Abstract
Streptococcus pyogenes (the group A streptococcus, GAS) is a Gram-positive bacterium responsible for a wide spectrum of diseases ranging from mild superficial infections (pharyngitis, impetigo) to severe often life-threatening invasive diseases (necrotizing fasciitis, streptococcal toxic shock syndrome) in humans. This unit describes molecular techniques for the genetic manipulation of S. pyogenes with detailed protocols for transformation, gene disruption, allelic exchange, transposon mutagenesis, and genetic complementation.
Keywords: Streptococcus pyogenes, plasmid, genomic DNA, transformation, transposon mutagenesis, genetic complementation, mariner
INTRODUCTION
This unit describes methods used for the genetic manipulation of Streptococcus pyogenes (the Group A Streptococcus, GAS). S. pyogenes is a strict human pathogen generally causing mild self-limiting infections of the upper respiratory tract (pharyngitis) or skin (impetigo). GAS has also re-emerged as a significant invasive pathogen of deeper tissues, leading to life-threatening diseases such as necrotizing fasciitis (flesh-eating disease) and streptococcal toxic shock syndrome (STSS). GAS strains vary widely in terms of antibiotic resistance, clinical severity and transmission rate. There are greater than 180 different serotypes of GAS based on the variable surface M protein (M-types) or its corresponding gene sequence (emm-types), and the serotype correlates with the site of infection at the throat or skin (Bessen, 2009).
GAS is a facultative anaerobic microorganism and is optimally grown at 37°C in 5% CO2 on TSA blood agar plates or in sealed screw-cap tubes in rich Todd-Hewitt liquid medium (for more information, see UNIT 9D.2: Growth and Maintenance of Streptococcus pyogenes). S. pyogenes is a member of the lactic acid bacteria and is homofermentative for lactic acid production from glucose fermentation.
Detailed methods will be presented for isolation of GAS genomic DNA (Basic Protocol 1), GAS transformation (Basic Protocol 2), gene disruption via plasmid integration (Basic Protocols 3 and 4), allelic exchange (Basic Protocol 5), current methods of transposon mutagenesis in GAS (Basic Protocols 6, 7, and 8), and genetic complementation (Basic Protocol 9).
CAUTION: GAS is a Biosafety Level 2 (BSL-2) pathogen. Follow all appropriate guidelines and regulations for the use and handling of pathogenic microorganisms as described in the Biosafety in Microbiological and Biomedical Laboratories (BMBL) manual from the Centers for Disease Control (CDC).
CAUTION: GAS remains sensitive to penicillin and this is a preferred treatment for infection in clinical settings. Therefore, the use of markers conferring resistance to β-lactam antibiotics for GAS genetic manipulations is both unethical and prohibited. Genes typically used for antibiotic selection during GAS genetic manipulations are: aad9 (spectinomycin resistance, Spr), cat194 (chloramphenicol resistance, Cmr), aphA3 (kanamycin resistance, Kmr) or ermC (erythromycin resistance, Emr).
NOTE: GAS serotypes and strains vary widely in terms of virulence as well as ease of genetic manipulation. This variability must be taken into account when studying GAS pathogenesis at a molecular level, as it will often influence the strains used and the available genetics. Typically, GAS laboratory strains (e.g., M49 NZ131 and M6 JRS4) are easy to manipulate genetically, but generally fail to produce strong disease phenotypes in models of GAS infection. In contrast, most clinical GAS isolates (e.g., M1T1 MGAS5005, M1T1 5448, M3 MGAS315) are more virulent in humans, and animal models, yet are generally harder to manipulate genetically.
BASIC PROTOCOL 1: LARGE-SCALE EXTRACTION OF GENOMIC DNA FROM S. PYOGENES (GAS) 1
This protocol describes an extended large-scale extraction of genomic DNA (gDNA) from a GAS liquid culture (ca. 18 h) adapted from Caparon and Scott (Caparon and Scott, 1991), resulting in gDNA suitable for downstream procedures requiring high yield and high quality (e.g., Southern blotting, library generation).
Materials
Todd-Hewitt Yeast (THY) broth for GAS growth (see recipe)
2 M glycine solution (see recipe)
Viable GAS isolated colony on the appropriate agar plate (UNIT 9D.2)
Sterile 250 ml Nalgene centrifugation bottles with inserts and screw caps
10 mM Tris pH 8 solution (see recipe)
Solution I (see recipe)
Lysozyme solution (see recipe)
Solution II (see recipe)
RNase A solution (see recipe)
Proteinase K solution (see recipe)
TE-saturated phenol
Phenol/chloroform/isoamyl alcohol (25:24:1)
Chloroform/isoamyl alcohol (24:1)
3 M sodium acetate (see recipe)
100% ethanol
Refrigerated centrifuges
Temperature-controlled (37°C) shaking water bath
Temperature-controlled (55°C) water bath
Spectrophotometer
Equipment for agarose gel electrophoresis
Inoculate and prepare culture
-
1
Inoculate 100 ml of THY broth containing 20 mM glycine and appropriate antibiotic, if needed, in a sealed 250 ml screw cap flask with an isolated GAS colony taken from an appropriate agar plate using sterile technique. Incubate static overnight at the proper temperature (30°C or 37°C).
Most GAS strains grow best at 37°C in 5% CO2 or in a sealed tube or flask. The addition of free glycine acts to reduce cell wall cross-bridges and make GAS easier to lyse.
Harvest and wash GAS cells
-
2
Transfer cells to a 250 ml centrifuge bottle and centrifuge 10 min at 10,000 x g, 4°C. Discard the supernatant.
-
3
Wash the cell pellet with 10 ml of 10 mM Tris pH 8.
-
4
Collect cells by centrifuging for 10 min at 10,000 x g, 4°C. Discard the supernatant.
Cell lysis
-
5
Resuspend the cells in 5.7 ml of Solution I, then transfer the suspension into a 50 ml polypropylene conical tube.
Polypropylene tubes are required since organic extractions will be performed. -
6
Add 3 ml of freshly prepared Lysozyme solution, then incubate for 2 h at 37°C with vigorous agitation in a shaking water bath.
-
7
Collect cells for 10 min at 10,000 x g, 4°C, then discard the supernatant.
-
8
Resuspend the cell pellet in 5.7 ml of Solution II, then incubate 15 min at 37°C with vigorous agitation in a shaking water bath.
-
9
Add 200 μl of RNase A solution and 130 μl of Proteinase K solution, then incubate for 1 h at 55°C. Invert tube often.
-
10
Transfer the lysate into a 15 ml polypropylene conical tube.
Genomic DNA extraction
-
11
Add 10 ml of TE-saturated phenol and invert several times to mix well (do not vortex), then centrifuge for 15 min at 10,000 x g, 4°C.
After centrifugation, two phases form, separated by a white interface. The aqueous phase (on top) contains the gDNA. -
12
Carefully transfer the aqueous phase, without disturbing the white interface, into a new 15 ml polypropylene conical tube.
-
13
Add 10 ml of phenol/chloroform/IAA solution and invert several times (do not vortex) to mix well, then centrifuge for 15 min at 10,000 x g, 4°C.
-
14
Transfer the aqueous phase (top) without disturbing the interface into a new 15 ml polypropylene conical tube, and repeat step 13 several times until the interface clears.
Initial extractions can be difficult due to the viscosity of the aqueous phase and the weak consistency of the interface. Do not disturb the interface during each extraction, and loss of volume of the aqueous phase is part of the process. -
15
Add 10 ml of chloroform/IAA (24:1) solution and invert several times to mix well (do not vortex), then centrifuge for 15 min at 10,000 x g, 4°C.
-
16
Transfer the aqueous phase (top) without disturbing the interface into a new 50 ml polypropylene conical tube. Bring volume to 10 ml with sterile distilled water.
Genomic DNA precipitation and purification
-
17
Add 1 ml of 3 M sodium acetate solution and 20 ml of 100% ethanol. Mix by inverting tube several times (do not vortex) and incubate the tube overnight at −20°C.
-
18
Centrifuge for 25 min at 10,000 x g, 4°C. Discard supernatant. Air-dry the DNA pellet. Resuspend the gDNA in 500 μl of sterile distilled water by pipetting (do not vortex). Store at 4°C.
Quality control
-
19
Check the quality of the gDNA both by agarose gel electrophoresis and concentration by spectrophotometry (OD260/280 = 1.8; 1 OD260 = 50 ug/ml).
Electrophoresis should show a single large band (>15 kb) with limited degradation. If the protein content is too high (poor restriction digestion), then return to step 11 and repeat organic extraction steps.
ALTERNATE PROTOCOL 1: MODERATE YIELD EXTRACTION OF GENOMIC DNA FROM S. PYOGENES (GAS)
This protocol describes the use of the MasterPure Complete DNA and RNA Purification Kit (Epicentre) adapted for rapid (ca. 4 h) extraction of GAS gDNA suitable for downstream procedures requiring moderate yield and high quality (e.g., Southern Blotting, library generation, arbitrary-primed-PCR [AP-PCR]).
NOTE: Kits for genomic DNA isolation are available from different manufacturers; however, optimization and troubleshooting steps might be required.
Additional materials (also see Basic Protocol 1)
MasterPure Complete DNA and RNA Purification Kit (Epicentre, Cat. No. MC85200)
Temperature-controlled thermomixer (Eppendorf, Cat. No. 5360 000.011)
Temperature-controlled water-bath
Isopropanol
70% ethanol
Inoculate and prepare culture
-
1
Inoculate 10 ml of THY broth containing 20 mM glycine and appropriate antibiotic, if needed, in a 15 ml conical tube with an isolated GAS colony taken from an appropriate agar plate using sterile technique. Incubate overnight at the proper temperature (30°C or 37°C).
Most GAS strains grow best at 37°C in 5% CO2 or in a sealed tube or flask.
Harvest GAS cells
-
2
Collect cells by centrifugation 10 min at 10,000 x g, 4°C. Discard the supernatant.
GAS cell lysis and genomic DNA purification
-
3
Follow the manufacturer’s protocol for genomic DNA isolation of Gram-positive bacteria using MasterPure Complete DNA and RNA Purification Kit.
Quality control
-
4
Check the quality of the gDNA both by agarose gel electrophoresis and concentration by spectrophotometry (OD260/280 = 1.8).
Electrophoresis should show a single large band (>15 kb) with limited degradation.
ALTERNATE PROTOCOL 2: QUICK EXTRACTION OF GENOMIC DNA FROM S. PYOGENES (GAS)
This protocol describes a rapid method (< 2 h) for GAS gDNA isolation using the FastDNA Spin Kit (MP Biomedicals). The yield and quality, although adequate for PCR, is not suitable for downstream procedures requiring intact, high quality gDNA.
Additional materials (also see Basic Protocol 1)
FastDNA Spin Kit (MP Biomedicals, Cat. No. 116540600)
FastPrep Cell Disruptor (MP Biomedicals, Cat. No. 116004500)
Inoculate and prepare culture
-
1
Inoculate 40 ml of THY broth containing 20 mM glycine and the appropriate antibiotic, if needed, in 50 ml conical tube with an isolated GAS colony taken from an appropriate agar plate using sterile technique. Incubate overnight at the proper temperature.
Most GAS strains grow best at 37°C in 5% CO2 or in sealed tube or flask.
Harvest and wash GAS cells
-
2
Pellet cells in same tube by centrifugation (6,000 x g, 20 min, 4°C).
-
3
Resuspend cells in 100 μl of distilled water.
GAS cell disruption and Genomic DNA extraction
-
4
Follow the manufacturer’s protocol for genomic DNA isolation of Gram-positive bacteria using the FastDNA Spin Kit and a FastPrep Cell Disruptor.
Quality control
-
5
Check the quality of the gDNA both by agarose gel electrophoresis and concentration by spectrophotometry (OD260/280 = 1.8).
Since this protocol is known to result in shearing, electrophoresis is not informative.
BASIC PROTOCOL 2: TRANSFORMATION OF S. PYOGENES (GAS) CELLS
This protocol describes the introduction of exogenous DNA (linear or plasmid) into GAS cells by means of electroporation. Electrocompetent GAS cells are prepared and then transformed according to the protocol first described by Caparon and Scott (Caparon and Scott, 1991).
Material
Todd-Hewitt Yeast (THY) broth for GAS growth (see recipe)
2 M glycine solution (see recipe)
Optimal: Hyaluronidase solution (see recipe)
Viable GAS isolated colony on the appropriate agar plate (UNIT 9D.2)
Refrigerated centrifuge
Spectrophotometer
Sterile Nalgene centrifugation bottles (500 ml, 250 ml) with inserts and screw caps
Bench-top centrifuge
Spectrophotometer
EP solution (see recipe)
−80°C freezer
0.025 μm membrane filter for plasmid dialysis (Millipore, Cat. No. VSWP02500)
Gene Pulser Xcell Microbial System apparatus (Bio-Rad, Cat. No. 165-2662)
Electroporation cuvettes (2 mm) (Bio-Rad, Cat. No. 165-2082)
Temperature-controlled CO2 incubator
THY agar plates with appropriate antibiotic (see recipe)
Preparation of electrocompetent GAS cells
-
1
Inoculate 10 ml of THY broth containing 20 mM glycine and the appropriate antibiotic, if needed, in 15 ml conical tube with an isolated GAS colony taken from an appropriate agar plate using sterile technique. Incubate overnight at the proper temperature (30°C or 37°C).
Most GAS strains grow best at 37°C in a sealed tube. The addition of free glycine acts to reduce cell wall cross-bridges and make the GAS envelope more permeable for DNA uptake during electroporation. Hyaluronidase solution should be added to the GAS culture if the strain expresses significant capsule. -
2
Inoculate 150 ml of THY broth containing 20 mM glycine and antibiotics, if needed, in a 200 ml screw-cap media bottles with 7.5 ml of the GAS overnight culture obtained in step 1.
-
3
Grow cells at 37°C until OD600 reaches 0.2 to 0.4. Check under microscope for contamination (presence of motile or non-cocci bacteria).
-
4
Store the GAS cells on ice for ~20 to 30 min.
For all subsequent steps, keep the cells on ice and chill all containers or tubes on ice before adding cells. -
5
Pellet GAS cells at 10,000 x g for 20 min, 4°C. Carefully pour off and discard the supernatant.
It is better to sacrifice yield by pouring off a few cells than to leave any supernatant behind. -
6
Wash cells 3 times with 20 ml ice-cold EP solution. Pellet as in step 5.
-
7
Resuspend the washed cells in 1 ml ice-cold EP solution. Transfer 200 μl aliquots (single reactions) into pre-chilled and labeled microfuge tubes and store at −80°C. Cells should be good for 6 months to 1 year.
GAS transformation by electroporation
Strains of GAS can vary significantly in their transformation efficiency by electroporation. Some strains (e.g., M49 NZ131) are highly transformable (over 2000 transformants/μg DNA), making them attractive genetic backgrounds. However, most clinical isolates are harder to transform (under 50 transformants/μg DNA) and require larger amounts of DNA.
-
8
Set up the electroporation experiment as follows:
(−) Control: negative control with competent cells only. No DNA.
-
(+) Control: positive control using a vector with same antibiotic marker as the experimental that will replicate in GAS (Use 5 μg DNA).
To determine the transformation efficiency of a GAS strain or a given competent cell preparation, perform a (+) control using 5 μg of the same replicating plasmid every time. This allows comparison between different experiments across time. -
Experimental:
Replicating Plasmid: origin replicates in GAS. Use 5 to 15 μg DNA for highly transformable GAS strains; whereas 30 to 40 μg DNA is needed for hard to transform clinical strains.
Linear DNA: Restriction digested or PCR-derived linear DNA. Only works with highly transformable GAS strains (e.g., NZ131). Use 10 to 25 μg DNA.
Conditional Temperature Sensitive Plasmid: See Basic Protocol #3 below.
-
Suicide Plasmid: See Basic Protocol #4 below.
Type of DNA used (circular versus linear), replication mode (replicating, conditional, suicide), and transformation efficiency of GAS strain used will each affect the amount of DNA needed for use. These are typical amounts in our hands and it may be necessary to test a range of concentrations.
Drop Dialyze DNA to remove salts
-
9
Dialyze the appropriate amount of DNA for electroporation by carefully placing as a large drop (30 to 50 μl maximum) in the center of a dialysis membrane (0.025 μm) floating on distilled water in a small 2″ petri dish. Incubate for 30 min at room temperature. Carefully remove dialyzed sample using a pipetteman without flooding membrane with distilled water.
Dialysis removes excess salts found in samples to prevent arcing during electroporation.
Electroporation and outgrowth of GAS
-
10
Place the required number of electroporation cuvettes on ice to chill.
-
11
Label the required number of sterile 15 ml conical tubes and add 10 ml THY broth to each.
-
12
Settings for electroporation using Gene Pulser system: voltage: 1.75 kV; resistance: 400 ohms; capacitance: 25 μF
-
13
Remove the required number of aliquotted competent GAS cells (200 μl per reaction) from −80°C and thaw quickly in your hand; place immediately on ice.
-
14
Add DNA to 200 μl aliquot of cells, mix by aspiration with a pipettor, and add all to chilled cuvette.
-
15
Tap lightly on bench to get cells to bottom and quickly electroporate sample. Record the time constant for each (expect 9.5 to 10.0 for cells only, No DNA).
Salt in DNA sample leads to lower time constant. Excess salt will result in arcing, where the pulse will cross the cuvette and bypass the sample. Arcing causes a loud audible popping noise and the reaction is not salvageable (discard). -
16
Quickly remove electroporated cells and place directly into 10 ml THY broth, containing no antibiotics, in a 15 ml conical tube. Wash electroporation cuvette once with 1 ml THY broth to collect all of the GAS cells from the cuvette and add to conical tube.
Outgrowth is done with no antibiotic selection to allow cells to recover. -
17
Grow cells static for 2 h at 37°C.
-
18
Pellet cells at 6,000 x g for 10 min at 4°C.
-
19
Resuspend cells in 300–500 μl of THY broth. Make serial dilutions (10−1 to 10−7) in saline solution.
-
20
Plate 100 μl of cells per plate onto:
Experimental: THY agar plates + appropriate antibiotic; 100 to 10−3 dilutions depending on expected efficiency
Viable counts: THY agar plates with no antibiotic; 10−6 to 10−7 dilutions
-
21
Incubate plates at appropriate temperature in 5% CO2 until colonies appear. For replicating plasmids at 37°C, colonies should be visible after overnight growth to 24 hours, whereas at 30°C it might require 1–2 days.
-
22
Determine the number of electroporants/μg DNA for your experimental samples. This can range from 102 to 106 depending on the strain and DNA used.
BASIC PROTOCOL 3: GENE DISRUPTION USING CONDITIONAL REPLICATIVE PLASMIDS
This protocol describes a method to generate mutations in GAS genes by insertional inactivation. The method relies on plasmids that are derived from pVE6002 (Maguin et al., 1992; Perez-Casal et al., 1993), which contain a pWV01-based thermosensitive origin of replication that replicates at 30°C (permissive) and not at 37°C or above (non-permissive). The plasmid must carry an antibiotic resistance marker that will function in the GAS strain to be mutagenized (i.e., aad9 [Spr]; cat194 [Cmr]; aphA3 [Kmr]; ermC [Emr]). To inactivate a gene, a fragment internal to the coding region for the gene is cloned into the conditional replicative plasmid. The resulting plasmid is then transformed into GAS under “permissive” growth conditions that allow the plasmid to replicate (30°C and appropriate antibiotic). The integration of the plasmid into the gene of interest by homologous recombination is obtained by growing the GAS transformants at the “non-permissive” temperature (37°C) with appropriate antibiotic selection. This results in the disruption of the gene and insertion of the plasmid into the genome of GAS.
If the promoter and start codon of the gene are included in the cloned fragment, the protocol can be used to integrate transcriptional or translational fusions into the GAS genome without interrupting the wild-type gene (for more details see Cho and Caparon (Cho and Caparon, 2006)). A strain that has lost the integration mutation (rescue) can be easily generated by passaging the integration mutant at the permissive temperature (30°C) and screening for loss of the plasmid antibiotic resistance. When targeting a gene within an operon, gene inactivation by plasmid insertion will result in the aberrant expression of genes located downstream of the mutation (polar effect) and should be taken into consideration when using this genetic approach.
Materials
Genomic DNA from GAS strain to mutagenize
Primers; design in step 1
High-fidelity DNA polymerase
Reagents and equipment for PCR
PCR purification kit (Promega)
DNA of a temperature-sensitive plasmid conditionally replicative in GAS
Restriction enzyme(s), DNA ligase and corresponding buffers
Electrocompetent cells of E. coli strain for plasmid propagation (see Unit --)
Electrocompetent cells of the GAS strain to mutagenize (see Basic Protocol 2)
Gene Pulser Xcell Microbial System apparatus (Bio-Rad, Cat. No. 165-2662)
Electroporation cuvettes (2 mm) (Bio-Rad, Cat. No. 165-2082)
Todd-Hewitt Yeast (THY) broth for GAS growth (see recipe)
Temperature-controlled CO2 incubator
THY agar plates with appropriate antibiotic (see recipe)
A set of primers targeting the gDNA surrounding the mutation
Plasmid construction
-
1
Design primers to amplify an internal fragment (within open reading frame) of the gene to mutagenize. The PCR product should be between 300 and 500 nucleotides and be located towards the 5′ end of the target gene. To facilitate the cloning of the PCR product, restriction sites can be added to the primers at both ends.
Integration of plasmid into the GAS genome results in duplication of the cloned homologous fragment. Since the 3′ end of the internal fragment represents the beginning of the mutation (site of plasmid insertion), the closer it is towards the 5′ end of the gene will maximize the gene disruption. -
2
Set up a PCR reaction using the gDNA from the GAS strain of interest, the two primers designed in step 1 and a high-fidelity DNA polymerase, following polymerase manufacturer’s instructions for amplification.
-
3
Purify the PCR product using a PCR purification kit.
-
4
Digest and clone the purified PCR product into a temperature-sensitive (pWVO1-derived) plasmid containing an antibiotic-resistance gene functional in GAS using standard techniques. Transform the resulting plasmid into E. coli for propagation. Confirm successful plasmid construct using DNA sequence analysis.
GAS transformation
-
5
Prepare the plasmid for electroporation using Drop Dialysis as described (see Basic Protocol 2). Plasmid DNA should be resuspended in water for a total volume less that 50 μl.
The amount of plasmid DNA required will be dependent on the transformation efficiency of the GAS strain being used. -
6
Prepare electrocompetent cells of the desired GAS strain (see Basic Protocol 2).
-
7
Transform the plasmid into the GAS cells using parameters for transformation of GAS with a replicating plasmid (see Basic Protocol 2).
-
8
Immediately after transformation, resuspend GAS cells in 10 ml of THY broth in a 15 ml screw-cap conical tube, seal tube, and outgrow for 3–4 h at the permissive temperature of 30°C.
-
9
Harvest GAS transformants by centrifugation at 6,000 x g for 20 min, 4°C. Resuspend GAS cells in 500 μl of saline. Also make serial dilutions in saline (10−1, 10−2).
-
10
Plate the following GAS transformation experiments (100 μl per plate) on THY agar containing the appropriate antibiotic.
(−) Control: No DNA, 2 plates of undiluted (100).
(+) Control: vector only, 2 plates of 10−1 and 10−2 dilution.
Experimental: 2–3 plates of undiluted (100) to 10−2 dilutions.
-
11
Incubate experimental plates at 30°C, 5% CO2 until colonies appear (1 to 2 days).
-
12
Streak resulting GAS transformants for isolated colonies on THY agar plates with appropriate antibiotic and incubate at 30°C, 5% CO2 until colonies appear.
Mutagenesis
-
13
Pick 4–6 isolated colonies to independently inoculate 10 ml of THY broth with appropriate antibiotic and incubate overnight at the non-permissive temperature of 37°C, 5% CO2.
-
14
Plate serial dilutions (10−5 to 10−7) of the overnight culture on THY agar plate with appropriate antibiotic and incubate overnight at 37°C, 5% CO2.
-
15
Use an isolated colony to start a culture in 10 ml of THY broth with the appropriate antibiotic and incubate overnight at 37°C, 5% CO2. Use culture for freezer stock.
Verification of the mutagenesis
-
16
Perform a gDNA extraction (see Basic Protocol 1.2 or 1.3).
-
17
Verify integration of the plasmid into the suitable locus by PCR.
Additional PCR primers should be designed to amplify the junctions between the chromosome and the integrated plasmid. Primers for the chromosome should target genome sequence outside the cloned internal fragment used for homology. Primers to the pWVO1ts origin or antibiotic resistance gene can be used to verify presence of the integrated plasmid; however, this does not confirm location in the genome.
Rescuing the mutation
Genetic complementation using the wild type gene of interest on a plasmid is important to demonstrate linkage. However, conditional integration mutants are reversible, so they can be rescued easily by passaging at the permissive temperature with no antibiotic selection to allow plasmid to excise and be lost. This should restore wild type function of target gene.
-
18
Inoculate a single colony of integration mutant from THY agar plate grown at 37°C, 5% CO2 with appropriate antibiotic into 10 ml of THY broth (no antibiotic) and grow overnight at 30°C. Repeat this overnight passage once.
-
19
Plate serial dilutions onto THY agar plates and incubate at 37°C, 5% CO2 overnight (no antibiotic).
-
20
Patch isolated colonies onto TSA blood agar plates and THY plates with the antibiotic used for integration plasmid selection. Rescue strains should be sensitive to antibiotic. Verify loss of integrated plasmid by PCR analysis of target gene. Test for wild type function of targeted gene.
BASIC PROTOCOL 4: GENE DISRUPTION USING SUICIDE PLASMIDS
Plasmids containing ColE1 origins will not replicate in GAS and can be used as ‘suicide’ vectors (Husmann et al., 1995; Okada et al., 1993) to integrate plasmid sequences into the GAS chromosome through homologous recombination. The only prerequisite is that the suicide plasmid must carry an antibiotic resistance marker that will function in GAS (i.e., aad9 [Spr]; cat194 [Cmr]; aphA3 [Kmr]; ermC [Emr]). Those successfully used in GAS include the Spr pUC-Spec (Husmann et al., 1995) and the Kmr pCIV2 (Okada et al., 1993). A fragment internal to the coding region of the gene of interest is cloned into a suicide vector for homology. Following transformation into GAS, integration of the plasmid following a single homologous crossover event will inactivate the gene. Because the plasmid cannot replicate to high numbers as with the conditional protocol, much larger amounts of plasmid DNA (300 and 500 μg) are required to compensate. Cloning larger regions of homology (>500 bp) may result in more efficient integration, but must still be within the coding region of gene of interest. Suicide integration is often challenging for producing GAS mutants and works best when using highly electrocompetent GAS strains (e.g., M49 NZ131 and M6 JRS4). When targeting a gene within an operon, gene inactivation by plasmid insertion will likely result in the aberrant expression of genes located downstream of the mutation (polar effect) and should be taken into consideration when using this genetic approach.
Materials
Genomic DNA from GAS strain to mutagenize
Primers; design in step 1
High-fidelity DNA polymerase
Reagents and equipment for PCR
PCR purification kit
DNA of a suicide plasmid (non-replicating in GAS)
Restriction enzyme(s), DNA ligase and corresponding buffers
0.025 μm membrane filter for plasmid dialysis (Millipore, Cat. No. VSWP02500)
Electrocompetent cells of E. coli strain for plasmid propagation (see APPENDIX 3L)
Electrocompetent cells of the GAS strain to mutagenize (see Basic Protocol 2)
Gene Pulser Xcell Microbial System apparatus (Bio-Rad, Cat. No. 165-2662)
Electroporation cuvettes (2 mm) (Bio-Rad, Cat. No. 165-2082)
Todd-Hewitt Yeast (THY) broth for GAS growth (see recipe)
Temperature-controlled CO2 incubator
THY agar plates with appropriate antibiotic (see recipe)
A set of primers targeting the gDNA surrounding the mutation
Plasmid construction
-
1
Design primers to amplify an internal fragment (within the open reading frame) of the gene to mutagenize. The PCR product should be between 300 and 500 nucleotides and be located towards the 5′ end of the target gene. To facilitate the cloning of the PCR product, restriction sites can be added to the primers at both ends.
-
2
Set up a PCR reaction using the gDNA from the GAS strain of interest, the two primers designed in step 1 and a high-fidelity DNA polymerase, following polymerase manufacturer’s instructions for amplification.
-
3
Purify the PCR product using a PCR purification kit.
-
4
Digest and clone the purified PCR product into a suicide vector containing an antibiotic-resistance marker functional in GAS using standard techniques. Transform the resulting plasmid into E. coli for propagation. Confirm clone using DNA sequence analysis.
GAS cells transformation
Much larger amounts of suicide plasmid DNA are required for transformation to compensate for lack of replication.
-
5
Set up transformations as follows:
(−) Control: No DNA, 2 plates of undiluted (100).
(+) Control: Use 5–10 μg DNA of a replicating plasmid.
Experimental: Use 300 to 500 μg DNA for ColE1-derived or other non-replicating plasmids. It will be necessary to concentrate DNA by ethanol precipitation. Resuspend in as small a volume as possible (20–50 μl). Prepare the plasmid for electroporation using Drop Dialysis as described (see Basic Protocol 2).
-
6
Electroporate the GAS strain of choice using normal parameters for transformation of Group A Streptococcus (see Basic Protocol 2).
-
7
Outgrow the 10 ml cultures 2–3 hour at 37°C.
-
8
Pellet cells at 6,000 x g for 20 min, 4°C.
-
9
Resuspend cells in 10 ml of THY broth with appropriate antibiotic. Grow cells static at 37°C overnight to allow outgrowth of suicide integrants.
-
10
Pellet cells at 6,000 x g for 20 min, 4°C.
-
11
Resuspend cells in 300 μl of Saline. Perform serial dilutions in saline (10−1, 10−2)
-
12
Plate the following GAS transformation experiments (100 μl per plate) on THY agar containing the appropriate antibiotic.
(−) Control: No DNA. 2 plates of undiluted (100).
(+) Control: Replicating vector. 2 plates of 10−1 and 10−2 dilution.
Experimental: 2–3 plates of undiluted (100), 1 plate of diluted (10−1).
Mutation
-
13
Incubate experimental plates at 37°C overnight or until colonies appear. Once integrants appear, streak them for isolated colonies and confirm chromosomal integration by PCR or Southern blotting analysis. PCR should target the junction between chromosome and integrated plasmid.
BASIC PROTOCOL 5: GENE REPLACEMENT BY ALLELIC EXCHANGE
This protocol describes a method to generate either polar or in-frame (non-polar) gene mutations by deleting the gene of interest and replacing it with an antibiotic resistance gene or cassette. Different genes encoding antibiotic-resistance cassette have been successfully used (i.e., aad9 [Spr]; cat194 [Cmr]; aphA3 [Kmr]; ermC [Emr]). If the native promoter of the deleted gene controls expression of the inserted antibiotic resistance gene, the deletion is in-frame, and translation signals are properly restored after the deletion, the resulting gene replacement will be non-polar. This is important for mutation of genes residing within an operon. The antibiotic-resistance gene flanked by the DNA regions surrounding the gene to delete is cloned into a conditional pWV01-based plasmid with a thermosensitive origin of replication (Maguin et al., 1992; Perez-Casal et al., 1993). The resulting construct is transformed into GAS cells and the allelic exchange occurs by double recombination between the two flanking regions of homologous DNA.
Materials
Genomic DNA from GAS strain to mutagenize
Plasmid DNA to amplify the desired antibiotic-resistance cassette
Two sets of primers to amplify DNA flanking the gene to mutagenize in step 1
A set of primers to amplify the desired antibiotic-resistance cassette in step 1
High-fidelity DNA polymerase
Reagents and equipment for PCR
PCR purification kit
DNA of a temperature-sensitive plasmid conditionally replicative in GAS
Restriction enzyme(s), DNA ligase and corresponding buffers
0.025 μm membrane filter for plasmid dialysis (Millipore, Cat. No. VSWP02500)
Electrocompetent cells of E. coli strain for plasmid propagation (see APPENDIX 3L)
Electrocompetent cells of the GAS strain to mutagenize (see Basic Protocol 2)
Gene Pulser Xcell Microbial System apparatus (Bio-Rad, Cat. No. 165-2662)
Electroporation cuvettes (2 mm) (Bio-Rad, Cat. No. 165-2082)
Todd-Hewitt Yeast (THY) broth for GAS growth (see recipe)
Temperature-controlled CO2 incubator
THY agar plates with appropriate antibiotic (see recipe)
A set of primers targeting the gDNA surrounding the mutation
Plasmid construction
-
1
Design three sets of primers to amplify (i) the gene conferring antibiotic-resistance to deletion (drug 1), (ii) the DNA region located upstream of the GAS gene to delete, (iii) the DNA region located downstream of the GAS gene to delete.
The size of PCR products for the DNA regions surrounding the GAS gene to delete should be at least 500 nucleotides. Subsequent ligation of the three PCR products together can use different cloning techniques, such as restriction digestion or the Splicing by Overlapping PCR technique (referred later as PCR SOE-ing in the text) (Warrens et al., 1997). If using PCR SOE-ing, the size of overlapping PCR products for the DNA regions surrounding the GAS gene to delete should be comparable to the size of the gene conferring the antibiotic resistance. -
2
Use genomic DNA from the GAS strain of interest (see Basic Protocol 1), a template with the gene conferring antibiotic-resistance, the sets of primers designed in step 1, and a high-fidelity DNA polymerase, to set up three PCR reactions to amplify the DNA region flanking the GAS gene to delete and the gene conferring antibiotic-resistance to the deletion, respectively, following polymerase manufacturer’s instructions.
For PCR SOE-ing, the three overlapping PCR products are first amplified individually. A PCR reaction is then set up by mixing identical amounts of the three PCR products, the primer located at the 5′ end of the DNA region located upstream of the GAS gene to delete, the primer located at the 3′ end of the DNA region located downstream of the GAS gene to delete, and a high-fidelity DNA polymerase, following the polymerase manufacturer’s instructions. Proper amplification and integration of the three DNA fragments should be verified by DNA sequencing prior continuing. -
3
Purify the PCR product using a PCR purification kit.
-
4
Digest and clone the purified PCR product into a pWV01-based plasmid containing a thermosensitive origin of replication using standard techniques. The plasmid should be chosen so it contains an antibiotic-resistance marker (drug 2) functional in GAS and different than antibiotic-resistance marker (drug 1) chosen in step 1. Transform the resulting plasmid into E. coli for propagation. Confirm clone using DNA sequence analysis.
Transformation of S. pyogenes (GAS)
-
5
Prepare the plasmid DNA and resuspend in a total volume of distilled water less that 50 μl. Prepare the plasmid for electroporation using Drop Dialysis as described (see Basic Protocol 2).
The amount of plasmid DNA required will be dependent on the transformation efficiency of the GAS strain being used. -
6
Prepare electrocompetent cells of the desired GAS strain (see Basic Protocol 2).
-
7
Transform the plasmid into the GAS cells using parameters for transformation of GAS with a replicating plasmid (see Basic Protocol 2).
-
8
Immediately after transformation, resuspend GAS cells in 10 ml of THY broth in a 15 ml screw-cap conical tube, seal tube, and outgrow for 3–4 h at the permissive temperature of 30°C.
-
9
Harvest GAS transformants by centrifugation at 6,000 x g for 20 min at 4°C. Resuspend GAS cells in 500 μl of saline. Perform serial dilutions (10−1, 10−2) in saline.
-
10
Plate the following GAS transformation experiments (100 μl per plate) on THY agar containing the appropriate antibiotic.
(−) Control: No DNA, 2 plates of undiluted (100).
(+) Control: vector only, 2 plates of 10−1 and 10−2 dilution.
Experimental: 2–3 plates of undiluted (100) to 10−2 dilution.
-
11
Incubate experimental plates at 30°C, 5% CO2 overnight or until colonies appear.
-
12
Streak resulting GAS transformants for isolated colonies on THY agar plates with appropriate antibiotic (drug 1 and/or 2) and incubate at 30°C, 5% CO2 until colonies appear.
Integration of the plasmid by single crossover
-
13
Pick 4–6 isolated colonies and independently inoculate 10 ml of THY broth with appropriate antibiotic (drug 1 and/or 2) and incubate overnight at the non-permissive temperature of 37°C, 5% CO2.
-
14
Plate serial dilutions (10−5 to 10−7) of overnight culture on THY agar with appropriate antibiotic (drug 1 and/or 2) and incubate overnight at 37°C, 5% CO2.
-
15
Use several isolated colonies to start independent cultures in 10 ml of THY broth with antibiotic (drug 1) and incubate overnight at 37°C, 5% CO2. Use cultures to produce individual freezer stocks.
Allelic exchange of the antibiotic-resistance cassette
-
16
Restreak several freezer stocks from step 15 for colony isolation onto THY plates with antibiotic (drug 1) and incubate at 30°C, 5% CO2. Colonies may take 1 to 2 days to appear.
-
17
Pick an isolated colony and inoculate 10 ml of THY broth containing drug 1 in a sealed 15-ml conical tube. Incubate at 30°C overnight, static.
-
18
Inoculate 10 ml of THY broth containing drug 1 with 10 μl of the overnight culture obtained in step 17 and grow static in sealed conical tube at 37°C overnight for second passage.
-
19
Repeat step 18 for third passage.
-
20
Plate different dilutions of the culture obtained in step 19 on THY agar plate with selecting drug 1.
Since gene-replacement by double crossover might be difficult to obtain, it is important to repeat step 18 to 20 while pursuing the following steps. -
21
Patch 200–300 integrants onto THY plates with drug 1 and THY agar plates with drug 2 and incubate overnight at 37°C, 5% CO2.
-
22
GAS cells with allelic exchange should be resistant to drug 1 and sensitive to drug 2, as the double crossover will have led to loss of the plasmid. If drug 1 resistant, drug 2 sensitive clones are not found during this first screen, repeat step 21.
If no allelic exchange mutants are found after 1,000 patches, return to step 16 and repeat from freezer stock. -
23
Perform a gDNA extraction (Basic Protocol 1).
-
24
Use PCR with primers designed to target the chromosome across the mutated region to verify potential gene-replacement strains.
BASIC PROTOCOL 6: TN4001 TRANSPOSITION IN S. PYOGENES (GAS)
This protocol is adapted from Lyon et al. (Lyon et al., 1998) and has proven successful for transposon mutagenesis of GAS. Tn4001 is a composite transposon from Staphylococcus aureus. Caparon and collaborators constructed a Spr mini-Tn4001.spc derivative and used it to identify protease-deficient mutants of GAS (Lyon et al., 1998). Since the transposon is introduced on a suicide vector (pMGC57) and transposants are selected for directly during each screen, it is not necessary to construct a stored library. The following protocol has been successfully used in highly transformable GAS strains, and optimizations might be required for use of Tn4001 in GAS strains that are more difficult to transform.
Material
DNA of the plasmid pMGC57
Electrocompetent cells of E. coli strain for plasmid propagation (see APPENDIX 3L)
Electrocompetent cells of the GAS strain to mutagenize (see Basic Protocol 2)
Gene Pulser Xcell Microbial System appatarus (Bio-Rad, Cat. No. 165-2662)
Electroporation cuvettes (2 mm) (Bio-Rad, Cat. No. 165-2082)
Todd-Hewitt Yeast (THY) broth for GAS growth (see recipe)
Temperature-controlled CO2 incubator
THY agar plates with appropriate antibiotic (see recipe)
GAS cell transformation
-
1
Prepare 2 batches of electrocompetent GAS cells using normal procedure (see Basic Protocol 2): use 300 ml cell culture for each batch and resuspend the electrocompetent cells for each batch in 1 ml of THY.
-
2
Transform 100 μl of competent GAS cells with 10 μg pMGC57 (Tn4001.spc).
-
3
Outgrow GAS transformants for 2 h at 37°C.
-
4
Pellet cells for 15 min at 6,000 x g in a 15-ml conical tube, and then resuspend cell pellets in 0.5 ml saline. All serial dilutions (10−1, 10−2) are performed in saline.
Selection of Tn4001 transposants
-
5
Plate 0.1 ml per plate on THYA Sp100. Use a dilution series to 10−2
-
6
Incubate plates under specific experimental condition (e.g., CO2 or temperature) until Spr colonies appear. Restreak to verify phenotype.
Identification of Tn4001 insertion sites
-
7
Locate insertion of Tn4001.spc. Direct DNA sequencing or cloning of insertion junction using spectinomycin resistance can be used as described elsewhere (Lyon et al., 1998). Alternatively, AP-PCR (see Basic Protocol 8) can be modified to work with Tn4001.spc.
-
8
Verify linkage of any phenotypes from Tn4001.spc mutations by recapitulating mutation in a clean background or via genetic complementation (see Basic Protocol 9).
BASIC PROTOCOL 7: MARINER TRANSPOSITION IN S. PYOGENES (GAS)
This section describes the use of the plasmid pOSKAR for in vivo transposition of the newly constructed mariner composite transposon osKaR (Le Breton et al., 2013) into GAS. The advantage of using a mariner transposon in the low G+C GAS is the potential saturation afforded by the frequent occurrence of the TA dinucleotide insertion site. pOSKAR contains the mariner-Himar1 transposase gene expressed in GAS from the P23 promoter, the temperature-sensitive pWV01 replicon, and a plasmid-encoded spectinomycin-resistance (Spr) cassette. The plasmid also includes the composite transposable element osKaR, consisting of a kanamycin-resistance (Kmr) cassette bracketed by Himar1-recognized inverse terminal repeats (ITR). This protocol has been successfully tested in a number of GAS serotypes. The pOSKAR plasmid is quite unstable in GAS as well as in E. coli and special measures are required for its preparation. pOSKAR is maintained in E. coli C43 (Miroux and Walker, 1996).
NOTE: For antibiotics, the superscript (Km300, Sp100) refers to the μg/ml final antibiotic concentration used for drug selection in that given experiment.
Material
DNA of the plasmid pOSKAR
Electrocompetent cells of E. coli strain for plasmid propagation (see APPENDIX 3L)
Plasmid Minipreps Kit (Wizard® Plus SV, Promega, Cat. No. A1460)
Plasmid Midi Kit (QIAGEN, Cat. No. 12143)
Electrocompetent cells of the GAS strain to mutagenize (see Basic Protocol 2)
Gene Pulser Xcell Microbial System apparatus (Bio-Rad, Cat. No. 165-2662)
Electroporation cuvettes (2 mm) (Bio-Rad, Cat. No. 165-2082)
Todd-Hewitt Yeast (THY) broth for GAS growth (see recipe)
Temperature-controlled CO2 incubator
THY agar plates with appropriate antibiotic (see recipe)
Production of pOSKAR
-
1
Transform pOSKAR into electrocompetent cells of E. coli C43 strain.
-
2
Outgrow E. coli transformants in 1 ml of LB for 1 h at 30°C, then plate 100 μl of serial dilutions 100 to 10−3 onto LB agar plates containing Sp100 and Km50. Incubate at 30°C overnight.
Plates with E.coli transformants containing pOSKAR can be stored at 4°C up to a week.
Culture of E. coli containing pOSKAR
-
3
Prepare 8 cultures of individual E. coli C43 clones containing pOSKAR by inoculating 250 ml of LB containing Sp100 and Km50. Incubate under gentle agitation overnight at 30°C.
Verification of the pOSKAR plasmid
-
4
Take 10 ml of each overnight culture. Cells are harvested by centrifugation and subsequently lysed for plasmid extraction performed with the Plasmid Miniprep Kit. Plasmid DNA is resuspended in 50 μl of dH2O.
In preparation for step 6, the remaining volume of each E. coli culture is either harvested by centrifugation and pellets frozen down until needed, or cultures stored at 4°C for several hours. -
5
Using 10 μl from each miniprep (and a stock pOSKAR control), set up two restriction digests (EcoRI and PstI) for each. Incubate reactions for 1h at 37°C and run the entire volume of each digest on an agarose gel. Cell pellets exhibiting the incorrect plasmid digestion profile compared to the control are discarded.
The expected pOSKAR digest profile should reveal 2 bands with EcoRI (5.86 kb and 1.32 kb) and 2 bands with PstI (5.57 kb and 1.61 kb). Instability of pOSKAR in E. coli can lead to rearrangements. Results can vary, with one experiment showing all 8 cultures correct and others showing only 1 to 2 correct.
Large-scale preparation of the pOSKAR plasmid
-
6
For each culture showing intact pOSKAR in step 5, harvest GAS cells by centrifugation at 8,000 x g for 10 min, 4°C. Purify pOSKAR plasmid using a Plasmid Midi Kit and the manufacturer’s protocol for isolation of very low-copy plasmids. Plasmid DNA is resuspended in 50 μl of distilled water.
-
7
Assay the quality of the pOSKAR plasmid preparation by 260/280 reading using a spectrophotometer.
-
8
Repeat the steps for preparation of the plasmid pOSKAR (see above, steps 1 to 7) until obtaining the total amount of pOSKAR DNA required for transformation of your GAS strain. Prepare the plasmid for electroporation using Drop Dialysis as described (see Basic Protocol 2).
Different amounts of pOSKAR may need to be used depending on the transformation efficiency of GAS strain used. For highly transformable GAS strains, 20 μg are generally sufficient whereas for less efficient GAS strains, as much as 300 μg may be necessary for transformation.
Introduction of the pOSKAR plasmid into GAS cells
-
9
Transform the required amount of pOSKAR into GAS cells as described above (see Basic Protocol 2).
-
10
Resuspend transformants in 10 ml of THY broth and outgrow the cells for 4 h at 30°C.
-
11
Pellet GAS cells by centrifugation at 6,000 x g for 20 min, and then resuspend them in 500 μl of saline and plate 100 μl aliquots (5 plates) onto THY agar plates containing Km300 and Sp100. Incubate at 30°C until GAS colonies appear.
Depending on the GAS strain used for the transformation, colonies are obtained after 2 to 3 days of incubation.
Verification of pOSKAR transformation and osKaR transposition
At least 60 to 80 individual GAS transformants should be screened by groups of 20 for resistance traits to verify (i) presence of pOSKAR, (ii) in vivo osKaR transposition.
-
12
Streak GAS colonies onto the following: three plates to incubate at 30°C: plate A (THY Km300 + Sp100), plate B (THY Sp100) and plate C (THY Km300) and three different plates to incubate at 37°C: plate D (THY Sp100), plate E (THY Km300) and plate F (TSA blood agar). Incubate plates at the proper temperature for 48 h in 5% CO2.
GAS clones containing the proper pOSKAR plasmid should grow on plates (A), (B), (C), (E) and (F), but not (D). For each clone containing the proper pOSKAR plasmid, proceed to step 13. -
13
Drop 3 ml of 25% glycerol in PBS onto the plate, and using a sterile cell spreader, scrape and resuspend the colonies that grew on the agar plate A (THY agar + Km300 + Sp100 incubated at 30°C). Transfer 1-ml aliquots of the cell suspension into three 2-ml screw-cap vials and freeze down at −80°C for GAS strain collection (pOSKAR source stock).
Quality control of osKaR transposition in the pOSKAR source stock
-
14
For each pOSKAR source stock obtained in step 13, thaw one of the three 2 ml screw-cap tubes and perform a serial dilution of the GAS cell suspension in saline (10−1 to 10−6). Plate a 100-μl aliquot of each dilution onto THY Km300 and THY Sp100 plates and incubate for 48 h at 37°C and 30°C, respectively.
-
15
Determine viable cell counts (CFU) from THY Km300 (37°C) and THY Sp100 (30°C) plates. The frequency of osKaR transposition is obtained by dividing the number of CFU from the plate THY Km300 at 37°C by the number of CFU from the plate THY Spec100 at 30°C.
The frequency of osKaR transposition usually varies from 10−2 to 10−3. -
16
Patch 100 to 200 GAS colonies from the plate THY Km300 (37°C) obtained in steps 13 and 14 onto plates containing THY Km300 or THY Sp100 and incubate the plates at 37°C overnight in 5% CO2. Determine the number of GAS transposants that are Spr.
Percentage of CFU Spr/CFU Kmr reflects the level of non-productive integration of pOSKAR into an osKaR transposon inserted into the GAS chromosome. Mutant libraries with a percentage greater than 5% are discarded.
Quality of the osKaR transposant library
-
17
Using clones from step 16 (THY Km300 plates at 37°C), inoculate 20 individual KmR SpS GAS clones in 10 ml of THY broth containing Km300 and incubate overnight at 37°C. Use 1 ml of each culture to produce a freezer stock and pellet cells from the remaining volume (9 ml) by centrifugation.
-
18
Prepare genomic DNA of each of the 20 osKaR transposants (see Basic Protocol 1.2).
-
19
Use AP-PCR assay to determine the location of each transposon and establish randomness of the library (see Basic Protocol 8).
Identical osKaR insertion sites within the mutant pool denote the presence of siblings, which are not desirable. Any library with a sibling percentage greater than 5% is discarded. -
20
To select for transposants in the complex library of osKaR mutants from the acceptable pOSKAR source stock (step 12) and lose the delivery plasmid, a 1 ml aliquot is amplified by culture in 100 ml of THY broth containing Km300 overnight at 37°C to generate transposon mutants and the resulting library is frozen down at −80°C in 15-ml aliquots until phenotype screening.
ALTERNATE PROTOCOL 3: MARINER TRANSPOSITION IN EASILY TRANSFORMABLE S. PYOGENES (GAS)
This protocol describes a faster variation of the Basic Protocol 7 for osKaR mariner transposition in GAS used in easily transformable GAS strains (e.g., NZ131).
Introduction of pOSKAR and osKaR mutagenesis
-
1
Prepare the pOSKAR plasmid as described in Basic Protocol 7 (steps 1 to 8).
-
2
Transform 20 μg of pOSKAR into electrocompetent GAS cells as described above (Basic Protocol 2). Resuspend transformants in 10 ml of THY broth and outgrow the cells for 4h at 30°C. Pellet GAS cells by centrifugation at 6,000 x g for 20 min and resuspend them in 500 μl of saline and plate 100-μl aliquots (5 plates) onto THY agar plates containing Km300. Incubate at 37°C until GAS colonies appear.
Depending on the GAS strain used for the transformation, colonies are obtained after 48h to 72h of incubation. The GAS colonies obtained should correspond to osKaR transposants.
Verification of osKaR mutagenesis
-
3
Verify the randomness of the osKaR mutant library by culturing at least 20 individual GAS colonies for AP-PCR analyses (see Basic Protocol 8) to identify the location of transposon insertions.
-
4
Steps 1 to 3 can be repeated and independent quality libraries combined until the total number of GAS osKaR transposants is reached in a Master Library. For instance, there are ca. 1,800 genes in the GAS genome. Thus, one would want a total of ca. 10,000 independent transposon mutants in a GAS Master Library to produce 5x coverage of the genome.
BASIC PROTOCOL 8: IDENTIFICATION OF OSKAR TRANSPOSON JUNCTIONS BY AP-PCR
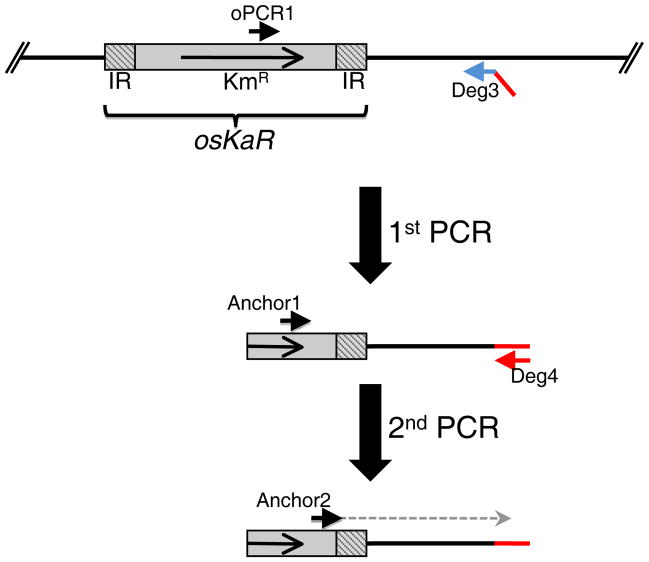
This section describes the use of arbitrary-primed PCR (AP-PCR) to identify the insertion point of transposons within the genome of a given GAS transposant. The protocol provided is designed specifically for osKaR insertions (Fig. 1); however, it can be adapted for Tn4001 or other transposons.
Figure 1. Identification of osKaR insertion site.
Arbitrary-primed PCR (AP-PCR) is a quick method to precisely identify the genomic region where a transposon has inserted. The following is specific for osKaR insertions: genomic DNA of the osKaR mutant is extracted (see Basic Protocol 1) and used for a semi-random PCR using the osKaR-specific primer oPCR1 and the random primer Deg3. The Deg3 primer consists of an 11-nucleotide random primer (in blue) with a 25-nucleotide specific tail (in red). The resulting PCR product is purified and used for a 2nd PCR using the osKaR-specific primer Anchor1 and the Deg3-tail specific primer Deg4. The resulting PCR product is purified and DNA sequencing is performed using the osKaR-specific primer Anchor2.
Materials
Genomic DNA of GAS osKaR mutant (see Basic Protocol 1)
Primers oPCR1, Deg3, Anchor1, Deg4 and Anchor2 (see recipe)
Reagents and equipment for PCR
Gel and PCR Clean-Up System kit (Wizard SV, Promega Cat. No. A9282)
Set up the first PCR round
-
1
Set up the first PCR (AP-PCR #1) using 1 μl of genomic DNA, 1 μl of primers oPCR1 and Deg3 (10 μM each) using Taq.
-
2
Set up the PCR using the following conditions:
Initial denaturation: 5 min at 95°C,
6 cycles: 95°C for 30 sec, 30°C for 30 sec, 72°C for 2min
30 cycles: 95°C for 30 sec, 45°C for 30 sec, 72°C for 2min
Final extension: 72°C for 5 min.
-
3
Purify the AP-PCR #1 products using the PCR Clean-Up kit and resuspend DNA in 50 μl of dH2O.
At this step, visualization of the AP-PCR product by agarose gel electrophoresis will reveal a smear.
Set up the second PCR round
-
4
Set up the second PCR (AP-PCR #2) using 1 μl of the AP-PCR#1, 1 μl of primers Anchor1 and Deg4 (10 μM each) using Taq.
-
5
Set up the PCR using the following conditions:
Initial denaturation: 5 min at 95°C, then
30 cycles: 95°C for 30 sec, 50°C for 30 sec, 72°C for 2 min
Final extension: 72°C for 5 min.
-
6
Purify the AP-PCR #2 products with the PCR Clean-Up kit and resuspend DNA in 50 μl of ddH2O.
At this step, a distinct band still may not be evident over a smear by agarose gel electrophoresis.
DNA sequencing and analysis
-
7
Set up a DNA sequencing reaction by mixing the appropriate amount of AP-PCR product #2 obtained from step 6 and primer Anchor2 as recommended for the DNA sequencing technique used.
-
8
Using a program for DNA sequence analyses, determine the osKaR insertion point.
DNA sequencing should reveal the partial sequence of the mariner ITR followed by the dinucleotide TA and the precise osKaR insertion point within the GAS genome.
BASIC PROTOCOL 9: MUTATION COMPLEMENTATION IN S. PYOGENES (GAS)
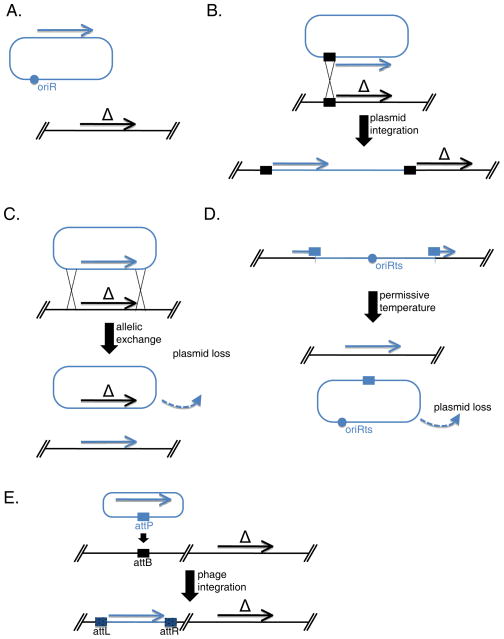
Basic Protocols 3, 4, 5 and 6 focused on mutating a gene of interest using different methods. To confirm the role of the studied gene in a particular GAS phenotypic trait, it is important to restore a wild-type copy of the allele and show the restoration of the wild-type phenotype. This section describes the use of different molecular approaches (Fig. 2) that can be used to perform genetic complementation or rescue.
Figure 2. Strategies for mutation complementation in S. pyogenes.
Complementation of a mutation (obtained using strategies described in Basic Protocols 3, 4, 5 and/or 6) is required to confirm the role of a gene of interest. The GAS strain with a mutated allele (depicted in black, with a Δ sign) of the gene of interest is manipulated to express a wild-type copy of that gene (depicted in blue) and additional tests are performed to see if the wild-type phenotype can be restored. (A) The wild-type allele of the gene of interest can be expressed in trans from a plasmid replicating in GAS. (B) The wild-type allele of the gene of interest can be re-introduced by ectopic integration at the location of mutation using single crossover integration of a conditionally replicative plasmid or a suicide vector. (C) The mutated allele of the gene of interest can be replaced by its wild-type gene using allelic exchange. (D) If the mutation of the gene of interest was carried out by single crossover using a conditionally replicating plasmid, wild-type allele of the gene of interest can be restored by rescuing the mutation by allowing the plasmid to excise from the chromosome and be lost in the absence of selection. (E) The wild-type allele of the gene of interest can be introduced by phage transduction (Cho and Caparon, 2006).
Option 1: Expression in trans from a replicating plasmid in GAS (Fig. 2A)
The wild-type allele of your gene of interest can be introduced on a plasmid in GAS. Several replicating plasmids are available possessing lactococcal (e.g., pWV01), staphylococcal (e.g., pC194, pE194, pUB110) and enterococcal (e.g., pAMβ1) replicons (Cho and Caparon, 2006; Perez-Casal et al., 1993; Trieu-Cuot et al., 1990). Use of plasmids is guided by compatibility with existing plasmids used for mutagenesis. If an antibiotic-resistance marker was used to inactivate the gene of interest, it is important to use a different antibiotic-resistance marker on the replicating plasmid to express the wild-type allele of the gene of interest. Plasmid copy number is also important.
Design primers to amplify the wild-type allele of the gene of interest along with its promoter region. To facilitate the cloning of the PCR product, restriction sites can be added to the primers at both ends. If necessary, a promoter may be provided for expression.
Set up a PCR reaction using the genomic DNA from the GAS strain of interest (see Basic Protocol 1), the two primers designed in step 1 and a high-fidelity DNA polymerase, following polymerase manufacturer’s instructions for amplification.
Purify the PCR product using a PCR purification kit.
Digest and clone the purified PCR product into a plasmid replicative in GAS and containing an antibiotic-resistance marker that is functional in GAS using standard techniques. Transform the resulting plasmid into E. coli for propagation. Confirm clone using DNA sequence analysis.
Transform the recombinant plasmid into the GAS mutant strain and plate transformants as described above (see Basic Protocol 2).
Option 2: Expression in trans by ectopic integration into GAS genome (Fig. 2B)
When a gene of interest has been disrupted by either allelic exchange (Basic Protocol 5) or by transposition (Basic protocol 6 or 7), the wild-type allele of the gene can be reintroduced by plasmid integration at the location of mutation using a conditionally replicative plasmid or a suicide vector.
Design primers to amplify the wild-type allele of the gene of interest along with its promoter region. To facilitate the cloning of the PCR product, restriction sites can be added to the primers at both ends.
Set up a PCR reaction using the genomic DNA from the GAS strain of interest, the two primers designed in step 1 and a high-fidelity DNA polymerase, following polymerase manufacturer’s instructions for amplification.
Purify the PCR product using a PCR purification kit.
-
Digest and clone the purified PCR product into a plasmid conditionally replicative in GAS and containing an antibiotic-resistance marker that is functional in GAS using standard techniques. Transform the resulting plasmid into E. coli for propagation. Confirm clone using DNA sequence analysis.
If an antibiotic-resistance marker was used to inactivate the gene of interest, it is important to use a different antibiotic-resistance marker on the plasmid used to express the wild-type allele of the gene of interest. -
Transform the recombinant plasmid into the GAS mutant strain. If using a conditionally replicative plasmid, see the steps 5 to 17 of Basic Protocol 3. If using a suicide vector, see the steps 5 to 13 of Basic Protocol 4.
Integration of the plasmid into the mutant chromosome results in a merodiploid mutant containing both the wild type and mutated allele of the gene of interest.
Option 3: Complementation of the mutation by allelic exchange (Fig. 2C)
When a gene of interest has been disrupted by either allelic exchange (Basic Protocol 5) or by transposition (Basic protocol 6 or 7), the native allele of the gene can be reintroduced by allelic exchange to replace the mutated allele of the gene of interest with its wild-type copy.
Design primers to amplify the wild-type allele of the gene of interest along with the flanking regions (at least 500 bp on both sides of the gene). To facilitate the cloning of the PCR product, restriction sites can be added to the primers at both ends.
Set up a PCR reaction using the genomic DNA from the GAS strain of interest, the two primers designed in step 1 and a high-fidelity DNA polymerase, following polymerase manufacturer’s instructions for amplification.
Purify the PCR product using a PCR purification kit.
-
Digest and clone the purified PCR product into a plasmid conditionally replicative in GAS and containing an antibiotic-resistance marker that is functional in GAS using standard techniques. Transform the resulting plasmid into E. coli for propagation. Confirm clone using DNA sequence analysis.
If an antibiotic-resistance marker was used to inactivate the gene of interest, it is important to use a second antibiotic-resistance marker on the plasmid used to clone the wild-type allele of the gene of interest. Transform the recombinant plasmid into the GAS mutant strain. Then proceed as described in the steps 5 to 17 of the Basic Protocol 3: mutation by double cross-over will result of the replacement of the mutated allele by the wild-type allele and therefore this event will be selected by the loss of antibiotic resistance.
Option 4: Rescue of plasmid integration mutant to restore native gene (Fig. 2D)
See steps 18 to 20 from the Basic Protocol 3.
Option 5: Phage transduction to introduce wild type allele into GAS genome (Fig. 2E)
For a detailed description for transduction of phage A25 in GAS, see (Caparon and Scott, 1991).
Integration vectors based on the attB site of GAS phage will only work if that location in your strain of interest is not already occupied with a prophage.
REAGENTS AND SOLUTIONS
Use deionized, distilled water in all recipes and protocols steps. For common stock solutions see APPENDIX 2A.
Chloramphenicol stock solution, 10 mg/ml
Dissolve 10 mg of chloramphenicol per ml of ethanol. Store at −20°C.
Chloramphenicol is used at a final concentration of 20 μg/ml for E. coli and 2 to 5 μg/ml for GAS.
EDTA stock solution, 0.5 M pH 8
Dissolve 186.1 g of disodium ethylenediaminetetraacetate.2H2O into 800 ml of water. Adjust pH to 8 with sodium hydroxide. Bring volume to 1 liter with water. Autoclave for 20 min. Store indefinitely at room temperature.
EP solution
Dissolve 100 ml of glycerol into water. Bring final volume to 1 liter. Autoclave for 20 min. Store indefinitely at 4°C.
Erythromycin stock solution, 250 mg/ml
Dissolve 250 mg of erythromycin per ml of ethanol. Store at −20°C.
Erythromycin is used at a final concentration of 250 μg/ml for E. coli and 1 to 2 μg/ml for GAS.
Glycine, 2 M
Dissolve 15 g of glycine per 100 ml of water. Sterilize by filtration. Store indefinitely at room temperature.
Hyarulonidase stock solution, 85 mg/ml
Dissolve 85 mg of hyaluronidase per ml of water. Sterilize by filtration. Prepare fresh for each use.
Kanamycin stock solution, 100 mg/ml
Dissolve 100 mg of kanamycin per ml of water. Sterilize by filtration. Store at 4°C up to a month.
Kanamycin is used at a final concentration of 50 μg/ml for E. coli and 1 to 300 μg/ml for GAS.
Lysozyme, 130 mg/ml
Dissolve 130 mg of lysozyme per ml of 10 mM Tris, pH 8. Prepare fresh for each use.
Primer Adapter-1
5′-AGTCTCGCAGATGATAAGGTGGTCGTGGT-3′
Primer Adapter-Tsp
5′-AATTACCACGACCACCTTATCATC-3′
Primer Anchor1
5′-CGCAACTGTCCATACTCTG-3′
Primer Anchor2
5′-GCCTACGAGGAATTTGTATCG-3′
Primer Deg3
5′-TAGAGTTATTAATGGAATTGCTGATNNNNNNNNNNN-3′
Primer Deg4
5′-TAGAGTTATTAATGGAATTGCTGAT-3′
Primer oID3
5′-TAATACGACTCACTATAGGGAG-3′
Primer oPCR1
5′-TACTGGATGAATTGTTTTAGTACC-3′
Proteinase K, 20 mg/ml
Dissolve 20 mg of proteinase K per ml of water. Store for 6 months at −20°C.
RNase A, 10 mg/ml
Dissolve 10 mg of RNase A per ml of water. Boil for 10 min. Store for 1 year at −20°C.
Saline solution
Dissolve 9.32 g of sodium hydroxide in 800 ml of water. Bring volume to 1 liter. Autoclave for 20 min. Store at room temperature.
SDS, 20%
Dissolve 20 g of SDS into 80 ml of water. Add water to 100 ml. Store indefinitely at room temperature.
Spectinomycin, stock solution, 100 mg/ml
Dissolve 100 mg of spectinomycin per ml of water. Sterilize by filtration and store at 4°C up to a month.
Spectinomycin is used at a final concentration of 100 μg/ml for E. coli and GAS.
Sodium acetate, 3 M
Dissolve 24.6 g of sodium acetate in 80 ml of water. Adjust pH to 5.2 with acetic acid. Bring volume to 100 ml. Store indefinitely at room temperature.
Solution I
Mix together 1.5 ml of 1 M Tris pH 8, 3 ml of 0.5 M EDTA pH 8, 15 ml of 50% Sucrose and complete with water to 30 ml.
Solution II
Mix together 0.3 ml of 1 M Tris pH 8, 0.3 ml of 0.5 M EDTA pH 8, 1.5 ml of 20% SDS and complete with water to 30 ml.
Sucrose, 50% (w/v)
Dissolve 50 g of sodium acetate in 80 ml of water. Adjust volume to 100 ml.
Filter sterilize. Store indefinitely at room temperature.
THY agar
Dissolve powdered Todd-Hewitt Broth extract (Alpha Biosciences, Cat. No. T20-106) (30 g/liter) and 2g/liter yeast extract (Fisher, Cat. No. BP-9727-500) in 800 ml of water. Bring volume to 1 liter with water and add 15 g/liter agar. Autoclave for 20 min, cool to 55°C, and add antibiotics if needed. Pour 25 ml into sterile Petri plates and allow to solidify. Store plates up to 2 months at 4°C.
THY broth
Dissolve powdered Todd-Hewitt Broth extract (Alpha Biosciences, Cat. No. T20-106) (30 g/liter) and 2g/liter yeast extract (Fisher, Cat. No. BP-9727-500) in 800 ml of water. Bring volume to 1 liter with water. Autoclave for 20 min. Store medium up to a month at room temperature.
Tris stock solution, 1 M pH 8
Dissolve 121.1 g of Tris base into 800 ml of water. Adjust pH to 8 with hydrochloric acid. Adjust volume to 1 liter with water. Autoclave for 20 min. Store indefinitely at room temperature.
COMMENTARY
Background Information
The Group A Streptococcus (GAS, Streptococcus pyogenes) is a major Gram-positive human pathogen that causes a wide array of diseases ranging from uncomplicated superficial infections (impetigo, Strep throat) to more rare but severe often life-threatening invasive manifestations (streptococcal toxic shock syndrome, necrotizing fasciitis, necrotizing pneumonia, sepsis syndrome) and post-infection complications (or sequelae) (Rheumatic fever, Acute proliferative glomerulonephritis). GAS strain diversity in terms of clinical manifestation, antibiotic resistance as well as transmission rate appears to be a key contributor to its broad pathogenesis (Bessen, 2009).
Individual gene mutagenesis and phenotypic screening remain the most common method to identify the function of any particular gene and therefore genetic manipulation of GAS appears of fundamental importance to shed lights upon the molecular mechanisms used by these different GAS strains to express their repertoire of virulence factors at different sites of infections (Cho and Caparon, 2006).
Unlike several Streptococcus species, GAS is not naturally competent for DNA uptake and limited options for genetic manipulations (mainly phage transduction) were available until the introduction of DNA transformation by electroporation (Dunny et al., 1991). Identification of plasmids able to replicate in GAS allowed the design of many genetic strategies for directed mutagenesis (insertional inactivation, in-frame deletion) and transposition (for review, see (Cho and Caparon, 2006)). These derive from plasmids initially isolated from Lactococci (pWV01 replicon) (Perez-Casal et al., 1993), Staphylococci (pC194, pE194, pUB110 replicons) (Cho and Caparon, 2006) and Enterococci (pAMβ1 replicon) (Trieu-Cuot et al., 1990). The more useful plasmids are able to replicate in both GAS and E. coli.
This unit gives protocols for the genetic tools that are currently used to study GAS pathogenesis at the molecular level.
Critical Parameters and Troubleshooting
Extraction of GAS genomic DNA (Basic Protocol 1.1)
The steps for the phenol/chloroform extractions require special attention when collecting the aqueous phase. Repeat the phenol/chloroform step until the interface is cleared.
Extraction of GAS genomic DNA (Basic Protocol 1.2)
The use of the MasterPure Complete DNA and RNA Purification Kit (Epicentre, Cat. No. MC85200) allows quick isolation of high quality gDNA from a small volume of GAS culture.
Quick extraction of GAS genomic DNA (Basic Protocol 1.3)
The use of the FastDNA Spin Kit (MP Biomedicals) allows quick isolation of gDNA; however it results in DNA shearing. Use this procedure for quick gDNA isolation for PCR analyses only.
Transformation of GAS cells (Basic Protocol 2)
Once GAS cultures reach the desired cell turbidity, it is critical to keep GAS cells on ice or chilled at 4°C during the centrifugation steps. Transformation efficiency varies significantly (i) among different batches of electrocompetent cells and (ii) among different GAS strains. The amount of DNA necessary for transformation varies between different GAS strains. Transforming a known amount of replicative plasmid will provide baseline for the transformation efficiency of different GAS competent cell batches.
Gene disruption using conditional replicative plasmids (Basic Protocol 3)
The amount of plasmid DNA necessary for transformation varies between different GAS strains. Chromosomal integration of the recombinant plasmid is reversible if grown under 37°C. Special attention is necessary to maintain the plasmid integration, i.e. growth of the GAS mutant at 37°C in the presence of the appropriate antibiotic. Gene insertional inactivation might result in aberrant expression of the genes downstream the mutation (polar effect) and additional precautions might be necessary to analyze mutation results.
Gene disruption using suicide plasmids (Basic Protocol 4)
The amount of plasmid DNA necessary for transformation varies between different GAS strains. This technique is usually more successful with highly transformable GAS strains. Gene insertional inactivation might result in aberrant expression of the genes downstream the mutation (polar effect) and additional precautions might be necessary to analyze mutation results.
Gene disruption by allelic exchange (Basic Protocol 5)
The amount of plasmid DNA necessary for transformation varies between different GAS strains. The event of gene replacement by double crossover might be difficult to obtain and several attempts to achieve it are usually necessary.
Tn4001 transposition (Basic Protocol 6)
The protocol presented is designed for highly transformable GAS strains, and optimization might be required when using GAS strains that are more difficult to transform.
Mariner transposition (Basic Protocol 7)
The plasmid pOSKAR is unstable and therefore difficult to maintain in E. coli. Freezer stocks are therefore not recommended. Efficiency of GAS strain transformation influences the osKaR transposition. If transposants can be directly selected after the outgrowth of highly transformable GAS strains, more precautionary measures are necessary for GAS strains difficult to transform.
Identification of osKaR transposon junctions by AP-PCR (Basic Protocol 8)
It is highly recommended to use a protocol resulting in high quality gDNA extraction.
Mutation complementation (Basic Protocol 9)
Among the different options presented the protocol describing the complementation by expressing the gene of interest from a plasmid is usually preferred. Special attention is required regarding plasmid compatibility as well as gene copy number.
Anticipated Results
Extraction of GAS genomic DNA (Basic Protocol 1.1)
This protocol generates high yield of high quality (not sheared) gDNA (100 to 1000 ng/μl) that is usually required for sensitive molecular biology techniques such as Southern blotting, AP-PCR or library generation.
Extraction of GAS genomic DNA (Basic Protocol 1.2)
This protocol quickly generates a high quality (not sheared) gDNA (100 to 1000 ng/μl).
Quick extraction of GAS genomic DNA (Alternate Protocol 1.3)
This protocol quickly generates gDNA (100 to 500 ng/μl) that can be used for PCR only.
Transformation of GAS cells (Basic Protocol 2)
This protocol generates GAS cells suitable for electroporation. Efficiency of the GAS transformation varies depending on the strain used. Initial tests might be necessary to optimize the transformation.
Gene disruption using conditional replicative plasmids (Basic Protocol 3)
This protocol to generate GAS mutant by gene insertional inactivation is generally efficient regardless of the GAS strain used. The possibility of the polar effect of the mutation needs to be taken into account.
Gene disruption using suicide plasmids (Basic Protocol 4)
This protocol to generate GAS mutant gene insertional inactivation by is generally more efficient when using highly-transformable GAS strains. The possibility of the polar effect of the mutation needs to be taken into account.
Gene disruption by allelic exchange (Basic Protocol 5)
This protocol generates GAS mutant where the allelic exchange does not affect the expression of the genes downstream the mutation. It is however a technique that might be time consuming.
Tn4001 transposition (Basic Protocol 6)
This protocol generates GAS transposon mutants in highly transformable GAS strains. The possibility of the polar effect of the mutation needs to be taken into account.
Mariner transposition (Basic Protocol 7)
This protocol has been successfully used to generate GAS osKaR mutants in different GAS strains. osKaR insertion can result in deregulation (polar effect) of the genes located downstream the mutation: this result needs to be taken into account. osKaR inserts in the dinucleotide TA which makes it ideal to potentially achieve near-saturation mutagenesis. osKaR has also been designed for whole-genome large-scale screens (TraSH, Tn-seq).
Time Considerations
Extraction of GAS genomic DNA (Basic Protocol 1.1)
The amount of time to achieve lysis of the GAS cells may vary by strain. The gDNA extraction can be quite time consuming (up to a day), especially if several phenol/chloroform extractions are needed.
Extraction of GAS genomic DNA (Basic Protocol 1.2)
The amount of time to achieve lysis of the GAS cells may vary by strain. The rest of the procedure takes up to 4 h.
Quick extraction of GAS genomic DNA (Basic Protocol 1.3)
The amount of time to achieve lysis of the GAS cells may vary by strain. The rest of the procedure takes up to 2 h.
Transformation of GAS cells (Basic Protocol 2)
Time required for the GAS culture to reach the required cell density may vary by strain. The preparation of electrocompetent cells may take about 1 h 30 to 2 h. The outgrowth following the electroporation may vary from 2 to 4 h depending on the incubation temperature.
Gene disruption using conditional replicative plasmids (Basic Protocol 3)
Time for the outgrowth following the electroporation may vary from 2 to 4 h depending on the incubation temperature. Much of the time required for the rest of the protocol is spent on agar plate incubation for colony formation, culture growth and clone patching.
Gene disruption using suicide plasmids (Basic Protocol 4)
Time for the outgrowth following the electroporation is about 2 h. Much of the time required for the rest of the protocol is spent on agar plate incubation for colony formation, culture growth and clone patching.
Gene disruption by allelic exchange (Basic Protocol 5)
Time for the outgrowth following the electroporation may vary from 2 to 4 h depending on the incubation temperature. Much of the time required for the rest of the protocol is spent on agar plate incubation for colony formation, culture growth and clone patching. If the step for single crossover of the plasmid is usually quite easily obtained, several attempts might be necessary to achieve the second cross-over step.
Tn4001 transposition (Basic Protocol 6)
Time for the outgrowth following the electroporation may vary from 2 to 4 h depending on the incubation temperature. Much of the time required for the rest of the protocol is spent on agar plate incubation for colony formation, culture growth and clone patching.
mariner transposition (Basic Protocol 7)
Time for the outgrowth following the electroporation requires a 4-h incubation. Much of the time required for the rest of the protocol is spent on agar plate incubation for colony formation, culture growth and clone patching. It is critical to spend the time necessary (sometimes several weeks) to construct the mutant libraries and verify their complexity.
Identification of osKaR transposon junctions by AP-PCR (Basic Protocol 8)
This protocol takes between 1 to 2 days for PCR.
Mutation complementation (Basic Protocol 9)
Time for the outgrowth following the electroporation may vary from 2 to 4 h depending on the incubation temperature. Much of the time required for the rest of the protocol is spent on agar plate incubation for colony formation, culture growth and clone patching.
Acknowledgments
We thank Kanika Gera and members of the McIver lab for critical review of this manuscript. This work was supported in part by funding to K.S.M. through the National Institutes of Health (NIH), National Institute for Allergy and Infectious Diseases (NIAID) for R01 AI47928 and through the American Heart Association for 10GRNT39000017. A UMB/UMCP Seed Grant awarded to K.S.M & Y.L.B also provided funding.
Literature Cited
- Bessen DE. Population biology of the human restricted pathogen, Streptococcus pyogenes. Infection, Genetics, and Evolution. 2009;9:581–593. doi: 10.1016/j.meegid.2009.03.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caparon MG, Scott JR. Genetic manipulation of pathogenic streptococci. Methods Enzymol. 1991;204:556–586. doi: 10.1016/0076-6879(91)04028-m. [DOI] [PubMed] [Google Scholar]
- Cho KH, Caparon M. Genetics of Group A Streptococci. In: Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI, editors. Gram–Positive Pathogens. 2. ASM Press; Washington, D.C: 2006. pp. 59–73. [Google Scholar]
- Dunny GM, Lee LN, LeBlanc DJ. Improved electroporation and cloning vector system for gram–positive bacteria. Appl Environ Microbiol. 1991;57:1194–1201. doi: 10.1128/aem.57.4.1194-1201.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husmann LK, Scott JR, Lindahl G, Stenberg L. Expression of the Arp protein, a member of the M protein family, is not sufficient to inhibit phagocytosis of Streptococcus pyogenes. Infect Immun. 1995;63:345–348. doi: 10.1128/iai.63.1.345-348.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Breton Y, Mistry P, Valdes KM, Quigley J, Kumar N, Tettelin H, McIver KS. Genome–wide identification of genes required for fitness of group a streptococcus in human blood. Infect Immun. 2013;81:862–875. doi: 10.1128/IAI.00837-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon WR, Gibson CM, Caparon MG. A role for Trigger Factor and an Rgg–like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. The EMBO Journal. 1998;17:6263–6275. doi: 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguin E, Duwat P, Hege T, Ehrlich D, Gruss A. New thermosensitive plasmid for gram–positive bacteria. J Bacteriol. 1992;174:5633–5638. doi: 10.1128/jb.174.17.5633-5638.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroux B, Walker JE. Over–production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- Okada N, Geist RT, Caparon MG. Positive transcriptional control of mry regulates virulence in the group A streptococcus. Mol Microbiol. 1993;7:893–903. doi: 10.1111/j.1365-2958.1993.tb01180.x. [DOI] [PubMed] [Google Scholar]
- Perez–Casal J, Price JA, Maguin E, Scott JR. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature–sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol Microbiol. 1993;8:809–819. doi: 10.1111/j.1365-2958.1993.tb01628.x. [DOI] [PubMed] [Google Scholar]
- Trieu–Cuot P, Carlier C, Poyart–Salmeron C, Courvalin P. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in gram–positive bacteria. Nucleic Acids Res. 1990;18:4296. doi: 10.1093/nar/18.14.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrens AN, Jones MD, Lechler RI. Splicing by overlap extension by PCR using asymmetric amplification: an improved technique for the generation of hybrid proteins of immunological interest. Gene. 1997;186:29–35. doi: 10.1016/s0378-1119(96)00674-9. [DOI] [PubMed] [Google Scholar]