem 4:1568–1582 [DOI] [PubMed] [Google Scholar]
- Spannhoff A, Sippl W, Jung M. (2009b) Cancer treatment of the future: inhibitors of histone methyltransferases. Int J Biochem Cell Biol 41:4–11 [DOI] [PubMed] [Google Scholar]
- Stefansson OA, Jonasson JG, Olafsdottir K, Hilmarsdottir H, Olafsdottir G, Esteller M, Johannsson OT, Eyfjord JE. (2011) CpG island hypermethylation of BRCA1 and loss of pRb as co-occurring events in basal/triple-negative breast cancer. Epigenetics 6:638–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolzenburg S, Bilsland A, Keith WN, Rots MG. (2010) Modulation of gene expression using zinc finger-based artificial transcription factors. Methods Mol Biol 649:117–132 [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. (2000) The language of covalent histone modifications. Nature 403:41–45 [DOI] [PubMed] [Google Scholar]
- Suganuma T, Workman JL. (2011) Signals and combinatorial functions of histone modifications. Annu Rev Biochem 80:473–499 [DOI] [PubMed] [Google Scholar]
- Sunami E, de Maat M, Vu A, Turner RR, Hoon DS. (2011) LINE-1 hypomethylation during primary colon cancer progression. PLoS ONE 6:e18884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpakowski S, Sun X, Lage JM, Dyer A, Rubinstein J, Kowalski D, Sasaki C, Costa J, Lizardi PM. (2009) Loss of epigenetic silencing in tumors preferentially affects primate-specific retroelements. Gene 448:151–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y. (2008) G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J 27:2681–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324:930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676 [DOI] [PubMed] [Google Scholar]
- Tan S, Guschin D, Davalos A, Lee YL, Snowden AW, Jouvenot Y, Zhang HS, Howes K, McNamara AR, Lai A, et al. (2003) Zinc-finger protein-targeted gene regulation: genomewide single-gene specificity. Proc Natl Acad Sci USA 100:11997–12002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. (2007) How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol 14:1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, et al. (2010) CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 464:1082–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran R.K., Henikoff J.G., Zilberman D., Ditt R.F., Jacobsen S.E., Henikoff S. (2005). DNA methylation profiling identifies CG methylation clusters in Arabidopsis genes. Curr Biol 15:154–159 [DOI] [PubMed] [Google Scholar]
- Turner BM. (2012) The adjustable nucleosome: an epigenetic signaling module. Trends Genet 28:436–444 [DOI] [PubMed] [Google Scholar]
- Uchimura Y, Ichimura T, Uwada J, Tachibana T, Sugahara S, Nakao M, Saitoh H. (2006) Involvement of SUMO modification in MBD1- and MCAF1-mediated heterochromatin formation. J Biol Chem 281:23180–23190 [DOI] [PubMed] [Google Scholar]
- Unoki M. (2011) Current and potential anticancer drugs targeting members of the UHRF1 complex including epigenetic modifiers. Recent Patents Anticancer Drug Discov 6:116–130 [DOI] [PubMed] [Google Scholar]
- van der Gun BT, Maluszynska-Hoffman M, Kiss A, Arendzen AJ, Ruiters MH, McLaughlin PM, Weinhold E, Rots MG. (2010) Targeted DNA methylation by a DNA methyltransferase coupled to a triple helix forming oligonucleotide to down-regulate the epithelial cell adhesion molecule. Bioconjug Chem 21:1239–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedadi M, Barsyte-Lovejoy D, Liu F, Rival-Gervier S, Allali-Hassani A, Labrie V, Wigle TJ, Dimaggio PA, Wasney GA, Siarheyeva A, et al. (2011) A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat Chem Biol 7:566–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeck J, Esteller M. (2010) Breast cancer epigenetics: from DNA methylation to microRNAs. J Mammary Gland Biol Neoplasia 15:5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. (2012) The epigenotype. 1942. Int J Epidemiol 41:10–13 [DOI] [PubMed] [Google Scholar]
- Wang L, Tang Y, Cole PA, Marmorstein R. (2008a) Structure and chemistry of the p300/CBP and Rtt109 histone acetyltransferases: implications for histone acetyltransferase evolution and function. Curr Opin Struct Biol 18:741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. (2008b) Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40:897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber B, Kimhi S, Howard G, Eden A, Lyko F. (2010) Demethylation of a LINE-1 antisense promoter in the cMet locus impairs Met signalling through induction of illegitimate transcription. Oncogene 29:5775–5784 [DOI] [PubMed] [Google Scholar]
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al. (2007) Epigenetic stem cell signature in cancer. Nat Genet 39:157–158 [DOI] [PubMed] [Google Scholar]
- Wigle TJ, Herold JM, Senisterra GA, Vedadi M, Kireev DB, Arrowsmith CH, Frye SV, Janzen WP. (2010a) Screening for inhibitors of low-affinity epigenetic peptide-protein interactions: an AlphaScreen-based assay for antagonists of methyl-lysine binding proteins. J Biomol Screen 15:62–71 [DOI] [PubMed] [Google Scholar]
- Wigle TJ, Provencher LM, Norris JL, Jin J, Brown PJ, Frye SV, Janzen WP. (2010b) Accessing protein methyltransferase and demethylase enzymology using microfluidic capillary electrophoresis. Chem Biol 17:695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. (2011) TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473:343–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RJ, Klimecki WT, Lau SS, Feinstein Y, Futscher BW. (2007) 5-Aza-2′-deoxycytidine-mediated reductions in G9A histone methyltransferase and histone H3 K9 di-methylation levels are linked to tumor suppressor gene reactivation. Oncogene 26:77–90 [DOI] [PubMed] [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. (2011) Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 473:389–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Min J, Lunin VV, Antoshenko T, Dombrovski L, Zeng H, Allali-Hassani A, Campagna-Slater V, Vedadi M, Arrowsmith CH, et al. (2010) Structural biology of human H3K9 methyltransferases. PLoS ONE 5:e8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Bian C., Lam R., Dong A., Min J. (2011). The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain. Nat Comm 2:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Chen P, Diao J, Cheng G, Deng L, Anglin JL, Prasad BVV, Song Y. (2011) Selective inhibitors of histone methyltransferase DOT1L: design, synthesis, and crystallographic studies. J Am Chem Soc 133:16746–16749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost JM, Korboukh I, Liu F, Gao C, Jin J. (2011) Targets in epigenetics: inhibiting the methyl writers of the histone code. Curr Chem Genomics 5 (Suppl 1):72–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Wang Q, Paulk J, Kubicek S, Kemp MM, Adams DJ, Shamji AF, Wagner BK, Schreiber SL. (2012) A small-molecule probe of the histone methyltransferase G9a induces cellular senescence in pancreatic adenocarcinoma. ACS Chem Biol 7:1152–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias N, Dougherty DA. (2002) Cation-pi interactions in ligand recognition and catalysis. Trends Pharmacol Sci 23:281–287 [DOI] [PubMed] [Google Scholar]
- Zakrzewski F, Weisshaar B, Fuchs J, Bannack E, Minoche AE, Dohm JC, Himmelbauer H, Schmidt T. (2011) Epigenetic profiling of heterochromatic satellite DNA. Chromosoma 120:409–422 [DOI] [PubMed] [Google Scholar]
- Zeng L, Zhang Q, Li S, Plotnikov AN, Walsh MJ, Zhou M-M. (2010) Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature 466:258–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE. (2011) Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet 12:7–18 [DOI] [PubMed] [Google Scholar]
- Ziller MJ, Müller F, Liao J, Zhang Y, Gu H, Bock C, Boyle P, Epstein CB, Bernstein BE, Lengauer T, et al. (2011) Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet 7:e1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Anisowicz A, Hendrix MJ, Thor A, Neveu M, Sheng S, Rafidi K, Seftor E, Sager R. (1994) Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science 263:526–529 [DOI] [PubMed] [Google Scholar]