Abstract
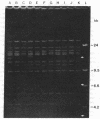
Examination of the genomes of 10 white-pock variants of cowpox virus strain Brighton red (CPV-BR) revealed that 9 of them had lost 32 to 38 kilobase pairs (kbp) from their right-hand ends and that the deleted sequences had been replaced by inverted copies of regions from 21 to 50 kbp long from the left-hand end of the genome. These variants thus possess inverted terminal repeats (ITRs) from 21 to 50 kbp long; all are longer than the ITRs of CPV-BR (10 kbp). The 10th variant is a simple deletion mutant that has lost the sequences between 32 and 12 kbp from the right-hand end of the genome. The limits of the inner ends of the observed deletions (between 32 and 38 kbp from the right-hand end of the CPV-BR genome) appear to be defined by the location of the nearest essential gene on the one hand and the location of the gene that encodes "pock redness" on the other. The genomes of the deletion/duplication white-pock variants appear to have been generated either by single crossover recombinational events between two CPV-BR genomes aligned in opposite directions or by the nonreciprocal transfer of genetic information. The sites where such recombination/transfer occurred were sequenced in four variants. In all of them, the sequences adjacent to such sites show no sequence homology or any other unusual structural feature. The analogous sites at the internal ends of the two ITRs of CPV-BR also were sequenced and also show no unusual features. It is likely that the ITRs of CPV-BR and of its white-pock variants, and probably those of other orthopox-virus genomes, arise as a result of nonhomologous recombination or by random nonreciprocal transfer of genetic information.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson R. A., Kato S., Camerini-Otero R. D. A pattern of partially homologous recombination in mouse L cells. Proc Natl Acad Sci U S A. 1984 Jan;81(1):206–210. doi: 10.1073/pnas.81.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archard L. C., Mackett M., Barnes D. E., Dumbell K. R. The genome structure of cowpox virus white pock variants. J Gen Virol. 1984 May;65(Pt 5):875–886. doi: 10.1099/0022-1317-65-5-875. [DOI] [PubMed] [Google Scholar]
- Archard L. C., Mackett M. Restriction endonuclease analysis of red cowpox virus and its white pock variant. J Gen Virol. 1979 Oct;45(1):51–63. doi: 10.1099/0022-1317-45-1-51. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay P. K., Watanabe S., Temin H. M. Recombination of transfected DNAs in vertebrate cells in culture. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3476–3480. doi: 10.1073/pnas.81.11.3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroudy B. M., Moss B. Sequence homologies of diverse length tandem repetitions near ends of vaccinia virus genome suggest unequal crossing over. Nucleic Acids Res. 1982 Sep 25;10(18):5673–5679. doi: 10.1093/nar/10.18.5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroudy B. M., Venkatesan S., Moss B. Incompletely base-paired flip-flop terminal loops link the two DNA strands of the vaccinia virus genome into one uninterrupted polynucleotide chain. Cell. 1982 Feb;28(2):315–324. doi: 10.1016/0092-8674(82)90349-x. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drillien R., Koehren F., Kirn A. Host range deletion mutant of vaccinia virus defective in human cells. Virology. 1981 Jun;111(2):488–499. doi: 10.1016/0042-6822(81)90351-2. [DOI] [PubMed] [Google Scholar]
- Esposito J. J., Cabradilla C. D., Nakano J. H., Obijeski J. F. Intragenomic sequence transposition in monkeypox virus. Virology. 1981 Mar;109(2):231–243. doi: 10.1016/0042-6822(81)90495-5. [DOI] [PubMed] [Google Scholar]
- FENNER F. The biological characters of several strains of vaccinia, cowpox and rabbitpox viruses. Virology. 1958 Jun;5(3):502–529. doi: 10.1016/0042-6822(58)90042-4. [DOI] [PubMed] [Google Scholar]
- GEMMELL A., FENNER F. Genetic studies with mammalian poxviruses. III. White (u) mutants of rabbitpox virus. Virology. 1960 May;11:219–235. doi: 10.1016/0042-6822(60)90063-5. [DOI] [PubMed] [Google Scholar]
- JOKLIK W. K. The purification fo four strains of poxvirus. Virology. 1962 Sep;18:9–18. doi: 10.1016/0042-6822(62)90172-1. [DOI] [PubMed] [Google Scholar]
- Kopchick J. J., Stacey D. W. Selective ligation of DNA molecules following microinjection. J Biol Chem. 1983 Oct 10;258(19):11528–11536. [PubMed] [Google Scholar]
- Lu A. L., Clark S., Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackett M., Archard L. C. Conservation and variation in Orthopoxvirus genome structure. J Gen Virol. 1979 Dec;45(3):683–701. doi: 10.1099/0022-1317-45-3-683. [DOI] [PubMed] [Google Scholar]
- Marko M. A., Chipperfield R., Birnboim H. C. A procedure for the large-scale isolation of highly purified plasmid DNA using alkaline extraction and binding to glass powder. Anal Biochem. 1982 Apr;121(2):382–387. doi: 10.1016/0003-2697(82)90497-3. [DOI] [PubMed] [Google Scholar]
- McFadden G., Dales S. Biogenesis of poxviruses: mirror-image deletions in vaccinia virus DNA. Cell. 1979 Sep;18(1):101–108. doi: 10.1016/0092-8674(79)90358-1. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Moyer R. W., Graves R. L., Rothe C. T. The white pock (mu) mutants of rabbit poxvirus. III. Terminal DNA sequence duplication and transposition in rabbit poxvirus. Cell. 1980 Nov;22(2 Pt 2):545–553. doi: 10.1016/0092-8674(80)90364-5. [DOI] [PubMed] [Google Scholar]
- Moyer R. W., Graves R. L. The mechanism of cytoplasmic orthopoxvirus DNA replication. Cell. 1981 Dec;27(2 Pt 1):391–401. doi: 10.1016/0092-8674(81)90422-0. [DOI] [PubMed] [Google Scholar]
- Moyer R. W., Rothe C. T. The white pock mutants of rabbit poxvirus. I. Spontaneous host range mutants contain deletions. Virology. 1980 Apr 15;102(1):119–132. doi: 10.1016/0042-6822(80)90075-6. [DOI] [PubMed] [Google Scholar]
- Nevins J. R., Joklik W. K. Isolation and properties of the vaccinia virus DNA-dependent RNA polymerase. J Biol Chem. 1977 Oct 10;252(19):6930–6938. [PubMed] [Google Scholar]
- Panicali D., Davis S. W., Mercer S. R., Paoletti E. Two major DNA variants present in serially propagated stocks of the WR strain of vaccinia virus. J Virol. 1981 Mar;37(3):1000–1010. doi: 10.1128/jvi.37.3.1000-1010.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickup D. J., Bastia D., Joklik W. K. Cloning of the terminal loop of vaccinia virus DNA. Virology. 1983 Jan 15;124(1):215–217. doi: 10.1016/0042-6822(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Pickup D. J., Bastia D., Stone H. O., Joklik W. K. Sequence of terminal regions of cowpox virus DNA: arrangement of repeated and unique sequence elements. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7112–7116. doi: 10.1073/pnas.79.23.7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Wilson J. H., Berget P. B., Pipas J. M. Somatic cells efficiently join unrelated DNA segments end-to-end. Mol Cell Biol. 1982 Oct;2(10):1258–1269. doi: 10.1128/mcb.2.10.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittek R., Moss B. Tandem repeats within the inverted terminal repetition of vaccinia virus DNA. Cell. 1980 Aug;21(1):277–284. doi: 10.1016/0092-8674(80)90135-x. [DOI] [PubMed] [Google Scholar]