Abstract
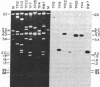
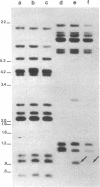
In earlier studies, we identified in Caenorhabditis elegans two informational suppressors sup-5 III and sup-7 X and recently showed that these suppressors acted via an altered tRNA to suppress translational termination at amber (UAG) stop codons. We now show that the sup-7 (st5) suppressor is a tRNATrpUAG amber suppressor. These studies utilized a radiolabeled purified tRNA fraction to identify hybridizing genomic sequences in a phage genomic library. DNA sequence analysis of the hybridizing segment of one clone showed that the probe recognized a tRNATrpUGG sequence. The sup-7 gene was shown to be one of an 11 or 12 member tRNATrp family by Southern blot analysis, taking advantage of an Xba I restriction site induced in the anticodon sequence by the mutational event to suppressor. Sequence analysis of a recombinant lambda clone containing sup-7 gene proved that sup-7(st5) is a tRNATrpUAG. This conclusive proof of the nature of sup7(st5) will permit unambiguous interpretation in genetic applications, and the availability of the cloned sequences may allow the sup-7 gene to be used to select for the reintroduction of DNA into C. elegans.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison D. S., Goh S. H., Hall B. D. The promoter sequence of a yeast tRNAtyr gene. Cell. 1983 Sep;34(2):655–664. doi: 10.1016/0092-8674(83)90398-7. [DOI] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974 May;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce A. G., Uhlenbeck O. C. Reactions at the termini of tRNA with T4 RNA ligase. Nucleic Acids Res. 1978 Oct;5(10):3665–3677. doi: 10.1093/nar/5.10.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciliberto G., Castagnoli L., Melton D. A., Cortese R. Promoter of a eukaryotic tRNAPro gene is composed of three noncontiguous regions. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1195–1199. doi: 10.1073/pnas.79.4.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H. Genetic studies of the lac repressor. IV. Mutagenic specificity in the lacI gene of Escherichia coli. J Mol Biol. 1977 Dec 15;117(3):577–606. doi: 10.1016/0022-2836(77)90059-6. [DOI] [PubMed] [Google Scholar]
- Files J. G., Hirsh D. Ribosomal DNA of Caenorhabditis elegans. J Mol Biol. 1981 Jun 25;149(2):223–240. doi: 10.1016/0022-2836(81)90299-0. [DOI] [PubMed] [Google Scholar]
- Galli G., Hofstetter H., Birnstiel M. L. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature. 1981 Dec 17;294(5842):626–631. doi: 10.1038/294626a0. [DOI] [PubMed] [Google Scholar]
- Goodman H. M., Olson M. V., Hall B. D. Nucleotide sequence of a mutant eukaryotic gene: the yeast tyrosine-inserting ochre suppressor SUP4-o. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5453–5457. doi: 10.1073/pnas.74.12.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorini L. Informational suppression. Annu Rev Genet. 1970;4:107–134. doi: 10.1146/annurev.ge.04.120170.000543. [DOI] [PubMed] [Google Scholar]
- Herman R. K., Madl J. E., Kari C. K. Duplications in Caenorhabditis elegans. Genetics. 1979 Jun;92(2):419–435. doi: 10.1093/genetics/92.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh D. Tryptophan transfer RNA as the UGA suppressor. J Mol Biol. 1971 Jun 14;58(2):439–458. doi: 10.1016/0022-2836(71)90362-7. [DOI] [PubMed] [Google Scholar]
- Hudziak R. M., Laski F. A., RajBhandary U. L., Sharp P. A., Capecchi M. R. Establishment of mammalian cell lines containing multiple nonsense mutations and functional suppressor tRNA genes. Cell. 1982 Nov;31(1):137–146. doi: 10.1016/0092-8674(82)90413-5. [DOI] [PubMed] [Google Scholar]
- Karn J., Brenner S., Barnett L., Cesareni G. Novel bacteriophage lambda cloning vector. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5172–5176. doi: 10.1073/pnas.77.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn L. J., Brown D. D. Nucleotide sequence of Xenopus borealis oocyte 5S DNA: comparison of sequences that flank several related eucaryotic genes. Cell. 1978 Dec;15(4):1145–1156. doi: 10.1016/0092-8674(78)90042-9. [DOI] [PubMed] [Google Scholar]
- Liebman S. W., Sherman F. Inhibition of growth by amber suppressors in yeast. Genetics. 1976 Feb;82(2):233–249. doi: 10.1093/genetics/82.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod A. R., Waterston R. H., Fishpool R. M., Brenner S. Identification of the structural gene for a myosin heavy-chain in Caenorhabditis elegans. J Mol Biol. 1977 Jul;114(1):133–140. doi: 10.1016/0022-2836(77)90287-x. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Coulondre C., Hofer M., Schmeissner U., Sommer H., Schmitz A., Lu P. Genetic studies of the lac repressor. IX. Generation of altered proteins by the suppression of nonsence mutations. J Mol Biol. 1979 Jun 25;131(2):191–222. doi: 10.1016/0022-2836(79)90073-1. [DOI] [PubMed] [Google Scholar]
- Page G. S., Hall B. D. Characterization of the yeast tRNA Ser genomic organization and DNA sequence. Nucleic Acids Res. 1981 Feb 25;9(4):921–934. doi: 10.1093/nar/9.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Robinson R. R., Davidson N. Analysis of a drosophila tRNA gene cluster: two tRNALeu genes contain intervening sequences. Cell. 1981 Jan;23(1):251–259. doi: 10.1016/0092-8674(81)90289-0. [DOI] [PubMed] [Google Scholar]
- SARABHAI A. S., STRETTON A. O., BRENNER S., BOLLE A. CO-LINEARITY OF THE GENE WITH THE POLYPEPTIDE CHAIN. Nature. 1964 Jan 4;201:13–17. doi: 10.1038/201013a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Gauss D. H. Compilation of sequences of tRNA genes. Nucleic Acids Res. 1982 Jan 22;10(2):r57–r81. [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Gauss D. H. Compilation of tRNA sequences. Nucleic Acids Res. 1982 Jan 22;10(2):r1–55. [PMC free article] [PubMed] [Google Scholar]
- Sulston J. E., Brenner S. The DNA of Caenorhabditis elegans. Genetics. 1974 May;77(1):95–104. doi: 10.1093/genetics/77.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston R. H. A second informational suppressor, SUP-7 X, in Caenorhabditis elegans. Genetics. 1981 Feb;97(2):307–325. doi: 10.1093/genetics/97.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston R. H., Brenner S. A suppressor mutation in the nematode acting on specific alleles of many genes. Nature. 1978 Oct 26;275(5682):715–719. doi: 10.1038/275715a0. [DOI] [PubMed] [Google Scholar]
- Waterston R. H., Fishpool R. M., Brenner S. Mutants affecting paramyosin in Caenorhabditis elegans. J Mol Biol. 1977 Dec 15;117(3):679–697. doi: 10.1016/0022-2836(77)90064-x. [DOI] [PubMed] [Google Scholar]
- Wills N., Gesteland R. F., Karn J., Barnett L., Bolten S., Waterston R. H. The genes sup-7 X and sup-5 III of C. elegans suppress amber nonsense mutations via altered transfer RNA. Cell. 1983 Jun;33(2):575–583. doi: 10.1016/0092-8674(83)90438-5. [DOI] [PubMed] [Google Scholar]
- Yaniv M., Folk W. R., Berg P., Soll L. A single mutational modification of a tryptophan-specific transfer RNA permits aminoacylation by glutamine and translation of the codon UAG. J Mol Biol. 1974 Jun 25;86(2):245–260. doi: 10.1016/0022-2836(74)90016-3. [DOI] [PubMed] [Google Scholar]