Summary
Enteric viruses, including poliovirus and reovirus, encounter a vast microbial community in the mammalian gastrointestinal tract, which has been shown to promote virus replication and pathogenesis. Investigating the underlying mechanisms, we find that poliovirus binds bacterial surface polysaccharides, which enhances virion stability and cell attachment by increasing binding to the viral receptor. Additionally, we identified a poliovirus mutant, VP1-T99K, with reduced lipopolysaccharide (LPS) binding. Although T99K and WT poliovirus cell attachment, replication and pathogenesis in mice are equivalent, following peroral inoculation of mice, VP1-T99K poliovirus was unstable in feces. Consequently, the ratio of mutant virus in feces is reduced following additional cycles of infection in mice. Thus, the mutant virus incurs a fitness cost when environmental stability is a factor. These data suggest that poliovirus binds bacterial surface polysaccharides, enhancing cell attachment and environmental stability, potentially promoting transmission to a new host.
Introduction
The gastrointestinal tract contains a diverse community of microbes, which play an essential role in host health. Imbalances in this microbial community have been linked to many human diseases including inflammatory bowl disease, diabetes, and obesity (Manichanh et al., 2012; Tremaroli and Backhed, 2012). While many studies have focused on bacterial-host interactions in the gastrointestinal tract, the impact of the microbiota on viruses is less clear.
Enteric viruses are a significant cause of disease worldwide and are transmitted by direct contact, fomites, and contaminated water or food. While many enteric viruses cause asymptomatic infection or mild symptoms, these viruses can cause serious to life-threatening illnesses. Poliovirus is a non-enveloped, single-stranded RNA virus in the Enterovirus genus of the Picornaviridae. Until the development of vaccines in the 1950s, poliovirus was a major cause of paralysis. Spread by the fecal-oral route, poliovirus can disseminate from the gastrointestinal tract and damage neurons.
Recent data suggest that intestinal microbiota enhance infection of three unrelated enteric viruses, poliovirus, reovirus, and mouse mammary tumor virus (MMTV) (Kane et al., 2011; Kuss et al., 2011). The retrovirus MMTV binds bacterial lipopolysaccharide (LPS) and induces immune tolerance through a Toll-like receptor 4-dependent mechanism (Kane et al., 2011). Reovirus replication and pathogenesis was enhanced by intestinal microbiota, although the mechanism is unknown (Kuss et al., 2011). Similarly, poliovirus replication and pathogenesis was enhanced by intestinal microbiota (Kuss et al., 2011). We determined that exposure to bacteria increased infectivity of poliovirus virions. After exposure to Gram-positive or Gram-negative bacteria, we recovered up to 5-fold more plaque forming units (PFU) than we started with. Since only ~1 of every 200 picornavirus particles are infectious, these data suggest that exposure to bacteria “resurrects” infectivity for some particles, reducing the particle:PFU ratio. However, the precise mechanisms and consequences of bacterial effects on poliovirus are unclear.
Here, we examined the mechanisms of bacterial-mediated enhancement of poliovirus. We demonstrate a dual role for bacterial polysaccharide-mediated enhancement of poliovirus infectivity through virion stability and cell attachment via the poliovirus receptor (PVR). Notably, we identified a single amino acid substitution in a viral capsid protein, VP1-T99K, which reduces LPS binding and limits virion stabilization by LPS. We determined that VP1-T99K poliovirus replicates and is pathogenic in perorally inoculated mice, but is unstable in feces. Our data reveal that microbiota play a critical role in stabilizing viral particles in the environment, and thus may promote transmission.
Results
Bacterial surface polysaccharides enhance poliovirus virion stability
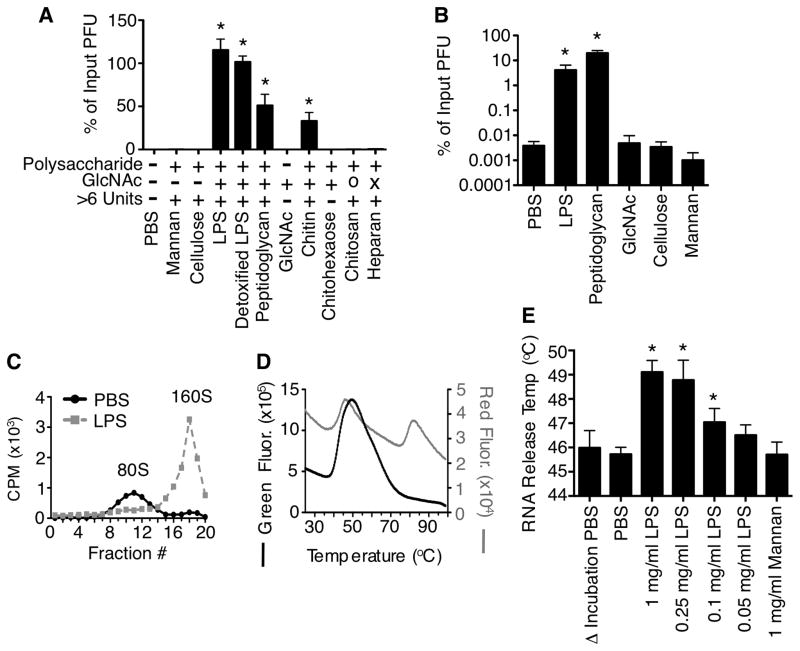
Previously we found that exposure of type 1 Mahoney poliovirus to bacteria or bacterial surface polysaccharides reduced thermal inactivation of viral particles at a variety of temperatures (Kuss et al., 2011). To define the polysaccharide requirements for stabilization we examined thermal inactivation of poliovirus in the presence of glycans. Poliovirus was incubated with various compounds at 42°C for 6 h followed by quantification of viral titers by plaque assay. We found that exposure to GlcNAc-containing polysaccharides, including the bacterial surface polysaccharides lipopolysaccharide (LPS) and peptidoglycan (PG), enhanced stability of poliovirus [Fig. 1A and (Kuss et al., 2011)]. Detoxified LPS lacking lipid A also enhanced poliovirus stability, suggesting that the polysaccharide component of LPS was sufficient for stabilization. Chitin, a long polysaccharide containing only GlcNAc, stabilized poliovirus. However, a 6-unit GlcNAc oligosaccharide (chitohexaose) or monomeric GlcNAc did not stabilize poliovirus. Similarly, poliovirus was not stabilized by deacetylated chitin (chitosan) or a GlcNAc-containing polysaccharide with sulfate groups (heparan). Therefore, viral stabilization required an acetylated GlcNAc-containing polysaccharide longer than 6 units.
Figure 1. Bacterial surface polysaccharides enhance poliovirus stability in vitro.
(A) GlcNAc-containing polysaccharides enhance poliovirus stability. WT Mahoney poliovirus (1×106 PFU) was incubated with indicated compounds (1 mg/ml) at 42°C for 6 h, followed by HeLa cell plaque assay. Input poliovirus titers were set to 100% and data are shown as % of input PFU. O=deacetylated chitin (chitosan); X=contains sulfate groups (heparan); n=5–9. (B) Bacterial polysaccharides enhance viral recovery following bleach treatment. WT Mahoney poliovirus was incubated with indicated compounds (1 mg/ml) at 37°C for 1 h followed by treatment with 0.1mg/ml bleach for 1 min and plaque assay; n=5–8. (C) Incubation with LPS limits virion inactivation as measured by gradient virion density analysis. 35S-labeled poliovirus was incubated with PBS or 1 mg/ml LPS at 42°C for 1 h prior to loading on 10–30% sucrose gradients. Gradients were subjected to ultracentrifugation and fractions were collected from the top of the gradient and scintillation counted. 160S=intact particles; 80S=empty particles. Graph is representative of 3 experiments. (D) Poliovirus thermostability profile using a cell-free Particle Stability Thermo Release assay (PaSTRy). Virus was added to SYBR Green or SYPRO Red and placed in a real-time PCR machine. Samples were heated from 25°C to 99°C on a 1% stepwise gradient with florescence monitoring. Peaks correspond to RNA release (SYBR Green) and capsid structural transitions (SYPRO Red). Graph is representative of three experiments. (E) Quantification of PaSTRy experiments revealing GlcNAc polysaccharides enhance thermostability of poliovirus by preventing premature RNA release. Poliovirus was incubated with indicated compounds at 37°C for 1 h prior to addition of SYBR Green and analysis with a real-time PCR machine; n=3–9. See also Fig. S1. Data are mean +/− SEM; * = P<0.05.
In addition to inactivation from heat, virus particles can be inactivated by disinfectants such as chlorine bleach through penetration of the viral capsid followed by RNA release and/or damage (Alvarez and O’Brien, 1982; O’Brien and Newman, 1979; Sharp et al., 1975; Taylor and Butler, 1982). To examine whether bacterial surface polysaccharides limit inactivation from bleach treatment, we pre-incubated poliovirus with various compounds for 1 h, exposed them to 1 mg/ml (0.001%) bleach for one min, neutralized with sodium bisulfite, and quantified viability by plaque assay. Viability of LPS and PG-exposed viruses was over 1000-fold higher than PBS- or control glycan-exposed viruses (Fig. 1B). These data suggest that GlcNAc-containing polysaccharides enhance poliovirus stability and increase bleach resistance of virions.
Plaque formation is influenced by several factors in addition to virion stability; therefore, we performed additional experiments to examine virion stability using physical/cell-free methods. First, we examined virion conformational changes using sucrose gradients. Poliovirus undergoes irreversible conformational changes that are part of the normal entry process (Fricks and Hogle, 1990; Hogle and Racaniello, 2002; Kaplan et al., 1990). Native virions (160S) transition to empty capsids (80S) after RNA release. 160S-80S transitions can also be triggered in solution by heating. Since bacterial polysaccharides enhance poliovirus thermostability and bleach resistance, we hypothesized that LPS exposure would limit the 160S-80S transition at elevated temperature. To test this, 35S-labeled poliovirus was incubated with LPS or PBS at 42°C for 1 h and loaded on a sucrose gradient. Following ultracentrifugation, radioactivity from gradient fractions revealed that polioviruses incubated with PBS were primarily 80S/empty particles while polioviruses incubated with LPS were primarily 160S/native particles (Fig. 1C).
As a second method to quantify virion stability using a physical/cell-free approach, we examined whether LPS influences viral RNA release using a Particle Stability Thermal Release assay (PaSTRy) (Walter et al., 2012). This calorimetry-type assay has been used to measure stability of several picornaviruses (Plevka et al., 2013; Walter et al., 2012). Poliovirus was pre-incubated with PBS, LPS, or mannan for 1 h, solutions were mixed with either SYBR Green (measures RNA release) or SYPRO Red (measures capsid denaturation via exposure of hydrophobic amino acids), and samples were heated in a real-time PCR machine with fluorescence monitoring. PBS-treated viruses generated a peak of fluorescence intensity for both fluorophores indicating RNA release and initial capsid denaturation at 46°C, consistent with results from Walter et al. (Walter et al., 2012) (Fig. 1DE). However, for LPS-treated poliovirus the peak of SYBR Green fluorescence was shifted to 49°C (Fig. 1D and Fig. S1), indicating that LPS exposure stabilized the capsid and delayed RNA release. LPS-mediated stabilization of poliovirus was concentration-dependent, as RNA release temperatures were similar to PBS controls when poliovirus was incubated in 0.05mg/mL of LPS instead of 1 mg/ml LPS (Fig. 1E). Mannan, a control polysaccharide, did not enhance virion thermostability. Overall, these data indicate that exposure to GlcNAc-containing bacterial polysaccharides enhanced viral stability by limiting premature RNA release.
Bacterial polysaccharides enhance poliovirus cell attachment by increasing binding to the viral receptor, PVR
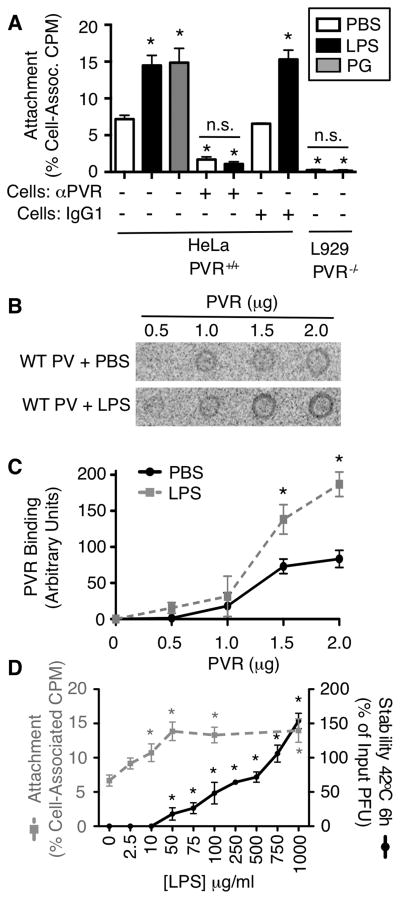
Previously we determined that exposure of poliovirus to bacteria enhanced cell attachment to PVR-expressing cells (Kuss et al., 2011). To determine whether bacterial polysaccharides are sufficient for the cell attachment enhancement, we examined attachment of 35S-labeled poliovirus treated with or without PG or LPS. We incubated 35S-labeled poliovirus with PBS or 1 mg/ml PG or LPS for 1 h at 37°C, incubated samples with HeLa cells at 4°C for 10 min to facilitate binding, washed, harvested cells, and quantified cell-associated virus by scintillation counting. We found that poliovirus pre-incubated with PG or LPS demonstrated enhanced binding to HeLa cells when compared to poliovirus pre-incubated with PBS (Fig. 2A). To determine whether PVR is required for the LPS-mediated enhancement of poliovirus attachment, we examined poliovirus attachment to HeLa cells treated with anti-PVR or isotype control antibody. Treatment with anti-PVR antibody reduced poliovirus binding to HeLa cells, regardless of whether virus was treated with LPS, indicating that LPS-mediated attachment enhancement is PVR dependent (Fig. 2A). Furthermore, negligible amounts of virus bound to non-PVR expressing mouse L929 cells, regardless of PBS or LPS pre-treatment (Fig. 2A). The PG- and LPS-mediated enhancement of poliovirus attachment to PVR-expressing cells could occur through a direct mechanism involving PVR or an indirect mechanism. To determine whether LPS-treated viruses have increased binding to PVR, we examined viral binding to purified PVR. PVR protein was immobilized on a nitrocellulose membrane and probed with 35S-labeled poliovirus pre-incubated with PBS or 1 mg/ml LPS. We found that poliovirus pre-incubated with LPS had significantly higher PVR binding when compared to poliovirus pre-incubated with PBS (Fig. 2BC). These data suggest that LPS enhances binding of poliovirus to PVR. Therefore, the LPS-mediated enhancement of poliovirus attachment to PVR-expressing cells is likely to occur through a direct mechanism.
Figure 2. LPS enhances poliovirus attachment to its cellular receptor, PVR.
(A) LPS and PG enhance poliovirus cell attachment in a PVR-dependent manner. Gradient purified 35S-labeled Mahoney poliovirus was incubated +/− 1 mg/ml PG or LPS at 37°C for 1 h, added to cells for 10 min (4°C), followed by washing and scintillation counting of cell-associated virus. As indicated, HeLa cells were incubated +/− PVR antibody or an isotype control antibody (IgG1) for 1 h prior to virus exposure; n=3–6. (C) Poliovirus binding to PVR. 35S-labeled poliovirus was incubated +/− 1 mg/ml LPS for 1 h at 37°C followed by incubation with nitrocellulose membranes containing immobilized PVR protein. After washing, bound viral CPM was used to quantify binding. Blot is representative of 3 experiments. (D) Quantification of poliovirus-PVR binding assays. Binding was quantified by phosphoimager analysis; n=3. (E) Differential LPS concentration requirements for poliovirus stabilization and cell attachment enhancement. 35S-poliovirus was incubated with different concentrations of LPS followed by HeLa cell attachment assay as described above (gray dashed line, left axis). Unlabeled poliovirus was incubated with different concentrations of LPS followed by thermal inactivation assay performed at 42°C for 6 h (black line, right axis) and statistical analysis compares data from each [LPS] to the 0 μg/ml LPS control; n=3–10. Data are mean +/− SEM; * = P<0.05, compared with PBS control.
Given that our results suggest that two mechanisms, stability and cell attachment, play a role in polysaccharide-mediated viral infectivity enhancement, we wondered whether the two mechanisms have similar polysaccharide concentration requirements. To test this, poliovirus was incubated with various concentrations of LPS and subjected to virion stability and cell attachment assays. We found that maximal thermal stabilization was achieved with 1 mg/ml LPS (Fig. 2D). However, maximal cell attachment enhancement was achieved with 0.05 mg/ml LPS. Therefore, maximal enhancement of viral attachment required 20-fold less LPS than maximal virion stabilization. Since poliovirus virions contain 60 copies of each capsid protein, there are up to 60 potential polysaccharide-binding sites per virion. Our results suggest that virion stabilization may require occupation of more binding sites than cell attachment enhancement.
Identification of a conditional mutant, VP1-T99K, with reduced LPS binding and virion stabilization at physiological temperatures
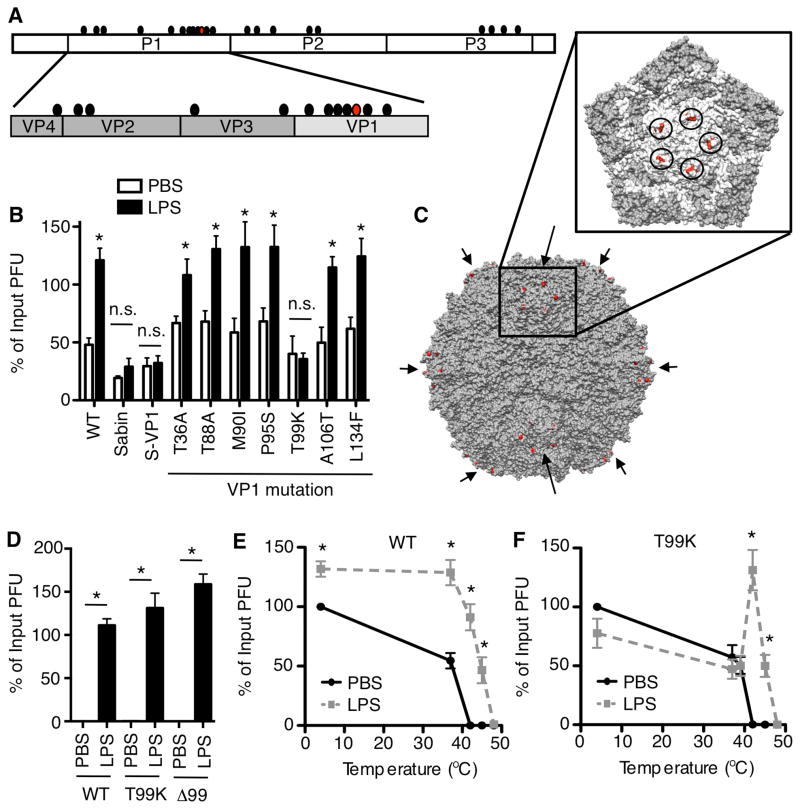
GlcNAc-containing polysaccharides such as LPS directly interact with the poliovirus capsid (Kuss et al., 2011); therefore, it may be possible to identify viral mutants with reduced LPS binding. We capitalized on the fact that many poliovirus capsid mutants have been characterized. One famous poliovirus mutant, the attenuated Sabin serotype 1 strain used in the oral vaccine, contains 12 non-synonymous mutations in the capsid-coding region in addition to other mutations in the non-structural coding region (Fig. 3A)(Nomoto et al., 1982). Several of these 12 capsid mutations are in surface-exposed regions. To screen for LPS effects on Sabin poliovirus, we examined virion stability following 6 h incubation at 37°C. Initially, we used 37°C rather than 42°C for these experiments since the Sabin strain is temperature sensitive. While LPS enhanced stability of wild-type poliovirus (virulent Mahoney, WT), LPS did not enhance stability of Sabin poliovirus at 37°C (Fig. 3B). Moreover, Mahoney poliovirus containing the Sabin VP1 capsid protein was not stabilized by LPS exposure at 37°C (Fig. 3B, S-VP1). Site-directed mutagenesis of the seven VP1 mutations identified the residue at 99 (T99K) as critical for the LPS- and PG-mediated stabilization at 37°C (Fig. 3B and Fig. S2A). Amino acid 99 is located in the highly exposed BC loop of VP1, near the 5-fold symmetry axis (Fig. 3C).
Figure 3. Identification of a conditional poliovirus mutant, VP1-T99K, with altered LPS-mediated virion stabilization.
(A) Amino acid differences (circles) between the wild-type (WT) serotype 1 Mahoney poliovirus and Sabin poliovirus. Red circle represents VP1-99. (B) LPS-mediated enhancement of viral stability is abrogated in Sabin and T99K polioviruses at 37°C. WT Mahoney, Sabin, and viral mutants were incubated with 1 mg/ml LPS at 37°C for 6 h and titers were quantified by plaque assay; n=3–11. (C) Mahoney poliovirus structure with the location of VP1-99 highlighted in red. Arrows indicate 5-fold symmetry axes (8 of 12 are visible). The inset shows one 5-fold symmetry axis and circles surround VP1-99. (D) LPS stabilizes T99K poliovirus and Δ99 poliovirus at high temperatures. Viruses were incubated with 1 mg/ml LPS at 42°C for 6 h prior to plaque assay; n=5–11. (E and F) Thermal stability profiles for WT vs. T99K polioviruses. WT or T99K viruses were incubated in PBS or 1 mg/ml LPS at various temperatures for 6 h prior to plaque assay; n=3–10. See also Fig. S2. Data are mean +/− SEM; * = P<0.05.
We next examined the temperature dependence of the T99K phenotype by performing virion stability experiments at various temperatures. To our surprise, T99K poliovirus was stabilized by LPS or PG exposure when incubations were performed at 42°C (Fig. 3D and Fig. S2B). To examine the precise temperature requirements for LPS-mediated virion stabilization, WT and T99K viruses were incubated with LPS at various temperatures from 4°C to 48°C for 6 h followed by plaque assay. LPS treatment enhanced WT infectivity at every temperature tested, when compared to PBS treatment (Fig. 3E). In contrast, LPS treatment only enhanced T99K viral infectivity at temperatures above 40°C (Fig. 3F).
We hypothesized that T99K is a conditional mutant for LPS binding and stabilization. In our model, the lysine at position 99 blocks polysaccharide binding to a nearby site at low temperatures but the binding site is open at high temperatures (see conceptual model in Fig. S2CDE). To test this, we deleted VP1 amino acid 99 in the Mahoney poliovirus strain and examined stability in the presence or absence of LPS at 42°C. The virus lacking VP1 amino acid 99 (Δ99) was viable and was stabilized by LPS at 42°C (Fig. 3D and Fig. S2). These results demonstrate that residue 99 is not the polysaccharide binding site, but influences binding.
Because LPS did not enhance T99K stability at physiological temperatures (Fig. 3B), we hypothesized that this mutant has reduced LPS binding. To test this, 35S-labeled WT or T99K virus was incubated with biotinylated LPS at 37°C for 1 h before precipitation with streptavidin beads. Binding of biotinylated LPS to poliovirus was saturable (Fig. 4A), and binding was reversible since addition of excess non-biotinylated LPS limited binding to biotinylated LPS (Fig. S3). While T99K virus bound biotinylated LPS, binding was reduced compared with WT virus (Fig. 4A). Since enhancement of viral attachment requires lower LPS concentrations than enhancement of virion stability (Fig. 2E), we wondered whether T99K virus binds enough LPS to enhance cell attachment but not virion stability. To examine cell attachment, WT and T99K viruses were pre-incubated with 1 mg/ml LPS at 37°C for 1 h and added to PVR-expressing HeLa cells. Pre-incubation with LPS enhanced T99K cell attachment to the same degree as for WT virus (Fig. 4B). These data suggest that the T99K mutation does not adversely affect LPS-mediated enhancement of cell attachment, but does impair LPS-mediated enhancement of virion stability. In summary, the T99K virus is a conditional mutant with reduced LPS-mediated virion stabilization at physiological temperatures.
Figure 4. The T99K mutant has reduced LPS binding but intact LPS-mediated cell attachment enhancement.
(A) Biotinylated LPS-35S-poliovirus binding assay. 35S-labeled viruses were incubated with biotinylated LPS or standard LPS for 1 h at 37°C followed by precipitation with streptavidin beads. The % of virus bound was quantified by scintillation counting; n=3–6. (B) T99K poliovirus cell attachment. 35S-labled WT or T99K poliovirus was incubated with PBS or 1 mg/ml LPS at 37°C for 1 h and added to HeLa cells at 4°C to facilitate viral binding. After washing, cell-associated radioactivity was quantified; n=5. See also Fig. S3. Data are mean +/− SEM; * = P<0.05.
T99K and WT poliovirus replication and pathogenesis are equivalent
Since T99K poliovirus has reduced LPS binding, which reduces LPS-mediated thermostability enhancement but not LPS-mediated cell attachment enhancement, we sought to use this virus as a tool to dissect the mechanisms and consequences of bacterial polysaccharide-mediated enhancement of poliovirus infectivity in vivo. First, to rule out general growth defects, we performed growth curves of WT and T99K viruses in HeLa cells. Quantification of viral titers by plaque assay revealed no growth defect of T99K compared with WT (Fig. 5A).
Figure 5. Viral replication, shedding, and pathogenesis are unaffected by the T99K mutation.
(A) T99K displays no in vitro growth defects compared to WT in growth curve assays. HeLa cells were infected with WT or T99K poliovirus (MOI 10) at 37°C and incubated at 2, 4, 8, and 16 h post-infection followed by plaque assay; n=3–4. (B) Fecal shedding kinetics of WT and T99K viruses. PVR-IFNAR−/− mice were inoculated perorally with either WT or T99K poliovirus (1×108 PFU) and feces were collected at 24, 48, and 72 h post-inoculation prior to plaque assay. (C) In vivo replication of WT and T99K viruses. Neutral red/light-sensitive WT or T99K poliovirus was used to perorally inoculate PVR-IFNAR−/− mice in the dark and feces were collected at 24, 48, and 72 h in the dark. Fecal virus was isolated and either exposed to light or kept in the dark. Viral titers were quantified and viral replication was determined by the ratio of light exposed virus to dark virus. (D) Survival of PVR-IFNAR−/− mice following peroral inoculation with WT or T99K poliovirus. Data in panels B, C, and D are from two independent experiments (18 mice per virus). In all panels, WT= black lines and T99K=gray dashed lines. Data are mean +/− SEM; * = P<0.05.
Next, we perorally inoculated susceptible PVR-transgenic (PVR) interferon α/β receptor knockout (IFNAR−/−) mice with WT or T99K viruses and examined viral shedding in feces, replication, and pathogenesis (Ida-Hosonuma et al., 2005; Kuss et al., 2011; Ohka et al., 2007). Mice were inoculated perorally with 108 PFU WT or T99K poliovirus. WT and T99K virus shedding in feces was approximately equivalent (Fig. 5B). Since a high amount of unreplicated inoculum virus is shed in the feces 1–3 days after peroral inoculation, viral titers in the feces do not necessarily reflect viral replication (Kuss et al., 2008). To overcome this problem, we used light-sensitive viruses. Virus propagated in the presence of neutral red dye is sensitive to light-induced inactivation by RNA cross-linking but loses light sensitivity upon replication, facilitating assessment of replication and differentiating inoculum virus from replicated virus (Brandenburg et al., 2007; Kuss et al., 2008; Kuss et al., 2011). Neutral red-labeled WT and T99K viruses were used to perorally inoculate PVR-IFNAR−/− mice in the dark and feces were collected in the dark. Virus was isolated from the feces and either exposed to light or dark. Viral replication was quantified by determining the ratio of light- vs. dark-exposed titers. At 24, 48, and 72 h post-inoculation no difference in viral replication was observed between WT and T99K viruses (Fig. 5C). Finally, we examined viral pathogenesis and found that mouse survival was equivalent for both viruses (Fig. 5D).
Overall these data suggest that T99K viral shedding, replication, and pathogenesis are similar to WT. These results were not particularly surprising, considering that the only defect observed for the T99K virus is lack of stabilization by bacterial polysaccharides due to reduced LPS binding. Since poliovirus initiates infection within 2–4 h following peroral inoculation of mice, and poliovirus is stable at 37°C for 2–4 h, virion stability is not a major selective pressure in this infection system (Kuss et al., 2008; Kuss et al., 2011).
Microbiota stabilize WT virus but not T99K virus in feces, revealing a consequence for viruses with reduced LPS binding
Given that T99K poliovirus is not stabilized by bacterial polysaccharides at physiological temperatures, we hypothesized that the T99K mutation may confer a fitness cost when selective pressure on virion stability is applied. This type of selective pressure happens routinely in nature, since virions must be stable in the environment to initiate infection in a new host. To examine environmental stability, we quantified the ratio of WT to T99K viruses in mixtures exposed to various selective pressures. To do this, we engineered a T99K mutant with a silent mutation creating a new NheI site to distinguish it from WT (Fig. 6A). First, we examined stability of the T99K-NheI virus in vitro to ensure that the virus was as stable as WT virus in PBS. WT or T99K-NheI virus was mixed with PBS and incubated at 22°C for 5 days. Aliquots were removed at 0, 72, and 120 h and quantified by plaque assay. Both WT and T99K-NheI viruses experienced slow but equivalent inactivation over time (Fig. S4). These results also demonstrate that T99K virions do not have an intrinsic stability defect in the absence of bacteria. Second, T99K-NheI and WT viruses were mixed 1:1 and incubated with PBS for 96 h at 22°C (Fig. 6B). Viruses were amplified in HeLa cells until cytopathic effects were observed, and RNA was isolated, reverse transcribed, and amplified by PCR with a radioactive primer. Amplified RT-PCR products were digested with NheI and analyzed by electrophoresis on an acrylamide gel. Following a 96 h incubation, the ratio of T99K:WT was similar to input suggesting that the decay of both viruses was similar (Fig. 6CD). These results confirm that T99K virions do not have stability defects in the absence of bacteria.
Figure 6. An environmental stability defect for T99K poliovirus.
(A) Schematic of the T99K virus marked with a silent NheI site. An additional NheI site in both WT and T99K PCR products serves as an internal digestion control. Star represents the 32P-labeled antisense primer location. For radioactive bands appearing on gels, undigested PCR products are 565 nt, NheI-digested WT PCR products are 465 nt, and NheI-digested T99K-Nhe PCR products are 170 nt. (B) In vitro stability experiment design. WT and T99K-NheI viruses were mixed 1:1 and incubated with PBS for 96 h at 22°C. Virus was amplified in HeLa cells, RNA was harvested, and RT-PCR products were digested with NheI. (C) Representative gel of NheI-digested RT-PCR products to quantify the ratio of WT:T99K viruses from the in vitro stability experiment. (D) Quantification of data from C;n =3. (E) In vivo environmental stability experiment design. WT and T99K-NheI viruses were mixed 1:1 and PVR-IFNAR−/− mice were perorally inoculated with 2×107 PFU total. Mice were euthanized at 24 h post-inoculation. Feces were harvested at 6 h or left in the cage bottom for 24 or 96 h at 22°C. Fecal viruses were amplified in HeLa cells, RNA was harvested, and RT-PCR products were digested with NheI. Viruses passaged exclusively in tissue culture (TC) served as a fitness control. (F) Representative gel of NheI digested RT-PCR products to quantify the ratio of WT:T99K viruses. Input=inoculum virus mixture. (G) Quantification of data from F; n=3–5. (H) In vivo environmental stability transmission experiment design. Performed as in 6E, except that primary mouse feces were aged for 96 h at 22°C, followed by amplification in HeLa cells, peroral inoculation of secondary mice, 96 h 22°C aging of secondary mouse feces, and analysis of virus ratios by digestion of RT-PCR products with NheI. (I) Quantification of data from (H). See also Fig. S4. Data are mean +/− SEM; * = P<0.05.
Finally, to examine environmental stability of WT vs. T99K viruses in a model mimicking natural transmission, we examined the ratio of the two viruses in feces from perorally inoculated mice. PVR-IFNAR−/− mice were inoculated with a 1:1 mixture of WT and T99K-NheI viruses. Fresh feces were collected at 6 h post-inoculation or left in the cage for 24 or 96 h at room temperature to impose environmental stability pressure (Fig. 6E). Feces were collected, fecal viruses were amplified in HeLa cells to generate viral RNA, and RT-PCR NheI digestion products were analyzed using an acrylamide gel. Samples from inoculum, 6 h feces, or 24 h feces revealed similar amounts of each virus (Fig. 6FG). In contrast, samples collected from 96 h feces revealed significantly lower ratios of T99K:WT indicating that the T99K-NheI was not as stable as WT in the environment (Fig. 6FG). These results indicate that viruses with reduced LPS binding and LPS-mediated stabilization incur a fitness cost when environmental stability is a factor. It is possible that even minor differences in transmission efficiency may have major consequences over repeated transmission cycles due to additive effects. To test this, we perorally inoculated primary mice with a 1:1 mixture of WT and T99K-NheI viruses, aged feces for 96 h at room temperature, amplified fecal viruses in HeLa cells to generate enough virus for secondary mouse inoculation, perorally inoculated naïve secondary mice, aged feces for 96 h at room temperature, and examined the ratio of T99K:WT virus at each step (Fig. 6H). As expected, samples from inoculum or cell culture-passaged viruses revealed similar amounts of each virus. However, T99K-NheI virus was reduced 2.6-fold in primary mouse feces and 3.9-fold in secondary mouse feces, confirming a fitness cost in the presence of selective pressure for virion stability (Fig. 6I). Overall, these data suggest that bacterial polysaccharides bind poliovirus particles and protect them from inactivation in the environment, potentially promoting transmission to the next host.
Discussion
Although microbiota-host interactions are widely studied, how microbiota interact with viruses is relatively unclear. Previously we found that microbiota enhance viral replication and pathogenesis for two enteric viruses, poliovirus and reovirus (Kuss et al., 2011). Here we examined mechanisms by which bacteria enhance poliovirus virion infectivity and we explored the consequences for a mutant virus with reduced LPS binding.
First, we found that GlcNAc-containing bacterial surface polysaccharides, including LPS, bind poliovirus virions and stabilize the particles by preventing premature RNA release. Poliovirus exposed to LPS or PG was stable at elevated temperatures and in the presence of dilute chlorine bleach (Fig. 1AB). Physical assays including density gradients and a calorimetry-type assay (PaSTRy)(Walter et al., 2012) demonstrated that LPS exposure prevents premature RNA release (Fig. 1CDE). Interestingly, LPS exposure shifted the temperature of RNA release by over 3°C (Fig. 1DE and Fig. S1). This 3°C shift is impressive considering that capsid stabilizing drugs that cause permanent virion stabilization and prevent uncoating of enterovirus 71 also shift RNA release by 3°C (Plevka et al., 2013). Interestingly, the polysaccharide-mediated stabilization could be a “double-edged sword”, as capsid plasticity is needed for uncoating during infection. It is unknown how polysaccharide-stabilized poliovirus retains the capsid flexibility required for uncoating following PVR binding.
Second, we found that LPS aids poliovirus attachment to host cells via a direct enhancement of binding to the viral receptor, PVR. Clearly, poliovirus can bind its receptor in the absence of LPS. However, LPS-bound viruses have enhanced PVR binding, likely explaining their enhanced attachment to HeLa cells (Fig. 2). Interestingly, we found that cell attachment enhancement required significantly less LPS than virion stabilization (Fig. 2D). Since each poliovirus virion contains 60 potential binding sites, it is possible that stabilization requires binding of several sites while cell attachment enhancement requires binding of fewer sites. While the concentration of bacterial polysaccharide present in the gastrointestinal tract is difficult to measure, it is likely to be in the mg/ml range (Bates et al., 2007; Freudenberg et al., 1991). Therefore, the gut environment likely contains enough bacterial polysaccharides needed for stabilization of virions and enhancement of host cell attachment.
A major goal of this study was to identify a mutant virus with reduced LPS binding and examine the consequences for viral fitness. To that end, we found that VP1-T99K-containing polioviruses, including the attenuated serotype 1 Sabin vaccine strain, have reduced LPS binding at physiological temperature. Our data suggest that while amino acid 99 is not the polysaccharide-binding site, it influences binding. We hypothesize that K99 viruses are in a “closed” conformation at low/physiological temperatures, but at high temperatures (>40°C) the structure “opens” to reveal the polysaccharide-binding site (Fig. S2CDE). In agreement with this hypothesis, we found that virus lacking amino acid 99 was stabilized by LPS exposure (Fig. 3D). Interestingly, many fecal/sewage isolates from the K99-containing Sabin serotype 1 vaccine have T, N, R, E, S, or G at position 99, suggesting that lysine at this position may confer a selective disadvantage (Kew et al., 2002; Kew et al., 1998; van der Sanden et al., 2009; Yakovenko et al., 2006). Residue 99 is located on the BC loop near the PVR binding site (Harber et al., 1995; He et al., 2003; Moss and Racaniello, 1991). This region of VP1 is especially antigenic and variable among picornaviruses (Page et al., 1988). In fact, the BC loop can be replaced and partially deleted without eliminating viral viability (Liao and Racaniello, 1997). Our data suggest that the selective pressure of environmental stability may be one reason for the variation at residue 99 often observed in environmental isolates of the Sabin vaccine.
Poliovirus is found in soil, sewage, drinking water, and shellfish, and virus remains infectious in the environment for varying lengths of time (Hovi et al., 2001; Jaykus, 1997; Li et al., 1998; Okoh et al., 2010; Tierney et al., 1977; Ward, 1977). Poliovirus in the environment typically originates from humans infected with circulating virulent poliovirus or the Sabin vaccine strains. Acutely infected individuals can excrete 105 to 1011 viral particles per gram of stool (Bosch, 1998). Since nearly 1013 to 1014 bacteria are found in feces (Gill et al., 2006), viruses remain in close contact with bacteria following excretion into the environment. Using an environmental stability assay, we found that the T99K virus had reduced viability when exposed to an environmental pressure as compared to WT (Fig. 6FGI). These results indicate that viral binding to bacteria or bacterial polysaccharides may offer a selective advantage by enhancing environmental stability.
Our work sheds light on how viruses interact with the microbiota in the gastrointestinal tract and in the environment. While microbiota are predominantly located in the gut, they are also found at numerous other sites on the human body including the skin, respiratory tract, and urogential tract. Since most viruses navigate one or more of these sites, interactions with the microbiota are likely to be common. While we and others have shown beneficial interactions for viruses with the microbiota (Kane et al., 2011; Kuss et al., 2011), this is not the rule for all viruses. For example, the intestinal microbiota antagonize rotavirus and influenza virus (Dolowy and Muldoon, 1964; Ichinohe et al., 2011; Varyukhina et al., 2012). Some strains of human norovirus bind extracellular glycans on certain strains of intestinal bacteria, although whether this influences infection is unclear (Miura et al., 2013). Future work into understanding how viruses interact with the microbiota may inform vaccine strategies and provide novel antiviral therapeutics.
Experimental Procedures
Viruses and Cells
Poliovirus work was performed in BSL2+ areas in accordance with practices recommended by the World Health Organization. Poliovirus (serotype 1 virulent Mahoney and serotype 1 attenuated Sabin) cell culture infections and plaque assays were performed using HeLa cells (Kuss et al., 2008). Construction of mutant viruses is described in Supplemental Information.
Poliovirus stability assays
For titer-based stability assays, 105 PFU of poliovirus was mixed with compounds and incubated at 37°C or 42°C. Before and after incubation, samples were quantified by plaque assay using HeLa cells. Poliovirus titers from the zero time point were compared to post-incubation titers to determine the % input PFU. For bleach experiments, 107 PFU Mahoney poliovirus was incubated with compounds at 37°C for 1 h followed by treatment +/− 0.1 mg/L (0.001%) bleach for 1 min. Samples were neutralized by diluting 10-fold into a solution of 0.01% sodium thiosulfate prior to plaque assay.
For sucrose gradient assays 35S-labeled Mahoney poliovirus was prepared and purified by CsCl gradient (Kuss et al., 2011). 35S-labeled virus (30,000 CPM) was incubated +/− LPS at 42°C for 1 h prior to loading on 10–30% sucrose gradients with a 60% cushion. Gradients were centrifuged at 25,000 RPM in a Fiberlite F50L-8×39 rotor for 1.5 h at 4°C. Fractions were collected and scintillation counted to determine CPM. 80S gradient marker controls consisted of 35S-labeled poliovirus heated at 56°C for 15 min and 160S gradient markers consisted of unheated 35S-labeled poliovirus.
For PaSTRy experiments the concentration of unlabeled CsCl-gradient purified virus was determined by optical density at 260 nm (Arita et al., 1998; Bibb et al., 1994). One microgram of virus was pre-incubated with compounds for 1 h at 37°C. Samples were mixed with SYPRO Red (3x final concentration) or SYBR Green II (10x final concentration) and buffer (10 mM HEPES pH 8.0, 200 mM NaCl) to a final volume of 50 μl (Walter et al., 2012). Samples were placed in an ABI 7500 real-time instrument and heated from 25°C to 99°C on a 1% stepwise gradient with fluorescence monitoring.
Poliovirus cell attachment and PVR binding assays
35S-labeled poliovirus (3000 CPM) was exposed to compounds at 37°C for 1 h and incubated with HeLa cells or murine L929 cells at 4°C for 10 min to facilitate viral binding. Cells were washed 3x with cold PBS, trypsinized, and 35S was quantified in the scintillation counter. In Fig. 2A experiments, HeLa cells were treated +/− 5 μg of mouse anti-PVR/CD155 antibody (Santa Cruz) or mouse IgG control antibody (BioLegend) for 1 h at 37°C followed by PBS washing and addition of virus. For PVR binding experiments, recombinant human PVR (R&D Systems, Minneapolis, MN) was spotted on a nitrocellulose membrane and dried at room temperature for 1 h. Wheat germ agglutinin protein, in identical concentrations to PVR, was spotted onto the membrane to control for non-specific viral binding (data not shown). To establish a standard curve, 35S-labeled poliovirus was quantified by scintillation counting and known CPM were also spotted to each membrane. Membranes were pre-hybridized in PBS+ (PBS supplemented with 100 μg/ml MgCl2 and 100 μg/ml CaCl2) with 3% BSA for 1 h at 37°C. 35S-labeled virus (20,000 CPM) was exposed to 1 mg/ml LPS or PBS at 37°C for 1 h before addition to the membranes in PBS+/3% BSA and incubated at 37°C for 3 h. Membranes were washed 3x in PBS+ and exposed to a phosphorimager screen for analysis on a STORM 860 Molecular Imager (Molecular Devices, Sunnyvale, California).
Biotin-LPS virus pull-down assay
35S-labeled poliovirus (107 PFU/7000 CPM) was mixed with biotinylated LPS (E.coli O111:B4; Invivogen, San Diego, CA) or non-biotinylated LPS (0127:B8; Sigma-Aldrich) at various concentrations. To each sample, 30 μl of streptavidin agarose resin (Thermo Scientific) was added and samples were incubated for 3 h at 37°C. Following incubation, streptavidin agarose resin was pelleted by centrifugation at 2,600 × g for 2 min. Resin was washed 3 times with PBS+ and resin-associated radioactivity was quantified by scintillation counting. Non-specific binding was subtracted using CPM from samples incubated with non-biotinylated LPS. Data are expressed as percent of input CPM (streptavidin agarose resin associated CPM/input CPM x 100). Prism version 6.0c was used for nonlinear regression (curve fit) analysis using a two site binding equation. For Fig. S3 experiments, 35S-labeled poliovirus was incubated at 37°C for 3 h with 150 μg/ml biotinylated LPS, followed by incubation for 3 h with streptavidin agarose resin +/− 1500 μg/ml non-biotinylated LPS and samples were processed as above.
Mouse experiments
All animals were handled according to the Guide for the Care of Laboratory Animals of the National Institutes of Health. All mouse studies were performed at UT Southwestern (Animal Welfare Assurance #A3472-01) using protocols approved by the local Institutional Animal Care and Use Committee in a manner designed to minimize pain and any animals that exhibited severe disease were euthanized immediately. C57BL/6 PVR-IFNAR−/− mice, expressing the human poliovirus receptor and deficient for the IFNα/β receptor, were obtained from S. Koike (Tokyo, Japan)(Ida-Hosonuma et al., 2005). Seven to nine week old PVR-IFNAR−/− mice were inoculated perorally with 1×108 PFU of poliovirus. Mice were euthanized upon severe disease onset, including paralysis, encephalitis, and severe lethargy. For shedding and replication experiments, mice were inoculated perorally with 1×108 PFU of standard poliovirus or neutral red/light-sensitive poliovirus (to measure replication), and feces were collected at indicated time points post-inoculation and processed as previously described (Kuss et al., 2011). For replication experiments, light-sensitive virus was processed in the dark under a red safety light and a portion was light exposed. Light-exposed and unexposed virus was quantified by plaque assay to determine the amount of replicating virus by dividing light-exposed PFU/ml by light unexposed PFU/ml and multiplying by 100.
In vivo environmental stability assay
For Fig. 6EFG experiments, WT and T99K-Nhe viruses were mixed (1×107 PFU each) and mice were inoculated perorally. Fresh feces were collected at 6 h post-inoculation. Mice were sacrificed at 24 h post-infection, but feces were incubated in the cage for 24 h or 96 h at room temperature. Fecal viruses were amplified in HeLa cells until cytopathic effects were observed and RNA was isolated. Viruses passaged only in cell culture served as a control. Fig. 6HI experiments were performed in a similar manner, except that primary mouse feces were incubated in the cage for 96 h at room temperature, followed by viral amplification in HeLa cells and peroral inoculation of naïve secondary mice with 2×107 PFU. Feces from secondary mice were incubated in the cage for 96 h at room temperature, followed by viral amplification in HeLa cells. In all cases, viral RNA was isolated with TRIzol as per manufacturers protocol (Invitrogen) and cDNA was prepared using SuperScript II Reverse Transcriptase (Invitrogen). A 32P end-labeled (Kuss et al., 2008) antisense oligonucleotide primer (5′GATTTAAGGCATGCCCATTGTTAG) was used for cDNA amplification by PCR with an unlabeled sense primer (5′TTTCGACACCCAGAGAGATGGA). RT-PCR products were digested with NheI overnight at 37°C and bands were separated by electrophoresis on a denaturing 5% acrylamide gel. Bands were quantified using phosphorimaging.
Statistical Analysis
The differences between groups were examined by two-tailed Student’s t tests. P<0.05 was considered statistically significant. WT and T99K mouse survival curves were not significantly different (P > 0.05 by log rank test, Fig. 5D).
Supplementary Material
Highlights.
Bacterial polysaccharides enhance poliovirus stability and cell attachment
VP1-T99K mutant poliovirus has reduced lipopolysaccharide (LPS) binding
Following peroral inoculation of mice, VP1-T99K poliovirus was unstable in feces
Microbiota-mediated stabilization promotes poliovirus fitness in the environment
Acknowledgments
We thank Gavin Best and Pamela de la Cruz for assistance with experiments, Satoshi Koike for mice, Stanley Lemon for advice on PaSTRy experiments, and Andrea Erickson and John Schoggins for comments on the manuscript. This work was funded by the World Health Organization Global Polio Eradication Initiative (J.K.P.), a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease award (J.K.P.), R01 AI74668 (J.K.P.), a Hartwell Foundation postdoctoral fellowship (C.M.R.), and T32 AI007520 (C.M.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez ME, O’Brien RT. Effects of chlorine concentration on the structure of poliovirus. Appl Environ Microbiol. 1982;43:237–239. doi: 10.1128/aem.43.1.237-239.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita M, Koike S, Aoki J, Horie H, Nomoto A. Interaction of poliovirus with its purified receptor and conformational alteration in the virion. J Virol. 1998;72:3578–3586. doi: 10.1128/jvi.72.5.3578-3586.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates JM, Akerlund J, Mittge E, Guillemin K. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe. 2007;2:371–382. doi: 10.1016/j.chom.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb JA, Witherell G, Bernhardt G, Wimmer E. Interaction of poliovirus with its cell surface binding site. Virology. 1994;201:107–115. doi: 10.1006/viro.1994.1270. [DOI] [PubMed] [Google Scholar]
- Bosch A. Human enteric viruses in the water environment: a minireview. International microbiology : the official journal of the Spanish Society for Microbiology. 1998;1:191–196. [PubMed] [Google Scholar]
- Brandenburg B, Lee LY, Lakadamyali M, Rust MJ, Zhuang X, Hogle JM. Imaging poliovirus entry in live cells. PLoS Biol. 2007;5:e183. doi: 10.1371/journal.pbio.0050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolowy WC, Muldoon RL. Studies of Germfree Animals. I. Response of Mice to Infection with Influenza a Virus. Proceedings of the Society for Experimental Biology and Medicine Society for Experimental Biology and Medicine. 1964;116:365–371. doi: 10.3181/00379727-116-29249. [DOI] [PubMed] [Google Scholar]
- Freudenberg MA, Meier-Dieter U, Staehelin T, Galanos C. Analysis of LPS released from Salmonella abortus equi in human serum. Microb Pathog. 1991;10:93–104. doi: 10.1016/0882-4010(91)90070-q. [DOI] [PubMed] [Google Scholar]
- Fricks CE, Hogle JM. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J Virol. 1990;64:1934–1945. doi: 10.1128/jvi.64.5.1934-1945.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harber J, Bernhardt G, Lu HH, Sgro JY, Wimmer E. Canyon rim residues, including antigenic determinants, modulate serotype-specific binding of polioviruses to mutants of the poliovirus receptor. Virology. 1995;214:559–570. doi: 10.1006/viro.1995.0067. [DOI] [PubMed] [Google Scholar]
- He Y, Mueller S, Chipman PR, Bator CM, Peng X, Bowman VD, Mukhopadhyay S, Wimmer E, Kuhn RJ, Rossmann MG. Complexes of poliovirus serotypes with their common cellular receptor, CD155. J Virol. 2003;77:4827–4835. doi: 10.1128/JVI.77.8.4827-4835.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogle JM, Racaniello VR. Poliovirus receptors and cell entry. In: Semler BL, Wimmer E, editors. Molecular Biology of Picornaviruses. Washington D.C: ASM Press; 2002. pp. 71–83. [Google Scholar]
- Hovi T, Stenvik M, Partanen H, Kangas A. Poliovirus surveillance by examining sewage specimens. Quantitative recovery of virus after introduction into sewerage at remote upstream location. Epidemiology and infection. 2001;127:101–106. doi: 10.1017/s0950268801005787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ida-Hosonuma M, Iwasaki T, Yoshikawa T, Nagata N, Sato Y, Sata T, Yoneyama M, Fujita T, Taya C, Yonekawa H, et al. The alpha/beta interferon response controls tissue tropism and pathogenicity of poliovirus. J Virol. 2005;79:4460–4469. doi: 10.1128/JVI.79.7.4460-4469.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaykus LA. Epidemiology and detection as options for control of viral and parasitic foodborne disease. Emerg Infect Dis. 1997;3:529–539. doi: 10.3201/eid0304.970418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane M, Case LK, Kopaskie K, Kozlova A, MacDearmid C, Chervonsky AV, Golovkina TV. Successful transmission of a retrovirus depends on microbiota. Science. 2011;334:245–249. doi: 10.1126/science.1210718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan G, Freistadt MS, Racaniello VR. Neutralization of poliovirus by cell receptors expressed in insect cells. J Virol. 1990;64:4697–4702. doi: 10.1128/jvi.64.10.4697-4702.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kew O, Morris-Glasgow V, Landaverde M, Burns C, Shaw J, Garib Z, Andre J, Blackman E, Freeman CJ, Jorba J, et al. Outbreak of poliomyelitis in Hispaniola associated with circulating type 1 vaccine-derived poliovirus. Science. 2002;296:356–359. doi: 10.1126/science.1068284. [DOI] [PubMed] [Google Scholar]
- Kew OM, Sutter RW, Nottay BK, McDonough MJ, Prevots DR, Quick L, Pallansch MA. Prolonged replication of a type 1 vaccine-derived poliovirus in an immunodeficient patient. J Clin Microbiol. 1998;36:2893–2899. doi: 10.1128/jcm.36.10.2893-2899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuss S, Etheredge C, Pfeiffer J. Mulitple Host Barriers Restrict Poliovirus Trafficking in Mice. PLoS Pathog. 2008;4:e1000082. doi: 10.1371/journal.ppat.1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, Hooper LV, Dermody TS, Pfeiffer JK. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 2011;334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JW, Wang XW, Rui QY, Song N, Zhang FG, Ou YC, Chao FH. A new and simple method for concentration of enteric viruses from water. J Virol Methods. 1998;74:99–108. doi: 10.1016/s0166-0934(98)00078-0. [DOI] [PubMed] [Google Scholar]
- Liao S, Racaniello V. Allele-specific adaptation of poliovirus VP1 B-C loop variants to mutant cell receptors. J Virol. 1997;71:9770–9777. doi: 10.1128/jvi.71.12.9770-9777.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nature reviews Gastroenterology & hepatology. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- Miura T, Sano D, Suenaga A, Yoshimura T, Fuzawa M, Nakagomi T, Nakagomi O, Okabe S. Histo-blood group antigen-like substances of human enteric bacteria as specific adsorbents for human noroviruses. J Virol. 2013;87:9441–9451. doi: 10.1128/JVI.01060-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG, Racaniello VR. Host range determinants located on the interior of the poliovirus capsid. EMBO J. 1991;10:1067–1074. doi: 10.1002/j.1460-2075.1991.tb08046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomoto A, Omata T, Toyoda H, Kuge S, Horie H, Kataoka Y, Genba Y, Nakano Y, Imura N. Complete nucleotide sequence of the attenuated poliovirus Sabin 1 strain genome. Proc Natl Acad Sci U S A. 1982;79:5793–5797. doi: 10.1073/pnas.79.19.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien RT, Newman J. Structural and compositional changes associated with chlorine inactivation of polioviruses. Appl Environ Microbiol. 1979;38:1034–1039. doi: 10.1128/aem.38.6.1034-1039.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohka S, Igarashi H, Nagata N, Sakai M, Koike S, Nochi T, Kiyono H, Nomoto A. Establishment of a poliovirus oral infection system in human poliovirus receptor (hPVR/CD155)-expressing transgenic mice that are deficient in interferon-{alpha}/{beta} receptor. J Virol. 2007;81:7902–7912. doi: 10.1128/JVI.02675-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoh AI, Sibanda T, Gusha SS. Inadequately treated wastewater as a source of human enteric viruses in the environment. International journal of environmental research and public health. 2010;7:2620–2637. doi: 10.3390/ijerph7062620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page GS, Mosser AG, Hogle JM, Filman DJ, Rueckert RR, Chow M. Three-dimensional structure of poliovirus serotype 1 neutralizing determinants. J Virol. 1988;62:1781–1794. doi: 10.1128/jvi.62.5.1781-1794.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plevka P, Perera R, Yap ML, Cardosa J, Kuhn RJ, Rossmann MG. Structure of human enterovirus 71 in complex with a capsid-binding inhibitor. Proc Natl Acad Sci U S A. 2013;110:5463–5467. doi: 10.1073/pnas.1222379110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DG, Floyd R, Johnson JD. Nature of the surviving plaque-forming unit of reovirus in water containing bromine. Appl Microbiol. 1975;29:94–101. doi: 10.1128/am.29.1.94-101.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GR, Butler M. A comparison of the virucidal properties of chlorine, chlorine dioxide, bromine chloride and iodine. J Hyg (Lond) 1982;89:321–328. doi: 10.1017/s0022172400070856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney JT, Sullivan R, Larkin EP. Persistence of poliovirus 1 in soil and on vegetables grown in soil previously flooded with inoculated sewage sludge or effluent. Appl Environ Microbiol. 1977;33:109–113. doi: 10.1128/aem.33.1.109-113.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- van der Sanden S, Pallansch MA, van de Kassteele J, El-Sayed N, Sutter RW, Koopmans M, van der Avoort H. Shedding of vaccine viruses with increased antigenic and genetic divergence after vaccination of newborns with monovalent type 1 oral poliovirus vaccine. J Virol. 2009;83:8693–8704. doi: 10.1128/JVI.02388-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varyukhina S, Freitas M, Bardin S, Robillard E, Tavan E, Sapin C, Grill JP, Trugnan G. Glycan-modifying bacteria-derived soluble factors from Bacteroides thetaiotaomicron and Lactobacillus casei inhibit rotavirus infection in human intestinal cells. Microbes and infection/Institut Pasteur. 2012;14:273–278. doi: 10.1016/j.micinf.2011.10.007. [DOI] [PubMed] [Google Scholar]
- Walter TS, Ren J, Tuthill TJ, Rowlands DJ, Stuart DI, Fry EE. A plate-based high-throughput assay for virus stability and vaccine formulation. J Virol Methods. 2012;185:166–170. doi: 10.1016/j.jviromet.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RL. Inactivation of poliovirus in wastewater sludge with radiation and thermoradiation. Appl Environ Microbiol. 1977;33:1218–1219. doi: 10.1128/aem.33.5.1218-1219.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovenko ML, Cherkasova EA, Rezapkin GV, Ivanova OE, Ivanov AP, Eremeeva TP, Baykova OY, Chumakov KM, Agol VI. Antigenic evolution of vaccine-derived polioviruses: changes in individual epitopes and relative stability of the overall immunological properties. J Virol. 2006;80:2641–2653. doi: 10.1128/JVI.80.6.2641-2653.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.