SUMMARY
Many visual animals have innate preferences for particular wavelengths of light, which can be modified by learning. Drosophila’s preference for UV over visible light requires UV-sensing R7 photoreceptors and specific wide-field amacrine neurons, called Dm8. Here we identify three types of medulla projection neurons downstream of R7 and Dm8, and show that selectively inactivating one of them (Tm5c) abolishes UV preference. Using a modified GRASP method to probe synaptic connections at the single-cell level, we reveal that each Dm8 neuron forms multiple synaptic contacts with Tm5c in the center of Dm8’s dendritic field, but sparse connections in the periphery. By single-cell transcript profiling and RNAi-mediated knockdown, we determine that Tm5c uses the kainate receptor Clumsy to receive excitatory glutamate input from Dm8. We conclude that R7s->Dm8->Tm5c form a hard-wired glutamatergic circuit that mediates UV preference by pooling ~16 R7 signals for transfer to the lobula, a higher visual center.
INTRODUCTION
Many visual animals, including vertebrates and invertebrates, have an innate ability to discriminate and to respond differentially to light of different wavelengths (Menzel, 1979). Zebrafish swim strongly towards ultraviolet, blue, and red light but weakly to green light while turtle hatchlings are more responsive to green relative to yellow and blue light (Orger and Baier, 2005; Young et al., 2012). Most flying insects, such as Drosophila, exhibit positive phototaxis toward short wavelengths of light when startled, preferring long ultraviolet (UV) to blue light, and blue light to green light (Gao et al., 2008; Yamaguchi et al., 2010). In contrast to true color vision, which discriminates visual stimuli based on wavelength independent of intensity, innate spectral preference is strongly intensity dependent and likely reflects each species’ ecological needs. In flower-visiting insects, such as butterflies, spectral preference differs within a family or even a genus (Ilse and Vaidya, 1956; Weiss, 1997), and such spectral preferences are thought to provide behavioral biases to facilitate initial recognition of flowers (Goyret et al., 2008). Because sunlight, but not reflected light, is rich in UV, Drosophila’s preference of UV over visible light is likely related to the fly’s attraction to open space (Hu and Stark, 1977). Butterflies, honeybees, and more recently Drosophila have been shown to have true color vision and are capable of associating colors with appetitive or aversive stimuli (Crane, 1955; Goyret et al., 2008; Koshitaka et al., 2008; Menne and Spatz, 1977; Menzel and Greggers, 1985; Schnaitmann et al., 2010). In Drosophila and butterflies, learning can significantly modify or even override innate spectral preference (Kelber, 1996; Goyret et al., 2008; Schnaitmann et al., 2010). To understand the neural basis of innate spectral preference and true color vision, it is essential to identify the neural circuits and synaptic mechanisms that underlie these processes. With a plethora of genetic tools for identifying neural circuit elements and for manipulating activity of targeted neurons, Drosophila is a well-suited model system for this task (reviewed in Borst, 2009; Meinertzhagen and Lee, 2012).
Drosophila vision is mediated by three classes of photoreceptors, defined by their stereotyped positions within the unit eye or ommatidia and their patterns of opsin gene expression. The outer photoreceptors R1-R6, which express the Rh1 opsin, respond to a broad spectrum of light and therefore are assumed to be achromatic. The inner, or chromatic, photoreceptors, R7 and R8, express opsins of narrow spectral sensitivity in a complex pattern (Hardie, 1979; Mikeladze-Dvali et al., 2005; O’Tousa et al., 1985). In the so-called “pale” (p) ommatidia, R7 expresses UV-sensitive Rh3 and the underlying R8 expresses blue-sensitive Rh5 opsin while in the “yellow” (y) ommatidia, R7 expresses Rh4, which is sensitive to longer wavelength of UV, and R8 contains the green-sensitive Rh6 opsin (Salcedo et al., 1999). The p and y types of ommatidia are stochastically distributed in the compound eye in a 30:70 ratio, and are thought to extend the spectral range of the eyes to mediate color vision (Morante and Desplan 2008). The chromatic photoreceptors R7 and R8 are functionally required for both spectral preference and color vision (Gao et al., 2008; Schnaitmann et al., 2010; Yamguchi et al., 2010; Melnattur and Lee, unpublished) while the achromatic R1-R6 mediate visual motion detection (Heisenberg and Buchner, 1977).
All visual information converges upon the medulla, the most complex optic neuropil. The medulla is organized in layers (M1-M10) and columns, which are innervated by afferents of different types in a retinotopic fashion. R7 and R8 axons directly innervate the M6 and M3 layers, respectively, while L1-L3 lamina neurons relay R1-R6 achromatic information to layers M1/5, M2, and M3, respectively (Figure 1A). Each medulla column receives inputs from individual R7, R8 and L1-3 neurons that view a single visual “pixel” (Meinertzhagen, 1976). Golgi studies have identified approximately 60 morphologically distinct types of medulla neurons (Fischbach and Dittrich, 1989). Using genetically-encoded reporters, a previous study identified a number of medulla neuron types that are postsynaptic to R7 and R8 and therefore are likely involved in spectral preference and color vision (Gao et al., 2008). Serial-section electron microscopy reconstruction revealed that each wide-field Dm8 amacrine neuron receives input from about 16 R7s. Targeted manipulation of neuronal activity further demonstrated that Dm8 is both required and sufficient for animals’ phototactic preference for UV over green light. However, the mechanisms enabling the wide-field Dm8 neurons to mediate UV preference remain unknown. Here we investigate the neural circuit that relays Dm8 signals to the lobula. Using a combination of histology, single-cell transcript profiling, RNAi-mediated knockdown and behavior assays, we identify and characterize the neural circuit and synaptic mechanism responsible for UV preference.
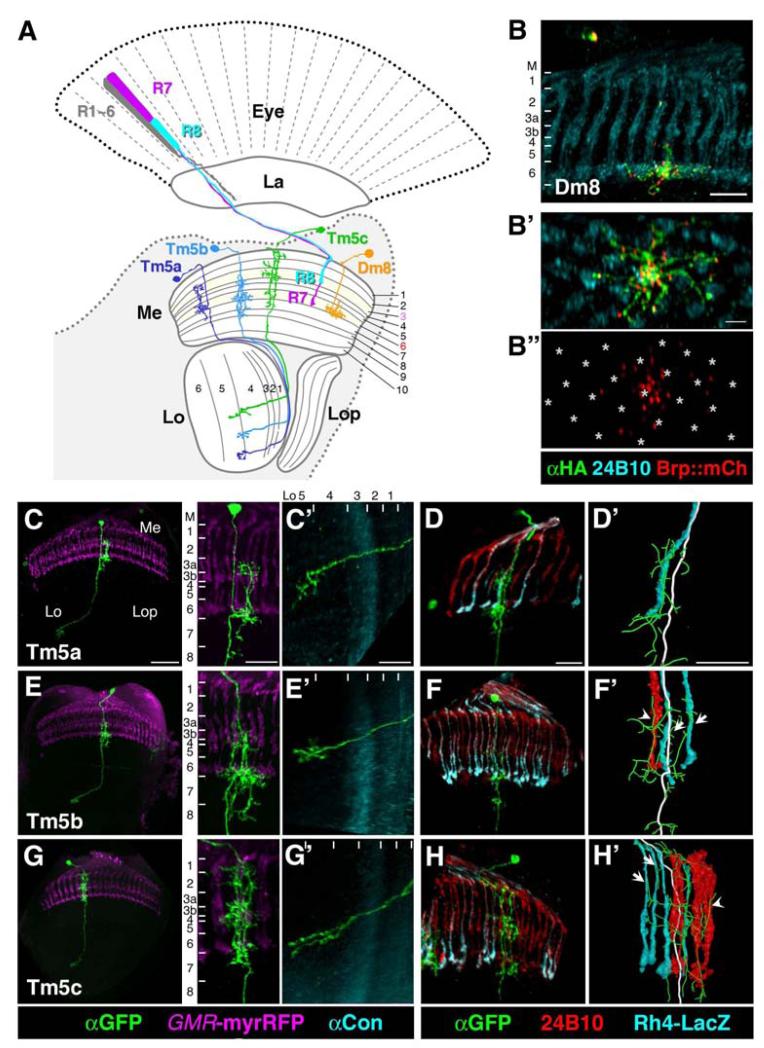
Figure 1. Three Distinct Subtypes of Tm5 Projection Neuron Relay Chromatic Information to the Lobula, a Higher Visual Center.
(A) A schematic illustration of the Drosophila visual system, including the eye (Eye), and four optic neuropils (lamina [La], medulla [Me], lobula [Lo], and lobula plate [Lop]). The inner photoreceptors, R7 (purple) and R8 (blue), project axons to the medulla layers M6 and M3, respectively, while the outer photoreceptors, R1-R6 (grey), terminate in the lamina. The amacrine neuron Dm8 (orange) extends dendrites in the M6 layer where they receive multiple R7 inputs. Three morphologically distinct types of Tm5 neurons, Tm5a (dark blue), Tm5b (cyan), and Tm5c (green), extend dendrites in multiple medulla layers, including M3 and M6, and project axons to the lobula layers Lo4 and Lo5.
(B-B″) Single Dm8 neurons were labeled using the single-cell flip-out technique. The dendritic arbors were labeled by a membrane-tethered marker (spGFP11::CD4::HA) and visualized by anti-HA antibody while their presynaptic terminals were labeled by the presynaptic marker Brp::mCherry (red). Photoreceptors were visualized using 24B10 antibody (cyan). (B) a dorsal-ventral view. (B′-B″) proximal-distal views. Dm8 dendrites span approximately 16 medulla columns in the M6 layer. The presynaptic sites of Dm8 cluster at the center of its dendritic field in the M6 layer. For clarity, B″ shows the presynaptic sites (red) of Dm8 with R7 terminals marked with asterisks.
(C-H′) The axonal and dendritic morphologies (C,C′,E,E′,G,G′) and dendritic arborization in the yellow/pale columns (D,D′,F,F′,H,H′) of Tm5a (C-D′), Tm5b (E-F′) and Tm5c (G-H′). Single Tm5a, Tm5b, or Tm5c clones were generated using the single-cell flip-out technique in flies carrying hs-Flp GMR-RFP; ortC1a-Gal4 UAS>CD2,y+>mCD8-GFP transgenes. R7 and R8 photoreceptors, visualized by GMR-RFP (red), served as landmarks for medulla columns. The medulla and lobula layers were identified using R7 and R8 terminals and anti-Connectin antibody (cyan), which labeled the Lo3 layer. (D, D′, F, F′, H, H′) The yellow-subtype R7 terminals (R7y), which occupied yellow-type medulla columns, were labeled by a Rh4-LacZ reporter and visualized by anti-β-galactosidase antibody (cyan). Left panels of (C, E, G): low magnification views showing the entire optic lobe; right panels of (C, E, G): high magnification views of the medulla; (C′, E′, G′): high magnification views of the lobula. (D, F, H): high magnification views of the medulla; (D′, F′, H′): close up of (D, F, H) showing skeletonized dendrites (green lines) and axons (white lines).
(C, C′) The Tm5a neuron extends a single primary dendrite along the photoreceptor axon and arborizes into many fine processes in the M3, M6, and M8 layers. Its axon makes a sharp turn and branches out in the Lo5 layer.
(D, D′) The main dendritic branch of Tm5a exclusively associates with R7y axons (cyan). (E-E′) The Tm5b neuron extends two or three main dendritic branches along the photoreceptor axons and arborizes into many fine processes in the M3, M6, and M8 layers. (E′) Its axonal terminal forms from multiple short branches in the Lo5 layer.
(F, F′) The main dendritic branches of Tm5b associate with either R7y (arrows) or R7p (arrow head) axons without apparent discrimination.
(G, G′) The Tm5c neuron extends multiple fine dendritic processes that span about eight medulla columns in the M1, M3 and M6 layers and the axon often bifurcates in the Lo4 layer and terminates in the Lo5/6 layers. The dendritic arborization in the M1 layer differentiates Tm5c from Tm5a and Tm5b.
(H, H′) The dendrites of Tm5c associate with either R7y (arrows) or R7p (arrowhead) axons. Scale bar: 30 μm in C (left panel); 5 μm in B′ and D′; 10 μm for the remaining panels.
RESULTS
Tm5 Neurons Are Divided Into Three Distinct Subtypes
A previous study revealed that Dm8 amacrine neurons mediate UV preference, but did not identify their downstream targets in the medulla (Gao et al., 2008). Using a flip-out genetic system (see Supplemental Experimental Procedures for details) to label single Dm8 neurons with an active zone marker, Brp::mCherry (Schmid et al., 2008), we found that most of the Dm8 presynaptic sites were localized in the M6 layer (Figures 1B-1B″). To identify the postsynaptic targets of Dm8 that mediate UV preference, we screened a collection of Gal4 lines for expression in medulla neurons that extend dendritic arbors in the M6 layer. We secondarily required that synaptic suppression of neurons within the Gal4 expression pattern eliminate UV preference behavior. One line, ortC1a-Gal4, in which Gal4 expression is driven by a highly conserved region of the ort (ora transientless; HisCl2) promoter (Figure S1A), labels a subset of ort(+) neurons that extend dendritic arbors in the M6 layer and project axons to the fifth layer of the lobula, Lo5 (Figure S1C). To assess whether ortC1a(+) neurons were required for UV preference, we used a quantitative spectral preference assay that tests phototaxis toward UV versus green light (Figure 2A; Gao et al., 2008). To quantify UV preference, we determined the UV/green intensity ratio at which flies found UV and green light equally “attractive”, and used the negative logarithm of the intensity ratio as a measure of attractiveness (AttrUV/G; Figures 2C and 2D). We found that driving the expression of tetanus toxin light chain (TNT, Sweeney et al., 1995) using ortC1a-Gal4, so as to block synaptic transmission in the ortC1a(+) neurons, significantly reduced UV attractiveness by approximately two orders of magnitude (AttrUV/G = 0.45 ± 0.29; mean ± SD, p<0.05), as compared with wild-type and control flies (AttrUV/G = 2.4 ± 0.24 for wild-type; 2.3 ± 0.31 for ortC1a-Gal4 control; 2.25 ± 0.30 for UAS-TNT control, Figures 2C and 2D). This reduction is comparable to that caused by inactivating Dm8 neurons (AttrUV/G = −0.11 ± 0.28), indicating that at least a subset of ortC1a(+) neurons lies in the pathway mediating UV preference.
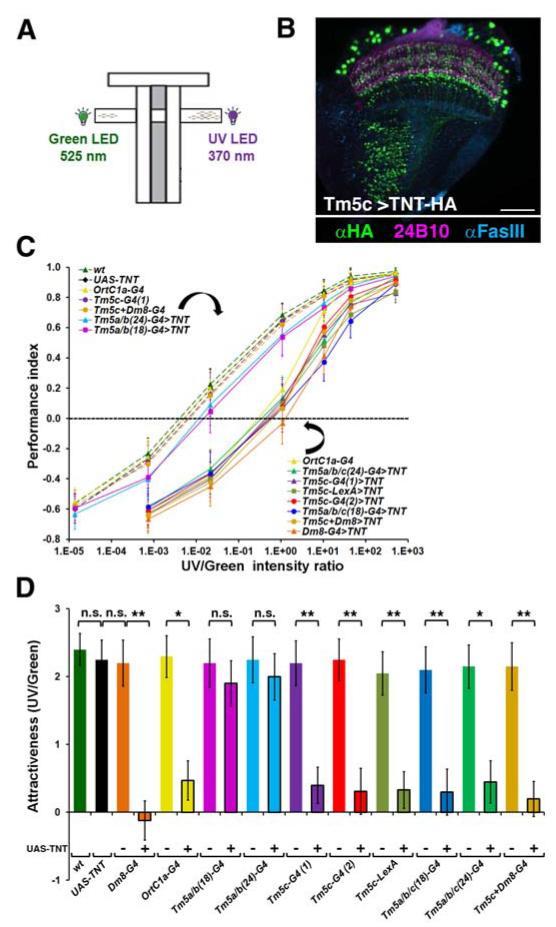
Figure 2. The Projection Neuron Tm5c Is Required for Normal UV Preference.
(A) A schematic diagram of the T-maze apparatus used in the spectral preference assay. (B) The expression of the HA-tagged TNT (tetanus toxin light chain) in Tm5c neurons was assessed in the flies carrying Tm5c-Gal4 UAS-TNT-HA. TNT-HA was visualized using anti-HA antibody (green). Photoreceptors were visualized by 24B10 antibody (magenta), and medulla and lobula layers were visualized by anti-FasIII antibody (cyan), serving as landmarks. TNT-HA was detected as punctate staining in axonal and dendritic terminals as well as cell bodies of Tm5c. Scale bar: 30 μm.
(C-D) Wild-type and various mutant flies were tested for phototactic preference to UV over green light. The intensity-response curves were measured at various UV/green intensity ratios (shown as a logarithmic scale). The performance index (P.I.) for each genotype was calculated from the numbers of flies choosing UV (NUV) or green (NG) light by the following formula: P.I. = [NUV - NG] / [NUV + NG]. The data were presented as mean ± standard deviation (C). Relative attractiveness (AttrUV/G) was calculated from the UV/green intensity ratio at which flies exhibited UV and green phototactic responses with equal frequency (i.e. P.I.=0) using the following formula: AttrUV/G= −log(UV/green ratio at P.I.=0). AttrUV/G values for all genotypes were plotted together as bar graphs (D).
(C) Inactivating ortC1a(+), Tm5c or Dm8 neurons, but not Tm5a/b, significantly reduced phototactic preference to UV light over green light. Wild-type (wt) flies exhibited phototactic preference to UV in an intensity-dependent fashion, resulting in a sigmoidal intensity-response curve. TNT expressed in ortC1a(+) neurons (ortC1a-G4>TNT) blocked the synaptic transmission of Tm5a/b/c and Tm20, and reduced flies’ phototactic preference to UV, resulting in a right-shifted intensity-response curve. Inactivating Tm5c alone using one of the three Tm5c-specific drivers to drive TNT expression (Tm5c-G4(1)>TNT, Tm5c-G4(2)>TNT, and Tm5c-LexA>TNT) caused reduced UV preference, comparable to that caused by inactivating Dm8 (Dm8-G4>TNT). In contrast, inactivating Tm5a/b (Tm5a/b(18)-G4>TNT and Tm5a/b(24)-G4>TNT) did not affect UV preference significantly. Inactivating Tm5a/b or Dm8 in addition to Tm5c (Tm5a/b/c(18)-G4>TNT, Tm5a/b/c(24)-G4>TNT, and Tm5c+Dm8-G4>TNT) did not enhance UV preference defects. Flies carrying Gal4 drivers or UAS-TNT alone had normal UV preference.
(D) Bar graph of the relative attractiveness of UV over green light (AttrUV/G) calculated from (C). n.s.: not significant (p>0.05); * and **: significant (p<0.05 and p<0.01, respectively).
To better characterize the ortC1a(+) neurons, we used the flip-out genetic method (Wong et al., 2002) to generate single cell clones and examine their morphologies. We found that ortC1a-Gal4 labeled four morphologically distinct types of transmedulla (Tm) neurons, including Tm20 and three Tm5 subtypes, Tm5a, Tm5b, and Tm5c (Figures 1C-1H′ and data not shown). Tm5a/b/c, but not Tm20, extend dendrites in the M6 layer (Figures 1C-1H′). Because Tm5a/b/c might be postsynaptic to Dm8 in the M6 layer, we chose to focus on these Tm5 subtypes. The three Tm5 subtypes have different axonal and dendritic morphologies (Figures 1C, 1E and 1G), suggesting that they have distinct functions. To identify subtype-specific drivers, we used the split-Gal4 system to refine the ortC1a expression pattern (Luan et al., 2006). Among approximately 200 dVP16AD enhancer trap lines, we identified two hemidriver lines, dVP16AD18K and dVP16AD24G, which in combination with the ortC1a-Gal4DBD hemidriver (ortC1aDBD∩18K [designated as Tm5a/b(18)-Gal4], ortC1aDBD∩24g [designated as Tm5a/b(24)-Gal4]) labeled Tm5a and Tm5b neurons (Figure S1D and data not shown). In addition, ortC1aDBD∩OK371 (designated as Tm5c-G4), the combination of the ortC1a-Gal4DBD and dVP16ADOK371 (a vesicular glutamate transporter enhancer trap) hemidrivers, labeled Tm5c neurons alone (Figure S1E). Using these subtype-specific drivers, we further examined whether the dendrites of Tm5 subtypes selectively arborize in pale or yellow medulla columns, which receive pale or yellow subtypes of R7/R8 terminals, respectively. We found that Tm5a arborized dendrites in single yellow columns (100%, n=57; Figures 1D and 1D′) while Tm5b and Tm5c dendrites populate multiple columns of mixed yellow and pale types (100%, n=21 and 25 for Tm5b and Tm5c, respectively; Figures 1F, 1F′, 1H and 1H′). Based on their selective targeting by type-specific drivers, distinct axonal and dendritic morphologies, and selective dendritic arborization of yellow and pale columns, we conclude that the three Tm5 subtypes are distinct neuronal classes and are likely to differ in connectivity and function.
Tm5c but not Tm5a/b Is Required for UV Preference
To assess which Tm5 subtypes are required for UV preference, we used the type-specific drivers to express TNT, so as to block synaptic transmission, and examined the behavioral consequences using the quantitative spectral preference assay. We found that inactivating Tm5a/b did not affect flies’ attraction to UV (AttrUV/G = 1.9 ± 0.32 for Tm5a/b(18)-Gal4>TNT, and 2.0 ± 0.34 for Tm5a/b(24)-Gal4>TNT; p>0.05; Figures 2C and 2D). In contrast, flies carrying Tm5c-Gal4>TNT, but not the corresponding Gal4 driver alone, exhibited UV preference defects comparable to those caused by inactivating Dm8 or ortC1a(+) neurons (AttrUV/G = 0.4 ± 0.27). Furthermore, flies carrying the combinatorial driver ortC1aLexADBD∩OK371dVP16AD (Tm5c-LexA) driving a HA(hemagglutinin)-tagged TNT had a reduced UV preference (AttrUV/G = 0.3 ± 0.29; Figures 2C and 2D), comparable to that of Tm5c-Gal4>TNT flies. By immunohistochemistry, we detected TNT-HA in Tm5c cell bodies, axonal terminals and dendrites but found no apparent defects in axonal or dendritic morphologies (Figure 2B). To confirm that synaptic transmission, rather than structural integrity, of Tm5c is essential for optimal UV preference, we used a temperature-sensitive allele of shibire (shits1) to block Tm5c’s synaptic transmission conditionally (Kitamoto, 2001). We found that Tm5c-Gal4 driving shits1 significantly reduced the UV attractiveness at non-permissive, but not permissive, temperatures (AttrUV/G = 0.2 ± 0.20 at 33°C and 1.9 ± 0.29 at 22°C for Tm5c>shits1; AttrUV/G = 1.6 ± 0.22 at 33°C and 2.0 ± 0.25 at 22°C for UAS-shits1 control; data not shown). To determine whether inactivating Tm5c causes any additional visual deficits, we examined optomotor behavior using a head-yaw assay (Figures S2A and S2B; Rister et al., 2007). We found that Tm5c-Gal4>TNT flies, like Dm8-Gal4>TNT flies, exhibited normal head-yaw responses to horizontally moving strips, indistinguishable from those of wild-type or control flies (Figure S2C). Thus, we conclude that Tm5c neurons, but not Tm5a/b, are functionally required for normal UV preference but not motion detection.
To test the possibility that Tm5a/b are partially redundant to Tm5c in the UV preference pathway, we inactivated all three Tm5 subtypes using both Tm5a/b and Tm5c-specific drivers (Tm5a/b/c(18)-Gal4 and Tm5a/b/c(24)-Gal4) to express TNT. We found that Tm5a/b/c(18)>TNT and Tm5a/b/c(24)>TNT flies had a reduced UV preference, indistinguishable from that of Tm5c>TNT flies (Figures 2C and 2D). Thus, inactivating Tm5a/b, in addition to Tm5c, did not exacerbate UV preference defects, suggesting that Tm5a/b are not functionally redundant to Tm5c for UV preference. Likewise, inactivating both Tm5c and Dm8 neurons (Tm5c+Dm8>TNT) did not further reduce UV preference (AttrUV/G = 0.2 ± 0.26), as compared with that caused by inactivating Tm5c or Dm8 alone (Figures 2C and 2D). These results suggest that Tm5c and Dm8 neurons function in the same pathway for UV preference.
Tm5c Receives Direct R8 Input to Mediate Phototaxis toward Green Light
Because Tm5c neurons were labeled by the ortC1a-Gal4 driver and extend dendrites in the M3 and M6 layers, we tested whether they express the histamine receptor Ort and receive direct histaminergic inputs from photoreceptors R7 and/or R8. We manually isolated GFP-labeled Tm5c cell bodies and confirmed ort expression by RT-PCR reactions (for details, see Supplemental Experimental Procedures). To examine the presumptive R7/R8->Tm5c synapses anatomically, we used a modified GRASP (GFP reconstitution across synaptic partners) method (Feinberg et al., 2008; Gordon and Scott, 2009). We expressed a vesicle-tethered split-GFP (syb::spGFP1-10) in R8s or R7s, and a membrane-tethered split-GFP (CD4::spGFP11) in Tm5c, and examined the native fluorescence signal of reconstituted GFP (see Experimental Procedures for details). Strong GRASP signal was observed at R8, but not R7, terminals, suggesting a functional synaptic connection between R8 and Tm5c (Figures 3B-C′). The GRASP signal was most prominent in the M3 layers but was also present in M1 and M2 layers, consistent with previous EM findings that R8’s presynaptic sites are localized in M1-M3 layers (Takemura et al., 2008).
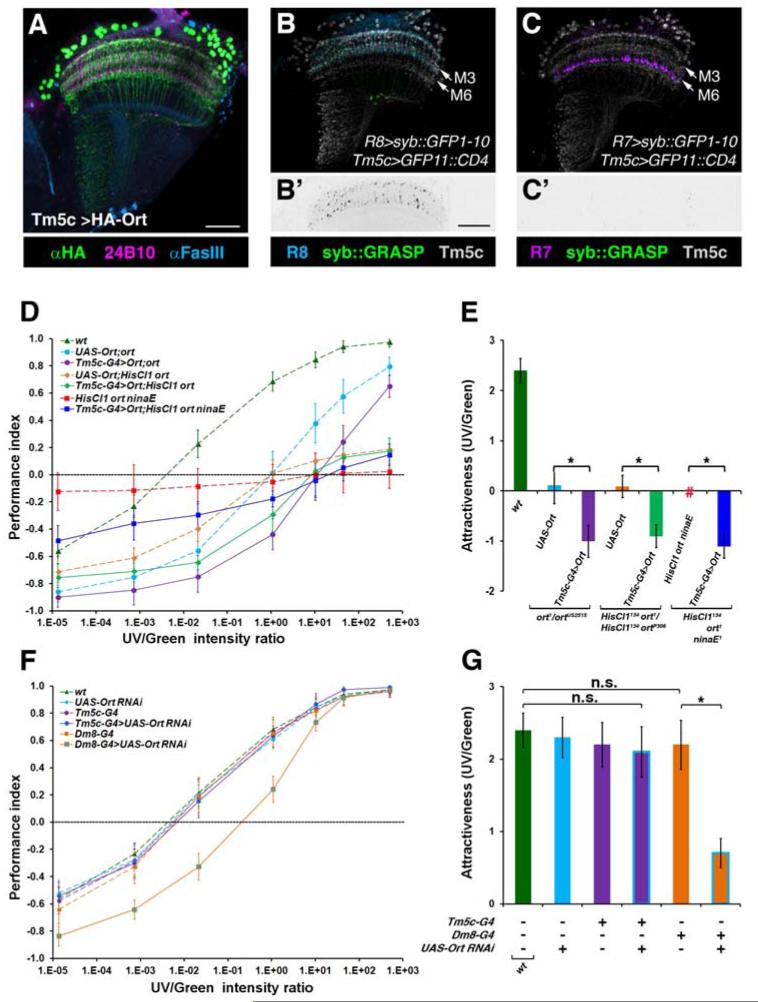
Figure 3. Tm5c Neurons Receive Direct Inputs from Photoreceptor R8s.
(A) The expression of the HA-tagged Ort in Tm5c neurons was assessed in the ort mutant flies carrying Tm5c-Gal4 UAS-2xHA-ORT transgenes. HA-Ort was concentrated in Tm5c dendritic arbors and cell bodies. (B-C′) GRASP revealed functional R8->Tm5c synapses (B), but not R7-> Tm5c (C). Flies expressing syb::spGFP1-10 in R8s/ R7s and CD4::spGFP11 in Tm5c were examined for native fluorescence of reconstituted GFP (green). Strong GRASP signal (green) was observed along with the R8 axons and their terminals (B-B′). On the contrary, no detectable GRASP signal was found in R7s (C-C′). (B′, C′) shows the green channel of (B, C), respectively. R8/R7 photoreceptors were labeled with αGFP antibody (pseudocolored in cyan for R8s; magenta for R7s). Anti-hCD4 immunolabeling (Grey) outlines the morphology of Tm5c neurons. Scale bar: 30 μm in A and B′. (D) Restoring Ort expression in Tm5c neurons in ort, HisCl1 ort double or HisCl1 ort ninaE triple mutant flies rendered a stronger green preference. ort mutants had a reduced UV preference and their intensity-response curve was shifted to the right, as compared to wild-type. HisCl1 ort double mutants exhibit some residual green phototactic responses, resulting in a flattened curve, as compared to ort mutants. HisCl1 ort ninaE triple mutants are near blind, choosing UV and green light indiscriminately. Ort expression in Tm5c in ort single, HisCl1 ort double or HisCl1 ort ninaE triple mutants resulted in a stronger green preference. (E) Bar graph of the relative attractiveness (AttrUV/G) calculated from (D). HisCl1 ort ninaE triple mutant flies are near blind and its AttrUV/G cannot be ascertained (#). (F) Blockage of the photoreceptors’ inputs to Tm5c neurons caused no detectable UV preference defect. RNAi-mediated knockdown of ort in Dm8, but not Tm5c, neurons significantly decreased UV preference. (G) Bar graph indicating the relative attractiveness of UV over green light (AttrUV/G) calculated from (F). n.s.: not significant (p>0.05); *: significant (p<0.05).
To determine whether the R8->Tm5c connections contribute to spectral preference, we prevented Tm5c neurons from receiving R8 input by knocking down the histamine chloride channel ort in Tm5c. We found that RNAi-mediated knockdown of ort in Tm5c had no detectable effects on flies’ UV preference (AttrUV/G = 2.1 ± 0.35; Figures 3F and 3G) as compared with wild-type and the UAS-ort RNAi control (AttrUV/G = 2.3 ± 0.28). In contrast, RNAi-mediated knockdown of ort in Dm8, hence blocking the reception of R7 inputs, significantly reduced UV preference (AttrUV/G = 0.7 ± 0.2; Figures 3F and 3G). These observations are consistent with the notion that the indirect pathway, R7->Dm8->Tm5c, is both required and sufficient for UV preference while the direct R8->Tm5c pathway plays, at best, a minor role in spectral preference under normal conditions.
To determine whether R8->Tm5c connections are functional, we examined whether expressing Ort in Tm5c affects spectral preference in various ort mutant, and thus sensitized, backgrounds. We found that expressing HA-tagged Ort in Tm5c enhanced green preference, as compared with the corresponding ort mutants (Figures 3A, 3D and 3E). To confirm these results, we tested flies in a fast phototaxis assay measuring their response to either UV or green light under dark-adapted conditions. We found that restoring Ort expression in Tm5c in HisCl1 ort ninaE triple mutants drove strong phototaxis toward green light but relatively weak phototaxis toward UV (Figures S3A-S3D), suggesting that Tm5c receives functional inputs directly from R8 photoreceptors.
Dm8 Provides Input for Tm5c in the Center of its Dendritic Field
To determine whether Tm5c neurons receive synaptic input from Dm8 to mediate UV preference, we examined potential membrane contacts between Dm8 and Tm5c using the GRASP method. In adult flies expressing one membrane-tethered split-GFP component (i.e. spGFP1-10::CD4) in Dm8 and the other (i.e. spGFP11::CD4) in Tm5c neurons (see Supplemental Experimental Procedures for details), we observed strong GFP fluorescence signals at the apparent contacts between Dm8 and Tm5c in the M6 layers (Figures 4A and 4A′), but no fluorescence upon expression of either split-GFP alone (data not shown). To determine whether these membrane contacts constitute genuine synapses, we developed a flip-out GRASP method that labels, in single neurons, the presynaptic sites with the presynaptic marker Brp::mCherry, in addition to an HA-tagged split-GFP, spGFP11::CD4::HA (Figure 4B). Using this method to mark single Dm8 neurons, we observed punctate GRASP signals at apparent contacts between single Dm8 neurons and multiple Tm5c neurons that expressed spGFP1-10::CD4 (Figures 4C and 4D). Most GRASP puncta co-localized with Brp::mCherry signals, indicating that these membrane contacts between Dm8 and Tm5c are on, or juxtaposed to, Dm8’s presynaptic sites. There were however some Brp::mCherry puncta devoid of GRASP signals, suggesting that Dm8 has other postsynaptic partners that do not share presynaptic sites with Tm5c (Figure 4D″, arrowheads). Interestingly, we observed clustered GRASP/Brp puncta at the center of the Dm8 dendritic field but only sparse puncta in the periphery. The dendritic field of Dm8 spans approximately 16 medulla columns but the area with clustered GRASP/Brp puncta corresponds to approximately one medulla column (Figures 4D′ and 4D″). We thus suggest that single Dm8 neurons form multiple synapses with Tm5c neurons in the center of Dm8’s dendritic field but only sparse synapses at the periphery.
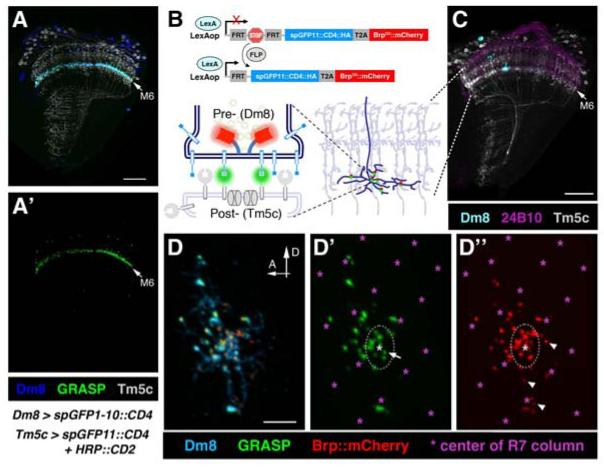
Figure 4. GRASP Visualization of Dm8->Tm5c Synapses.
(A, A′) Membrane contacts between Dm8 and Tm5c were assessed by the GRASP method. The split-GFP1-10 component was expressed in the Dm8 neurons and the split-GFP11 component was expressed in the Tm5c neurons. Strong green fluorescence signal from reconstituted GFP (green) was observed at the apparent contacts between Dm8 dendrites and Tm5c dendrites in the M6 layer. Top panel shows Dm8 (blue) and Tm5c (grey) neurons visualized by anti-GFP and anti-HRP antibodies, respectively. A′ shows the green channel (GRASP) alone.
(B) Schematic illustration of the single-cell GRASP method. Dm8-LexA flies carrying LexAop>FRT-Stop-FRT>spGFP11::CD4::HA::T2A::Brp::mCherry and hs-Flp transgenes allow dual-labeling with the split-GFP (spGFP11) and the active zone marker, Brp::mCherry (separated by the ribosomal skipping sequence T2A) in single Dm8 neurons. A mild heat shock was applied to 3rd instar larvae to remove the FRT-Stop-FRT cassette and place both markers under LexAop control. The same flies also carried Tm5c-Gal4 UAS-spGFP1-10::CD4 transgenes to express the other split-GFP (spGFP1-10) in all Tm5c neurons. Functional GFP molecules were reconstituted at the membrane contacts between Tm5c and single Dm8. The active zone marker labels the presynaptic sites of Dm8, differentiating synaptic contacts (labeled by both GRASP and Brp::mCherry) from mere membrane contacts (GRASP alone).
(C) A single Dm8 neuron labeled with both spGFP11::CD4::HA and Brp::mCherry was visualized by anti-HA antibody (cyan). Tm5c neurons co-expressing spGFP1-10 and HRP::CD2 were visualized by anti-HRP antibody (grey). Photoreceptor axons visualized by 24B10 antibody were used as landmarks (magenta).
(D-D″) Presynaptic sites of a single Dm8 neuron and its membrane contacts with Tm5c were visualized by native fluorescence of Brp::mCherry (red) and reconstituted GFP (GRASP, green), respectively. Most GRASP puncta co-localized with Brp::mCherry signal, suggesting they are synaptic contacts. Dm8 dendrites, which span about 14 medulla columns, were visualized by anti-HA antibody (cyan). GRASP/Brp::mCherry puncta cluster in the center of the Dm8 dendritic field. D′ and D″ show the green (GRASP) and red (Brp::mCherry) channels of D, respectively. Arrows mark the puncta with GRASP signal but not Brp::mCherry and arrow heads mark the puncta with Brp::mCherry alone. The dotted line circles the area corresponding to one medullar column in the center of the Dm8 dendritic field. Photoreceptor terminals (marked by asterisks), visualized by 24B10 antibody, were used as landmarks for medulla columns.
Scale bar: 30 μm for A and C; 5 μm in D for D-D″.
Glutamatergic Output of Dm8 and Tm5c Is Required for UV Preference
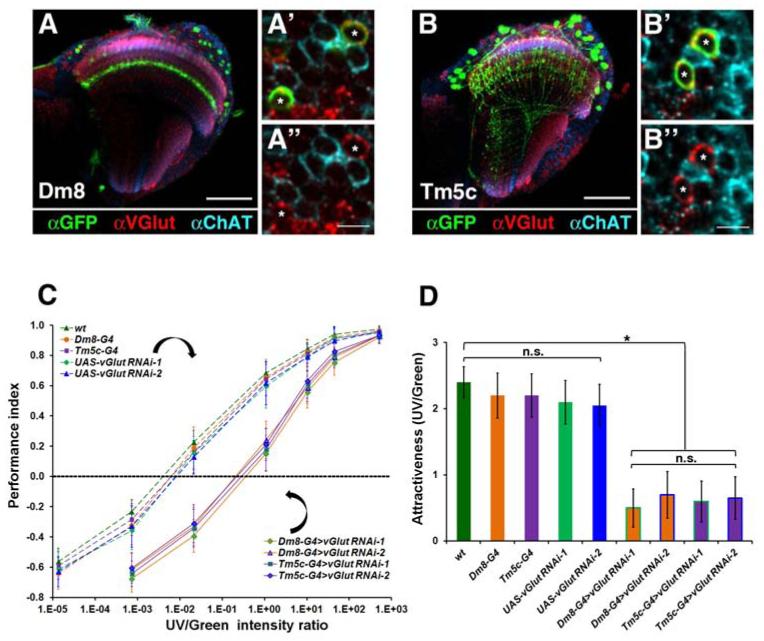
To further characterize the synapses between Dm8 and Tm5c, we determined their neurotransmitter usage. We manually isolated GFP-labeled Tm5c and Dm8 neurons and used RT-PCR to assess mRNA expression of diagnostic transporters or biosynthetic enzymes for all known fly neurotransmitters (Table S1). While RT-PCR revealed the presence of transcripts for all diagnostic genes in the optic lobes (Figure S4A), only the transcript of the vesicular glutamate transporter (VGlut) was detected in Dm8 and Tm5c (data not shown). We quantified VGlut transcript levels using real-time PCR and found that Dm8 and Tm5c expressed high levels of VGlut transcript (5,805 ± 1054 and 6,122 ± 1031 copies per cell, respectively), comparable to those of Rp49, a ribosomal protein (Dm8: 8,754 ± 345; Tm5c: 8,927 ± 717 copies per cell) (Figures S4B-S4E). The expression of VGlut protein in Dm8 and Tm5c was further confirmed by immunohistochemistry using anti-VGlut antibody (Figures 5A-5B″). In contrast, both Tm5a and Tm5b express choline acetyltransferase (ChAT) but not VGlut, as assessed by immunohistochemistry (Figure S5). Thus, Dm8 and Tm5c express VGlut and therefore are likely glutamatergic while Tm5a and Tm5b are likely cholinergic.
Figure 5. Glutamatergic Output of Dm8 and Tm5c Is Required for Normal UV Preference.
(A-B″) Dm8 and Tm5c express the vesicular glutamate transporter (VGlut). The expression of VGlut and choline acetyl transferase (ChAT) in Dm8 (A-A″) and Tm5c (B-B″) neurons was assessed using anti-VGlut (red) and anti-ChAT (cyan) antibodies in adult flies. Dm8 and Tm5c (asterisks) neurons were labeled with the mCD8GFP membrane marker (green) using the Dm8-Gal4 and Tm5c-Gal4 drivers, respectively. For both Dm8 and Tm5c, anti-GFP staining co-localized with anti-VGlut, but not anti-ChAT, staining. (A′, B′) are high magnification views of the medulla cortex in (A, B). (A″, B″), GFP staining was omitted from (A′, B′) for clarity.
Scale bar: 50 μm for A and B; 5 μm for A′ and B′.
(C, D) RNAi-mediated knockdown of VGlut in Dm8 or Tm5c neurons significantly reduced UV preference, as compared with wild-type and UAS-RNAi controls. Wild-type and various VGlut RNAi-knockdown flies were tested for spectral preference to UV light over green light as described in Figure 2. Dm8-G4 and Tm5c-G4 drivers were used to drive two different UAS-VGlut RNAi transgenes (VGlut RNAi-1 [Bloomington] and VGlut RNAi-2 [VDRC]) in Dm8 and Tm5c, respectively.
(D) Bar graph indicating the relative attractiveness of UV over green light (AttrUV/G) calculated from (C). n.s.: not significant (p>0.05); *: significant (p<0.05).
To determine whether VGlut function, hence the glutamatergic outputs, of Dm8 and Tm5c are required for UV preference, we knocked down VGlut transcripts in either Dm8 or Tm5c neurons by targeted expression of VGlut RNAi and examined the behavioral consequence. We found that RNAi-mediated knockdown of VGlut in Dm8 neurons significantly reduced UV preference (AttrUV/G = 0.5 ± 0.29 for Dm8>VGlut-RNAi-1 and 0.7 ± 0.35 for Dm8>VGlut-RNAi-2), as compared with their matched controls (AttrUV/G = 2.2 ± 0.33 for Tm5c-Gal4, 2.1 ± 0.33 for UAS-VGlut-RNAi-1 and 2.05 ± 0.32 for UAS-VGlut-RNAi-2; p<0.05) (Figures 5C and 5D). Similarly, RNAi-mediated knockdown of VGlut in Tm5c significantly reduced but did not completely abolish the UV preference seen in controls (AttrUV/G = 0.6 ± 0.31 for Tm5c>VGlut-RNAi-1 and 0.65 ± 0.32 for Tm5c>VGlut-RNAi-2 vs. AttrUV/G = 2.2 ± 0.33 for Tm5c-Gal4, 2.1 ± 0.33 for UAS-VGlut-RNAi-1 and 2.05 ± 0.32 for UAS-VGlut-RNAi-2). In contrast, expressing ChAT RNAi in either Dm8 or Tm5c failed to cause any UV preference defects (data not shown). These results suggested that the glutamatergic outputs of Dm8 and Tm5c are required for normal UV preference. We note that RNAi-mediated VGlut knockdown in either Dm8 or Tm5c caused somewhat weaker UV preference defects than those caused by TNT-mediated synaptic blockage. To determine whether partial efficacy of RNAi-knockdown might account for this difference, we quantified the VGlut transcript levels in Dm8 and Tm5c neurons expressing VGlut-RNAi. We found that expressing VGlut RNAi in Dm8 and Tm5c neurons reduced VGlut transcript levels by about 60% (VGlut transcript level for Dm8-Gal4>VGlut RNAi-1: 2,062 ± 259 and RNAi-2: 2,460 ± 498 copies per cell; Tm5c-Gal4>VGlut RNAi-1: 2011 ± 455 and RNAi-2: 2,233 ± 205 copies per cell; Figure S4B), without affecting Rp49 transcript level (Dm8-Gal4>VGlut RNAi-1: 8,622 ± 409 and RNAi-1: 8,433 ± 586 copies per cell; Tm5c-Gal4>VGlut RNAi-1: 8,719 ± 620 and RNAi-1: 8,583 ± 530 copies per cell; Figure S4D). Thus, the effects of RNAi knockdown are partial, consistent with its effects on UV preference behavior. We conclude that VGlut, and hence glutamate neurotransmission, is required in Dm8 and Tm5c for UV preference.
Tm5c Expresses Kainate Receptors to Receive Excitatory Glutamate Input
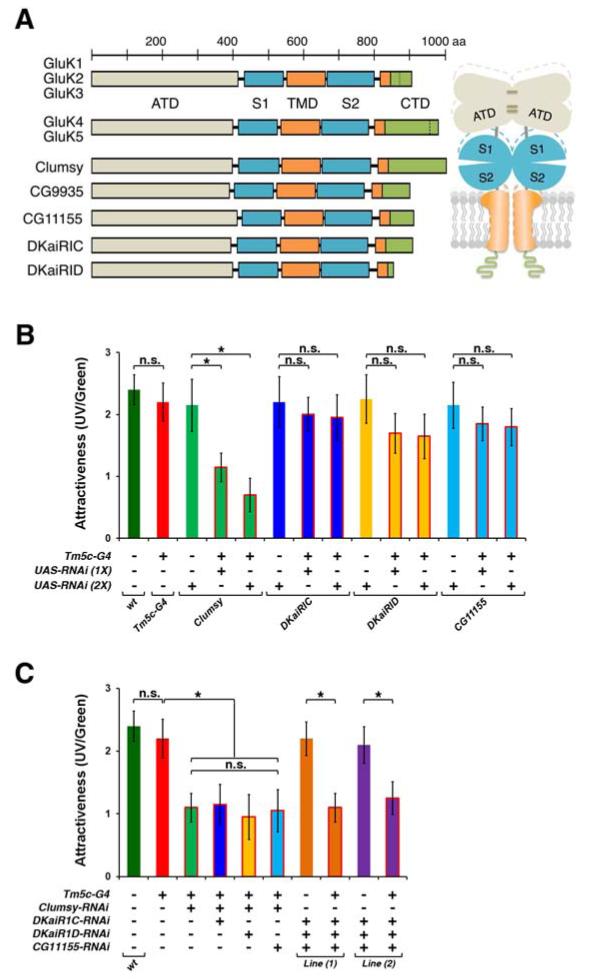
The preceding evidence suggested that Dm8 provides glutamatergic inputs to Tm5c in the UV preference pathway. To determine the nature of the Dm8->Tm5c connections, we extended our transcript profile analysis of Tm5c to include all known glutamate receptor subunits. The Drosophila genome encodes fifteen known glutamate-gated ionotropic receptors (iGluR), including three conserved classes (Kainate, AMPA, and NMDA types) of cation iGluR and one chloride channel (GluClα), as well as five metabotropic receptors (mGluR) (Table S1; Littleton and Ganetzky, 2000; Meinertzhagen and Lee, 2012). In addition, a large family of ionotropic receptors (IR), distantly related to iGluR, has been identified recently and at least three family members are expressed in the CNS (Abuin et al., 2011). We performed RT-PCR and confirmed their expression in adult flies (Figure S6A). Single cell transcript profiling revealed that Tm5c expresses four Kainate iGluR subunits, Clumsy, DKaiRIC, DKaiRID, and CG11155 (Figure 6A). By quantitative RT-PCR, we found that these iGluR were expressed at a modest level (Clumsy: 1904 ± 324; DKaiRIC: 2484 ± 368; DKaiRID 3078 ± 625; CG11155: 2155 ± 530 copies per cell; Figures S6B-S6I), as compared to that of VGlut. Insofar as Tm5c expressed Kainate-type iGluR but not GluClα or metabotropic receptors, we concluded that Tm5c receives fast excitatory glutamatergic inputs from Dm8.
Figure 6. The Kainate iGluR Clumsy Is Required in Tm5c Neurons for Normal UV Preference.
(A) Domain structures of the subunits of kainate ionotropic glutamate receptors in vertebrates (GluK1-5) and Drosophila (Clumsy, CG9935, CG11155, DKaiRIC, DKaiRID). The Drosophila kainate iGluR subunits share the same domain organization as those of vertebrates. Clumsy has a long cytoplasmic terminal domain (CTD), which shares limited sequence homology with that of the vertebrate receptor subunit GluK5. ATD: amino-terminal domain; S1 and S2: two parts of ligand binding domain; TMD: transmembrane domain; CTD: cytoplasmic terminal domain.
(B) Wild-type and various iGluR RNAi-knockdown flies were tested for spectral preference to UV over green light as described in Figure 2. Tm5c-Gal4 was used to drive RNAi expression of four kainate iGluRs subunits that are normally expressed in Tm5c neurons. RNAi-mediated knockdown of the Clumsy iGluR subunit (Tm5c-G4>Clumsy-RNAi [1X]), but not DKaiRIC, DKaiRID, or CG11155, caused a significant reduction of UV preference. The performance of wild-type flies and matched controls is shown for comparison. Increasing the copy number of UAS-Clumsy RNAi transgenes (Tm5c-G4>Clumsy-RNAi [2X]) further reduced flies’ UV preference. n.s.: not significant (p>0.05); *: significant (p<0.05).
(C) RNAi-knockdown any of the three subunits (i.e. DKaiRIC, DKaiRID and CG11155), in addition to Clumsy, caused no further reduction of UV preference, as compared with Clumsy RNAi knockdown. n.s.: not significant (p>0.05); *: significant (p<0.05). RNAi-knockdown of the three subunits, DKaiRIC, DKaiRID and CG11155, reduced UV preference close to the level caused by RNAi clumsy alone, suggesting these subunits are functionally redundant. n.s.: not significant (p>0.05); *: significant (p<0.05).
To determine whether Tm5c function requires iGluR for normal UV preference, we knocked down each iGluR subunit in Tm5c neurons by RNAi and examined the behavioral consequence. We found that knocking down Clumsy, but not the other three subunits, significantly reduced UV preference (AttrUV/G= 1.15 ± 0.23, p<0.05), as compared with wild-type or matched controls (Figures 6B and S7A). To increase the efficacy of RNAi-mediated knockdown, we increased the number of UAS-RNAi transgenes. We found that doubling Clumsy RNAi transgenes further reduced UV preference (AttrUV/G= 0.7 ± 0.27) to levels comparable to those caused by inactivating Tm5c (AttrUV/G = 0.4 ± 0.27). In contrast, Tm5c-Gal4 driving two copies of UAS-DKaiRIC, DKaiRID or CG11155 RNAi transgene failed to cause any significant UV preference deficit, as compared to their matched controls (Figures 6B and S7A). We quantified the efficacy of RNAi knockdown in Tm5c using quantitative RT-PCR and found no significant differences in the knockdown efficacy (Clumsy: 841 ± 103 [44.2% of wild-type], DKaiRIC: 1197 ± 175 [48.2% of wild-type], DKaiRID 1425 ± 231 [46.3% of wild-type], CG11155: 1065 ± 406 [49.4% of wild-type], Rp49: 8195 ± 614 [94.8% of wild-type] copies per cell; Figures S6B-S6I). These results suggest that the unique effect of Clumsy RNAi on UV preference was due to Clumsy’s function rather than RNAi knockdown efficacy.
To determine potential functional redundancy of iGluRs, we further knocked down these iGluRs in various combinations in Tm5c and examined the behavioral consequences. We found that RNAi-mediated knockdown of any one of the other three iGluRs in addition to Clumsy did not reduce UV preference below levels caused by Clumsy RNAi knockdown alone, suggesting that Clumsy does not compensate for the loss of either DKaiRIC, DKaiRID or CG11155 (AttrUV/G= 1.1 ± 0.32 for Clumsy+DKaiRIC; 1.0 ± 0.36 for Clumsy+DKaiRID; 1.05 ± 0.34 for Clumsy+CG1115; Figure 6C). Interestingly, simultaneous RNAi knockdown of these three iGluRs (DKaiRIC, DKaiRID, and CG11155) in Tm5c reduced UV preference (AttrUV/G= 1.1 ± 0.23 for DKaiRIC+DKaiRID+CG11155[1]; 1.25 ± 0.26 for DKaiRIC+DKaiRID+CG11155[2]; Figures 6C and S7B) close to the level caused by Clumsy RNAi knockdown alone, suggesting that these iGluRs are functionally redundant. We conclude that the kainate iGluR subunit Clumsy is required in Tm5c for normal UV preference while the other three iGluRs, DKaiRIC, DKaiRID and CG11155, play redundant roles in Tm5c.
DISCUSSION
Understanding how visual systems translate light impulses into adaptively tuned percepts to guide behavior is a central goal in neurobiology. The Drosophila visual system, with its amenability to genetic manipulation, has enabled increasingly deep investigation of the molecular and cellular basis of visual-driven behaviors, including the spectral preference for UV light examined here (Borst, 2009; Clark et al., 2013). UV spectral preference has previously been shown to require first-order interneurons in the visual medulla (i.e. the wide-field amacrine Dm8 neurons) that receive inputs from multiple UV-sensing photoreceptors (Gao et al., 2008). Here, we show that a subclass of Tm5 neurons, called Tm5c, receive excitatory glutamatergic input from Dm8 neurons through the kainite-type receptor, Clumsy. We demonstrated that glutamatergic signaling, both to and by Tm5c, is necessary for normal UV preference. Together our results define not only critical elements of the molecular and cellular machinery underlying UV preference, but also patterns of connectivity and information flow at the first several processing stations of this important visual circuit.
Establishing the Neuronal Connectivity and Synaptic Mechanisms Underlying UV Preference by Single-cell GRASP and Transcript Profiling
By sparse reconstruction of serial-section transmission electron microscopy (ssTEM), we previously identified Dm8 as the major postsynaptic partner for R7. In this study, we show that Tm5c interneurons are required for transducing Dm8’s signal to the lobula, a higher visual center, in the UV preference pathway. The thin and complex dendrites of Tm5c make them challenging to resolve by ssTEM, due to the limited axial resolution (~50 nm) of this method (Denk et al., 2012; Meinertzhagen and Lee, 2012; Takemura et al., 2013). To visualize Dm8->Tm5c and R8->Tm5c synapses, we therefore resorted to the GRASP technique. To differentiate synaptic contacts from mere membrane contacts and to visualize the spatial distribution of Dm8->Tm5c synapses, we adapted the GRASP method to permit single-cell identification of presumptive presynaptic neurons in which active zones were fluorescently tagged. By applying this single-cell GRASP method, we demonstrated that each Dm8 neuron has multiple synaptic contacts to one (or, at most a few) Tm5c neuron in the center of its dendritic field but sparse synaptic contacts in the periphery. The nature of synaptic signaling at the R8->Tm5c and Dm8->Tm5c contacts was established by single-cell transcript profiling and functional studies. Tm5c expresses the histamine-gated chloride channel Ort, and restoring Ort expression in Tm5c in an ort mutant background drove strong green preference. Dm8 expresses VGlut, and Tm5c correspondingly expresses glutamate-gated ionotropic receptors. RNAi-mediated knockdown of VGlut in Dm8, or the Clumsy iGluR in Tm5c, abolished UV preference. We believe that the approach taken here, which combines single-cell GRASP, transcript profiling, RNAi-mediated knockdown and behavioral assays, could be profitably applied to the dissection and characterization of other complex neural circuits.
Kainate Receptors Mediate Excitatory Glutamatergic Transmission in the UV Preference Circuit
Using single-cell transcript profiling, we identified four kainate-type glutamate receptor subunits (Clumsy, CG11155, DKaiRIC, and DKaiRID) expressed in Tm5c. These four iGluRs and CG9935 share sequence homology and domain structures with vertebrate kainate-type iGluRs (GluK1/2/3 and GluK4/5) (Figure 6A; Littleton and Ganetzky, 2000). RNAi-mediated knockdown further revealed that Clumsy is functionally required in Tm5c for UV preference. This is, to our knowledge, the first demonstration of kainate-type iGluR function in the Drosophila CNS. In vertebrates, functional kainate receptors assemble tetramerically as dimers of dimers; GluK1-3 are capable of forming functional homotetramers while GluK4/5 are obligatory heteromers (Sobolevsky et al., 2009; Kumar et al., 2011, Mayer, 2011). Fly kainate receptor subunits share with vertebrates’ the key hydrophobic resides at the dimerization interfaces, suggesting that they assemble in a similar way to the vertebrate receptors (Figure 6A). RNAi-knockdown of CG11155, DKaiRIC, or DKaiRID did not enhance UV preference defects caused by RNAi-knockdown of Clumsy. However, simultaneous RNAi-knockdown of all three iGluR subunits significantly reduced UV preference (Figure 6C), suggesting that they are functionally redundant. We thus suggest that Clumsy forms functional heterotetramers with one of the other kainate receptors in Tm5c to mediate UV preference. Further in vitro assembly and electrophysiological studies will be needed to determine the exact subunit composition of the functional iGluRs. Ionotropic glutamate receptors in flies have been exclusively studied at the neuromuscular junction, in part as a surrogate model for CNS glutamate synapses. Our functional identification of kainate-type iGluRs in the fly visual system, in combination with the robust UV preference behavior they mediate, opens the door to studying the assembly, function, and regulation of this important class of glutamate receptors in the Drosophila CNS.
A Hard-wired Pooling Circuit Is Superimposed on a Retinotopic Circuit to Mediate UV Preference
As the wide-field Dm8 neurons have no presynaptic sites or axonal projections outside of the external medulla, they depend on medulla projection neurons to transduce signals to higher visual centers. While all three subtypes of Tm5 neurons appear to be postsynaptic to Dm8 and therefore are capable of transducing the Dm8 signal to the lobula (Gao et al., 2008; Takemura et al., 2013), we show here that only Tm5c is functionally required for UV preference. Inactivating Tm5a/b or Dm8 in addition to Tm5c did not enhance UV preference defects, suggesting that Dm8’s function in UV preference is solely communicated through Tm5c. Tm5c differs from Tm5a/b in axonal morphology and neurotransmitter usage. We thus suggest that Tm5c has unique synaptic functions and/or targets in that visual compartment that account for its role in UV preference.
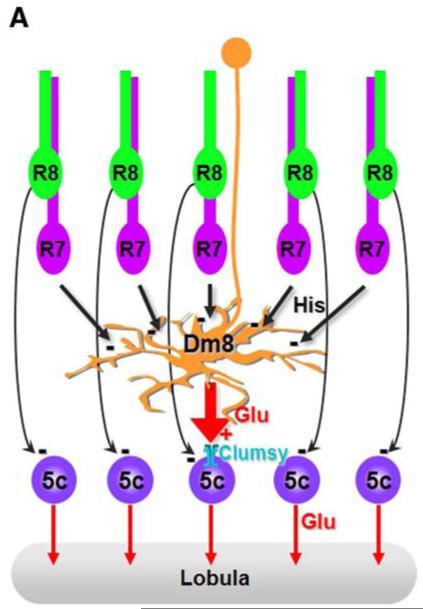
The spatial organization of the R7s->Dm8->Tm5c circuit suggests a neural pooling mechanism for UV preference. Each Dm8 amacrine neuron has a large dendritic field that receives ~16 R7 inputs (Gao et al., 2008), while a single Tm5c is present in most, if not all, medulla columns and receives direct retinotopic inputs from R8s (Figure 7). Our single-cell GRASP experiments reveal that each Dm8 forms multiple synaptic contacts with one, or at most a few, Tm5c neurons in the center of Dm8’s dendritic field but forms few synapses with Tm5c outside of the center. By pooling 16 R7 inputs to a single Tm5c, Dm8 could increase UV sensitivity by up to 16-fold at some cost in spatial resolution. It is interesting to note that the amplification magnitude of the R7s->Dm8->Tm5c circuit depends primarily on the size of the Dm8’s dendritic field, which is negatively regulated by R7-derived Activin during development: excess Activin reduces Dm8’s dendritic field size while lack of Activin enhances it (Ting and Lee, unpublished observation). It is thus tempting to speculate that size of the Dm8 arbor, and thus the trade off between UV sensitivity and spatial resolution, has been adjusted in the course of insect evolution to meet each insect’s ecological needs.
Figure 7. A Hard-wired Glutamatergic Pooling Circuit Mediates UV Preference.
Summary diagram of R7->Dm8->Tm5c pathway. The amacrine neuron Dm8 receives inhibitory histaminergic inputs from approximately 16 R7 photoreceptors and provides excitatory glutamatergic inputs to one (or at most a few) Tm5c projection neurons. Kainate iGluR receptors expressed in Tm5c are required for receiving glutamatergic inputs from Dm8 and for flies’ UV preference. In addition to the indirect pathway via Dm8, Tm5c neurons also receive direct photoreceptor inputs from R8.
In addition to the excitatory glutamate input from Dm8, Tm5c neurons also receive inhibitory histaminergic inputs directly from R8 photoreceptors. Thus, the R7s->Dm8->Tm5c pooling circuit is superimposed on the retinotopic circuit R8->Tm5c. Tm5c expresses Ort, and restoring Ort expression in Tm5c in various ort mutant backgrounds rescued green phototaxis. Thus, both direct (R8->Tm5c) and indirect (R7s->Dm8->Tm5c) pathways transduce sign-inverting signals to Tm5c and both pathways are capable of driving phototaxis. However, RNAi-mediated knockdown of ort in Tm5c, which prevents the reception of R8 inputs, did not affect normal UV preference (Figures 3F and 3G). This observation is consistent with our previous conclusions that the indirect pathway through R7s->Dm8, is both required and sufficient for optimal UV preference, at least under the test condition, and that multiple redundant pathways mediate green phototaxis (Gao et al., 2008). While the direct pathway is not involved in UV preference, it might play a role in true color vision. Notably, chloride ions are a known modulator for kainate receptor (Chaudhry et al., 2009) and the direct pathway signals through the histamine-gated chloride channel Ort. Given that multiple pathways function redundantly in true color vision, differentiating these possibilities must await single-unit electrophysiological recording and/or new genetic techniques to segregate their functions.
EXPERIMENTAL PROCEDURES
Fly Stocks and Transgenesis
Fly stocks were maintained on standard fruit fly medium at 23 °C-25 °C. Transgenic flies were generated using standard P-element or PhiC31-mediated transformation protocols by Rainbow Transgenic Flies, Inc. Fly stocks used in this study are described in the Supplemental Experimental Procedures.
Molecular Biology
Transgenic constructs were generated by general subcloning, PCR, and In-Fusion cloning. Detailed procedures are given in Supplemental Experimental Procedures.
Immunohistochemistry
Immunohistochemistry and confocal imaging was performed as described previously (Ting et al., 2007). Detailed procedures are given in Supplemental Experimental Procedures.
GFP Reconstitution Across Synaptic Partners (GRASP)
GRASP for detecting synaptic contacts between R7/R8 and Tm5c were performed in flies carrying Rh4-Gal4 Rh3-Gal4 ortC1a-LexADBD, OK371-dVP16AD UAS-syb::spGFP1-10 LexAop-spGFP11::CD4 and PanR8-Gal4 ortC1a-LexADBD, OK371-dVP16AD UAS-syb::spGFP1-10 LexAop-spGFP11::CD4, respectively. Flies were kept in a constant light condition for five days before dissection. The construction of UAS-syb::spGFP1-10 will be reported elsewhere.
GRASP for detecting membrane contacts between Dm8 and Tm5c was performed as described previously (Gordon and Scott, 2009), with the addition of a LexAop-HRP::CD2 transgene for labeling LexA expressing neurons.
Single-cell GRASP was carried out using the transgene LexAop>FRT-Stop-FRT> spGFP11::CD4::HA::T2A::Brp::mCherry, to express split-GFP (spGFP11::CD4::HA) and the active zone maker Brp::mCherry in single Dm8 neurons. The ribosomal skipping sequence T2A allows co-translational cleavage of the polyprotein (Diao and White, 2012). A brief heat shock (38°C for 3.5 min) was applied at the third larval instar to express flipase, which removes the FRT-Stop-FRT cassette and allows Dm8-LexA to drive the expression of both spGFP11::CD4::HA and Brp::mCherry. These flies also carried Tm5c-Gal4 UAS-spGFP1-10::CD4 UAS-HRP::CD2 transgenes, which label all Tm5c neurons with the other split-GFP component (spGFP1-10::CD4) and the HRP::CD2 membrane marker. Functional GFP molecules were reconstituted at the membrane contacts between Tm5c and single Dm8 neurons and detected by native GFP fluorescence (GRASP signal). The active zone marker labeled the presynaptic sites of Dm8 to differentiate synaptic contacts (GRASP and Brp::mCherry) from mere membrane contacts (GRASP alone).
To visualize the morphologies of Dm8, Tm5c and photoreceptors, in addition to the GRASP and Brp::mCherry signals, we carried out imaging in two consecutive steps. First, we visualized single Dm8 neurons by HA-staining, and visualized GRASP and Brp::mCherry by their native fluorescence signals. Second, we visualized photoreceptors and Tm5c by immunolabeling the same brain sample with 24B10 and anti-HRP antibodies, respectively. Two confocal image stacks from these two steps were superimposed using Imaris software (Bitplane).
UV/Green Spectral Preference and Phototaxis Assays
The phototaxis assay was carried out by using a T-maze device made of transparent plexiglass as previously described (Gao et al., 2008). Student’s t test was used for comparisons between two groups. Comparisons between multiple groups were made by one-way ANOVA with Tukey post hoc test. Asterisks indicate levels of significant differences (*p < 0.05; **p < 0.01).
Head Yaw Optomotor Response Assay
Rotary visual stimuli and corresponding head yaw response measurements were conducted with an automated system. The basis for the assay custom instrumentation, including control, video acquisition, and processing software, development has been described (Rister et al., 2007; Gao et al., 2008). Detailed procedures are given in Supplemental Experimental Procedures.
Single-Cell Transcript Profiling
Single Dm8, Tm5b, and Tm5c neurons labeled with GFP were dissociated from adult medulla and single GFP-positive cells were isolated using a custom-made microcapillary system as previously described (Takemura et al., 2011). Single-cell PCR analyses were carried out to determine the presence of specific transcripts (Table S1) as described previously (Takemura et al., 2011). Real-time PCR was carried out to quantify the transcript levels of VGlut, Clumsy, DKaiRIC, DKaiRID and CG11155 in control and RNAi-knockdown flies. Rp49, which encodes a ribosomal protein, was used as an internal reference gene. PCR primer sequences are provided in the Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Shinya Takemura and Dmitri Chklovskii for communicating results prior to publication and Tzumin Lee, Kristin Scott, Stephan Sigrist, Aaron DiAntonio, Gerald Rubin, and Claude Desplan for providing critical reagents, Mark Mayer and Alan Hinnebusch for helpful discussion. This work was supported by the Intramural Research Programs of the NIH, Eunice Kennedy Shriver National Institute of Child Health and Human Development (grant Z01-HD008776 to C.-H.L), National Institute of Mental Health (Project 1ZIA-MH-002800-07, B.H.W.) and Center for Information Technology (R.P. and T.P.). L.J.M. and M.G.’s research was supported by NIH grants (RC1-NS069014 and R01-NS076774) to Charles Zuker.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information includes seven figures, one table, and Supplemental Experimental Procedures.
REFERECNES
- Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, Benton R. Functional architecture of olfactory ionotropic glutamate receptors. Neuron. 2011;69:44–60. doi: 10.1016/j.neuron.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst A. Drosophila’s view on insect vision. Curr. Biol. 2009;19:R36–R47. doi: 10.1016/j.cub.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Chaudhry C, Plested AJ, Schuck P, Mayer ML. Energetics of glutamate receptor ligand binding domain dimer assembly are modulated by allosteric ions. Proc. Natl. Acad. Sci. USA. 2009;106:12329–12334. doi: 10.1073/pnas.0904175106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DA, Freifeld L, Clandinin TR. Mapping and cracking sensorimotor circuits in genetic model organisms. Neuron. 2013;78:583–595. doi: 10.1016/j.neuron.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane J. Imaginal behavior of a Trinidad butterfly, Heliconius erato hydara Hewitson, with a special reference to the social use of color. Zoologica. 1955;40:167–196. [Google Scholar]
- Denk W, Briggman KL, Helmstaedter M. Structural neurobiology: missing link to a mechanistic understanding of neural computation. Nat. Rev. Neurosci. 2012;13:351–358. doi: 10.1038/nrn3169. [DOI] [PubMed] [Google Scholar]
- Diao F, White BH. A novel approach for directing transgene expression in Drosophila: T2A-Gal4 in-frame fusion. Genetics. 2012;190:1139–1144. doi: 10.1534/genetics.111.136291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg EH, Vanhoven MK, Bendesky A, Wang G, Fetter RD, Shen K, Bargmann CI. GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron. 2008;57:353–363. doi: 10.1016/j.neuron.2007.11.030. [DOI] [PubMed] [Google Scholar]
- Fischbach KF, Dittrich AP. The optic lobe of Drosophila melanogaster. I. A Golgi analysis of wild-type structure. Cell Tissue Res. 1989;258:441–475. [Google Scholar]
- Gao SY, Takemura SY, Ting CY, Huang SL, Lu ZY, Luan HJ, Rister J, Thum AS, Yang ML, Hong ST, Wang JW, Odenwald WF, White BH, Meinertzhagen IA, Lee CH. The neural substrate of spectral preference in Drosophila. Neuron. 2008;60:328–342. doi: 10.1016/j.neuron.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon MD, Scott K. Motor control in a Drosophila taste circuit. Neuron. 2009;61:373–384. doi: 10.1016/j.neuron.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyret J, Pfaff M, Raguso RA, Kelber A. Why do Manduca sexta feed from white flowers? Innate and learnt colour preferences in a hawkmoth. Naturwissenschaften. 2008;95:569–576. doi: 10.1007/s00114-008-0350-7. [DOI] [PubMed] [Google Scholar]
- Hardie RC. Electrophysiological analysis of fly retina. I: Comparative properties of R1-6 and R7 and 8. J. Comp. Physiol. 1979;129:19–33. [Google Scholar]
- Heisenberg M, Buchner E. Role of retinula cell-types in visual behavior of Drosophila melanogaster. J. Comp. Physiol. 1977;117:127–162. [Google Scholar]
- Hu KG, Stark WS. Specific receptor input into spectral preference in Drosophila. J. Comp. Physiol. 1977;121:241–252. [Google Scholar]
- Ilse D, Vaidya VG. Spontaneous feeding response to colours in Papilio demoleus L. Proc. Ind. Acad. Sci. 1956;43:23–31. [Google Scholar]
- Kelber A. Colour learning in the hawkmoth Macroglossum stellatarum. J. Exp. Biol. 1996;199:1127–1131. doi: 10.1242/jeb.199.5.1127. [DOI] [PubMed] [Google Scholar]
- Kitamoto T. Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J. Neurobiol. 2001;47:81–92. doi: 10.1002/neu.1018. [DOI] [PubMed] [Google Scholar]
- Koshitaka H, Kinoshita M, Vorobyev M, Arikawa K. Tetrachromacy in a butterfly that has eight varieties of spectral receptors. Proc. Biol. Sci. 2008;275:947–954. doi: 10.1098/rspb.2007.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J, Schuck P, Mayer ML. Structure and assembly mechanism for heteromeric kainate receptors. Neuron. 2011;71:319–331. doi: 10.1016/j.neuron.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littleton JT, Ganetzky B. Ion channels and synaptic organization: analysis of the Drosophila genome. Neuron. 2000;26:35–43. doi: 10.1016/s0896-6273(00)81135-6. [DOI] [PubMed] [Google Scholar]
- Luan H, Peabody NC, Vinson CR, White BH. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron. 2006;52:425–436. doi: 10.1016/j.neuron.2006.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML. Structure and mechanism of glutamate receptor ion channel assembly, activation and modulation. Curr. Opin. Neurobiol. 2011;21:283–290. doi: 10.1016/j.conb.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinertzhagen IA. The organization of perpendicular fibre pathways in the insect optic lobe. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1976;274:555–594. doi: 10.1098/rstb.1976.0064. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA, Lee CH. The genetic analysis of functional connectomics in Drosophila. Adv. Genet. 2012;80:99–151. doi: 10.1016/B978-0-12-404742-6.00003-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menne D, Spatz HC. Color vision in Drosophila melanogaster. J. Comp. Physiol. 1977;114:301–312. [Google Scholar]
- Menzel R. Spectral sensitivity and colour vision in invertebrates. In: Autrum H, editor. Vision in Invertebrates (Handbook of Sensory Physiology. VII/6A. Springer Verlag; New York: 1979. pp. 503–580. [Google Scholar]
- Menzel R, Greggers U. Natural phototaxis and its relationship to color vision in honeybees. J. Comp. Physiol. 1985;157:311–321. [Google Scholar]
- Mikeladze-Dvali T, Desplan C, Pistillo D. Flipping coins in the fly retina. Curr. Top. Dev. Biol. 2005;69:1–15. doi: 10.1016/S0070-2153(05)69001-1. [DOI] [PubMed] [Google Scholar]
- Morante J, Desplan C. The color-vision circuit in the medulla of Drosophila. Curr. Biol. 2008;18:553–565. doi: 10.1016/j.cub.2008.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orger MB, Baier H. Channeling of red and green cone inputs to the zebrafish optomotor response. Vis. Neurosci. 2005;22:275–281. doi: 10.1017/S0952523805223039. [DOI] [PubMed] [Google Scholar]
- O’Tousa JE, Baehr W, Martin RL, Hirsh J, Pak WL, Applebury ML. The Drosophila ninaE gene encodes an opsin. Cell. 1985;40:839–850. doi: 10.1016/0092-8674(85)90343-5. [DOI] [PubMed] [Google Scholar]
- Rister J, Pauls D, Schnell B, Ting CY, Lee CH, Sinakevitch I, Morante J, Strausfeld NJ, Ito K, Heisenberg M. Dissection of the peripheral motion channel in the visual system of Drosophila melanogaster. Neuron. 2007;56:155–170. doi: 10.1016/j.neuron.2007.09.014. [DOI] [PubMed] [Google Scholar]
- Salcedo E, Huber A, Henrich S, Chadwell LV, Chou WH, Paulsen R, Britt SG. Blue- and green-absorbing visual pigments of Drosophila: ectopic expression and physiological characterization of the R8 photoreceptor cell-specific Rh5 and Rh6 rhodopsins. J. Neurosci. 1999;19:10716–10726. doi: 10.1523/JNEUROSCI.19-24-10716.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid A, Hallermann S, Kittel RJ, Khorramshahi O, Frolich AM, Quentin C, Rasse TM, Mertel S, Heckmann M, Sigrist SJ. Activity-dependent site-specific changes of glutamate receptor composition in vivo. Nat. Neurosci. 2008;11:659–666. doi: 10.1038/nn.2122. [DOI] [PubMed] [Google Scholar]
- Schnaitmann C, Vogt K, Triphan T, Tanimoto H. Appetitive and aversive visual learning in freely moving Drosophila. Front. Behav. Neurosci. 2010;4:10. doi: 10.3389/fnbeh.2010.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney ST, Broadie K, Keane J, Niemann H, O’Kane CJ. Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron. 1995;14:341–351. doi: 10.1016/0896-6273(95)90290-2. [DOI] [PubMed] [Google Scholar]
- Takemura SY, Lu Z, Meinertzhagen IA. Synaptic circuits of the Drosophila optic lobe: the input terminals to the medulla. J. Comp. Neurol. 2008;509:493–513. doi: 10.1002/cne.21757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura SY, Karuppudurai T, Ting CY, Lu Z, Lee CH, Meinertzhagen IA. Cholinergic circuits integrate neighboring visual signals in a Drosophila motion detection pathway. Curr. Biol. 2011;21:2077–2084. doi: 10.1016/j.cub.2011.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura SY, Bharioke A, Lu Z, Nern A, Vitaladevuni S, Rivlin PK, et al. A visual motion detection circuit suggested by Drosophila connectomics. Nature. 2013;500:175–181. doi: 10.1038/nature12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting CY, Herman T, Yonekura S, Gao S, Wang J, Serpe M, O’Connor MB, Zipursky SL, Lee CH. Tiling of R7 axons in the Drosophila visual system is mediated both by transduction of an activin signal to the nucleus and by mutual repulsion. Neuron. 2007;56:793–806. doi: 10.1016/j.neuron.2007.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss MR. Innate colour preferences and flexible colour learning in the pipevine swallowtail. Anim. Behav. 1997;53:1043–1052. [Google Scholar]
- Wong AM, Wang JW, Axel R. Spatial representation of the glomerular map in the Drosophila protocerebrum. Cell. 2002;109:229–241. doi: 10.1016/s0092-8674(02)00707-9. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Desplan C, Heisenberg M. Contribution of photoreceptor subtypes to spectral wavelength preference in Drosophila. Proc. Natl. Acad. Sci. USA. 2010;107:5634–5639. doi: 10.1073/pnas.0809398107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M, Salmon M, Forward R. Visual wavelength discrimination by the loggerhead turtle, Caretta caretta. Biol. Bull. 2012;222:46–55. doi: 10.1086/BBLv222n1p46. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.