Abstract
Fecal microbiota transplantation (FMT) has emerged as a highly effective therapy for refractory, recurrent Clostridium difficile infection (CDI), which develops following antibiotic treatments. Intestinal microbiota play a critical role in the metabolism of bile acids in the colon, which in turn have major effects on the lifecycle of C. difficile bacteria. We hypothesized that fecal bile acid composition is altered in patients with recurrent CDI and that FMT results in its normalization. General metabolomics and targeted bile acid analyses were performed on fecal extracts from patients with recurrent CDI treated with FMT and their donors. In addition, 16S rRNA gene sequencing was used to determine the bacterial composition of pre- and post-FMT fecal samples. Taxonomic bacterial composition of fecal samples from FMT recipients showed rapid change and became similar to the donor after the procedure. Pre-FMT fecal samples contained high concentrations of primary bile acids and bile salts, while secondary bile acids were nearly undetectable. In contrast, post-FMT fecal samples contained mostly secondary bile acids, as did non-CDI donor samples. Therefore, our analysis showed that FMT resulted in normalization of fecal bacterial community structure and metabolic composition. Importantly, metabolism of bile salts and primary bile acids to secondary bile acids is disrupted in patients with recurrent CDI, and FMT corrects this abnormality. Since individual bile salts and bile acids have pro-germinant and inhibitory activities, the changes suggest that correction of bile acid metabolism is likely a major mechanism by which FMT results in a cure and prevents recurrence of CDI.
Keywords: Clostridium difficile, bile acids, fecal microbiota transplantation
currently, Clostridium difficile infection (CDI) is one of the most common nosocomial infections in the United States, affecting >500,000 people annually in health care facilities and many more in the community (1, 7, 23, 26, 32). Unfortunately, antibiotic therapy alone commonly fails to cure this disease; in fact, the use of antibiotics is the primary risk factor for CDI, and antibiotic treatment of CDI can perpetuate its recurrence (3, 23, 32, 42). Approximately 20–30% of patients experience recurrence of the disease after antibiotic treatment of the initial infection (22, 27). The risk of recurrence continues to rise with each relapse, and some patients ultimately develop a recurrent CDI (R-CDI) syndrome, where the cycle of infections becomes indefinite (3, 42). Fecal microbiota transplantation (FMT), which restores the normal composition of fecal bacteria, is a powerful emerging therapy that successfully treats >90% of patients with R-CDI (3, 13, 46). However, the mechanisms by which this transplanted microbiota prevents recurrence of CDI remain poorly understood.
One known function of the normal human gut microbiota that which may affect C. difficile physiology is the metabolism of primary bile acids to secondary bile acids in the colon. The primary bile acids cholic acid and chenodeoxycholic acid are produced in the liver and conjugated to taurine or glycine to form bile salts before secretion into bile to assist in lipid digestion in the small intestine (21). While ∼95% of secreted bile salts are reabsorbed from the small intestine via the enterohepatic circulation pathway, ∼5% reach the colon, where, by the actions of intestinal bacteria, they are deconjugated and dehydroxylated at C-7 to form deoxycholic and lithocholic acids from cholic and chenodeoxycholic acids, respectively (21). Because of the cytotoxic effects of antibiotics on microbiota, including normal gut microflora, it is not surprising that a variety of antibiotics, including ciprofloxacin, neomycin, and β-lactams, significantly alter the fecal bile acid pool in rodents and in vitro human fecal cultures, reducing the proportion of secondary relative to primary bile acids (5, 12, 18, 45). These findings suggest that antibiotic treatment is likely to substantially alter intestinal bacteria-mediated fecal bile acid metabolism in R-CDI patients.
Bile acids substantially affect germination and growth of C. difficile. The primary bile acid taurocholic acid, the taurine-conjugated form of cholic acid secreted by the liver, is a potent germinant of the organism and is even used as a key component of C. difficile growth media (39, 47). Recently, a putative C. difficile bile acid germinant receptor was identified to be a protease CspC, and a cspC mutant of C. difficile had decreased pathogenicity in a hamster model (11). Notably, the secondary bile acids lithocholic and ursodeoxycholic acids have been shown to inhibit C. difficile germination in vitro (40). Fecal samples from mice treated with clindamycin, which have a significantly lower proportion of secondary bile acids than untreated mice, are better able to stimulate colony formation from C. difficile spores than are controls, suggesting that secondary bile acids may also inhibit germination and/or growth in vivo (12). These studies suggest that secondary bile acids in feces, which decrease following antibiotic treatment, may inhibit germination and/or growth of C. difficile in the colon.
To examine the influence of FMT on the gut microflora-mediated metabolism in R-CDI patients, we conducted general metabolomic and targeted bile acid analyses on fecal extracts from 12 R-CDI patients before and after treatment with FMT. Using high-throughput 16S rRNA gene sequencing, we also analyzed the composition of fecal microbiota in these patients. Here we demonstrate that restoration of normal bacterial composition of fecal microbiota is accompanied by rapid normalization of fecal bile acid composition in these patients. These results offer a potential mechanism by which FMT treats R-CDI and suggest novel pathways of therapeutics development for this frustrating clinical condition.
EXPERIMENTAL PROCEDURES
Patients and donors.
All patients recruited into this study had R-CDI syndrome and failed to clear the infection, despite multiple rounds of antibiotic treatments (Table 1). Two standard donors were used in the preparation of fecal microbiota material, as described previously (16). The Institutional Review Board at the University of Minnesota approved prospective collection of fecal specimens and their analysis (Protocol 1303M29781).
Table 1.
Clinical characteristics of the patient cohort
| Patient No. | Age, yr | Sex | Total No. of CDI Episodes Prior to FMT | Duration of R-CDI Prior to FMT, mo | Original Trigger Event | Trigger Antibiotic | History of Hospitalization for Severe or Complicated CDI |
|---|---|---|---|---|---|---|---|
| 1 | 68 | F | 6 | 6 | Sinusitis | Not documented | No |
| 2 | 66 | F | 3 | 3 | Spinal surgery | Cephalosporin | Yes |
| 3 | 29 | F | 7 | 8 | C-section | Cephalosporin | No |
| 4 | 59 | M | 4 | 4 | Bronchitis | Azithromycin Ciprofloxacin | Yes |
| 5 | 56 | F | 5 | 6 | Knee surgery | Cephalosporin | No |
| 6 | 87 | F | 8 | 9 | Duodenal ulcer perforation | Not documented; Multiple antibiotics | Yes |
| 7 | 65 | F | 5 | 7 | Vaginal infection | Clindamycin | No |
| 8 | 52 | F | 8 | 11 | Urinary tract infection | Ciprofloxacin | No |
| 9 | 47 | F | 4 | 4 | Sinusitis | Not documented | Yes |
| 10 | 52 | F | 9 | 7 | Pelvic reconstruction surgery | Clidamycin | No |
| 11 | 72 | M | 6 | 7 | Diarrheal illness of unclear cause | No identified antibiotic trigger | Yes |
| 12 | 83 | F | 3 | 5 | Urinary sepsis | Not documented; Multiple antibiotics | No |
| Patient No. | Metronidazole Courses | 2-Week Vancomycin Courses | Vancomycin Taper/Pulse | Fidaxomicin | Rifaximin Chaser | Intercurrent Antibiotics | Immunosuppression |
|---|---|---|---|---|---|---|---|
| 1 | 2 | 1 | 2 | 1 | 0 | None | None |
| 2 | 1 | 1 | 1 | 0 | 0 | None | None |
| 3 | 2 | 3 | 2 | 0 | 1 | None | None |
| 4 | 1 | 1 | 1 | 1 | 0 | None | None |
| 5 | 1 | 1 | 1 | 1 | 1 | None | None |
| 6 | 2 | 2 | 1 | 1 | 1 | None | None |
| 7 | 2 | 1 | 2 | 0 | 0 | None | None |
| 8 | 2 | 3 | 2 | 2 | 0 | None | None |
| 9 | 1 | 1 | 1 | 0 | 1 | None | None |
| 10 | 1 | 2 | 4 | 1 | 1 | None | None |
| 11 | 2 | 2 | 2 | 0 | 0 | None | None |
| 12 | 1 | 1 | 1 | 0 | 0 | None | None |
| Patient No. | Probiotics Prior to FMT | Medications Potentially Relevant to Bile Acid Metabolism | Past GI Surgical History | Findings at Colonoscopy | Type of Fecal Microbiota Preparation | Donor | Resolution of CDI With One FMT |
|---|---|---|---|---|---|---|---|
| 1 | Florastor | Simvastatin; omeprazole | None | Normal | Frozen | 1 | Yes |
| 2 | None | Pantoprazole | Partial gastrectomy | Severe diverticulosis | Frozen | 1 | Yes |
| 3 | None | None | None | Normal | Frozen | 1 | Yes |
| 4 | Lactobacillus rhamnosus (Culturelle) | None | None | Moderate diverticulosis | Fresh | 1 | Yes |
| 5 | None | None | None | Terminal ileum ileitis (Crohn's disease) | Frozen | 1 | Yes |
| 6 | Florastor | None | None | Normal | Frozen | 1 | Yes |
| 7 | None | Simvastatin | Appendectomy | Normal | Fresh | 1 | Yes |
| 8 | None | None | None | Normal | Frozen | 1 | Yes |
| 9 | None | None | Cholecystectomy | Lymphocytic colitis | Fresh | 1 | Yes |
| 10 | Acidophilus probiotic blend | None | None | Mild diverticulosis | Frozen | 1 | Yes |
| 11 | None | Atorvastatin | None | Severe diverticulosis | Fresh | 1 | No |
| 12 | None | Simvastatin; glipizide | None | Nondiagnostic Inflammation in lamina propria | Frozen | 2 | Yes |
FMT, fecal microbiota transplantation; C-section, cesarean section; CDI, Clostridium difficile infection; R-CDI, recurrent CDI; severe CDI, albumin <3 and one of the following: ≥15,000 white blood cells/mm3, abdominal tenderness, documentation of colitis on CT scan; complicated CDI, one of the following attributed to CDI: ICD admission, ≥35,000 white blood cells/mm3, albumin <2.5; hypotension, fever >38.5°C, mental status changes, evidence of end organ failure, serum lactate ≥2.2 mmol/l; intercurrent antibiotics, antibiotics used to treat non-CDI after CDI diagnosis.
FMT procedure.
The fecal microbiota were infused via colonoscopy, as previously described (16). All patients took oral vancomycin until 2 days prior to the procedure. The patients received a polyethylene glycol-based colonoscopy preparation (GoLYTELY or MoviPrep) on the day prior to FMT. Fecal microbiota were prepared from one of two standard donors, selected as described previously, by successive filtration to separate the microbial fraction from the rest of the fecal material (16). Ten patients received preparations, which were frozen in 10% (vol/vol) glycerol and stored at −80°C until used, as previously described (16), while six patients received freshly processed (nonfrozen) material. Recovery from CDI was defined by resolution of clinical diarrhea (≤3 bowel movements per day and normalization of stool consistency) over 2 mo. All patients continue to be followed clinically since their FMT.
Sample collection.
Fecal samples were collected by patients prior to FMT and at 7–22 days following FMT. All patients were taking vancomycin when pre-FMT samples were collected. Samples were stored at −80°C within 24 h of collection until DNA extraction. An unprocessed portion of each donor sample was also collected at the time of donation and stored immediately at −80°C.
DNA extraction.
DNA was extracted from each patient and donor sample (0.25–0.50 g) using the PowerSoil DNA Isolation Kit (MO BIO, Carlsbad, CA) according to the manufacturer's instructions. Samples with high water content (Bristol stool scale types 5–7) were centrifuged at 12,000 rpm for 3 min to pellet solids, which were used for DNA extraction. Each sample was extracted in triplicate, and each replicate was eluted in 50 μl of 10 mM Tris·HCl buffer (pH 8.0) and pooled. DNA concentrations of extracted samples were measured with a QuBit DNA quantification system (Invitrogen, Carlsbad, CA) using QuBit high-sensitivity assay reagents. All extracted DNA samples were stored at −20°C until amplification.
PCR amplification.
Fecal DNA samples (25 ng) were used as template for PCR amplification of the V6 region of the 16S rRNA gene. Degenerate primer sets (Table 2) were designed with 6-bp Illumina index sequences on the 5′ end of the reverse primer, which were specific to each fecal DNA sample and allowed for multiplexed sequencing. Primers also contained Illumina PCR primer sequences (reverse primer) and Illumina TruSeq Universal Adapter sequences (forward primers) for library creation. Triplicate reactions were electrophoresed on a 2% agarose gel and then extracted using the QIAquick gel extraction kit (Qiagen, Valencia, CA), eluted in 30 μl of 10 mM Tris·Cl buffer, pH 8.0, and pooled. DNA concentrations were measured using the QuBit DNA quantification system and high-sensitivity assay reagents. Samples were stored at −20°C until pooled for sequencing.
Table 2.
Primers for V6 16S PCR
| Sequence (5′—3′) | |
|---|---|
| Forward primer 1 | [*]NCNACGCGAAGAACCTTANC |
| Forward primer 2 | [*]NNCAACGCGAAAAACCTTACC |
| Forward primer 3 | [*]NNNCAACGCGCAGAACCTTACC |
| Forward primer 4 | [*]NNNNATACGCGARGAACCTTACC |
| Forward primer 5 | [*]NNNNNCTAACCGANGAACCTYACC |
| Reverse primer | [†][6-bp index sequence][‡][4–7 N] CGACRRCCATGCANCACCT |
Illumina TruSeq Universal Adapter sequence;
Illumina PCR primer;
Illumina multiplexing PCR primer 2.0.
DNA sequencing.
Up to 24-equimolar aliquots of each product were pooled to give 3 samples of ∼1 μg of DNA in 100 μl of total volume. DNA concentration in the final pooled solutions was measured using the Quant-IT PicoGreen quantitation system (Invitrogen). Amplicon size was analyzed using an Agilent DNA 1000 chip and 2100 BioAnalyzer (Agilent, Santa Clara, CA). Sequencing was performed at the University of Minnesota BioMedical Genomics Center. Paired-end sequences were generated on the Illumina MiSeq personal sequencer (2 × 150 nt read length). Reads in each pair for each sequencing run overlapped, and paired ends were merged. The hamming distance (number of substitutions) was calculated for sliding overlaps of the two reads in a pair to find the best overlap (lowest hamming distance with ≥25-nt overlap and 98% identity). Merged sequences were binned according to barcode sequence, and barcode and amplicon primer sequences were trimmed using custom Perl scripts.
Sequence processing and analysis.
Sequence data were processed and analyzed using the MOTHUR program (36). To ensure high-quality data for analysis, sequence reads containing ambiguous bases, >7-bp homopolymers, >1 mismatch in the primer sequence, or an average per base quality score <35 within each 50-bp window were removed. Sequences that only appeared once in the total set were assumed to be a result of sequencing error and were removed from the analysis. Chimeric sequences were removed from the data set using the UCHIME algorithm within the MOTHUR program (10). A random subset of 22,353 sequences per sample was used to balance read numbers and clustered into operational taxonomic units (OTUs). Taxonomy was assigned at a cutoff of >90% (17) using a 16S rRNA database prepared from Ribosomal Database Project 9, using the Bayesian method with a bootstrap algorithm (100 iterations) and a probability cutoff of 0.60 (8). Samples were clustered using the Fast UniFrac algorithm to generate trees and principal coordinate analysis (PCoA) plots (14). The UniFrac algorithm was run using the Fast UniFrac program available at www.bmf2.colorado.edu/fastunifrac/.
Reagents.
All solvents and chemicals were purchased from Sigma-Aldrich (St. Louis, MO), unless stated otherwise. Liquid chromatography (LC)-mass spectrometry (MS)-grade acetonitrile was purchased from Fisher Scientific (Pittsburgh, PA); cholic acid, cholesterol, and diethyl phosphorocyanidate from Alfa Aesar (Ward Hill, MA); and isodeoxycholic acid from Steraloids (Newport, RI).
LC-MS analysis of fecal extracts.
Fecal samples were suspended in 1 ml of 50% acetonitrile (wt/vol) and extracted by vortexing and sonication for 10 min. The suspension was centrifuged twice at 18,000 g for 10 min, and after passage of the supernatant through a 2-μm filter, the filtrate was transferred to a sample vial and subjected to LC-MS analysis. A 5-μl aliquot of fecal extract was injected into an Acquity ultraperformance LC system (Waters, Milford, MA) and separated by a mobile phase gradient ranging from H2O to 95% aqueous acetonitrile containing 0.1% formic acid over a 10-min run. The LC eluant was introduced into a SYNAPT QTOF mass spectrometer (Waters) for accurate mass measurement and ion counting. Capillary and cone voltage for electrospray ionization were maintained at −3 kV and −35 V, respectively, for negative-mode detection. Source temperature and desolvation temperature were 120°C and 350°C, respectively. Nitrogen was used as a cone gas (50 l/h) and desolvation gas (600 l/h) and argon as the collision gas. For accurate mass measurement, the mass spectrometer was calibrated with sodium formate solution [mass-to-charge ratio (m/z) = 50–1,000] and monitored by the intermittent injection of the lock mass leucine enkephalin ([M-H]− = 554.2615 m/z) in real time. Mass chromatograms and mass spectral data were acquired and processed by MassLynx software (Waters) in centroided format. Additional structural information was obtained via tandem MS (MS/MS) fragmentation, with collision energies ranging from 15 to 30 electron volts. The concentration of bile acids in fecal samples was determined on the basis of the peak areas of individual bile acids and external standards.
Chemometric analysis and biomarker identification.
Chromatographic and mass spectral data of fecal samples were deconvoluted using MarkerLynx software (Waters). Each detected ion was represented by its retention time in the LC system and its m/z. A multivariate data matrix comprising sample identity, ion identity (retention time and m/z), and relative ion abundance was generated through centroiding, deisotoping, filtering, peak recognition, and integration and further exported into SIMCA-P+ software (Umetrics, Kinnelon, NJ) (6). Unsupervised principal component analysis and supervised orthogonal partial least squares analysis were used to analyze data from fecal extracts. Major latent variables in the data matrix were described in a scores scatter plot of the established multivariate model. Fecal metabolites affected by FMT were identified by analysis of ions contributing to the separation of pre- and post-FMT samples in the multivariate models (4, 6). The chemical identities of metabolites of interest were determined by accurate mass measurement, elemental composition analysis, database search [Lipid Maps (http://www.lipidmaps.org/), Human Metabolome Database (http://www.hmdb.ca/), MS/MS fragmentation, and comparisons with authentic standards].
Statistics.
Experimental values for fecal metabolites are expressed as means ± SD. Statistical analysis was performed using ANOVA followed by Tukey's honestly significant difference test on significant groups at α = 0.05. P < 0.05 was considered statistically significant.
RESULTS
Complete patient recovery following one or two FMT procedures.
Eleven of the 12 patients (91.6%) treated with FMT and analyzed in this study achieved clinical recovery with no recurrence of CDI following a single FMT over a ≥1-yr period of follow-up. Interestingly, while the patients were maintained on vancomycin prior to FMT, they continued to experience mild diarrheal symptoms (4.1 ± 1.3 stools per day, median type 5 on the Bristol stool scale). After the FMT procedure, the frequency of stools decreased (2.0 ± 1.4 stools per day, median type 4 on the Bristol stool scale). After a second FMT, the single patient with an initial recurrence of the disease also recovered. He was documented to be C. difficile toxin B-negative and remained free of diarrhea for 6 mo, at which point he experienced a relapse of CDI. This relapse was associated with a course of laxatives taken for constipation associated with his Parkinson's disease. The infection was successfully treated with a single 10-day course of fidaxomicin. Two patients were incidentally discovered to have underlying inflammatory bowel disease at the time of their FMT: one had Crohn's terminal ileitis and the second had lymphocytic colitis (Table 1). Neither patient required additional pharmacological therapy.
Composition of fecal communities dramatically changes following FMT.
The composition of fecal microbiota in 12 patients before and after FMT, as well as each donor sample, was examined by sequencing the V6 region of the 16S rRNA gene. A total of 13,873,260 sequences were generated, and 8,175,859 sequences remained after quality filtering, removal of chimeras, and preclustering. Sequences were subsampled to 22,535 to obtain the same number of reads for each sample, clustered into OTUs at 90% sequence identity as used previously (17), and classified using the Ribosomal Database Project (8).
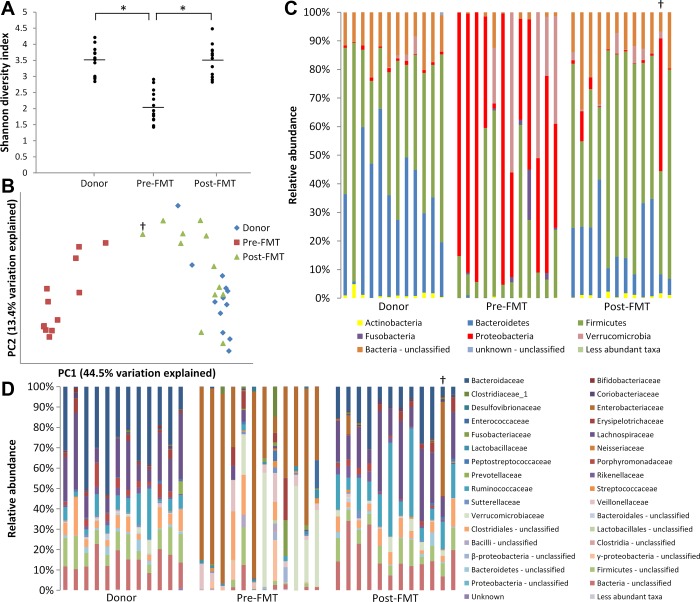
As we previously showed for a small set of patients (17), fecal bacterial communities changed dramatically following FMT. Initially, α diversity (i.e., diversity within each sample) was examined using the Shannon diversity index, which indicated that donor and post-FMT samples were significantly more diverse than pre-FMT samples (Fig. 1A). To examine β diversity (i.e., diversity between samples), PCoA was performed on the basis of UniFrac distances (Fig. 1B). The PCoA demonstrated that while donor and post-FMT samples clustered together along principal coordinate 1, pre-FMT sample communities were distinct.
Fig. 1.
Changes in fecal bacterial communities following fecal microbiota transplantation (FMT). A: Shannon diversity indexes of fecal samples. *P < 0.05. B: principal coordinate analysis of UniFrac distances between bacterial communities. PC1, principal coordinate 1; PC2, principal coordinate 2. C: relative abundance of operational taxonomic units (OTUs) from bacterial phyla in fecal samples. Colors correspond to phyla (see key at bottom). D: relative abundance of OTUs from bacterial families in fecal samples. Colors correspond to families (see key at right). †Patient who failed initial FMT.
These shifts in diversity following FMT were related to changes in the relative abundance of the bacterial phyla Bacteroidetes, Firmicutes, and Proteobacteria (Fig. 1C). Donor samples, representative of a healthy fecal microbiome (2, 48), were dominated by Bacteroidetes and Firmicutes (together accounting for ∼80–90% of OTUs in each sample), with a very low abundance of Proteobacteria (∼2% or fewer OTUs per sample). In contrast, patient samples prior to FMT tended to be dominated by the Proteobacteria, comprising ≥30% of OTUs in 13 of 14 samples (92.8%) and >50% of OTUs in 7 samples. After FMT, microbiota in patient samples more closely resembled the donors' composition, with increased abundance of Firmicutes and Bacteroidetes and decreased abundance of Proteobacteria. Overall, the number of Actinobacteria, Bacteroidetes, Firmicutes, and unclassified bacterial OTUs significantly increased following FMT (P = 0.044, P = 0.0015, P = 0.0016, and P < 0.0001, respectively), while the number of Proteobacteria and Verrucomicrobia significantly decreased (P < 0.0001 and P = 0.035, respectively; Table 3).
Table 3.
Abundance of major bacterial phyla in patient and donor samples
| Average No. of OTUs (of 22,535 total) |
||||
|---|---|---|---|---|
| Phylum | Donor | Pre-FMT | Post-FMT | P (pre- vs. post-FMT) |
| Actinobacteria | 275 | 24 | 219 | 0.044 |
| Bacteroidetes | 8,316 | 36 | 4,402 | 0.0015 |
| Firmicutes | 10,223 | 5,412 | 12,443 | 0.0016 |
| Fusobacteria | 0 | 480 | 2 | 0.177 |
| Proteobacteria | 207 | 12,689 | 1,234 | <0.0001 |
| Verrucomicrobia | 189 | 3,469 | 449 | 0.035 |
| Bacteria-unclassified | 3,302 | 424 | 3,785 | <0.0001 |
OTUs, operational taxonomic units.
Dramatic differences in community composition were also seen at the family level (Fig. 1D). Together, the bacterial families Lachnospiraceae, Bacteroidaceae, and Ruminococcaceae comprised ≥50% of the OTUs in each donor sample. In contrast, these families were found in much lower abundance in the majority of pre-FMT patient samples, with Lachnospiraceae at <2% abundance in 11 of 14 samples (78.6%), Bacteroidaceae at <1% abundance in each sample, and Ruminococcaceae at <1% abundance in 13 of 14 samples (92.8%). Instead, Enterobacteriaceae, Veillonellaceae, and Verrucomicrobiaceae, minor constituents of donor communities, together represented ≥50% of OTUs in 13 of 14 pre-FMT samples (92.8%). After FMT, these patterns reversed in most patient samples, with increased Lachnospiraceae, Bacteroidaceae, and Ruminococcaceae and decreased Enterobacteriaceae, Veillonellaceae, and Verrucomicrobiaceae. Bacterial families with significantly altered OTU abundance after FMT compared with pre-FMT samples are shown in Table 4.
Table 4.
Significantly changed bacterial families in patient and donor samples
| Average No. of OTUs (of 22,535 total) |
||||
|---|---|---|---|---|
| Family | Donor | Pre-FMT | Post-FMT | P (pre- vs. post-FMT) |
| Bacteroidaceae | 6,451 | 25 | 4,348 | 0.0009 |
| Coriobacteriaceae | 150 | 12 | 150 | 0.034 |
| Enterobacteriaceae | 13 | 11,572 | 1,083 | <0.0001 |
| Lachnospiraceae | 4,002 | 286 | 4,737 | <0.0001 |
| Lactobacillaceae | 2 | 255 | 12 | 0.0019 |
| Rikenellaceae | 388 | 2 | 455 | 0.0014 |
| Ruminococcaceae | 1,749 | 112 | 3,797 | 0.0002 |
| Veillonellaceae | 117 | 1261 | 81 | 0.0006 |
| Verrucomicrobiaceae | 189 | 3469 | 449 | 0.035 |
| Bacteroidetes-unclassified | 2,185 | 501 | 1,809 | 0.0064 |
Changes in fecal bile acid composition following FMT.
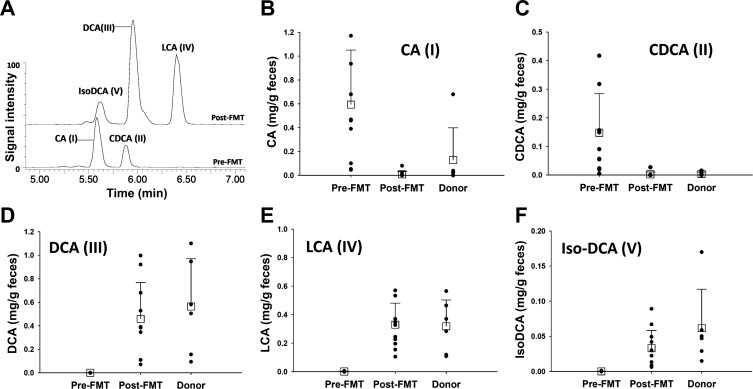
To explore changes in fecal bile acid composition induced by FMT, fecal samples collected from 12 CDI patients before and after the FMT procedure and 6 fecal samples from 2 donors were analyzed via LC-MS together. Microbial composition in these patient and 4 donor samples (1 from donor 1 and 3 from donor 2) was analyzed as described above (Fig. 1). The profile of primary and secondary bile acids in fecal extracts was defined by examination of the extracted ion chromatograms of deprotonated bile acids in the pre- and post-FMT samples. The results showed that FMT induced dramatic changes in fecal bile acid composition (Fig. 2A). Two primary bile acids, cholic acid (I) and chenodoxycholic acid (II), were present in significant amounts in most pre-FMT samples but were absent or existed in low abundance in most post-FMT and donor samples (Fig. 2, B and C). In contrast, three secondary bile acids, lithocholic acid (III), deoxycholic acid (IV), and isodeoxycholic acid (V), were present in the post-FMT and donor samples but were absent in the pre-FMT samples (Fig. 2, Fig. 2D–Fig. 2F).
Fig. 2.
Bile acid concentrations before and after FMT. A: overlay of representative chromatograms of bile acid metabolites in pre- and post-FMT fecal extracts. Chromatograms were generated by extraction of mass spectrometry signals within 20 ppm of calculated exact masses (407.2797, 391.2848, and 375.2899 mass-to-charge ratio in negative mode) of primary and secondary bile acids of sterol metabolites [cholic acid (CA, I), chenodeoxycholic acid (CDCA, II), deoxycholic acid (DCA, III), lithocholic acid (LCA, IV), and IsoDCA (V)]. Signal intensity of deoxycholic acid (III) in the post-FMT sample was arbitrarily set as 100%. B–F: concentration of bile acids (I–V) in pre-FMT, post-FMT, and donor samples.
Metabolomic analysis of FMT-induced metabolic changes in the fecal extracts of R-CDI patients.
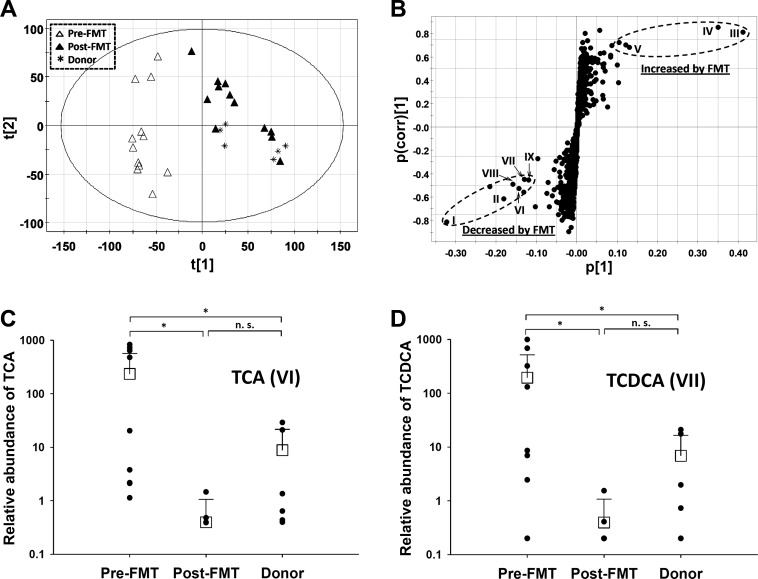
To examine overall metabolic changes following FMT, the same fecal samples analyzed for bile acid composition were also analyzed via LC-MS-based metabolomics. Unsupervised principal components analysis of chromatographic and mass spectral data acquired from LC-MS analysis revealed a clear separation of pre-FMT samples from the donor and post-FMT samples in a multivariate model (Fig. 3A). These findings suggest that FMT transformed the fecal metabolome of CDI patients to that consistent with the donors. The fecal metabolites, which differed among pre-FMT, post-FMT, and donor samples, were revealed in an S plot of the supervised orthogonal partial least squares model (Fig. 3B, Table 5). Besides confirmation of the primary and secondary bile acids (I–V) as the fecal metabolites that distinguish pre- and post-FMT samples, bile salts, including taurine conjugates (VI and VII; Fig. 3, C and D) and glycine conjugates of cholic acid and chenodeoxycholic acid (VIII and IX; data not shown), were identified as major contributors to sample separation in the multivariate models. The presence of a significant amount of bile salts in many pre-FMT samples suggested their incomplete hydrolysis in these patients.
Fig. 3.
Identification and characterization of FMT-responsive sterol metabolites in fecal extracts of Clostridium difficile infection (CDI) patients. A: scores plot of a principal components analysis model on pre-FMT (▲), post-FMT (△), and donor (∗) samples. The t[1] and t[2] values represent scores of each sample in principal components 1 and 2, respectively. B: loadings plot of principal components analysis model. Sterol metabolites contributing to separation of pre- and post-FMT samples (I–IX) were labeled, and their chemical identities are listed in Table 3. C and D: relative abundances of taurine conjugates of primary bile acids [taurocholic acid (TCA, VI) and taurochenodeoxycholic acid (TCDCA, VII)] in pre-FMT, post-FMT, and donor samples. NS, not significant. *P < 0.05.
Table 5.
Fecal metabolite markers of FMT
| ID | [M-H]− | Formula | Identity | Effect of FMT |
|---|---|---|---|---|
| I | 407.2798 | C24H40O5 | Cholic acid | ↓ |
| II | 391.2848 | C24H40O4 | Chenodeoxycholic acid | ↓ |
| III | 391.2848 | C24H40O4 | Deoxycholic acid | ↑ |
| IV | 375.2899 | C24H40O3 | Lithocholic acid | ↑ |
| V | 391.2848 | C24H40O4 | Isodeoxycholic acid | ↑ |
| VI | 514.2839 | C26H45NO7S4 | Taurocholic acid | ↓ |
| VII | 498.2889 | C26H45NO6S | Taurochenodeoxycholic acid | ↓ |
| VIII | 464.2817 | C26H43NO6 | Glycocholic acid | ↓ |
| IX | 448.3063 | C26H43NO5 | Glycochenodeoxycholic acid | ↓ |
Major primary and secondary bile acids and conjugates of primary bile acids (I–IX) were identified as metabolite markers contributing to separation of pre-FMT samples from post-FMT and donor samples in metabolomic analysis. [M-H]−, mass-to-charge ratio of deprotonated metabolite; ↑, increased after FMT; ↓, decreased after FMT.
DISCUSSION
The appearance of hypervirulent strains of CDI has led to an increased incidence of recurrent and severe forms of the disease refractory to antibiotic therapies alone (19, 31, 42). FMT has emerged as a solution to this unmet clinical need, and its use is becoming increasingly widespread in clinical practice (3, 42, 43). However, the mechanisms behind the success of this therapy remain largely unknown. Previous work by our group and others has demonstrated dramatic changes in the fecal microbiota of patients undergoing FMT, with fecal microbial communities becoming more similar to that of the donor, including an increased abundance of Bacteroidetes and Firmicutes, a decreased abundance of Proteobacteria, and an increase in overall microbial diversity (17, 24, 37, 46). The results presented in this study indicate that these microbiome changes also hold true for a larger sample size of patients. Consistent with the previous reports, FMT resulted in an increase in OTUs corresponding to Bacteroidetes and Firmicutes and a decrease in the abundance of Proteobacteria. The PCoA analysis also indicated that these changes represent shifts in the fecal bacterial community toward the composition of the donor and a shift from a highly altered community dominated by Proteobacteria toward the community typically found in healthy individuals (2, 48). The dramatic dysbiosis documented in the pre-FMT samples is likely induced and perpetuated by continuous use of an antibiotic, mainly vancomycin, that the patients were taking at the time of collection.
Because gut microbiota in the colon have multiple metabolic functions, targeted metabolite profiling and untargeted metabolomics were adopted to examine the metabolic events induced by FMT. The results showed that after FMT the overall metabolic composition of samples shifted to one similar to that of the donors, mimicking our findings for taxonomic microbial composition. The bile acids and primary bile salts were by far the dominant metabolites contributing to the separation of pre-FMT samples from post-FMT and donor samples revealed by the multivariate model on the fecal metabolomes, suggesting that the changes in bile acid composition account for much of the overall change in the fecal metabolome in R-CDI patients treated with FMT. Strikingly, secondary bile acids were simply not detectable in any of the pre-FMT fecal samples. Because of the well-known functions of gut microbiota in the hydrolysis of bile salts (taurine and glycine conjugates) to bile acids and the biotransformation of primary bile acids to secondary bile acids in the colon (20, 21, 28, 41, 44), this observation suggests that pre-FMT patients in this study were deficient in microbiota that are capable of metabolizing secondary bile salts and bile acids, possibly due to the depletion or decrease of bile acid-metabolizing microbiota after extensive antibiotic treatment (5, 18, 45). Therefore, FMT likely rehabilitated bile acid-metabolizing microbiota in these same patients. It is even plausible that normalization of bile acid metabolism contributed to resolution of mild diarrheal symptoms experienced by patients while they were maintained on vancomycin prior to FMT. Indeed, increased content of primary bile acids in stool is correlated with diarrheal symptoms, while increased content of secondary bile acids correlates with constipation (38).
Increased concentrations of primary bile acids and their conjugates in R-CDI patients are consistent with relentless relapses of the infection each time suppressive antibiotics are discontinued. Taurocholic acid, which existed in high abundance in many pre-FMT samples, is a potent germinant for C. difficile, transforming inert spores to toxin-producing, free-living vegetative cells (39). In fact, taurocholic acid has been used for decades as a key component of laboratory medium used to germinate C. difficile spores (47). In contrast, lithocholic acid and other secondary bile acids, which were deficient in pre-FMT samples but abundant in post-FMT and donor samples, inhibit C. difficile germination and colony growth (12, 40). These findings suggest that changes in fecal bile acid composition in FMT patients following the procedure may generate an environment unsuitable for C. difficile germination or growth. Prompt restoration of normal fecal bile acid composition following FMT fits this hypothesis, as clinical recurrence of CDI would otherwise peak during the 2nd and 3rd wk after discontinuation of antibiotics.
The changes in fecal bile acid composition of FMT patients may be directly related to changes in fecal bacterial composition. Many of the bacteria known to have 7α-dehydroxylation activity, which transforms primary bile acids to secondary bile acids, are members of the Lachnospiraceae and Ruminococcaceae families (i.e., Clostridium clusters XIVa and IV), including the best-studied of these species, Clostridium scindens (9, 20, 25, 35, 41, 44). By increasing the relative abundance of members of these bacterial families, it is possible that FMT increases 7α-dehydroxylation activity, leading to increased secondary bile acids and decreased primary bile acids. This may directly inhibit germination and, thus, toxin production by C. difficile.
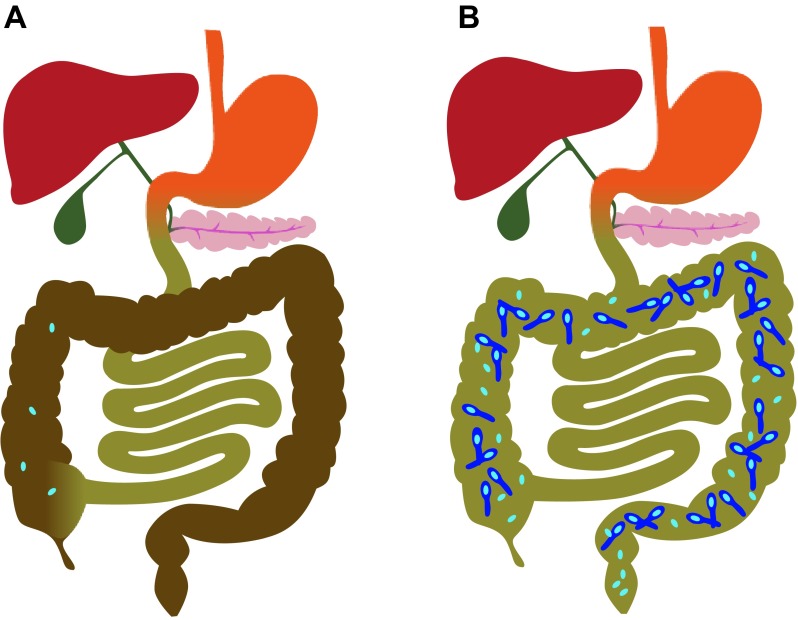
In conclusion, our results support a mechanistic model where FMT prevents recurrence of CDI by restoring normal bile acid composition in the colon (Fig. 4). In this model, antibiotics used to treat CDI also inhibit normal members of the microbiota that hydrolyze bile salts to bile acids and then convert primary bile acids to secondary bile acids. The lack of bile acid metabolism in antibiotic-treated R-CDI patients generates a local environment that promotes germination of C. difficile spores and growth of new vegetative bacteria that actively produce toxins once antibiotic treatment is completed. Restoration of normal colonic microbial ecology by FMT restores bile acid metabolism and normal bile acid composition in the colon, producing an unfavorable environment for C. difficile spore germination and allowing clinical recovery of R-CDI patients. Therefore, microbiological or pharmacological strategies to manipulate the intestinal bile acid composition may become effective approaches for the treatment of R-CDI.
Fig. 4.
Antibiotics promote recurrence of CDI by decreasing hydrolysis of bile salts and conversion of primary bile acids to secondary bile acids (the model). A: normal bile acid composition in the colon (shown in brown) prevents germination of C. difficile spores. B: antibiotics allow increased levels of bile salts and primary bile acids in the colon (shown in green), which promote germination of C. difficile spores and growth of vegetative forms of bacteria.
GRANTS
This research was supported, in part, by National Institute on Aging Grant R21 AI-091907 (A. Khoruts and M. J. Sadowsky). A. R. Weingarden was supported by funding from the University of Minnesota BioTechnology Institute.
DISCLOSURES
A. Khoruts and M. J. Sadowsky received funding from CIPAC, LLC, to carry out research on FMT using frozen fecal microbiota. A. Khoruts and M. J. Sadowsky have provided consulting services for CIPAC; the University of Minnesota Conflicts of Interest Program is managing conflicts of interest.
AUTHOR CONTRIBUTIONS
A.R.W., C.C., M.J.S., and A.K. are responsible for conception and design of the research; A.R.W., C.C., A.B., D.Y., Y.L., V.M.N., and A.K. performed the experiments; A.R.W., C.C., M.J.S., and A.K. analyzed the data; A.R.W., C.C., M.J.S., and A.K. interpreted the results of the experiments; A.R.W., C.C., and A.K. prepared the figures; A.R.W. and C.C. drafted the manuscript; A.R.W., C.C., M.J.S., and A.K. edited and revised the manuscript; A.R.W., C.C., A.B., D.Y., Y.L., V.M.N., M.J.S., and A.K. approved the final version of the manuscript.
ACKNOWLEDGMENTS
We thank the nursing staff, volunteer donors, and recipient patients; we also thank Katerina Helebrantova for help with the artwork on Fig. 4.
REFERENCES
- 1.Ananthakrishnan AN. Clostridium difficile infection: epidemiology, risk factors and management. Nat Rev Gastroenterol Hepatol 8: 17–26, 2011 [DOI] [PubMed] [Google Scholar]
- 2.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Dore J, Antolin M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Merieux A, Melo Minardi R, M'Rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P. Enterotypes of the human gut microbiome. Nature 473: 174–180, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borody TJ, Khoruts A. Fecal microbiota transplantation and emerging applications. Nat Rev Gastroenterol Hepatol 9: 88–96, 2011 [DOI] [PubMed] [Google Scholar]
- 4.Bylesjo M, Rantalainen M, Nicholson JK, Holmes E, Trygg J. K-OPLS package: kernel-based orthogonal projections to latent structures for prediction and interpretation in feature space. BMC Bioinformatics 9: 106, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carman RJ, Simon MA, Petzold HE, 3rd, Wimmer RF, Batra MR, Fernandez AH, Miller MA, Bartholomew M. Antibiotics in the human food chain: establishing no effect levels of tetracycline, neomycin, and erythromycin using a chemostat model of the human colonic microflora. Regul Toxicol Pharmacol 43: 168–180, 2005 [DOI] [PubMed] [Google Scholar]
- 6.Chen C, Ma X, Malfatti MA, Krausz KW, Kimura S, Felton JS, Idle JR, Gonzalez FJ. A comprehensive investigation of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) metabolism in the mouse using a multivariate data analysis approach. Chem Res Toxicol 20: 531–542, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chitnis AS, Holzbauer SM, Belflower RM, Winston LG, Bamberg WM, Lyons C, Farley MM, Dumyati GK, Wilson LE, Beldavs ZG, Dunn JR, Gould LH, Maccannell DR, Gerding DN, McDonald LC, Lessa FC. Epidemiology of community-associated Clostridium difficile infection, 2009 through 2011. JAMA Intern Med: 1–9, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37: D141–D145, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doerner KC, Takamine F, LaVoie CP, Mallonee DH, Hylemon PB. Assessment of fecal bacteria with bile acid 7α-dehydroxylating activity for the presence of bai-like genes. Appl Environ Microbiol 63: 1185–1188, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francis MB, Allen CA, Shrestha R, Sorg JA. Bile acid recognition by the Clostridium difficile germinant receptor, CspC, is important for establishing infection. PLoS Pathog 9: e1003356, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giel JL, Sorg JA, Sonenshein AL, Zhu J. Metabolism of bile salts in mice influences spore germination in Clostridium difficile. PLos One 5: e8740, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gough E, Shaikh H, Manges AR. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis 53: 994–1002, 2011 [DOI] [PubMed] [Google Scholar]
- 14.Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4: 17–27, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. The role of butyrate on colonic function. Aliment Pharmacol Ther 27: 104–119, 2008 [DOI] [PubMed] [Google Scholar]
- 16.Hamilton MJ, Weingarden AR, Sadowsky MJ, Khoruts A. Standardized frozen preparation for transplantation of fecal microbiota for recurrent Clostridium difficile infection. Am J Gastroenterol 107: 761–767, 2012 [DOI] [PubMed] [Google Scholar]
- 17.Hamilton MJ, Weingarden AR, Unno T, Khoruts A, Sadowsky MJ. High-throughput DNA sequence analysis reveals stable engraftment of gut microbiota following transplantation of previously frozen fecal bacteria. Gut Microbes 4: 125–135, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hashimoto S, Igimi H, Uchida K, Satoh T, Benno Y, Takeuchi N. Effects of β-lactam antibiotics on intestinal microflora and bile acid metabolism in rats. Lipids 31: 601–609, 1996 [DOI] [PubMed] [Google Scholar]
- 19.He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, Connor TR, Harris SR, Fairley D, Bamford KB, D'Arc S, Brazier J, Brown D, Coia JE, Douce G, Gerding D, Kim HJ, Koh TH, Kato H, Senoh M, Louie T, Michell S, Butt E, Peacock SJ, Brown NM, Riley T, Songer G, Wilcox M, Pirmohamed M, Kuijper E, Hawkey P, Wren BW, Dougan G, Parkhill J, Lawley TD. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet 45: 109–113, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirano S, Nakama R, Tamaki M, Masuda N, Oda H. Isolation and characterization of thirteen intestinal microorganisms capable of 7α-dehydroxylating bile acids. Appl Environ Microbiol 41: 737–745, 1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofmann AF, Hagey LR. Bile acids: chemistry, pathochemistry, biology, pathobiology, therapeutics. Cell Mol Life Sci 65: 2461–2483, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly CP, LaMont JT. Clostridium difficile—more difficult than ever. N Engl J Med 359: 1932–1940, 2008 [DOI] [PubMed] [Google Scholar]
- 23.Khanna S, Pardi DS, Aronson SL, Kammer PP, Orenstein R, St Sauver JL, Harmsen WS, Zinsmeister AR. The epidemiology of community-acquired Clostridium difficile infection: a population-based study. Am J Gastroenterol 107: 89–95, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J Clin Gastroenterol 44: 354–360, 2010 [DOI] [PubMed] [Google Scholar]
- 25.Kitahara M, Takamine F, Imamura T, Benno Y. Assignment of Eubacterium sp. VPI 12708 and related strains with high bile acid 7α-dehydroxylating activity to Clostridium scindens and proposal of Clostridium hylemonae sp. nov, isolated from human faeces. Int J Syst Evol Microbiol 50: 971–978, 2000 [DOI] [PubMed] [Google Scholar]
- 26.Lessa FC, Gould CV, McDonald LC. Current status of Clostridium difficile infection epidemiology. Clin Infect Dis 55 Suppl 2: S65–S70, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Louie TJ, Miller MA, Mullane KM, Weiss K, Lentnek A, Golan Y, Gorbach S, Sears P, Shue YK. Fidaxomicin versus vancomycin for Clostridium difficile infection. N Engl J Med 364: 422–431, 2011 [DOI] [PubMed] [Google Scholar]
- 28.Mahony DE, Meier CE, Macdonald IA, Holdeman LV. Bile salt degradation by nonfermentative Clostridia. Appl Environ Microbiol 34: 419–423, 1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchesi JR, Dutilh BE, Hall N, Peters WH, Roelofs R, Boleij A, Tjalsma H. Towards the human colorectal cancer microbiome. PLos One 6: e20447, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.May T, Mackie RI, Fahey GC, Jr, Cremin JC, Garleb KA. Effect of fiber source on short-chain fatty acid production and on the growth and toxin production by Clostridium difficile. Scand J Gastroenterol 29: 916–922, 1994 [DOI] [PubMed] [Google Scholar]
- 31.Merrigan M, Venugopal A, Mallozzi M, Roxas B, Viswanathan VK, Johnson S, Gerding DN, Vedantam G. Human hypervirulent Clostridium difficile strains exhibit increased sporulation as well as robust toxin production. J Bacteriol 192: 4904–4911, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller BA, Chen LF, Sexton DJ, Anderson DJ. Comparison of the burdens of hospital-onset, healthcare facility-associated Clostridium difficile infection and of healthcare-associated infection due to methicillin-resistant Staphylococcus aureus in community hospitals. Infect Control Hosp Epidemiol 32: 387–390, 2011 [DOI] [PubMed] [Google Scholar]
- 33.Nagai F, Morotomi M, Watanabe Y, Sakon H, Tanaka R. Alistipes indistinctus sp. nov and Odoribacter laneus sp. nov, common members of the human intestinal microbiota isolated from faeces. Int J Syst Evol Microbiol 60: 1296–1302, 2010 [DOI] [PubMed] [Google Scholar]
- 34.Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett 217: 133–139, 2002 [DOI] [PubMed] [Google Scholar]
- 35.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47: 241–259, 2006 [DOI] [PubMed] [Google Scholar]
- 36.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. Introducing MOTHUR: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shahinas D, Silverman M, Sittler T, Chiu C, Kim P, Allen-Vercoe E, Weese S, Wong A, Low DE, Pillai DR. Toward an understanding of changes in diversity associated with fecal microbiome transplantation based on 16S rRNA gene deep sequencing. MBio 3, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin A, Camilleri M, Vijayvargiya P, Busciglio I, Burton D, Ryks M, Rhoten D, Lueke A, Saenger A, Girtman A, Zinsmeister AR. Bowel functions, fecal unconjugated primary and secondary bile acids, and colonic transit in patients with irritable bowel syndrome. Clin Gastroenterol Hepatol 11: 1270–1275, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol 190: 2505–2512, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sorg JA, Sonenshein AL. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J Bacteriol 192: 4983–4990, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stellwag EJ, Hylemon PB. Characterization of 7α-dehydroxylase in Clostridium leptum. Am J Clin Nutr 31: S243–S247, 1978 [DOI] [PubMed] [Google Scholar]
- 42.Surawicz CM, Alexander J. Treatment of refractory and recurrent Clostridium difficile infection. Nat Rev Gastroenterol Hepatol 8: 330–339, 2011 [DOI] [PubMed] [Google Scholar]
- 43.Surawicz CM, Brandt LJ, Binion DG, Ananthakrishnan AN, Curry SR, Gilligan PH, McFarland LV, Mellow M, Zuckerbraun BS. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am J Gastroenterol 108: 478–499, 2013 [DOI] [PubMed] [Google Scholar]
- 44.Takamine F, Imamura T. Isolation and characterization of bile acid 7-dehydroxylating bacteria from human feces. Microbiol Immunol 39: 11–18, 1995 [DOI] [PubMed] [Google Scholar]
- 45.Toda T, Ohi K, Kudo T, Yoshida T, Ikarashi N, Ito K, Sugiyama K. Antibiotics suppress Cyp3a in the mouse liver by reducing lithocholic acid-producing intestinal flora. Yakugaku Zasshi 129: 601–608, 2009 [DOI] [PubMed] [Google Scholar]
- 46.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, Speelman P, Dijkgraaf MG, Keller JJ. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 368: 407–415, 2013 [DOI] [PubMed] [Google Scholar]
- 47.Wilson KH. Efficiency of various bile salt preparations for stimulation of Clostridium difficile spore germination. J Clin Microbiol 18: 1017–1019, 1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zoetendal EG, Rajilic-Stojanovic M, de Vos WM. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 57: 1605–1615, 2008 [DOI] [PubMed] [Google Scholar]