Abstract
Mechanisms of molecular self-replication have the potential to shed light upon the origins of life. In particular, self-replication through RNA-catalysed templated RNA synthesis is thought to have supported a primordial ‘RNA World’. However, existing polymerase ribozymes lack the capacity to synthesise RNAs approaching their own size. Here we report the in vitro evolution of such catalysts directly in the RNA-stabilising medium of water-ice, which yielded RNA polymerase ribozymes specifically adapted to sub-zero temperatures and able to synthesise RNA in ices at temperatures as low as −19°C. Combination of cold-adaptive mutations with a previously described 5′ extension operating at ambient temperatures enabled the design of a first polymerase ribozyme capable of catalysing the accurate synthesis of an RNA sequence longer than itself (adding up to 206 nucleotides), an important stepping stone towards RNA self-replication.
Introduction
Compelling evidence, ranging from aspects of modern metabolism to the central informational and catalytic roles of RNA in translation and splicing, supports the hypothesis that in a distant evolutionary past, biology used RNA for both genetic information storage and metabolism1. Central to this “RNA world”2, arising from the products of prebiotic chemistry3-5, would have been a ribozyme catalyst able to perform templated RNA synthesis to enable replication and expression of the emerging RNA genomes6,7.
RNA-catalysed RNA replication is a complex process, requiring iterative cycles of accurate substrate selection, elongation and translocation along the RNA template. While any primordial replicase appears to have been lost, functional aspects of life’s first genetic system may be studied using modern-day analogues obtained through directed evolution8 the best of which are a family of ribozymes based upon the R18 RNA polymerase ribozyme9-11.
R18 is a ribozyme, approximately 200 nucleotides (nt) long, derived from a ligase ribozyme generated de novo from a random RNA sequence pool12,13, and can catalyse the templated extension of RNA primers by up to 14 nucleotides (nts) using nucleoside triphosphates (NTPs)9. However, the activity of R18 falls short of that required for self-replication in both its limited synthetic capacity (exacerbated by a low affinity for substrate primer/template duplex14) as well as its poor stability in the presence of the high concentration of Mg2+ ions (200 mM) required for polymerase activity9,15,16.
We had previously shown that ice, a biphasic medium, could enhance both polymerase ribozyme activity and stability15 - despite the poor adaptation of the R18 polymerase ribozyme (evolved at 22°C) to the low temperatures of the ice phase (−7°C). Ice is also one of a range of heterogeneous prebiotic media (including aerosols, lipidic membranes and porous rock17-20) with the potential to effect RNA compartmentalisation, a prerequisite for evolution6.
In an attempt to better understand the potential of ice as a prebiotic medium we have investigated the directed evolution of RNA in this frozen environment. Harnessing the compartmentalised bead-tagging (CBT) RNA selection strategy11 directly in ice, we isolated RNA polymerase ribozymes adapted to frozen conditions, and derived a ribozyme - tC9Y - able to accurately synthesise RNA oligomers longer than the ribozyme itself using a favourable RNA template sequence at 17°C. Furthermore, we discovered traits that characterize such favourable RNA templates, defining the upcoming challenges on the road towards RNA self-replication.
Results
In-ice selection of ribozyme polymerase activity
Nucleic acid enzymes are commonly studied in homogenous media such as aqueous solutions at ambient temperatures. However, the primordial ribozymes of the RNA world are likely to have arisen and/or functioned in a range of heterogeneous media at variable ionic and substrate concentrations19. Here we have investigated the effects of one such non-uniform environment - water-ice15,21 - on RNA evolution.
Ice formation from an aqueous solution comprising solutes such as ions and RNA results in a non-uniform, biphasic system whereby the solutes, including RNA, are excluded from the growing ice crystals and are concentrated in an interstitial brine, called the eutectic phase. Having previously shown that freezing could enhance, prolong and compartmentalise RNA polymerase ribozyme activity15, we sought to explore the accessibility of novel ribozyme phenotypes by direct in-ice evolution using a modification of the previously described CBT (compartmentalised bead-tagging) in vitro selection strategy11.
During in-ice CBT, the selection step of primer extension by ribozyme variants is carried out directly in the eutectic phase of water ice at −7°C, where ice-active ribozyme variants tag the microbead to which they are tethered by extending RNA primer/template duplexes on the same bead (Fig. 1a). After thawing, primer extension initiates rolling circle amplification (RCA) and is scored by fluorescent staining of RCA products, permitting flow cytometric sorting and isolation of beads carrying active ribozymes (Supplementary Fig. S1a, b).
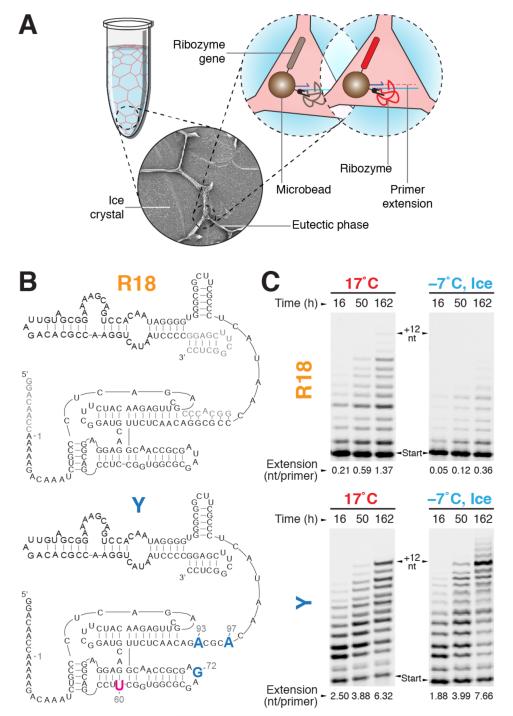
Figure 1. In-ice selection for polymerase ribozyme activity.
a, Short scheme of in-ice compartmentalised bead-tagging (CBT) selection (full scheme in Supplementary Fig. S1a). Solutes and microbeads are concentrated into the channels of the liquid eutectic phase (pale red) surrounding ice crystals (pale blue). Inset: scanning electron micrograph of ice selection (diameter, 0.15 mm). Ice-active ribozymes (red) extend primers linked to the same bead as themselves and their encoding gene, enabling recovery by flow cytometry. b, Secondary structures of the wild-type R18 ribozyme construct (residues not mutagenised in the starting library are shown in grey) and ribozyme Y, with mutations derived from the in-ice selected ribozyme C8 in blue. c, Polymerase activities of R18 and the evolved ribozyme Y (denaturing PAGE of extension time-courses (primer A /template I)) at 17°C and in ice at −7°C. The average number of nucleotides added per primer is indicated below each lane.
We initiated in-ice selection for RNA polymerase ribozyme activity starting from a random mutant library (7.6 mutations/gene) of the original R18 polymerase ribozyme9. To effectively explore sequence space using our modestly sized mutant repertoire (5×107) we exploited the strategies of neutral drift and recombination using three initial rounds of in-ice CBT at low stringency (requiring primer extension by 3-5 nucleotides for RCA signal, to deplete the sequence pool of detrimental mutations), followed by recombination and five additional rounds of in-ice CBT selection with increasing stringency (requiring primer extension by 10-12 nucleotides for RCA signal, Supplementary Table S1).
Generic and ice-specific adaptations
We screened this enriched pool for ribozymes with improved in-ice RNA polymerase activity, using a modification of the ribozyme polymerase plate assay (RPA)11. RPA screening identified two polymerase ribozyme clones with improved in-ice primer extension activity, C30 and C8, both of which outperformed R18 on two different primer/template combinations (Supplementary Fig. S1c, d). Engineering C30 yielded the ribozyme ‘W’ which displayed a phenotype related to that of ‘Z’ (an R18 ribozyme variant with increased sequence generality evolved independently at 17°C11), but which possessed an additional set of moderately cold-adaptive mutations (Supplementary Fig. S2). C8, however, showed a more striking improvement of in-ice activity, and we focused further investigations upon this phenotype. C8 exhibited only three mutations with respect to the parent R18 sequence (U72G, G93A, C97A, Supplementary Fig. S3a), which together provided superior in-ice polymerase activity. Introducing the previously described C60U mutation11 from Z gave a moderate further boost to activity (Supplementary Fig. S3b, c) yielding the ribozyme ‘Y’ (Fig. 1b). Y outperformed the parent R18 ribozyme on a range of templates (Supplementary Fig. S3d), but in particular displayed substantial polymerase activity in ice at −7°C (Fig. 1c). Indeed, Y synthesised more RNA in ice (at −7°C) than at 17°C, after just two days’ incubation. During incubation for one week in ice, Y extends over 35% of primers by 12 nts, before pausing at a challenging template sequence block16.
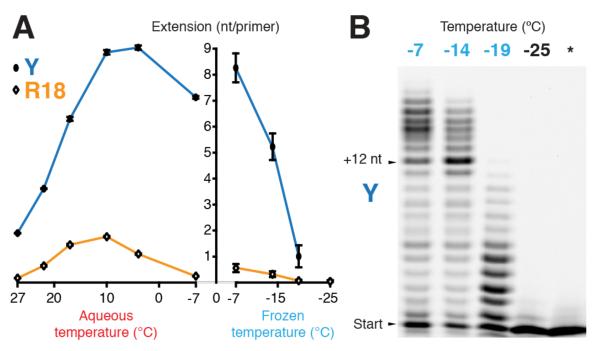
Comparison of RNA synthesis by R18 and Y over a range of temperatures revealed a lower temperature optimum for Y that supported significant activity at −7°C in ice and in supercooled solutions (Fig. 2a). While Y outperformed R18 by fourfold at 17°C (as judged by gel densitometry of average extension per primer15,22), at −7°C the differential rose to more than 15-fold in ice, and more than 30-fold in a supercooled solution. These results indicate both generic as well as ice-specific improvements in RNA polymerase ribozyme activity with the latter arising primarily from an adaptation to cold temperatures rather than to ice surfaces or structural features. Substantial RNA synthesis by the ribozyme could be observed down to temperatures as low as −19°C (Fig. 2b).
Figure 2. Cold adaptation of ribozyme activity.
a, Influence of temperature upon RNA synthesis by ribozymes Y and R18: quantification of average extension (primer A / template I, 7 days) in aqueous (left panel) and frozen (right panel) reactions (error bars represent s.d. of 3 repeats, except for −7°C frozen (6 repeats)). b, Primer extension by Y in ices at different temperatures (primer A / template I; 40 days; * = −25°C, no NTPs).
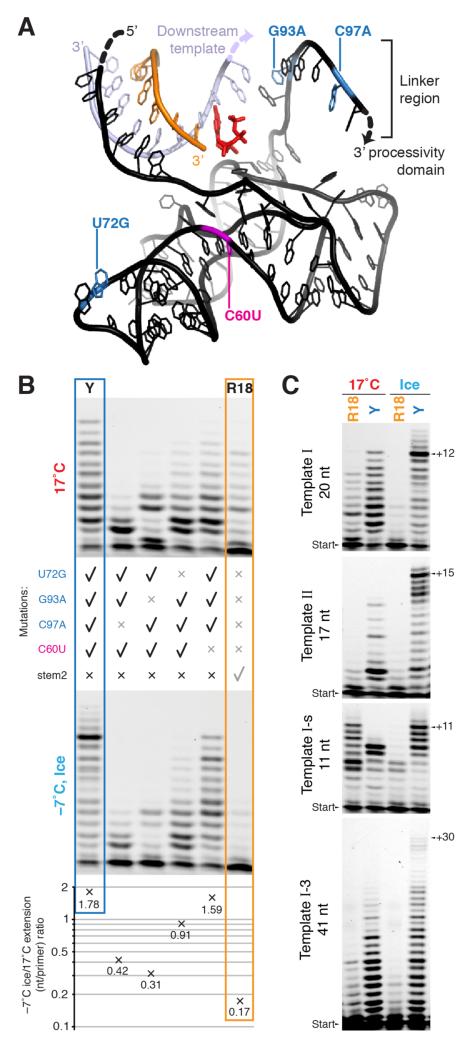
We sought to disentangle the contributions of the different selected mutations in Y (approximate positions in Fig. 3a) to both generic as well as to cold-specific adaptations of polymerase ribozyme activity by reverting individual Y mutations and measuring the activity of the resulting polymerase ribozymes under different conditions. Reversion greatly reduced polymerase activity, but its impact differed at different temperatures: ratios of quantified extensions at ambient temperatures and in ice indicate that G93A and C97A are the key mutations responsible for boosting cold-specific activity, with U72G providing a minor contribution (Fig. 3b). As expected, the C60U mutation, derived from the Z ribozyme evolved at ambient temperatures, did not improve activity in ice any more than at ambient temperatures.
Figure 3. Basis of cold adaptation.
a, Representation of the relative positions of primer (orange), template (lilac) and catalytic core of Y (black), modelled on shared regions in the crystal structure of the progenitor class I ligase ribozyme30. The incoming NTP (red) is positioned at the 3′ end of the primer, and the locations of the four mutations comprising Y are highlighted. The indicated single-stranded downstream template and the linker region leading to the processivity domain (not shown) are likely flexible in the polymerase. b, Backmutation analysis of Y to uncover contributions to cold adaptation. Denaturing PAGE of extension (3 days, primer A / template I) at 17°C (top panel) and at −7°C in ice (lower panel), by R18, Y, and four reversion mutants of Y as indicated. These extensions were quantified (average nucleotides added/primer), allowing calculation of the ratio of observed extension in ice versus 17°C for each ribozyme, varying ~10-fold between Y and R18 (bottom panel). c, Influence of template length (indicated) and sequence on extension by R18 and Y (denaturing PAGE of extensions of primer A upon different templates at 17°C and at −7°C in ice (7 days)).
G93A and C97A may be involved in low-temperature interactions with the RNA template, improving primer/template duplex binding. This is unlikely to be mediated by sequence-specific hybridisation, as Y performed even more proficient primer extension on a template (II) with a different downstream sequence (Fig. 3c), continuing up to a couple of bases from its 5′ end. Furthermore, the parental R18 ribozyme shows a preference for a shorter template (I-s) on which Y performs worse than on the longer template I, suggesting that Y may be able to productively interact with downstream single-stranded template RNA to achieve enhanced polymerisation in ice.
Synthesis of long RNAs in ice and at ambient temperatures
To explore the potential of this apparent enhancement of downstream template interactions, we examined ribozyme activity upon long templates consisting of repeats of the template I sequence (I-3, Fig. 3c). Whereas R18 could only achieve limited polymerisation upon such templates, Y was able to synthesise long extension products up to +30 nt in ice.
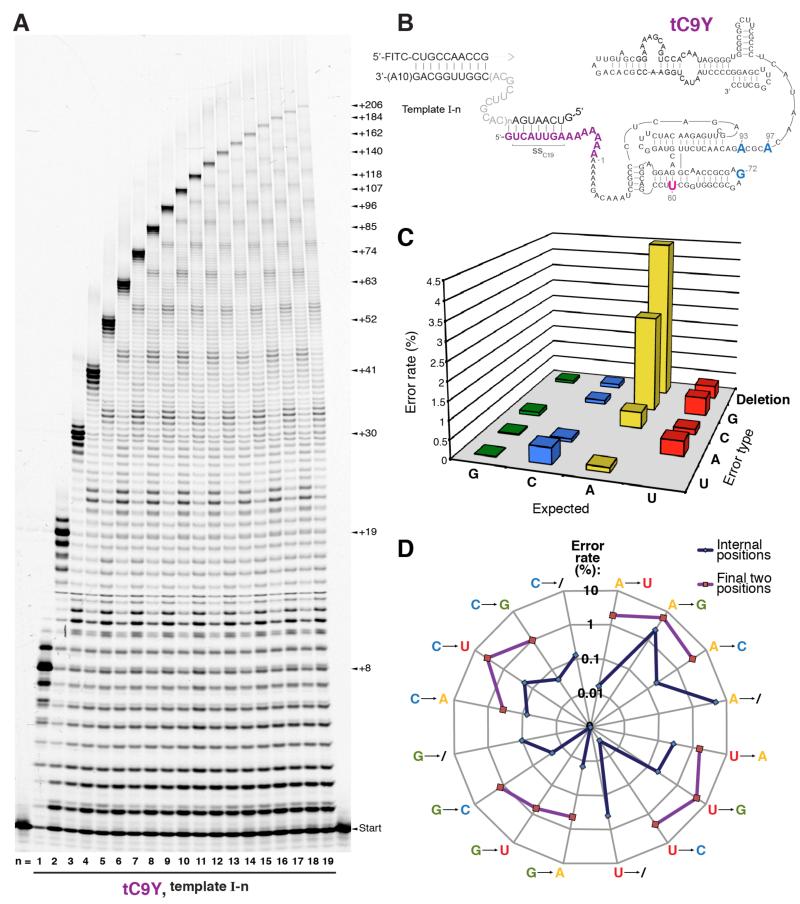
We attempted to fully leverage these benefits of Y upon extension by adding to Y, via a flexible A8 linker sequence, the 5′-‘ssC19’ sequence extension (5′-GUCAUUG-), previously shown to enhance synthesis of long RNAs by facilitating ribozyme tethering to the 5′ end of the template11. This new engineered tC9Y polymerase ribozyme proved exceptionally good at synthesising very long RNAs on these favourable templates, yielding RNA products up to 206 nucleotides long (Fig. 4a, b) at 17°C.
Figure 4. Long range RNA synthesis by ribozyme tC9Y.
a, Denaturing PAGE showing the tC9Y-ctalysed extension of primer BioFITC-A upon the (I-n series of templates (17°C, 7 days). Size of synthesised products (nucleotides added to primer) is indicated. b, Secondary structure of the tC9Y ribozyme, shown hybridised to the 5′ end of the template (I-n, where n is the number of central 11-nucleotide repeats) via the 5′ ssC19 sequence (purple). c, The full spectrum of error rates within all sequenced products synthesised by tC9Y upon template (I-10 at 17°C. The total fidelity (97.4%) is calculated using a geometric mean of the total error rates opposite A, C, G and U. d, The errors shown in (c) were subdivided into rates of each error type in the final two bases of all sequenced extension products versus the rates in all preceding internal bases up to the final two. Note that deletions of a penultimate base would appear as and are assigned as substitutions, as further extension would be required to manifest their identity; thus, the displayed substitution rates in the final two bases represent overestimates. The geometric mean between the total substitution rates at A, C, G, and U is 7.1% in the final two bases of each extension product compared to 0.8% in the preceding bases.
Due to the influence of the Y mutations, tC9Y is also the fastest and most efficient polymerase ribozyme described so far, yielding 63 nt products after only 16 hours and 206 nt products after 60 hours (Supplementary Fig. S4). With ~97.5% of primers extended beyond each successive nucleotide by the end of the week-long incubation (Supplementary Fig. S5a), tC9Y exhibits a termination probability per incorporation on these favourable templates of 0.012-0.025. This is an approximately 2-fold improvement on tC19 (0.019-0.05) and 4-fold improvement on tC19Z (0.03-0.1) (Supplementary Fig. S6), previously the most efficient polymerase ribozymes described11.
In ice (−7°C), long-range synthesis by tC9Y only yielded products up to 118 nucleotides long (Supplementary Fig. S5b). As well as proceeding much more slowly (Supplementary Fig. S4a), initiation of frozen extension in this setup was inefficient. Furthermore, termination patterns indicated that some primers were extended in an ssC19-independent manner (Supplementary Fig. S5c). This may be due to altered dynamics and a reduced functionality of the ssC19 tethering function - optimized for ambient conditions - under the altered temperature and salt conditions in ice, greatly reducing overall synthetic efficiency.
Long range synthesis allowed enhanced precision when determining the fidelity of RNA synthesis, a critical parameter of RNA replication23. Sanger sequencing of full length products11 (Supplementary Table S2) indicated that the Y mutations conferred a modest fidelity improvement at 17°C to 98.3% compared to 97.3% for tC1911, and in ice to 94.8% compared to 93.4% for R1815. We next performed Illumina sequencing of the entire length spectrum of tC9Y (17°C) extension products, to gain a global picture of the fidelity of ribozyme-catalysed RNA synthesis and to uncover potential biases generated by restricting analysis to full length products. Analysis of 1.17×105 positions revealed a 97.7% fidelity for full length products, and a slightly lower overall fidelity of 97.4% when including all incomplete extension products (Supplementary Fig. S7a, Table S2). In-detail examination of polymerase errors across all extension products revealed both the ribozyme’s global error spectrum (dominated by deletions and transition substitutions of A, Fig. 4c) and an enrichment of certain types of errors towards the 3′ ends of incomplete extension products: the last two bases showed a 7.1% substitution rate, compared to a 0.8% substitution rate (plus a 1.5% deletion rate) in all preceding residues (Fig. 4d). This increase may reflect misincorporations that prevented further extension by the ribozyme (Supplementary Fig. S7b), a phenomenon that is also observed in templated nonenzymatic replication24. However, the majority (>85%) of incomplete extension products terminated in the correct residues, suggesting that non-processive synthesis as well as degradation of the ribozyme and single-stranded template in the Mg2+-rich buffer were likely responsible for the majority of the observed terminations. Indeed, the total 2.5% termination rate per position (Supplementary Fig. S5a) suggests just a ~1 / 300 chance of misincorporation-induced termination per position during these tC9Y-catalysed RNA syntheses.
In-ice selection of RNA polymerase ribozyme templates
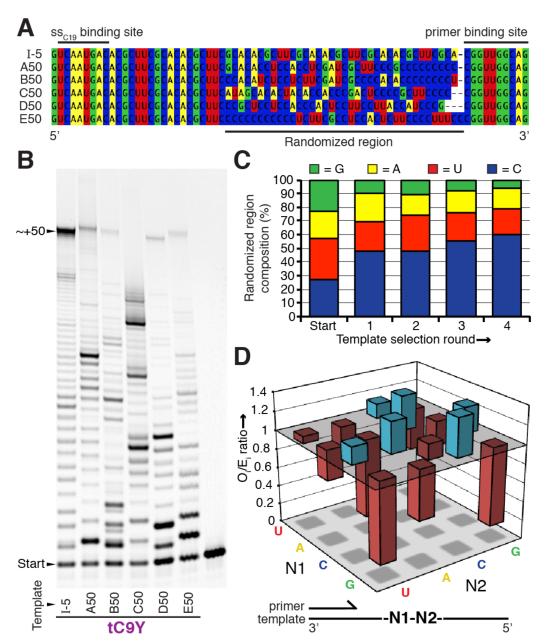
In early RNA replication, adaptive pressures would have acted not only on the replicase ribozyme itself, but on the replication system as a whole, including the replication templates. We complemented our studies of the directed evolution of RNA polymerase ribozyme activity in ice by investigating the effects of in-ice replication upon template sequence evolution. To this end we selected for long (50 nt) templates that could be copied in ice by the RNA polymerase ribozyme, from a pool of random RNA sequences. While an earlier template selection experiment at ambient temperatures had led to a rapid collapse of diversity to a single template (I-5)11, in ice the template sequence pool remained diverse over four rounds of selection, during which templates capable of directing full-length syntheses emerged (Supplementary Fig. S8a). From this pool (Supplementary Table S3) we randomly selected five ~50 nt template sequences (A50, B50, C50, D50 and E50, Fig. 5a), of which four (A50, B50, D50, E50) could be extended to full length (~+50 nts) both at ambient temperatures (Fig. 5b) and in ice (Supplementary Fig. S8b). tC9Y copied these templates with an overall fidelity comparable to the I-n repeat template (Supplementary Fig. S8c, Table S2). It is notable that although these template selections harnessed the ssC19 tag, the templates’ beneficial properties extended to both the Y as well as the parental R18 RNA polymerase ribozyme (Supplementary Fig. S8d).
Figure 5. Template selection.
a, Sequences of five in-ice evolved templates (round 4) alongside template I-5. b, Extension of primer A by ribozyme tC9Y on ice-selected templates compared to favourable template I-5. (17°C, 7 days). c, Base composition of the central 36-nt evolved region of sequences from template pools over the course of selection (Positions sequenced: Starting pool = 271, Round 1 = 232, Round 2 = 582, Round 3 = 739, Round 4 = 3086). d, Ratio of the observed dinucleotide frequencies (Oi) to those expected (Ei) in the pool of round 4 sequenced templates (based on nucleotide composition). Those overrepresented are in blue, those underrepresented in red.
Having discovered a pool of efficient template sequences for the RNA polymerase ribozyme, we sought to describe traits that distinguished these templates from the many RNA sequences that are poor templates25. Analysis of 95 selected template sequences revealed a consistent set of attributes that appear to critically influence the replicability of a given RNA sequence. Firstly, the majority of the template sequences exhibited a low propensity to form stable secondary structure elements (as judged by mfold26). Secondly, we observed a clear and progressive trend of the template nucleotide composition away from parity towards a reduced G and an increased C content (Fig. 5c). As CTP is efficiently incorporated opposite template G9, this trend does not reflect the elimination of inefficient template bases but rather the tendency of G-rich templates to promote the formation of highly stable secondary structures including G-rich hairpin motifs (through both G-C and G·U pairing) and potentially G-quadruplexes. Thirdly, we found a striking deviation of the dinucleotide frequencies from those expected (correcting for the biased nucleotide composition) (Fig. 5d). All dinucleotides lacking C (except -UU-) were underrepresented, as the abundant C residues appeared to function to space out A, U and G, suggesting an important role for intermittent interactions to unpaired template C for productive template binding and/or translocation. Indeed, extension ‘blocks’ are frequently observed preceding template stretches lacking C are frequently observed16. Conversely, RNA polymerisation often proceeds efficiently on template regions with regularly spaced C residues (such as template I-n).
Discussion
Compartmentalisation promotes the linkage of genotype and phenotype, a prerequisite for Darwinian evolution. Prior to the advent of membranous protocells, emerging organisms of the RNA world might have used a range of alternative compartmentalisation strategies, including porous rock cavities, aerosol droplets, aqueous phase separation, micelles or eutectic ice phases. Of these, some have been investigated for compatibility with ribozyme activity18,22,27,28, but none have been examined for the emergence of adaptive, environment-specific RNA phenotypes. In this study we have demonstrated the in-ice directed evolution of RNA function, showing adaptation of RNA polymerase ribozyme activity to the conditions of the eutectic phase of water ice (Fig. 1). The novel in-ice selected RNA polymerase ribozymes (Supplementary Fig. S9) displayed both cold-specific adaptations, allowing ribozyme-catalysed RNA synthesis to proceed in ices at temperatures as low as −19°C (Fig. 2), and generic i.e. temperature- and medium-independent activity improvements, highlighting the potential of water-ice (and perhaps other heterogeneous media) as a favourable environment for the emergence of ribozyme phenotypes that may not be accessible under ambient conditions.
The G93A and C97A mutations that provide the main cold-specific activity boost are located in the single-stranded linker region of the ribozyme between the catalytic core (1-90) and the processivity domain (105-187). Examination of the crystal structure of the Class I RNA ligase29,30, from which the catalytic core of the ribozyme polymerase is derived, suggests this sequence is well-positioned to interact with the downstream RNA template (Fig. 3a), and could augment the sequence-general contacts the ribozyme makes with the upstream duplex16,29,31 to enhance primer/template binding. Indeed, Y performs better when a long downstream single-stranded template is available, without requiring specific base-pairing with a template sequence motif (Fig. 3c).
Combining these three mutations from the in-ice ribozyme selection with the previously identified C60U mutation and the 5′ terminal ssC19 tag11 yielded a novel polymerase ribozyme tC9Y that could synthesise RNA polymers of up to 206 nts in length (Fig. 4a), longer than itself (202 nts), illustrating the synthetic potential of polymerase ribozymes.
Although synthesis of long RNA oligomers might in itself be of adaptive value - for example as a form of chemical storage, capturing nucleotides and ions into a less mobile form - the strong sequence dependence of RNA synthesis in general remains an obstacle to self-replication. To better understand this, we isolated a diverse repertoire of novel long (50 nt) RNA sequences by in-ice selection that proved efficient templates for enzymatic RNA synthesis both in ice and at ambient temperatures. Features of the selected sequences allowed us to make inferences about template sequence features generally favourable to RNA synthesis. These (likely interdependent) characteristics include a low tendency for secondary structure formation, a reduced G-content as well as bias against -DD- (D= A, G, U) dinucleotides (with the exception of -UU-) (Fig. 5). Such compositional limitations are reminiscent of those observed in nonenzymatic contexts32, highlighting the analogous constraints upon both processes arising from the distinct stacking and hydrogen bonding properties of the natural nucleobases.
Discovery of these biases underscores the formidable challenges ahead in the development of an RNA replicase. The need to copy both its complementary minus and plus strands during a replication cycle points to conflicting pressures with regards to nucleotide composition and secondary/tertiary structure formation needed for ribozyme activity. Of key importance will therefore be strategies to overcome template secondary structures as well as improving sequence generality of ribozyme-template interactions. This might include the engineering or evolution of an RNA unfolding or strand displacement activity on the polymerase ribozyme, the deconstruction of polymerase ribozymes into shorter RNA oligomers, and/or the definition of reaction environments and conditions that modulate RNA structure without compromising ribozyme activity. Generalising the ribozyme polymerase activity described here to a wider range of sequences and RNA structures, including that of the ribozyme itself, will be a key advance towards realising molecular self-replication and evolution.
Materials and methods
Oligonucleotides
NTPs were purchased from Promega and mutagenic dNTPs from TriLink. Ribozyme, RNA template and primer sequences, sources, and purification methods are listed in detail in Supplementary Table S3.
Polymerisation assay
Primer extension reactions were set up similar to as described in15, typically by annealing ribozyme, 5′-FITC-labelled primer and template together in 2 μl H2O (80°C for 2 min, 17°C for 10 min), before adding 18 μl chilled extension buffer (final concentrations: 200 mM MgCl2, 50 mM Tris·HCl pH 8.3, 0.5 μM each RNA). To enhance activity in some frozen reactions, MgCl2 was replaced by MgSO4, where described15. A 4 mM concentration of each NTP was provided for reactions in aqueous solution, and 1 mM of each NTP in ice (unless otherwise indicated). Reactions in ice were set up by freezing at −25°C for 10 min to induce ice crystal formation, before incubation in a Techne RB-5 refrigerated bath, maintained by a Techne Tempette TE-8D thermostat at −7°C to allow eutectic phase formation; omitting the −25°C step allowed the preparation of supercooled reactions. Reactions at −14°C or −19°C were instead transferred to freezers maintained at these temperatures. Reactions were stopped by addition of 0.5 volumes 0.5 M EDTA, heated to 94°C for 5 min in 6 M urea in the presence of a 10× excess of RNA competing oligonucleotide able to hybridise to the template sequence, and resolved by urea-PAGE (20% polyacrylamide, 8 M urea). Gels were analysed using a Typhoon Trio scanner (GE Healthcare), and quantitation of extension products was performed as previously described15,22. Gel image brightness and contrast were adjusted to illustrate banding patterns.
In ice ribozyme selection
The ice-CBT protocol employed is described in depth in Supplementary Fig. S1a. Briefly, a mutagenised R18 library was prepared and microbead-bound clonal ribozyme/primer/template repertoires (Supplementary Fig. S1a(iii)) were generated as for the selection of Z described previously11. However, for selections in ice beads were resuspended in 600 μl final volume chilled extension buffer (50 mM MgCl2, 12.5 mM Tris·HCl, pH 8.3, 125 μM each NTP, 0.125 μM RNA template, 0.125 μM stem2) and frozen at −25°C (10 min) before transfer and incubation at −7°C to allow eutectic phase formation. The concentration effect of eutectic phase formation restores an optimal eutectic phase composition from this diluted extension buffer, reducing bead aggregation within the eutectic phase. Ribozymes remained ligated to beads during extension, improving local primer extension. Beads were recovered by thawing with 75 μl 0.5 M EDTA pH 7.5 and Tween-20 (to 0.1%). Detection of ice-synthesised extension products by RCA, staining with PicoGreen (Invitrogen), and sorting of bead-bound active ribozyme genes were conducted as for the Z selection described previously11; recombination via StEP shuffling of libraries is described in Supplementary Materials and Methods.
Analysis of in-ice ribozyme selection
Polyclonal selection pools were transcribed and quantified by agarose gel electrophoresis by comparison with purified standards and added to annealed BioFITCU10-A/template I in extension buffer, before freezing, incubation at −7°C and determination of primer extension activity by urea-PAGE and densitometric quantitation (Supplementary Table S1). Screening of the final output pool (Supplementary Fig. S1c, Table S1) was performed using the ribozyme polymerase plate assay (RPA) as described previously11, except ribozyme extension (of BioU10-A on template I) was assayed under frozen conditions (200 mM MgCl2, 50 mM Tris·HCl, pH 8.3, 0.5 mM each NTP, frozen on dry ice prior to −7°C incubation for 334 h).
Scanning electron microscopy
A sample with the same composition as a selection extension mixture (Supplementary Fig. S1a(iv)) was frozen and imaged as previously15, including a 10 min incubation at −90°C under vacuum to sublimate ice prior to gold-coating and imaging.
Sequencing of extension products and template selection are described in detail in Supplementary Materials and Methods.
Supplementary Material
Acknowledgements
We thank J. N. Skepper (University of Cambridge) for help with SEM imaging, S. James and S. Brunner for help with MiSeq sequencing and analysis and Maria Daly (MRC LMB) for help with FACS. This work was supported by a Homerton College, Cambridge junior research fellowship (J.A.) and by the Medical Research Council (programme number U105178804).
Footnotes
Additional information. Supplementary information is available online. Correspondence and requests for materials should be addressed to P.H.
Competing financial interests. The authors declare no competing financial interests.
References
- 1.RNA Worlds . In: Atkins JF, Gesteland RF, Cech TR, editors. Cold Spring Harbor Laboratory Press; 2011. [Google Scholar]
- 2.Gilbert W. Origin of life: The RNA world. Nature. 1986;319:618–618. [Google Scholar]
- 3.Powner MW, Gerland B, Sutherland JD. Synthesis of activated pyrimidine ribonucleotides in prebiotically plausible conditions. Nature. 2009;459:239–242. doi: 10.1038/nature08013. [DOI] [PubMed] [Google Scholar]
- 4.Bowler FR, et al. Prebiotically plausible oligoribonucleotide ligation facilitated by chemoselective acetylation. Nature Chem. 2013;5:383–389. doi: 10.1038/nchem.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engelhart AE, Powner MW, Szostak JW. Functional RNAs exhibit tolerance for non-heritable 2′-5′ versus 3′-5′ backbone heterogeneity. Nature Chem. 2013;5:390–394. doi: 10.1038/nchem.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szostak JW, Bartel DP, Luisi PL. Synthesizing life. Nature. 2001;409:387–390. doi: 10.1038/35053176. [DOI] [PubMed] [Google Scholar]
- 7.Robertson MP, Joyce GF. The Origins of the RNA World. Cold Spring Harb. Perspect. Biol. 2010 doi: 10.1101/cshperspect.a003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellington AD, Chen X, Robertson M, Syrett A. Evolutionary origins and directed evolution of RNA. Int. J. Biochem. Cell Biol. 2009;41:254–265. doi: 10.1016/j.biocel.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 9.Johnston WK, Unrau PJ, Lawrence MS, Glasner ME, Bartel DP. RNA-catalyzed RNA polymerization: accurate and general RNA-templated primer extension. Science. 2001;292:1319–1325. doi: 10.1126/science.1060786. [DOI] [PubMed] [Google Scholar]
- 10.Zaher HS, Unrau PJ. Selection of an improved RNA polymerase ribozyme with superior extension and fidelity. RNA. 2007;13:1017–1026. doi: 10.1261/rna.548807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wochner A, Attwater J, Coulson A, Holliger P. Ribozyme-catalyzed transcription of an active ribozyme. Science. 2011;332:209–212. doi: 10.1126/science.1200752. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP, Szostak JW. Isolation of new ribozymes from a large pool of random sequences. Science. 1993;261:1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- 13.Ekland EH, Szostak JW, Bartel DP. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science. 1995;269:364–370. doi: 10.1126/science.7618102. [DOI] [PubMed] [Google Scholar]
- 14.Lawrence MS, Bartel DP. Processivity of ribozyme-catalyzed RNA polymerization. Biochemistry. 2003;42:8748–8755. doi: 10.1021/bi034228l. [DOI] [PubMed] [Google Scholar]
- 15.Attwater J, Wochner A, Pinheiro VB, Coulson A, Holliger P. Ice as a protocellular medium for RNA replication. Nat. Commun. 2010;1:76. doi: 10.1038/ncomms1076. [DOI] [PubMed] [Google Scholar]
- 16.Attwater J, et al. Chemical fidelity of an RNA polymerase ribozyme. Chem. Sci. 2013;4:2804–2814. [Google Scholar]
- 17.Dobson CM, Ellison GB, Tuck AF, Vaida V. Atmospheric aerosols as prebiotic chemical reactors. Proc. Natl. Acad. Sci. USA. 2000;97:11864–11868. doi: 10.1073/pnas.200366897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen IA, Salehi-Ashtiani K, Szostak JW. RNA catalysis in model protocell vesicles. J. Am. Chem. Soc. 2005;127:13213–13219. doi: 10.1021/ja051784p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Budin I, Szostak JW. Expanding roles for diverse physical phenomena during the origin of life. Annu. Rev. Biophys. 2010;39:245–263. doi: 10.1146/annurev.biophys.050708.133753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monnard PA. Catalysis in abiotic structured media: an approach to selective synthesis of biopolymers. Cell. Mol. Life. Sci. 2005;62:520–534. doi: 10.1007/s00018-004-4342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vlassov AV, Kazakov SA, Johnston BH, Landweber LF. The RNA world on ice: a new scenario for the emergence of RNA information. J. Mol. Evol. 2005;61:264–273. doi: 10.1007/s00239-004-0362-7. [DOI] [PubMed] [Google Scholar]
- 22.Muller UF, Bartel DP. Improved polymerase ribozyme efficiency on hydrophobic assemblies. RNA. 2008;14:552–562. doi: 10.1261/rna.494508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kun A, Santos M, Szathmary E. Real ribozymes suggest a relaxed error threshold. Nat. Genet. 2005;37:1008–1011. doi: 10.1038/ng1621. [DOI] [PubMed] [Google Scholar]
- 24.Rajamani S, et al. Effect of stalling after mismatches on the error catastrophe in nonenzymatic nucleic acid replication. J. Am. Chem. Soc. 2010;132:5880–5885. doi: 10.1021/ja100780p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence MS, Bartel DP. New ligase-derived RNA polymerase ribozymes. RNA. 2005;11:1173–1180. doi: 10.1261/rna.2110905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strulson CA, Molden RC, Keating CD, Bevilacqua PC. RNA catalysis through compartmentalization. Nature Chem. 2012;4:941–946. doi: 10.1038/nchem.1466. [DOI] [PubMed] [Google Scholar]
- 28.Vlassov AV, Johnston BH, Landweber LF, Kazakov SA. Ligation activity of fragmented ribozymes in frozen solution: implications for the RNA world. Nucleic Acids Res. 2004;32:2966–2974. doi: 10.1093/nar/gkh601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shechner DM, et al. Crystal structure of the catalytic core of an RNA-polymerase ribozyme. Science. 2009;326:1271–1275. doi: 10.1126/science.1174676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shechner DM, Bartel DP. The structural basis of RNA-catalyzed RNA polymerization. Nat. Struct. Mol. Bio. 2011;18:1036–1042. doi: 10.1038/nsmb.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller UF, Bartel DP. Substrate 2′-hydroxyl groups required for ribozyme-catalyzed polymerization. Chem. Biol. 2003;10:799–806. doi: 10.1016/s1074-5521(03)00171-6. [DOI] [PubMed] [Google Scholar]
- 32.Joyce GF, Orgel LE. Non-enzymatic template-directed synthesis on RNA random copolymers. Poly(C,A) templates. J. Mol. Bio. 1988;202:677–681. doi: 10.1016/0022-2836(88)90297-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.