Abstract
Recently, several structural genomics centers have been established and a remarkable number of three-dimensional structures of soluble proteins have been solved. For membrane proteins, the number of structures solved has been significantly trailing those for their soluble counterparts, not least because over-expression and purification of membrane proteins is a much more arduous process. By using high throughput technologies, a large number of membrane protein targets can be screened simultaneously and a greater number of expression and purification conditions can be employed, leading to a higher probability of successfully determining the structure of membrane proteins.
This unit describes the cloning, expression and screening of membrane proteins using high throughput methodologies developed in our laboratory. Basic Protocol 1 deals with the cloning of inserts into expression vectors by ligation-independent cloning. Basic Protocol 2 describes the expression and purification of the target proteins on a miniscale. Lastly, for the targets that express at the miniscale, basic protocols 3 and 4 outline the methods employed for the expression and purification of targets at the midi-scale, as well as a procedure for detergent screening and identification of detergent(s) in which the target protein is stable.
Keywords: Escherichia coli, high-throughput, cloning/expression, recombinant protein expression, membrane protein, ligation-independent cloning (LIC), membrane protein purification
Introduction
Integral membrane proteins are of special biological interest as they are involved in a variety of cellular processes including signal transduction, transport and cell-to-cell communication (Tan et al., 2008). As such, they are of great interest as drug targets. Thus, solving the three-dimensional structures of integral membrane proteins is not only a boon for commercial companies but would also add a tremendous amount of knowledge to our understanding of some of the most important molecular mechanisms of the cell.
Although membrane proteins represent between 20-30% of all the genes in a genome (Wallin and von Heijne, 1998), relatively few structures have been solved. In fact, only around 2% of all the structures in the Protein Data Bank (PDB) are of membrane proteins (Mao et al., 2009; see also http://blanco.biomol.uci.edu/mpstruc/listAll/list and http://pdbtm.enzim.hu/). The reasons for this scarcity of data are manifold, but can mainly be ascribed in part to the fact that the level of expression of recombinant membrane proteins (in heterologous, or homologous, systems) is significantly lower than that of cytosolic proteins and in many cases they are toxic to the host cell (Wagner et al., 2007, 2006). As large amounts of protein are required for structure determination (X-ray, nuclear magnetic resonance), the typically low yields of membrane proteins in recombinant systems can severely hinder this process.
In addition, due to the hydrophobic nature of integral membrane proteins, detergents are required during the extraction and the purification procedures. Although a variety of detergents are available for this purpose (Speers and Wu, 2007; Eshaghi, 2009), detergents are poor substitutes of a lipid bilayer, and indeed many, if not most of them, are relatively harsh and can impact the structural stability of the protein in a negative way (i.e. conformational aberration, unfolding, or incorrect folding). Another level of difficulty is added when dealing with eukaryotic membrane proteins as many of them fail to fold properly in bacterial expression systems. Nor are they correctly modified post-translationally, as bacteria lack the mechanisms to do so (Wagner, 2006). In such cases, switching to a eukaryotic host (i.e. yeast, insect, mammalian cells) is preferable, although yields are typically considerably lower than in bacterial systems (Sahdev et al., 2008).
Recently, several structural genomics networks, with interests in integral membrane proteins, have been established (Lundstrom, 2006b; Stroud et al., 2009; Graslund et al., 2008; see also Kloppmann et al., 2012 for a review). These networks are able to simultaneously screen large numbers of targets and rapidly identify proteins that can be successfully purified and crystallized. These structural genomics centers are also capable of handling multiple kinds of expression and purification systems, which can further increase the success rate. Another advantage of such kinds of networks is that statistical and bioinformatics-based approaches can be applied to their extensive target databases. This could lead to improved prediction analyses of which types of membrane proteins may constitute better targets for expression and/or purification (or conversely, which membrane proteins would be poor targets).
As part of the NIH-supported Protein Structure Initiative (PSI-2 and PSI-Biology; www.nigms.nih.gov/Research/FeaturedPrograms/PSI/; see also Dessailly et al., 2009), the New York Consortium on Membrane Protein Structure (NYCOMPS) is one of the specialized centers with focus on developing new methodologies and technologies for membrane protein structure determination.
Target selection, the first step in the screening process, has been described in great detail elsewhere (Punta et al., 2009; Mancia and Love, 2010). A brief summary of our target selection procedure is provided here. First, a model organism, whose genome has been completely sequenced and annotated, must be selected. For earlier NYCOMPS efforts, Escherichia coli was used as the model organism, for our more recent work, focus has shifted to Homo sapiens. Next, using algorithms commonly employed to predict transmembrane domains, such as TMHMM2 (Sonnhammer et al., 1998; Krogh et al., 2001) and PolyPhobius (Käll et al., 2005; Käll et al., 2007), all integral membrane proteins of that selected model organism are identified. These algorithms, while excellent, are not perfect. So, in order to assure that only true integral membrane proteins are pursued, several filters are applied. For example, any proteins predicted to contain just one transmembrane domain are discarded. Next, the remaining proteins are grouped into families. The exact requirements for creation of each family are somewhat complex, but the main criterion is amino acid sequence similarity to one another (Punta et al., 2009). Then, a single member of each family (typically the longest in sequence) is chosen as the ‘seed protein’. Using the same criteria as that used for the creation of families in the model organism, this ‘seed protein’ is then used to identify related proteins from among the collection of 96 bacterial genomes currently employed by NYCOMPS.
This approach of creating families and selection of homologues from other organisms serves two important purposes. First, by grouping the integral membrane proteins from the selected model organism into families, one can then expect to have complete coverage of the model organism's integral membrane ‘proteome’, without having to clone and test for expression every integral membrane protein from that organism. Second, since one cannot predict which proteins will be successfully expressed and purified in our expression system, using homologues from other organisms improves the chances of identifying a well-behaved member of each protein family. We have decided to focus our efforts on bacterial alpha helical bundle membrane proteins because they are significantly more abundant than β barrel integral membrane proteins, they are found throughout the bacterial kingdom and they have biological and medical relevance (see Punta et al., 2009; von Heijne, 2007; Overington et al., 2006). Currently, our database contains in excess of 12,000 bacterial targets (see TargetTrack at www.sbkb.org/tt/). Overviews of the NYCOMPS protein expression pipeline have appeared elsewhere (Mancia and Love, 2010; Love et al., 2010). Here we describe, in much more significant detail, the high-throughput cloning, expression and purifications of these targets.
Strategic Planning
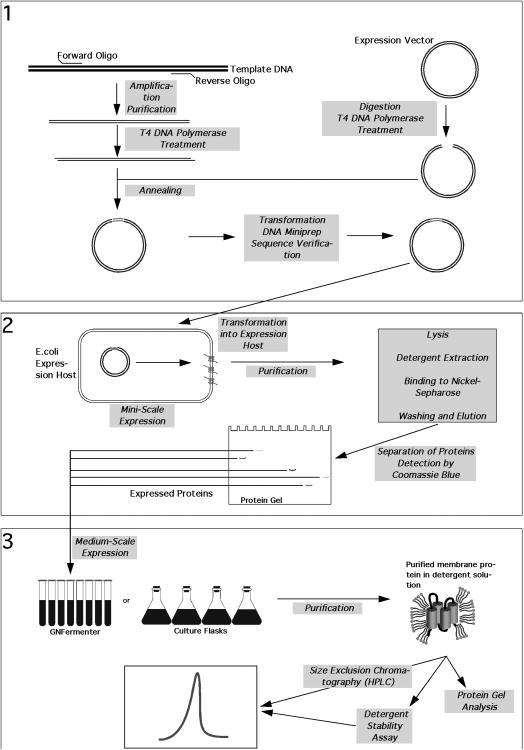
An outline of the methods described in this unit is shown in the flowchart in Figure 1.
Figure 1.
Flowchart outlining the methods described in this unit.
Part 1: Amplification and cloning of targets. Part 2: Expression and miniscale purification of target proteins. Part 3: Scale-up and detergent screening.
The entire screening procedure can be divided into three parts:
Part 1 deals with the cloning of the target DNA in bacterial expression vectors. This includes PCR amplification, purification of the amplified products and insertion of the targets into the appropriate expression vectors. These steps are performed in 96- or 384-well plates, depending upon the total number of reactions and available equipment. All subsequent steps, (i.e. transformation into a cloning strain and preparation of plasmid minipreps) are performed in 96-well plates. Using SBS-format 96- and 384-well plates for these procedures allows the use of automation, without which, although possible, would be tedious. Basic Protocol 1 describes our cloning procedure, including treatment of both vector and insert to create single-stranded overhangs, annealing of the two and transformation of E. coli with the resulting mixture. Support Protocol 1 describes the large-scale preparation of vector for the subsequent cloning of hundreds, or even thousands, or targets. Support protocol 2 provides details for PCR amplification and purification of targets at the 96- or 384-well scale. Support Protocol 3 describes the robotic purification of plasmid DNA in a 96 well format, using the CosMCPrep kit from Beckman Coulter.
In part 2, plasmid DNA is transformed into an expression host and recombinant protein is expressed and purified at the miniscale. These steps are performed in a 96-well platform. Proteins are then separated on SDS PAGE gels and visualized by Coomassie staining. Basic Protocol 2 describes our procedures for transformation of an E. coli expression host with the constructs prepared in Basic Protocol 1, the conditions for expression and purification of the recombinant proteins at the small-scale.
Part 3 describes the scale-up of targets that express at the miniscale and analysis of the affinity-purified proteins by size exclusion chromatography on an HPLC. In our laboratory, for scale-up of targets in high-throughput fashion, we employ the GNF Fermenter, a 96-channel airlift fermenter developed by the Genomics Institute of the Novartis Research Foundation. Basic Protocol 3 describes our procedures for growth and expression in the GNF Fermenter. As this is a relatively expensive and specialized piece of equipment, not available to all laboratories, an Alternate Protocol is provided that describes procedures for growth and expression of a smaller number of targets, at a comparable scale. While this procedure is much more laborious and time-consuming, particularly if a large quantity of targets are to be processed simultaneously, it allows laboratories without such dedicated, specialized equipment to still screen a reasonable number of targets.
In Basic Protocol 4 we provide details of our procedures for the purification of His-tagged proteins from the resulting cell pellets recovered following growth in the GNF Fermenter, or in traditional shake flasks. A small portion of the purified protein is reserved for analysis by SDS-PAGE, while the remaining majority of the purified protein is used for size exclusion chromatography. An Alternate Protocol provides details of our Detergent Stability Assay, which allows to rapidly identify detergents more suitable for crystallization of the expressed and purified targets.
Basic Protocol 1
High-Throughput Cloning of Open Reading Frames Encoding Integral Membrane Proteins Into E. Coli Expression Vectors
High-throughput cloning is most easily accomplished using the method of Ligation Independent Cloning (LIC; Aslanidis and de Jong, 1990, Eschenfeldt et al., 2009). Aside from its lower cost, as compared to traditional restriction enzyme digestion/ligation-mediated cloning, no targets are eliminated due to the presence of restriction enzyme sites located within the open reading frame. Inserts are prepared by Polymerase Chain Reaction (PCR), which easily permits just the open reading frame, or other region(s) of interest to be cloned directly into the desired expression vector, without the need to go through a cloning vector intermediate.
When cloning genes from prokaryotes, genomic DNA is the most convenient template, due to the absence of introns. (In our laboratory we use bacterial genomic DNA from ATCC [http://www.atcc.org] and have found them to be a suitable source for template DNA). For cloning of genes from eukaryotic organisms, the presence of introns necessitates the use of cDNA clones, such as those from the I.M.A.G.E consortium (www.imageconsortium.org), or synthetic genes as templates for PCR. While cDNA libraries are a potential alternative, in our experience, amplification of targets from these, especially in large numbers, is problematic. We believe this is for several reasons. First, one must have prior knowledge of the tissue(s) in which their target is expressed, so that the correct library can be purchased. However, commercially available libraries are typically expensive, which becomes even more daunting when one considers that it may be necessary to have available several different libraries from different animals/tissues. Homemade libraries are an alternative, but preparation of a quality cDNA library of suitable quality is not trivial and one must still know in which tissue their target is expressed. Further, one now is also faced with the task of obtaining the tissue of choice, fresh, or carefully and quickly frozen to prevent degradation of the RNA. Last, transcripts from many eukaryotic genes undergo alternative splicing, potentially producing multiple bands following PCR amplification. The researcher must then decide which of the amplified products to clone, if not all of them. It is for these reasons that we strongly encourage purchasing sequence-verified cDNAs, especially since they are only ∼$100 each. The only downside is that they are available from only about six different organisms. However, with the cost of gene synthesis becoming less and less expensive, this may be the best option for obtaining gene sequences from less common organisms. Gene synthesis has the added advantage of codon optimization and removal of any predicted RNA secondary structure, which together may prevent expression of the target to the levels required for subsequent analysis.
Vectors used for the expression of recombinant targets in E. coli can be obtained commercially, designed by the individual lab to suit their specific needs, or obtained inexpensively from one of many centers for deposition of materials that exist. The four prokaryotic expression vectors most commonly used by the NYCOMPS protein production facility are all available through the materials repository of the Protein Structure Initiative, DNASU (http://psimr.asu.edu). They are based on the pET series of expression vectors (Novagen) and contain a T7 bacteriophage promoter under the control of the lac operator (Studier et al., 1990). These vectors and their key features are provided in Table 1 and Figure 2.
Table 1.
Brief description of the four expression vectors used by NYCOMPS. See also figure 2.
| Vector | Antibiotic | Backbone | Fusion Tag | Position |
|---|---|---|---|---|
| pNYCOMPS-N-term. | Kanamycin | pET24 | FLAG-10×His-TEV | N-term. |
| pNYCOMPS-C-term. | Kanamycin | pET24 | TEV-10×His | C-term. |
| pMCSG7-10× His | Ampicillin | pET21 | 10× His-TEV | N-term. |
| pMCSG28-10× His | Ampicillin | pET21 | TEV-10×His | C-term. |
Figure 2.
Schematic outline of the four expression vectors used by NYCOMPS.
Only relevant features are shown. lacI: lac repressor. RBS: ribosomal binding site. FLAG: DYKDDDDK epitope. 10×His: decahistidine tag. TEV: tobacco etch virus protease recognition site (ENLYFQS). 5′ and 3′ LIC: overhang sites for ligation-independent cloning. ccdB: CcdB toxin gene for plasmid maintenance. Restriction enzymes for linearizing the vectors for LIC treatment are also shown. Features are not to scale.
Other expression systems, such as the arabinose-induced araBAD promoter (Guzman et al., 1995), the hybrid trc and tac promoter (Brosius and Holy, 1984; Amann and Brosius, 1985) or the bacteriophage-based pL(λ) system (Bernard and Helinski, 1979), might also be suitable but they are not as well-established as the T7-based system. They also rely on E.coli RNA polymerase for transcription whereas T7 RNA polymerase is fully dedicated to the expression of the target gene. Other than using a LysS-containing expression strain, to dampen expression of the integral membrane protein target and possibly avoiding the deleterious effects of expression of a foreign protein in the host cell, one can carry out expression of integral membrane proteins under pretty much the same conditions as those used for any soluble/non-integral membrane proteins. Many suitable expression controls are readily available and if a laboratory would like to include such a control in their studies, we would suggest obtaining one, or more, of the constructs successfully used for structural determination from the PSI materials repository, DNASU.
Regardless of the vector(s) selected for use by the individual laboratory, there are several features that should be considered. Any vector used for expression of targets in high-throughput fashion should have a simple tag that permits purification of any target by the same method (see also Graslund et al., 2008; Stevens, 2000; Xie et al., 2009). Although one can choose from several available tags, the most commonly used one, by far, is a polyhistidine tag (6-10 histidines in length). Since they are small tags, their presence often will not interfere with downstream applications such as crystallization, biochemical studies, or antibody development. Proteins tagged with histidines can be easily purified using nickel- or cobalt-chelating resin. Other tags such as glutathione S-transferase (GST), maltose-binding protein (MBP) or SUMO proteins can also be used (Stevens, 2000), but these tags have several drawbacks, most notably, their large size. While such tags may initially appear to improve solubility of the target protein, other genomics consortia have found that quite often the targets fused to these large proteins are no longer well-behaved once the fusion partner is removed, leading to a considerable number of false positives is their screening pipeline. (Donnelly, et al., 2006)
Each expression vector should also contain a protease cleavage site for removal of the tag used for purification, if desired. For several reasons, the protease from Tobacco Etch Virus (TEV) is commonly employed: The protease remains active under a variety of conditions; a vector that allows recombinant expression in E. coli of a version of TEV protease with increased stability and activity is readily available; and the enzyme expressed from this plasmid can be purified in large quantities by well-established methods (Parks et al., 1994; Blommel and Fox, 2007). Other characteristics of the expression vectors, most notably the position of the fusion tag, the backbone of the parent vector and the antibiotic resistance conferred by the plasmid, can have dramatic influence with regard to the successful expression and/or purification of the target. Cloning of the same target into multiple vectors significantly increases the probability for successful expression of the target of interest.
Since the expression vector is to be used for the insertion of hundreds, if not thousands, of targets for expression, it is worthwhile to prepare large quantities of vector. While gel extraction and purification is inherently inefficient (we typically recover only ∼75% of the starting material), the resulting purified, linearized vector from 100 μg circular plasmid is still sufficient for cloning almost 2000 targets. Therefore, we strongly believe that the time and effort expended to prepare high quality vector, free of salt, enzymes, or other contaminants, is worthwhile. (See Support Protocol 1 – Digestion and Gel Purification of Vector for LIC Cloning).
Materials
10× Buffer 4 (New England Biolabs): 10mM Tris-HCl, pH 7.9 50mM NaCl 10mM MgCl2 1mM DTT 100× Bovine Serum Albumin (BSA) (10 mg/ml; New England Biolabs)
T4 DNA Polymerase (New England Biolabs M0203)
Deoxynucleotides (100 mM) from Invitrogen
25 mM EDTA
SOC medium (see recipe)
E. coli DH10B-T1R competent cells
96 well PCR plates (VWR 82006-704)
96 well PCR plate (Eppendorf 951020401)
384 well PCR plates (Eppendorf 951020702
Adhesive-backed foil seals (VWR 60941-076)
Adhesive-backed porous seals (VWR 60941-086)
24 well blocks (Qiagen 19583)
Luria Broth (LB) Lennox (see recipe)
LB-Agar (see recipe)
Antibiotics
Glass beads (Sigma Z273627)
New Brunswick Innova 44 shaking incubator
Shel Lab shaking incubator (SI6R-HS)
37°C oven
We have found that the specific brand of plates or seals is not terribly important and we have provided catalog numbers here primarily for reference. By all means, laboratories should try to find such disposables at the best price possible, especially since these items will be used in considerable quantities.
We routinely re-use our 24 well blocks multiple times by scraping out old agar, running them through a standard laboratory glassware washer and autoclaving.
We have found that 96- and 384-well plates can become misshapen following repeated heating and cooling (as in a PCR amplification), or after freezing. For this reason, we strongly suggest using high-quality, hard-shell plates with a full skirt for any such applications, especially those that will later be handled by a robot.
For applications such as plating and storage of competent cells, where potentially large numbers of plates could be consumed, we recommend using less expensive 96 well plates (i.e. VWR 82006-704).
Preparation of vector
Linearize expression vector by digestion with appropriate restriction enzyme and gel purify according to Support Protocol 1.
-
Assemble reaction to create single-stranded overhangs on expression vector:
For 500 cloning reactions: 100 μl 10× Buffer 2 (New England Biolabs) 10 μl 100× BSA linearized and gel-purified expression vector (20 ng/μl final) 25 μl 100 mM dNTP (2.5 mM final; see table 2 for nucleotides) 12.5 μl T4 DNA Polymerase (0.0375 Units/μl final) H2O to 1 ml final volume Depending upon the anticipated number of targets to be expressed, vector preparation volumes can be scaled up or down, accordingly. For preparation of large quantities of expression vector, it is best to prepare a reaction mix and divide among the wells of an 8-tube strip, or a 96 well PCR plate. LIC-treated vector can be stored at -20°C, where it remains stable for months.
Incubate at 22°C for 60 minutes. Heat inactivate enzyme at 75°C for 20 minutes.
Table 2.
Oligonucleotides used for addition of LIC overhangs to PCR products.
Oligonucleotides are shown in 5′to 3′ direction. Deoxynucleotides required for the creation of single-stranded overhangs for each vector and insert combination are provided.
| Vector | Oligonucleotides (top: Forward; bottom: Reverse) | dNTP | |
| Vector | Insert | ||
| pNYCOMPS-N-term. | TATTTTCAATCCTACGTA (ATG or first codon) | dGTP | dCTP |
| CCCTCAATATTATACGGG (stop codon) | |||
| pNYCOMPS-C-term. | TTAAGAAGGAGATATACT (ATG) | dCTP | dGTP |
| TGAAAATAGAGGTTTTCGGC (last codon) | |||
| pMCSG7-10×His | TACTTCCAATCCAATGCC (ATG or first codon) | dGTP | dCTP |
| TTATCCACTTCCAATG (stop codon) | |||
| pMCSG28-10×His | GTCTCTCCC (ATG) | dATP | dTTP |
| GGTTCTCCCCAGC (last codon) | |||
Preparation of inserts
Inserts for cloning into expression vectors are prepared by PCR, using KOD Hot Start DNA Polymerase and plasmid, or genomic DNA, as template. (See Support Protocol 2 -°C PCR Amplification and Purification of PCR Products). Small numbers of PCR products can be purified free from enzyme and unincorporated nucleotides and primers using the Qiagen PCR clean-up kit, as per the manufacturer's instructions. For PCR reactions at the 96- or 384-well scale, PCR products may be purified robotically using the AMPure PCR purification kit from Beckman Coulter Genomics (see Support Protocol 2 - PCR Amplification and Purification of PCR Products).
-
Prepare a reaction mix for inserts:
For 120 reactions: 120 μl 10× Buffer 2 12 μl 100× BSA 30 μl 100 mM dNTP (2.5 mM final; see table 2) 15 μl T4 DNA Polymerase (0.0375 Units/μl) H2O to a final volume of 960 μl -
Combine 8 μl of the above mixture with 2 μl of each purified PCR product (See Support Protocol 2).
Depending on the plate format used for the PCR reactions and purifications, the LIC reactions are performed in 96- well or 384-well plates.
Incubate at 22°C for 60 minutes. Heat inactivate enzyme at 75°C for 20 minutes.
Annealing and transformation
-
Combine 2 μl of vector with 4 μl of each insert. Incubate at 22°C for 60 minutes.
Mixing of vector and insert can be performed by hand, using a multichannel pipettor, or with a liquid handling robot. Establishing protocols for robotic assembly is worthwhile, particularly when assembling large numbers of annealing reactions. In our laboratory, we can assemble 384 annealing reactions in less than five minutes with the use of a Biomek FX liquid handling robot (Beckman Coulter).
There are numerous liquid handling robots currently available. At the time the NYCOMPS Protein Production Facility was being set up, a thorough comparison was made of many of the instruments then on the market. The Biomek FX was selected as if was felt that it would both satisfy our needs at the time and also allow for potential future applications. Now, more than six years later, we are extremely pleased with our choice, as the Biomek FX still meets all of our liquid handling needs.
Add 2 μl 25 mM EDTA and incubate at 22°C for 5 minutes.
-
Using a multichannel pipettor, combine 2 μl of each LIC reaction with 20 μl DH10B-T1R competent cells in a 96 well PCR plate and leave on ice for 30 minutes.
We recommend using only bacteriophage T1-resistant (T1R) bacterial strains for cloning and expression. Several companies now offer T1R strains (Sigma, Invitrogen, New England Biolabs). In our laboratory, we routinely use DH10B-T1R for cloning purposes, but any other commercially available strains will be appropriate. We suggest preparing competent cells in-house, using a standard protocol, as the cost of repeatedly purchasing them can be prohibitive.
-
Heat shock at 42°C for 45 seconds. Add 90 μl pre-warmed SOC, cover with a porous seal and allow cells to recover in 37°C shaker (Shel Lab), at 400-500 rpm, for one hour.
Heat shock of cells in a 96 well PCR plate can be conveniently done in a thermal cycler.
-
Plate transformations on to LB agar, containing appropriate antibiotic, in 24 well blocks containing two to four glass beads per well. Shake plates in orbital shaker until all liquid is absorbed into plate.
Transformations can be plated using a liquid handling robot, or a multichannel pipettor with an expandable head. Four 24 well blocks recreate a standard 8 × 12 96 well plate.
Incubate inverted blocks overnight at 37°C.
Isolate miniprep plasmid DNA robotically, as described in Support Protocol 3 – Robotic Isolation of Plasmid DNA.
Basic Protocol 2
Small-Scale Expression and Purification
Expression of target proteins is under the control of the T7 promoter (see figure 2), necessitating the use of bacterial strain expressing the T7 RNA polymerase (Studier and Moffat, 1986; Rosenberg et al., 1987; Studier et al., 1990; Dubendorff and Studier, 1991). In our laboratory, we primarily use BL21(DE3)-pLysS-T1R cells, a lambda DE3 lysogenic E.coli strain expressing T7 RNA polymerase under the control of the lacUV5 promoter.
Expression of T7 RNA polymerase is induced by IPTG, which, in turn, induces the expression of the target protein. Additionally, the pLysS plasmid, encoding the T7 RNA polymerase-specific lysozyme, reduces the level of “leaky” expression of the target protein by degrading the T7 RNA polymerase in the un-induced state (Moffat and Studier, 1987; Studier, 1991). This tight control of T7 RNA polymerase expression is critical, especially when expressing proteins that are potentially toxic to the host (e.g. membrane proteins).
Other BL21 derivatives such as the Tuner and Origami strains from Novagen may also be used for expression. The Tuner strain and its derivatives permit tighter control of the level of protein expression by adjusting the amount of IPTG used for induction, possibly avoiding the toxic effects of the expressed protein to the host cell. The Origami strain contains the pRARE2 plasmid, which encodes seven tRNAs for codons not commonly used by E. coli. This can be a faster, cheaper and more practical alternative to gene synthesis and codon optimization when attempting to express large number of eukaryotic targets in bacteria. We don't have much experience the Tuner strains, but some laboratories may find that there are particular applications where their use is beneficial. The majority of the targets selected by our laboratory are from prokaryotes. However, when we do attempt to express targets of eukaryotic origin, we always use expression strains containing the pRARE plasmid.
When first determining the conditions to be used for our high-throughput expression pipeline, we invested considerable effort comparing expression strains, media, IPTG concentration used for induction of protein expression and temperature post-induction. We now use a growth and induction protocol that has been found to be optimal, in the sense that we found the greatest number of targets expressing from a given test set of 96 unique targets. This is this protocol that we describe below. Nonetheless, these procedures can easily be modified to improve the expression of any specific target (or targets). For instance, the time of induction (e.g. A600 ranges from 0.5-1.0), duration of induction, temperature used for growth (e.g. overnight induction at 18°C, 25°C or 30°C), or IPTG concentration used for induction (typically 0.4-1.0 mM final concentration), can all be customized to optimize the expression of certain targets. These factors are discussed in greater detail in the Commentary section at the end of this chapter.
After growth and induction, bacterial pellets are frozen at -80°C for at least one day (the freeze/thaw promotes cell lysis). Pellets are then resuspended in buffer and lysed with a robotic sonicator. A hand-held sonicator can also be used, although this is more labor-intensive.
For protein purification, we routinely use N-dodecyl-β-D-maltopyranoside (DDM) for extraction. We have found this detergent to be the most useful in our hands, although other detergents such as N-decyl-β-D-maltopyranoside (DM) or N-octyl-β-D-glucopyranoside (OG) can also be used.
After extraction, proteins are bound to nickel chelating resin, washed and eluted from the resin with Imidazole. The affinity-purified proteins are separated by SDS-PAGE and visualized by staining with Coomassie blue.
Materials
2×TY media (see recipe)
E. coli BL21(DE3)-pLysS-T1R competent cells
Antibiotics
Isopropyl-β-D-1-thiogalactopyranoside (IPTG, 100 mM)
2× Glycerol Storage Solution (see recipe)
Resuspension Buffer (see recipe)
Benzonase nuclease (EMD Millipore 70664-250KUN)
4-(2-aminoethyl)-benzenesulfonyl fluoride HCl (AEBSF, Bio-Research Products #401)
Resuspension Buffer containing 12% (w/v) N-dodecyl-β-D-maltopyranoside (DDM, Affymetrix D310)
Nickel-chelating resin (GE Healthcare, Qiagen, G Biosciences and others)
Large-orifice pipet tips
Wash Buffer B (see recipe)
Elution Buffer (see recipe)
5× SDS-PAGE loading buffer (see recipe)
96 well deep-well blocks (Costar 3960)
Shel Lab shaking incubator (SI6R-HS)
Sonicator Robot-small probe (ST Robotics, Cambridge), 30% maximum amplitude (or a hand-held sonicator)
Titramax 1000 shaking platform (Heidolph Instruments, Elk Grove Village, IL)
Adhesive-backed porous (VWR 60941-086) and foil (VWR 60941-076) seals
96 well, 2ml filter plates (Thompson Instrument Company 931919)
96 well bottom plate seal (Thompson Instrument Company 982005)
Vacuum manifold
Centrifuge with plate rotor
96 well U-bottomed plates (Greiner 650101)
Criterion Precast 4-20% (w/v) acrylamide Tris-HCl Gels, 26 wells (Biorad 345-0034)
Growth and expression
Combine 1 μl of miniprep plasmid DNA with 20 μl BL21(DE3)pLysS-T1R competent cells in a 96 well PCR plate and incubate on ice for 30 minutes.
Heat shock cells at 42°C for 45 seconds and recover exactly as done for transformation of the cloning strain.
-
Transformations are used to directly inoculate 500 μl 2×TY (plus antibiotics) in a 96 well deep-well block. Incubate overnight 37°C, shaking at 850 RPM (Shel Lab)
For growth of pLysS versions of the BL21 expression strains, two antibiotics are included in the media – one for the resistance conferred by the vector and Chloramphenicol, at a final concentration of 34 μg/ml, to maintain the pLysS plasmid.
-
The next morning, use 30-40 μl of each saturated, overnight culture to inoculate 1 ml pre-warmed 2×TY (plus antibiotics, added immediately before use) in a 96 well deep-well block.
Use the remaining overnight culture to prepare a glycerol stock in a 96 well deep-well block by combining equal volumes of the overnight culture and 2× glycerol storage solution. Seal with an adhesive-backed foil seal and mix by inverting several times. Store the block at -80°C. This stock can be used to repeat small-scale expression, or for inoculating larger cultures for scale-up (see Basic Protocol 3).
-
Incubate in 37°C shaker (Shel Lab), at 850 rpm until A600 is ∼1.0.
Absorbance at 600 nm can be monitored during growth by removing a small portion of a well chosen at random and diluting in media. After several growths at the small-scale, one can simply use time to determine when to induce. In our laboratory, it usually takes about 1.5 hours to reach an A600 of ∼1.0
Add 10 μl 100 mM IPTG (1 mM final concentration) and return deep well block to 37°C shaker for four hours. Harvest cells by centrifugation at 4500 × g for 10 minutes, pour off the supernatant, seal blocks with foil and store cell pellets at -80°C.
Cell lysis and solubilization
-
Place 500 μl of resuspension buffer, containing 0.5 mM TCEP, 4 μl/25 ml (4 units/ml final) Benzonase and 600 mg/500 ml (5 mM final) AEBSF, into each well
Benzonase is a genetically engineered endonuclease from Serratia marcescens, which degrades both DNA and RNA, while having no proteolytic activity. AEBSF is a serine protease inhibitor with a specificity similar to that of phenylmethylsulfonyl fluoride (PMSF). Prepare enough extra Resuspension Buffer to make Resuspension Buffer with 12% (w/v) DDM for solubilization.
Lyse cells with two rounds of sonication using a sonicator robot.
Add 100 μl of resuspension buffer containing 12% (w/v) DDM, to obtain a final DDM concentration of 2% (w/v). Incubate blocks on a shaking platform (Titramax 1000) at 4°C for one hour to complete the solubilization of membrane proteins.
Nickel affinity protein purification
Equilibrate Ni2+ resin with Resuspension Buffer by pouring desired amount of Ni2+ resin into a 50 ml centrifuge tube. Centrifuge at 500 × g for two minutes and pour off supernatant. Repeat. Add equal amount of Resuspension Buffer to the pelleted Ni2+ resin, resulting in a 50% (v/v) slurry. Prepare at least 50 ml at a time. It can be stored at 4°C and is enough for several hundred small-scale purifications. We have used Ni2+ resin from several different manufacturers and have never noticed any difference in performance. We suggest obtaining resin from the least expensive source available.
-
Transfer lysate to a 96 well Thomson filter plate, sealed with a Thomson bottom seal and add 50 μl of a 50% (v/v) slurry of Ni2+ resin to each well.
Use tips with large orifices when pipetting the nickel slurry.
Place the filter plate on a shaking platform (Titramax 1000) at 4°C and shake at 600 rpm overnight.
The following day, unseal filter plate and place it on a vacuum manifold to remove the lysate.
Reseal filter plate and add 1 ml of Wash Buffer B to each well.
Shake at 600 rpm for 30 minutes on a shaking platform at 4°C
Unseal filter plate and place it on a vacuum manifold to remove wash buffer. Repeat wash step.
Place filter plate on top of a 96 well U-bottomed plate and centrifuge for two minutes at 500 × g to completely remove wash buffer.
Replace Thomson bottom seal on filter plate and add 35 μl of elution buffer into each well. Shake at 600 rpm for 30 minutes at 4°C on a shaking platform
Remove bottom seal and place filter plate on top of a clean 96 well U-bottomed plate. Centrifuge for two min at 500 × g to collect eluates.
-
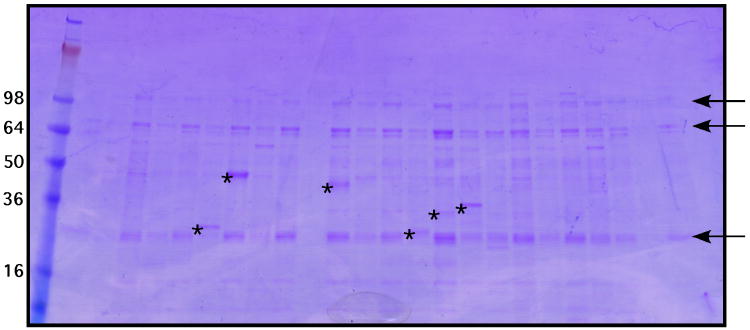
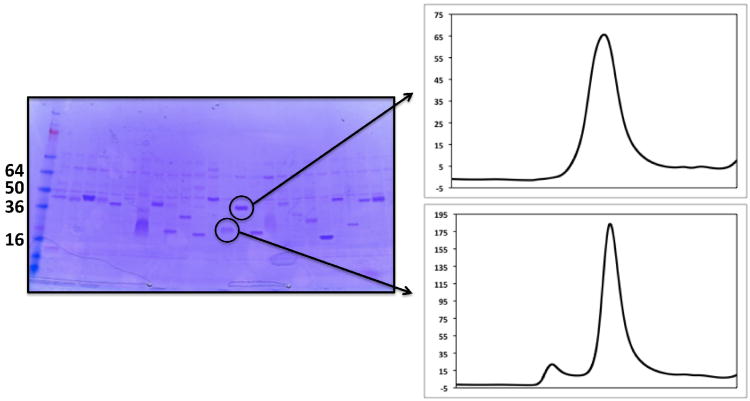
Add 7 μl 5× SDS-PAGE loading buffer and load 15-20 μl of each eluate on to a Bio-Rad Criterion gel. Visualize purified proteins by staining with Coomassie blue. Typical results from the small-scale expression and purification from 24 unique targets is shown if Figure 3.
Unlike traditional sample preparation for SDS-PAGE, membrane protein samples are not heated prior to loading on to the gel. It has been reported that this often leads to precipitation of the membrane protein, them not to migrate into the gel. We frequently observe membrane proteins migrating at an apparent molecular weight approximately 10% smaller than its predicted molecular weight. This could be due to the fact that without heating, the membrane protein sample is not completely denatured.
Targets that produce a visible band of the expected size are being expressed at ∼0.5 mg/ml.
Figure 3.
Coomassie-stained gel of proteins expressed and purified at the small scale.
24 unique targets were grown in 1 ml media in a 96 well deep-well block and purified by Ni2+ affinity chromatography. A portion of each purified protein was separated by SDS-PAGE and visualized by staining with Coomassie blue. Sizes of molecular weight standards are indicated along the left hand side of the gel. Contaminating proteins are indicated by arrowheads. Expressed proteins are indicated by asterisks.
Basic Protocol 3
Mid-Scale Expression of 96 Proteins in the GNF Fermenter
Once a set of targets that express at the small-scale have been identified, it is useful to re-test these on a slightly larger (e.g. 500 ml) scale. The quantity of protein purified at this scale is then used for further analysis, such as size exclusion chromatography and/or a detergent stability assay. In our laboratory we are able to simultaneously grow 36 500 ml cultures, in 2-liter baffled Erlenmeyer flasks, using a set of three stacked Innova shakers. Even at this capacity, which is already greater than that of most laboratories, it would require three days to grow an entire set of 96 targets. Although we have used this method previously (see Alternate Protocol), we now employ a much more efficient and less time-consuming approach with the GNF Fermenter (Genomics Institute of the Novartis Research Foundation, www.gnf.org). The GNF Fermenter is a programmable, 96-channel, airlift fermenter, which uses tubes arrayed in an 8 × 12 grid. Each tube holds approximately 65 ml of media and cells can be grown to a very high density, as no shaking is involved. The mass of the cell pellet obtained from each tube of the GNF fermenter is roughly equivalent to that from a 500 ml culture grown in a shake flask.
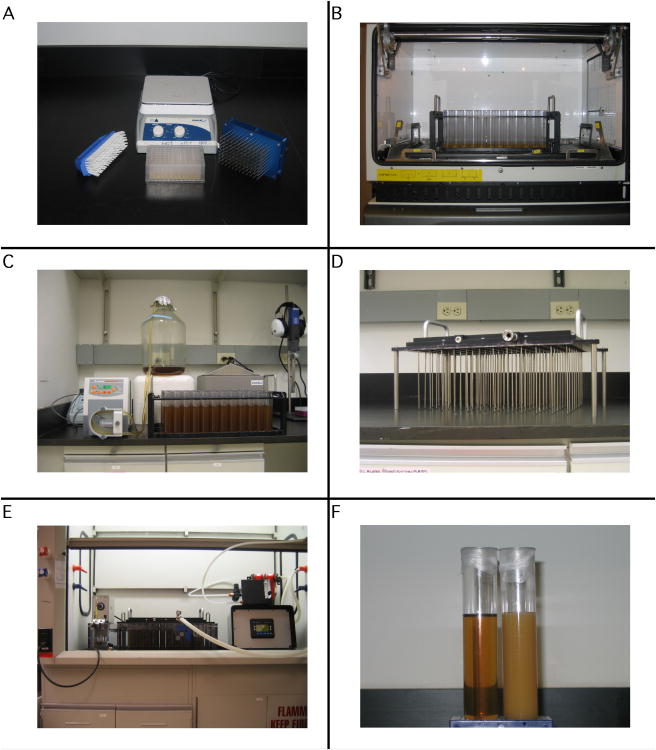
To initiate a growth and expression protocol in the GNF Fermenter, we use the glycerol stocks of transformed BL21 cells from the previous step (Basic Protocol 2) to start a fresh culture. By using a liquid handling robot and a “rearray” file, all clones that showed expression at the small scale are transferred to a single 96 well deep-well block. After a short recovery, the cultures are used to inoculate 10 ml of media in each of the corresponding fermenter tubes. The GNFermenter tube rack is then placed in an Innova 44 shaking incubator and incubated overnight. The next day, each tube is filled with additional media and the cultures are allowed to grow to high density by using a mixture of compressed air and oxygen. At the end of the growth and expression protocol, the cells are collected by centrifugation and stored at -80°C for future purification (see Basic Protocol 4). Details of the assembly of the GNF Fermenter and an example of a typical culture at the end of the expression protocol are shown in Figure 4.
Figure 4.
Assembly of the GNF Fermenter.
The photo in Panel A shows the 96-pronged replicating tool from Enzyscreen. The tips of the tool are washed with the brush under deionized water and sterilized by heating to 300°C on a hot plate for 5 minutes. 8-10 ml 2×TY in each of the 96 tubes of the GNF Fermenter are inoculated from glycerol stocks and the entire rack is loaded into an Innova 44 shaker and grown to saturation overnight at 37°C as shown in Panel B. The following morning, fresh media is added to each tube with the aid of a peristaltic pump as shown in Panel C. Panel D shows the gas manifold. The manifold sits on top of the fermenter tubes, with each of the 96 cannulae delivering air and O2 directly into each culture. This provides exceptional aeration and mixing. Panel E shows the completely assembled fermenter inside a fume hood. The rack of tubes is lowered into a circulating water bath for temperature control. The gas mixing chamber and controller are to the right of the fermenter. Panel F shows the media before (left) and after (right) the 5.5 hour growth and induction protocol. Optical densities, measured at 600 nm, of greater than 20, are routinely obtained.
Materials
2×TY
Terrific Broth (TB, see recipe)
Antibiotics
1 M 4-morpholinopropanesulfonic acid (MOPS), pH 7.6 (see recipe)
1 M MgSO4
1 M Isopropyl-β-D-1-thiogalactopyranoside (IPTG)
Cryo-replicator (EnzyScreen #CR1000, Haarlem, The Netherlands)
Liquid handling robot
GNF Fermenter (Genomics Institute of the Novartis Research Foundation, San Diego, CA)
Compressed air and bottled O2
Starting the overnight cultures
Fill the required number of 96 well deep well blocks with 1 ml 2×TY (plus antibiotics) per well.
Set a hot plate to 300°C
Wash EnzyScreen cryo-replicator under deionized water with included brush and sterilize by heating on the hot plate for five minutes. Remove from hot plate, place on its side and allow it to cool for 12 minutes.
Place cryo-replicator on glycerol stock for five seconds and transfer tool to deep well block containing media to inoculate. Repeat for all blocks to be inoculated.
Place deep well block in shaker incubator (Shel Lab) and allow cells to recover for at least 30 minutes, at 37°C, shaking at 850 rpm.
Meanwhile, prepare at least one liter 2×TY, containing the required antibiotics. Transfer 8-10 ml of media (plus antibiotics) to each tube of the GNF Fermenter, held in the fermenter tube rack.
Use liquid handling robot to re-array cultures from step 5, so that each of the clones to be tested is now in a single 96 well deep well block.
Transfer each 1 ml of inoculated media from the 96 well deep well block to the corresponding GNF fermenter tubes, held in the tube rack. Transfer rack to Innova shaker with the appropriate shelf installed.
Incubate cultures overnight at 37°C, shaking at 250 rpm.
Growth and Induction Protocol for GNF Fermenter
The next morning, prepare Terrific Broth, supplemented with 100 mM MOPS, pH 7.6, 2 mM MgSO4 and antibiotic(s). This is the media to be used for growth and expression of the target proteins.
Use a peristaltic pump to transfer 55 ml of growth media to each tube of the GNF Fermenter containing the overnight culture.
Lower rack of tubes into the GNF Fermenter water bath, pre-warmed to 37°C. Install the gas manifold, connect the tubing and set the programmer to the desired growth protocol.
-
In our laboratory, our standard growth protocol is one hour in the presence of house air, followed by a 90 minute-long period, during which the oxygen concentration is increased linearly to 100%. We then induce protein expression by the addition of IPTG to a final concentration of 1 mM. Protein expression is for three hours in the presence of pure oxygen.
There can be a considerable amount of evaporation that takes place during the growth and expression protocol. Typically, we dilute our 1 M IPTG stock in 5% (v/v) glycerol to a concentration of 16 mM. Four ml of the IPTG/glycerol mixture is then added to each culture at the point of induction. This results in an IPTG concentration of 1 mM final and helps replace some of the culture volume lost as a result of evaporation. It may be helpful to do this at one additional time (without IPTG) during the 5.5-hour procedure, to ensure that one recovers at least 50 ml culture at the time of harvest. The additional feeding can be media instead of dilute glycerol and we find it is often convenient to save any leftover TB media used for filling the tubes at the beginning of the procedure for this purpose.
-
At the end of the growth and expression protocol, pour 50 ml of each culture into a 50 ml conical centrifuge tube. Pellet cells by centrifugation, pour off supernatant and store cell pellets at -80°C.
For simplicity, tubes can be labeled with well position and date as long as a record is kept separately so that one knows which clone is in each well/tube.
Alternate Protocol
Mid-Scale Expression Using Erlenmeyer Flasks
This is a more conventional method for growing cells for a medium scale-up, but does not have the high throughput capacity of the GNF Fermenter. A single deep well block, containing 96 different targets that have been shown to express at the small scale can be prepared as above, but the saturated overnight cultures are used to inoculate 500ml culture media in a 2-liter Erlenmeyer flask. Growth and induction is similar to the small-scale expression (Basic Protocol 2).
Materials
2×TY (see recipe)
Antibiotics
1 M Isopropyl-β-D-1-thiogalactopyranoside (IPTG)
Antifoam 204 (Sigma A6426)
2 Liter baffled Erlenmeyer flasks
Innova 44 incubator (Eppendorf)
Shel Lab incubator
Shaking incubator containing a rack for holding 50 ml tubes
Preparing the overnight cultures
-
Re-array targets from glycerol stocks as in steps 1-5 above (Starting the overnight cultures) and use them to inoculate 10 ml of 2×TY (containing appropriate antibiotics) in 50 ml conical centrifuge tubes.
Alternatively, if only a small number of cultures are to be grown, cultures can be directly recovered in 10 ml of 2×TY by using sterile plastic inoculating loops to remove some of the frozen cells from the deep-well blocks.
-
Incubate the 50 ml tubes overnight at 37°C, shaking at 250 rpm. Pre-warm the required number of 2-liter flasks containing 500 ml of 2×TY at 37°C.
If space permits, it is convenient to use the same incubator for pre-warming the media as will be used for expression. Do not shake the flasks. We have observed that growth is substantially better when media is pre-warmed.
Growth and Induction
The following day, add the appropriate antibiotics and the 10 ml saturated overnight cultures to the pre-warmed media in the flasks. Incubate flasks in an Innova 44 incubator at 37°C, shaking at 250 rpm, until A600 is ∼1.0 (typically 1.5-2 hours).
Add 0.5 ml of 1 M IPTG (final concentration 1 mM) to each flask and continue shaking for another 4 hours at 37°C.
-
Harvest cells by centrifugation and store pellets at -80°C.
We recommend using a spatula to transfer the bacterial cell pellet from the centrifugation bottles to 50 ml conical tubes for long-term storage. An ordinary rubber, plastic, or silicone kitchen spatula works quite well.
Basic Protocol 4
Mid-Scale Purification and Size Exclusion Chromatography
The mid-scale purification protocol is essentially a scaled-up version of the small-scale purification protocol (Basic Protocol 2), with an additional washing step that includes ATP. This wash step is included to remove a potential chaperone (heat-shock protein GroEL) frequently found bound to over-expressed proteins (Thain et al., 1996). ATP achieves this by stimulating substrate release from GroEL. This step is not included in the small-scale purification protocol, as the proteins recovered from those purifications are not used for further analytical steps (such as size exclusion chromatography) where the presence of the GroEL contaminant might interfere and make conclusive analysis more difficult. It should be noted that we have never performed a thorough comparison of purification results with and without the ATP wash. But neither have we ever observed anything that suggests that inclusion of the wash had a negative effect. The necessity of including this step should be determined by the individual laboratory.
Lysis, solubilization and binding are all performed in 50 ml conical tubes. For convenience, all wash steps and elution are done in 96 well filter plates. After proteins are eluted, a small portion of the eluate is resolved by SDS-PAGE and the gel stained with Coomassie blue, to confirm that the protein of the expected size is expressed in the larger culture volume. The remainder of the sample is used to screen the aggregation status of the membrane proteins by size exclusion chromatography. Proteins that are in a soluble, monodisperse state (i.e. not in an aggregated form) will produce single and relatively sharp peaks by size exclusion chromatography, whereas proteins that form nonspecific aggregates will produce broad and numerous peaks, or appear in the void volume of the column. Only those proteins that are soluble and monodisperse are useful for further use (large-scale purification, crystallization etc). For size exclusion chromatography, we use an Agilent Technologies 1200 Series HPLC system with a Superdex S200 column, although any chromatography system capable of doing size exclusion chromatography may be used.
Materials
Benzonase nuclease (EMD Millipore 70664-250KUN)
4-(2-aminoethyl)-benzenesulfonyl fluoride HCl (AEBSF, Bio-Research Products #401)
Nickel-chelating resin (GE Healthcare, Qiagen, G Biosciences and others)
Resuspension Buffer (see recipe)
ATP Wash Buffer (see recipe)
Wash Buffer B (see recipe)
Elution Buffer (see recipe)
2× SDS-PAGE loading buffer (see recipe)
96 well, 2ml filter plates (Thompson Instrument Company 931919)
96 well bottom plate seal (Thompson Instrument Company 982005)
Vacuum manifold
50 ml conical centrifuge tubes
Sonicator Robot-large probe (ST Robotics, Cambridge), 60% maximum amplitude (or a hand-held sonicator)
Titramax 1000 shaking platform (Heidolph Instruments, Elk Grove Village, IL)
Roto-Shake Genie (VWR 58815-176; or similar rotating platform)
Beckman Coulter Avanti J26-XP centrifuge (or similar)
96 well U-bottomed plates (Greiner 650101)
Criterion Precast 4-20% (w/v) acrylamide Tris-HCl Gels, 26 wells (Biorad 345-0034)
Agilent Technologies 1200 Series HPLC system
-
TSK-Gel Super SW3000 size exclusion column, 4.6 × 300 mm (Tosoh 18675)
Alternatively, 5/150 Superdex S200 column (GE Healthcare 28-9065-61)
Resuspension and Lysis
-
Remove cell pellets from -80°C freezer, and resuspend each pellet in 25 ml resuspension buffer, containing 0.5 mM TCEP, 4 μl/25 ml (4 units/ml final) Benzonase and 600 mg/500 ml (5 mM final) AEBSF.
Benzonase is a genetically engineered endonuclease from Serratia marcescens, which degrades both DNA and RNA, while having no proteolytic activity. AEBSF is a serine protease inhibitor with a specificity similar to that of phenylmethylsulfonyl fluoride (PMSF). Prepare enough extra Resuspension Buffer to make Resuspension Buffer with 12% (w/v) DDM for solubilization.
Vortex and mix vigorously to dislodge the cell pellet.
Lyse cells with five rounds of sonication, using robotic sonicator (or hand-held sonicator).
Add 5 ml Resuspension Buffer, containing 12% (w/v) DDM, 0.5 mM TCEP and 600 mg/500 ml (AEBSF) to each 50 ml conical tube.
Rotate 50 ml conical tubes end-over-end on a Roto-Shake Genie at 4°C for one hour, or until lysate is clear.
Remove insoluble material by centrifugation at 6800 × g for 20 minutes at 4°C.
Purification of fusion proteins
In separate, clean, 50 ml conical tubes add 200 μl of a 50% (v/v) Ni2+ slurry.
Pour clarified lysates into clean 50 ml conical tubes containing the Ni2+ slurry.
Rotate 50 ml conical tubes end-over-end for one hour at 4°C to allow proteins to bind to the Ni2+ resin.
-
Centrifuge the tubes at 500 × g for 5 minutes at 4°C. Pour off all but ∼1 ml of the supernatant.
Ni2+ resin will form a loose pellet, be careful not to pour off the resin along with the supernatant. Leaving some buffer with the resin will make transferring the resin to the filter plates easier and more efficient.
Pipette the pelleted resin into an unsealed 96 well Thompson filter plate and allow buffer to drain by gravity. Seal the filter plate with a Thompson 96 well bottom plate seal.
Add one 1 ml ATP wash buffer, containing 5 mM ATP, 0.1 mM TCEP, 0.05% (w/v) DDM and shake at 600 rpm on a Titramax 1000 shaking platform for 30 minutes at 4°C.
Remove the seal and remove wash buffer by vacuum.
Add 1 ml Wash Buffer B, containing 0.1 mM TCEP and 0.05 (w/v) DDM, to the filter plate. Shake at 600 rpm for 30 minutes. Remove wash buffer by vacuum. Repeat wash step.
Elution and analysis
Unseal the filter plate and remove Wash Buffer B by vacuum.
Place a 96 well round bottom plate beneath the unsealed filter plate and centrifuge at 500 × g for two minutes to remove any remaining wash buffer.
Reseal filter plate and add 100 μl Elution Buffer, containing 0.1 mM TCEP and 0.05% (w/v) DDM. Shake filter plate at 600 rpm for 30 minutes at 4°C.
-
Place a clean 96 well round bottom plate beneath the filter plate and centrifuge at 500 × g for two minutes to collect eluates containing purified proteins.
Optional - Add another 100 μl Elution Buffer to the filter plate, seal and shake at 600 rpm at 4°C for 30 minutes. Remove seal, place a clean 96 well round bottom plate beneath the filter plate and centrifuge at 500 × g for two minutes. Combine eluates. Whether or not you choose to perform this step depends upon what is planned for the sample. An eluate volume of 100 μl is sufficient for loading a gel and analysis by gel filtration in a single detergent. If one wishes to perform the Alternate Protocol – Detergent Stability Assay, then a total eluate volume of 200 μl is required.
Combine 10 μl eluate with 10 μl 2× SDS-PAGE loading buffer. Load entire volume on to a Bio-Rad Criterion gel. Visualize proteins by staining with Coomassie Blue.
-
Use the remainder of the sample to perform a gel filtration analysis on an automated Agilent HPLC system with a Superdex S200 column at a flow rate of 0.2 ml/min, coupled to a variable wavelength detector. Protein absorbance is measured at 280 nm.
We prefer using a Superdex column for screening, instead of a silica-based HPLC column, for the simple reason that a larger sample volume can be injected, as per the manufacturer's specifications. We typically inject 50 μl of sample on the Superdex column, whereas the maximum sample volume for the silica-based column is just 5 μl. Moreover, almost all membrane proteins purified at a scale suitable for obtaining enough protein for crystallographic studies are performed with the same two-step purification procedure - Ni2+ affinity chromatography and gel filtration. Therefore, our analysis better reflects the types of results our colleagues might expect after scaling up.
Alternate Protocol
Detergent Stability Assay
An alternative method for analyzing the aggregation status of membrane proteins is to test whether they are stable in an environment where there is an excess of detergent (2-10 times the critical micelle concentration, or CMC). This system can also be conveniently used to test the stability of a protein in different detergents (it is well-established that membrane proteins that remain stable in different detergents are more likely to crystallize). In our laboratory, we routinely test four of the most commonly-used detergents for membrane protein crystallization (DDM, LDAO, C8E4 and βOG), but additional detergents can also be assayed (such as DM and OM).
After incubation of the protein with the detergents, samples are filtered to remove insoluble material, and the aggregation status of the protein is analyzed by size exclusion chromatography as described above except that a Superdex column must to be used as the Tosoh TSK-Gel Super SW3000 columns (and most silica-based columns) will often perform poorly in the presence of LDAO.
It has been reported that silica can adsorb the detergent LDAO (Moller and le Maire, 1993; Matsson et al., 2005), so using LDAO-containing buffers for size exclusion chromatography on silica-based columns should be done with caution, or avoided altogether. Equilibrating the column with several column volumes of LDAO-containing buffer prior to sample injection, or using greater than 2× CMC LDAO in the mobile phase may help avoid any problems. Alternatively, avoid any complications altogether by employing a Superdex S200 column, instead of a silica-based column, as described above.
Materials
Gel Filtration Buffer (see recipe)
N-dodecyl-β-D-maltopyranoside (DDM)
n-Dodecyl-N, N-Dimethylamine-N-Oxide (LDAO),
2-[2-[2-(2-octoxyethoxy)ethoxy]ethoxy]ethanol (C8E4)
-
n-Octyl-β-D-Glucopyranoside (βOG)
All detergents are from Affymetrix and are dissolved in Gel Filtration Buffer. Affymetrix (formerly Anatrace) is by far the largest supplier of detergents used for the purification and analysis of integral membrane proteins. The quality of their products is superb. For crystallization, it is of utmost importance to use detergents of the highest quality available. Among the members of our consortium, only detergents from Affymetrix are utilized. If the purified proteins are to be used for other purposes, less expensive (and likely lower quality) sources of detergents may possibly be used, but advise caution.
96 well U-bottom plates (Greiner 650101)
Adhesive-backed foil (VWR 60941-076)
384 well filter plates (Pall 5070)
384 well receiver plates (Nunc 264573)
96 well plates for HPLC samples (Agilent 5042-8502, or Eppendorf 951020401)
Pierceable silicone sealing mats (Agilent 521-01-151)
Agilent Technologies 1200 Series HPLC system
-
TSK-Gel Super SW3000 size exclusion column, 4.6 × 300 mm (Tosoh 18675)
Alternatively, 5/150 Superdex S200 column (GE Healthcare 28-9065-61)
Incubate purified protein in excess quantities of detergents to be tested
-
In a 96 well U-bottom plate, combine 4 × 40 μl of each eluate with the volume, concentration and type of detergent indicated below.
2 μl of 5% (w/v) DDM (0.25% (w/v) final)
2 μl of 5% (w/v) LDAO (0.25% (w/v) final)
4 μl of 5% (w/v) C8E4 (0.5% (w/v) final)
3 μl of 10% (w/v) βOG (0.75% (w/v) final)
Seal the plate with adhesive-backed foil and incubate at room temperature for two hours.
-
Place a Pall 384 well filter plate on top of a Nunc 384 well receiver plate. Transfer eluate/detergent mixes to the filter plate. Centrifuge the plate at 2500 × g for 10 minutes to remove insoluble material.
If the full volume of sample is not recovered, due to precipitation and clogging of the filter membrane, repeat centrifugation until enough volume is obtained for gel filtration chromatography.
Transfer flow through to an Agilent 96 well plate, cover with a piercable silicone-sealing mat and proceed with gel filtration
-
Gel filtration analysis is performed on an automated Agilent HPLC system, using a Tosoh TSK-Gel Super SW3000 size exclusion column at a flow rate of 0.25 ml/min, coupled to a variable wavelength detector. Protein absorbance is measured at 280 nm.
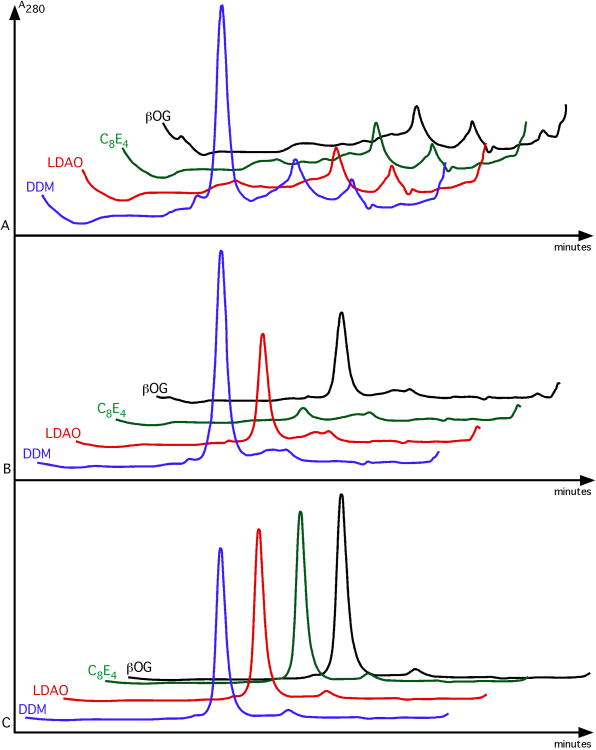
Representative gel filtration results of poor-, fair- and well-behaved proteins are shown in Figure 6.
If using a Superdex S200 column instead of a silica column for SEC, reduce flow rate to 0.2 ml/min.
Figure 6.
Detergent stability assay results.
Three different proteins were purified in DDM, divided into four portions and mixed with a large excess of LDAO, C8E4, or βOG. The addition of an excess quantity of the same detergent used for purification, DDM, serves as a control. Panel A shows a protein that is well behaved in DDM, but no others. The protein in Panel B tolerates LDAO and βOG, although there is likely some protein loss, as seen by the lower protein peak height as compared to that seen in DDM. The same protein is completely lost when combined with C8E4. It most likely precipitated during incubation at room temperature and was removed by filtration prior to size exclusion chromatography. Panel C is well behaved in all four detergents and is an excellent candidate for scale-up and crystallographic studies.
Support Protocol 1
Digestion and Gel Purification of Vector for LIC Cloning
Before vectors can be treated for LIC (see Basic Protocol 1), they need to be linearized by digestion with a unique restriction enzyme. That is, the enzyme should cut between the two regions that will be treated to produce single-stranded overhangs, but not elsewhere in the vector. Refer to figure 2 for the appropriate restriction enzyme to be used for any given vector.
Materials
10× NEB Buffers
100× BSA (10 mg/ml)
Restriction enzymes SnaBI, BfuAI, SspI and SmaI (NEB)
Incubators set at 25°C, 37°C, 50°C and 65°C
Agarose gel electrophoresis system
Gel extraction kit (Qiagen 28706) or similar
-
Digest 50-100 μg of plasmid DNA in a total volume of 100-150 μl, using the conditions recommended by the manufacturer (New England Biolabs).
In order to achieve complete digestion of large quantities of plasmid DNA, we find that overnight digests are acceptable, as long as one avoids using a large excess of enzyme. If in doubt, refer to the manufacturer's recommendations for the specific enzyme.
Separate DNA fragments on a 0.8-1.0% (w/v) agarose gel and isolate the linearized vector.
-
Purify the linearized vector using the Qiagen Gel Extraction Kit (or similar) according to the conditions recommended by the manufacturer.
One may make a compromise between the number of columns suggested by the manufacturer, given the weight of the agarose gel slice and the number of columns one can reasonably handle at the same time. However many columns are used, it is important that the concentration of the purified, linearized plasmid be greater than 40 ng/μl for the next step. To gel purify 100 μg linearized plasmid, we typically use 12 columns, regardless of the weight of the gel slice, without any noticeable problems with generation of the single-stranded overhangs, or annealing.
Measure the DNA concentration of the purified vector and store at -20°C.
Support Protocol 2
Pcr Amplification and Purification of PCR Products
Target DNA is amplified from bacterial genomic DNA or plasmid DNA using oligonucleotides containing the appropriate overhangs for a given vector (see Basic Protocol 1 and Table 2). When designing the oligonucleotides, care should be taken in regards to the native start and stop codon (i.e. N-terminally tagged constructs should contain the native stop codon (inclusion of the start codon is optional), whereas C-terminally tagged constructs should contain the native start codon and omit the stop codon).
For generating oligonucleotide sequences with correct overhangs, especially for large numbers of targets, we recommend using a freely available web-based program such as Primer Prim'er (Everett et al., 2004; http://www-nmr.cabm.rutgers.edu/bioinformatics/Primer_Primer/). For oligonucleotide synthesis we use Integrated DNA Technologies (www.idtdna.com). IDT will assemble oligos in 384- or 96-well plates with forward and reverse primers pre-mixed in the same well and diluted to working concentrations. For amplification, we routinely use KOD Hot Start DNA Polymerase (Novagen). Although other thermostable DNA Polymerases can be used, we have found KOD Hot Start DNA Polymerase to be superior to several others we have tested (for very long targets we recommend KODXL Polymerase [also from Novagen]). PCR reactions (15 μl) are assembled in 384-well PCR plates according to the manufacturer's specification (alternatively, reactions can also be done in 96-well plates (50 μl).
If amplifying from plasmid DNA, the PCR reactions should subsequently be treated with DpnI to remove template DNA (this step is not essential if the antibiotic resistance marker of the template DNA is different from the one carried on the cloning vector and in such circumstance can be avoided).
Following amplification, PCR products are purified to remove primers, nucleotides, salts, enzymes, etc. We use the Agencourt AMPure XP PCR purification kit (Beckman Coulter), which is amenable to automated platform (i.e. liquid handling robots). Alternatively, for manual applications, a high throughput 96-well purification kit should be considered (i.e. Qiagen MinElute 96 UF PCR Purification Kit).
Materials
KOD Hot Start DNA Polymerase Kit (Novagen #71086)
Template DNA (genomic DNA at 10 ng/μl, plasmid DNA at 0.5 ng/μl, cDNA etc)
Sense and anti-sense primer (5 μM each) assembled in 384 or 96 well plates
Dimethyl Sulfoxide (DMSO)
-
Restriction enzyme DpnI (New England Biolabs): 20mM Tris-Acetate, pH 7.9
10× Buffer 2 (New England Biolabs): 20mM Tris-Acetate, pH 7.9 50mM Potassium Acetate 10mM Magnesium Acetate 1mM DTT AMPure XP PCR purification kit (Beckman Coulter A63880)
96 well PCR plates (Eppendorf 951020401)
384 well PCR plates (Eppendorf 951020702)
Adhesive foil for microplates (VWR # 60941-076)
96 well silicone sealing mat (Axygen CM-96-RD)
Thermal cycler (Eppendorf Mastercycler, Eppendorf Mastercycler ep384, or similar)
Liquid handling robot
E-gel 96 2% (w/v)agarose (Invitrogen G7008-02)
Mother E-Base™ (Invitrogen EB-M03)
We have found that 96- and 384-well plates can become misshapen following repeated heating and cooling (as in a PCR amplification), or after freezing. For that reason, we strongly suggest using high-quality, hard-shell plates with a full skirt for any such applications, especially those that will later be handled by a robot.
On our newer Eppendorf 384-well thermal cycler the motor-driven lid comes down tightly on to the top of the plate. We find that an adhesive-backed foil seal is sufficient to prevent evaporation. However, evaporation can be a problem when using older thermal cyclers with mechanical lids that don't come down as tightly (e.g. older Eppendorf Mastercycler models). In these cases, we would recommend using a silicone seal.
Assembling and performing the PCR reactions
-
For 384 amplifications, assemble the following master mix (enough for 420 reactions, to allow for losses):
- 630 μl 10× Buffer for KOD Hot Start Polymerase
- 630 μl of dNTPs (0.2 mM each final)
- 378 μl of 25 mM MgSO4 (1.5 mM final)
- 126 μl of KOD Hot Start Polymerase (0.3 U/reaction)
- 3318 μl of water
- ------
- 5.082 ml
Optional: Add DMSO to a final concentration of 10%. This might improve amplification success, especially when using genomic DNA as template.
For more, or fewer PCR reactions, scale reaction mixture above accordingly. For 96 or fewer reactions, reactions can be performed in a 96 well PCR plate.
Keep on ice until ready.
-
Using a liquid handling robot, or a multi-channel pipettor, dispense 12.1 μl of the reaction mixture above, 2 μl of template DNA and 0.9 μl of primers to each well in a 384 well PCR plate.
We suggest using a 96 well plate containing the template DNA stocks (genomic DNA or plasmid DNA). Using a liquid handling robot and a “rearray” file, DNAs can be transferred from the 96 well plate to a 384 well PCR plate. Final amounts for genomic DNAs is 20 ng/reaction and for plasmid DNA 1 ng/reaction. Primers are at a final concentration of 0.3 μM.
Seal plate with an adhesive-backed aluminum foil. Vortex the plate and give it a short spin to collect liquid at the bottom of the well.
-
Perform amplification using a 384 well thermal cycler using cycling conditions recommended by the supplier of the thermostable DNA polymerase.
Our standard cycling conditions for KOD Hot Start Polymerase are as follows:
Following an initial denaturing step of 2 minutes at 95°C, then perform 45 amplification cycles of 25 seconds at 95°C, 20 seconds at 55°C and 1.5minutes at 70°C. These conditions are for targets with average sizes (i.e. 700-3000 bps). For amplification of longer targets, the extension step should be lengthened accordingly. Alternatively, for very long targets (over 5 Kbps) we recommend using KODXL Polymerase (Novagen).
-
Optional: Add DpnI to amplification reactions to remove plasmid templates and incubate at 37°Cfor 1-2 hours.
Since very little template DNA is used in each reaction, very little restriction enzyme is needed to remove it. We usually dilute DpnI 1:40 in 1× Buffer 4 (final conc. 0.5 U/μl) and add 1 μl per well.
Purification and analysis of PCR products
Purify the PCR products using the Agencourt AMPure XP PCR purification kit, using a liquid handling robot. Alternatively, split the 384 well PCR plate into four quadrants of 96 wells each and use a manual 96-well purification kit to clean up the products.
-
Analyze PCR products on a 96 well E-gel system (or similar high throughput agarose gel electrophoresis system)
The E-Gel® agarose gel system is a bufferless system for gel electrophoresis of DNA samples. Although the resolution is not very high, bands of the expected size are clearly visible and allow one to get an indication of the overall success of the amplification. We find that cloning by LIC to be so reliable, that if amplification of the target is successful, then cloning will be, as well.
Support Protocol 3
Robotic Isolation of Plasmid DNA
Plasmid DNA is isolated using the CosMCPrep plasmid purification system and a liquid handling robot. Although the CosMCPrep kit could be used manually, it is not recommended due to the numerous pipetting steps. For non-automated 96 well plasmid purification, we recommend PureLink 96 HQ Mini Plasmid DNA Purification Kit (Invitrogen), or the QIAprep 96 Turbo Miniprep Kit from Qiagen.
We typically pick just a single colony per construct and have found that our cloning success rate (i.e. percentage of correct clones as verified by sequencing) to be acceptable (usually in the 80% pass/total number of targets range). Picking more than one colony will increase the passing rate marginally, but is also more labor-intensive and expensive.
For insert verification, we recommend sequencing each construct. Other methods, such as restriction enzyme analysis, or amplification of inserts with vector-specific primers, can be used. However, the results can often be ambiguous and difficult to interpret.
Materials
2×TY (see recipe)
Antibiotics
Sterile toothpicks
70% (v/v) ethanol, prepared fresh
Isopropanol
96 well deep-well blocks (Costar 3960)
Shel Lab shaking incubator
CosMCPrep plasmid DNA isolation kit (Beckman Coulter A29174)
Liquid Handling Robot
96 well round bottom plates (Greiner 650101)
Using sterile toothpicks, pick a single colony from each well of the 24 well blocks and use to inoculate 1 ml 2×TY, containing appropriate antibiotic, in a 96 well deep-well block. Incubate overnight in 37°C incubator, shaking at 850 rpm.
-
Recover cells by centrifugation at 4500 × g for 10 minutes. Decant media.
A small aliquot of the overnight culture should be used to inoculate another culture for preparation of a glycerol stock and/or for sequencing. For sequencing, we use single pass sequencing services from Beckman Coulter Genomics using the T7 Universal primer (beckmangenomics.com). Sequences are verified by running BLAST against a target database (Altschul et al., 1990).
Isolate plasmid DNA using the CosMCPrep plasmid DNA isolation kit from Beckman Coulter. Elute DNA into 100 μl buffer RE1 into a 96 well round bottom plate. Store DNA at -20°C
Reagents and Solutions
Luria Broth (LB) Lennox Medium
1% (w/v) Tryptone
0.5% (w/v) Yeast Extract
0.5% (w/v) NaCl
LB-Agar
1% (w/v) Tryptone
0.5% (w/v) Yeast Extract
0.5% (w/v) NaCl
1.5% (w/v) Agar
SOC Medium
2% (w/v) Tryptone
0.5% (w/v) Yeast Extract
10 mM NaCl
2.5 mM KCl
10 mM MgCl2.
20 mM MgSO4
20 mM Glucose
2×TY
1.6% (w/v) Tryptone
1.0% (w/v) Yeast Extract
0.5% (w/v) NaCl
Terrific Broth (TB)
1.2% (w/v) Tryptone
2.4% (w/v) Yeast Extract
0.4% (w/v) Glycerol
17 mM KH2PO4
72 mM K2HPO4
2× Glycerol Storage Solution
65% (v/v)glycerol
0.1 M MgSO4
25 mM Tris-Cl pH 8.0
Resuspension Buffer
50 mM Hepes pH 7.8
300 mM NaCl
20 mM Imidazole pH 7.8
5% (v/v)glycerol
-
1 mM MgCl2.
The protease inhibitor AEBSF is added fresh to a final concentration of 1.2 mg/ml
The reducing agent TCEP is added fresh to a final concentration of 0.5 mM
ATP Wash Buffer
50 mM Hepes pH 7.8
300 mM NaCl
40 mM Imidazole pH 7.8
5% (v/v) glycerol
-
5 mM MgCl2.
NaATP is added fresh to a final concentration of 5 mM. Detergent is added fresh at a concentration of five times its critical micelle concentration (CMC). TCEP is added fresh to a final concentration of 0.1 mM
Wash Buffer B
25 mM Hepes pH 7.8
500 mM NaCl
75 mM Imidazole pH 7.8
-
5% (v/v) glycerol
Detergent is added fresh to a final concentration of twice its CMC. TCEP is added fresh to a final concentration of 0.1 mM
Elution Buffer
25 mM Hepes pH 7.8
200 mM NaCl
500 mM Imidazole pH 7.8
-
5% (v/v) glycerol
Detergent is added fresh to a final concentration of twice its CMC. TCEP is added fresh to a final concentration of 0.1 mM
5× SDS-PAGE loading buffer
200 mM Tris pH 6.8
10% (w/v) SDS
25% (v/v) glycerol
5% (v/v) β-Mercaptoethanol (added fresh)
0.05 (w/v) Bromophenol Blue
2× SDS-PAGE loading buffer
100 mM Tris pH 6.8
4% (w/v) SDS
20% (v/v)glycerol
0.2% (w/v) Bromophenol Blue
200 mM DTT (added fresh)
Gel Filtration Buffer
40 mM Hepes pH 7.8
200 mM NaCl
-
5% (v/v) glycerol
Detergent is added fresh to a final concentration of twice its CMC. TCEP is added fresh to a final concentration of 0.1 mM
All bacteriological growth media, once sterilized by autoclaving, can be stored indefinitely at room temperature. Media should be discarded if it becomes cloudy, or contamination is otherwise suspected.
All other buffers and solutions can be stored at 4°C for up to six months.
Commentary
Background Information
Escherichia coli are ideally suited for high-throughput expression of membrane proteins. They are inexpensive to grow and maintain, have short generation times and do not require sophisticated, specialized equipment for their culture. Furthermore, E. coli are especially well-suited for high throughput protein expression studies, as they are amenable to miniaturization. Vectors for expression of recombinant proteins in bacteria are widely available or can be easily constructed to suit particular purposes.
Even without the availability of automated liquid handling equipment, it is entirely possible to perform cloning and protein expression studies on the order of hundreds of targets with just a multichannel pipet. The advantages of a high throughput approach are that many different genes, many different constructs of a particular gene (i.e. deletions, mutations), or some combination of both can be tested simultaneously. This becomes especially useful when trying to determine the three-dimensional structures of membrane proteins. For instance, by testing homologues and orthologues for a desired membrane protein family, one increases the likelihood of resolving the structure of a member of that family. In fact, this high throughput approach has proven to increase dramatically the structures of membranes proteins to be deposited in the Protein Data Bank (PDB; see Kloppmann et al., 2012 for a summary) including depositions from our center (Cao et al., 2011a; Cao et al., 2011b; Chen et al., 2010; Czyzewski and Wang, 2012; Levin et al., 2009, 2012; Shaffer et al., 2009; Waight et al., 2010; Mancusso et al., 2012).
Moreover, there is a significant amount of additional data, protocols and reagents that can be collected from these structural genomics enterprises. This can facilitate other research efforts in the field of protein structures including drug development, functional analyses and antibody development (Skolnick et al., 2000; Lundstrom, 2006a, 2007; Weigelt, 2010).
Critical Parameters and Troubleshooting
Amplification of targets by PCR
After target selection, perhaps the most important step in establishing a high throughput pipeline is the first one - amplification of the target genes. The successful amplification depends on several parameters. And as no single set of conditions will lead to the amplification of all targets, it is imperative to adjust the various parameters until a satisfactory percentage of targets can be amplified. Optimization of PCR amplification is discussed in numerous other sources, including in the materials provided with the polymerase. As such, it won't be addressed here. Using the PCR conditions described above, we have found that a ≥90% of the total number of selected targets are successfully amplified, as determined by agarose gel electrophoresis (Support Protocol 2). In our experience, if we successfully amplify the target, we will almost certainly clone it successfully into one of our collection of expression vectors.
Optimization of protein expression-background
If a target has been successfully cloned, then how does one get it expressed? The expression and purification conditions described above (Basic Protocols 2 and 3) are those that we determined empirically to produce the greatest number of expressed proteins for a randomly chosen test set. Regardless of whether you are trying to obtain suitable levels of expression of a single protein for your particular needs, or obtain the greatest number of expressed proteins from a large set of targets (as in our case), modifying the expression conditions can have a significant effect.
Once a target has been cloned into a given expression vector and used to transform a particular expression host, the easiest variable to change is temperature. Over-expression of a heterologous protein in E. coli can be stressful and in extreme cases, toxic, to the host cell (Wagner et al., 2007). In many cases, expression of the foreign protein overwhelms the cell such that the protein is either misfolded, ends up in inclusion bodies or is proteolytically cleaved (Baneyx and Mujacic, 2004.) While it is possible to denature, purify and refold proteins from the inclusion bodies, it is best if this can be avoided.
Optimization of protein expression-temperature
One simple alternative is to induce protein expression at lower temperatures. If one lowers the temperature of the culture following induction, all of the metabolic processes of the cell, including transcription and translation of the induced protein, are slowed down. Lowering the temperature, therefore, lowers the expression level of the target protein, but in turn, these lowered protein levels may be less stressful to the cell and permit more properly folded protein to be purified as a result (Baneyx and Mujacic, 2004). The only concession of lower temperatures post-induction is that induction is typically done for a longer time. It is common to allow expression of the target protein to proceed at 22°C upwards of 24 hours. We are now in the habit of testing expression of all of our constructs at both 37°C and 22°C, sometimes with dramatic results. While it is quite common for a protein to be satisfactorily expressed at both temperatures, we find numerous examples of proteins that express at only one temperature, or the other.
Optimization of protein expression-expression strain
Once a construct (or set of constructs) has been prepared, the next variable worth exploring is the host cell strain. In our work, we routinely use BL21(DE3)pLysS. LysS is a T7 RNA polymerase-specific lysozyme, the presence of which helps to reduce the level of basal T7 RNA polymerase present in the uninduced state, in order to prevent any “leaky” expression of the heterologous protein (Moffatt and Studier, 1987). Additionally, LysS dampens the maximum level of protein expression by degrading some portion of the T7 RNA polymerase following induction with IPTG. If higher levels of protein expression are desired and the expressed protein does not appear to harm the host strain, then the pLysS-less version of BL21 should be tested. And conversely, if over-expression of the target protein in the absence of LysS seems to affect the growth and/or health of the host strain, then the pLysS-containing derivative of BL21 should be tried.
Other expression strains worth trying are the C41 and C43 strains, also known as the “Walker strains” (Miroux and Walker, 1996). In these strains, expression of the T7 RNA polymerase is under the control of a wild-type lacUV5 promoter, rather than the hybrid trp/lacUV5 promoter of the BL21 strains. In the C41 and C43 strains, both the basal and induced level of expression of the T7 RNA polymerase is lower than in the BL21 strain. Almost counter-intuitively, like lower temperature, lower levels of target protein expression as a result of lower levels of T7 RNA polymerase, often results in higher yields of recombinant protein that can be purified (Miroux and Walker, 1996).
One drawback, however, is that the C41 and C43 strains are not T1/T5 phage-resistant. Many high-throughput laboratories, ours included, are very reluctant to use non phage-resistant strains due to the potential of phage infection and loss of considerable time and effort. Fortunately, several phage-resistant strains, which are phenotypically, if not genotypically, identical to the Walker strains, are now available. These include the T7 Express and Lemo(DE3) strains sold by New England Biolabs (see also Wagner et al., 2008). We have only limited experience with these strains, but when a test set of 96 targets was expressed in the BL21, BL21pLysS, T7 Express and Lemo(DE3) strains, anywhere from 3-6 targets were expressed in one strain, but not in any of the other three (data not shown). This finding suggests these newer expression strains are potentially quite valuable and certainly worthy of additional testing. If one is handling a relatively small set of targets, and/or if suitable personnel resources are available, then testing each construct in each of the different expression strains, in parallel, is certainly worthwhile.
Optimization of protein expression-media
We have tested a variety of E. coli growth media and have found, unsurprisingly, that richer media tends to produce higher yields of protein per volume of culture. This is almost certainly due to the fact that the cells can grow to higher densities in rich media, leading to greater cell mass and consequently, larger quantities of protein. We use 2×TY almost exclusively, although we sometime use Terrific Broth (TB, sometimes with addition of buffer, see Basic Protocol 3), with comparable results. In our experience, LB does not give satisfactory results. One could consider using autoinduction media (Studier, 2005), which is much more convenient than traditional methods that induce protein expression by the addition of IPTG. However, from our experience, however, we have noted that a small but not negligible percentage of targets that don't express in autoinduction media are expressed at detectable levels in 2×TY, or TB when induced by IPTG.
Detergents for extraction and purification
As described in Basic Protocol 2 above, extraction and purification of membrane proteins expressed in E. coli is done in the presence of DDM. This detergent, like all maltose-based ones, is extremely mild, yet very efficient in solubilizing E. coli inner membranes, This, combined with its low critical micelle concentration (CMC), make DDM the preferred detergent for initial attempts at membrane protein purification. LDAO is an alternative, as its CMC is similar to that of DDM (for CMCs and other physical properties of detergents consult the Affymetrix website, http://www.affymetrix.com). Since its micelle size is small compared to that of DDM, it has the added advantage of being able to be easily removed and/or exchanged for another detergent by dialysis, SEC, or ultrafiltration. We have found a small number of examples from amongst our targets that actually seem to prefer being extracted and purified with LDAO, but the vast majority of success at the crystallization step has come with proteins extracted in DDM or DM.
Recently, a new class of maltose-neopentyl glycol (MNG) amphiphiles with very low CMCs has been developed for solubilization and purification of membrane proteins (Chae et al., 2010). We have used them in a limited number of purifications and have found their performance to be slightly inferior to that of DDM at the solubilization step. However, these are apparently excellent detergents for maintaining the biophysical properties of membrane proteins (Chae et al., 2010).
Anticipated Results
With a few subtle modifications over the past few years, we have produced and tested for expression more than 20,000 constructs using the procedures described above. We find that we typically successfully amplify by PCR greater than 90% of our targets. Our cloning success rate, as determined by sequencing of the resulting construct, is around 80%. This cloning success is the result of picking just a single colony for each construct.
We can typically increase our cloning success rate to ∼85% simply by picking a second colony. Of course, picking a second colony adds to the cost of the procedure, mainly due to the cost of DNA sequencing. It is up to the individual to determine if the additional cost incurred is worthwhile. We have found cloning of open reading frames into several different expression vectors to be extremely beneficial, as one can greatly increase the success of expression of a set of targets by trying them in a variety of different vectors. When we clone a given set of targets into the four different vectors described in Basic Protocol 1, we find we can obtain detectable levels of expression for upwards of 30% of the targets. With the use of the GNF Fermenter expression procedure described in Basic Protocol 3, we find that almost all of the targets that express at the small-scale are reproducibly expressed at the mid-scale, so that there are no losses of targets at this stage. How many targets make it all the way through the gel filtration stage to produce a good chromatographic profile, indicating a well-behaved protein, is highly dependent upon the nature of the targets selected for cloning and expression. However, we find that slightly less than half of all the targets to make it to the mid-scale purification stage produce a good gel filtration profile. This represents about 7.5% of the total number of targets originally selected for cloning. While this is a considerable attrition rate, we feel strongly that the targets that do make it through this rigorous screening procedure are indeed worth the effort to pursue. Since we now have a great deal of expression, purification and crystallization available to us, considerable effort has been invested by our bioinformatics group in hopes of better understanding what features make for a successful target. After considering qualities such as GC content, amino acid content, predicted molecular weight, predicted number of transmembrane domains, isoelectric point, etc., efforts were made to create predictive models that would allow us to focus our efforts only on those targets determined to more likely to express and behave well. Unfortunately, after creating test sets of proteins predicted to fail and others predicted to be successful, the predictor was found to be no better than our current approach (M. Punta, personal communication).
Time Considerations
The amount of time required for taking a set of selected targets all the way through the cloning, expression and characterization procedures described above is highly dependent upon the number of targets and the degree of automation available to the individual laboratory. With the aid of a liquid handling robot and two 384-well thermal cyclers, we can easily amplify upwards of 800 targets in a single afternoon. Purification of the PCR products and generation of the single-stranded overhangs required for cloning by LIC takes the better part of a day. Preparation of the gel-purified vector, containing the complementary single-stranded overhangs also requires about one day. Annealing of the PCR products to the vector, transformation of the cloning strain and plating of the transformed cells can be done in a day. Colonies can be picked the following day and grown overnight for isolation of plasmid DNA the following morning. The isolated plasmids can be used to transform the expression strain that afternoon and small-scale expression performed the next 1-2 days. Purification of the heterologously expressed proteins by nickel affinity and analysis of the recovered proteins by SDS-PAGE and staining with Coomassie blue can take 2-3 days. Mid-scale growth in the GNF fermenter is accomplished in a single day. Purification at the mid-scale is done in batches of two dozen/day, such that it takes four days to purify and analyze the entire set of 96 cultures from a single fermenter run. Assuming 2-3 researchers coordinate their efforts, approximately 800 targets could be processed, start to finish, in under a month.
Figure 5.
Coomassie-stained gel of proteins expressed and purified at the mid-scale.
24 unique targets were grown in the GNF Fermenter and purified from 50 ml of fermenter culture. Approximately 10% of each purified protein was loaded on to a gel and visualized by staining with Coomassie blue. Molecular weight standards are indicated along the left side of the gel. Half of each purified protein was immediately analyzed by size exclusion chromatography in DDM. Gel filtration traces of two of the 24 purified proteins are shown. Both proteins are monodisperse and are excellent candidates for further analysis.
Acknowledgments
We would like to thank the members of NYCOMPS, in particular Dr. James Love, who was instrumental in developing many of the procedures described here and Dr. Wayne Hendrickson, the Principal Investigator on the Protein Structure Initiative grants that supported this work. We would like to acknowledge the efforts of all of our past and present Research Technicians who are the ones performing much of the work described here on a daily basis. We would like also to thank Dr. Filippo Mancia for critical review of the manuscript. We would also like to acknowledge the bioinformatics group of Dr. Burkhard Rost, especially Drs. Marco Punta and Edda Kloppmann, who are responsible for creating our lists of targets for cloning and expression. This work was supported by NIGMS GM075026 (Wayne Hendrickson P.I.).
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amann E, Brosius J. ATG vectors' for regulated high-level expression of cloned genes in Escherichia coli. Gene. 1985;40:183–190. doi: 10.1016/0378-1119(85)90041-1. [DOI] [PubMed] [Google Scholar]
- Aslanidis C, de Jong PJ. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic Acids Res. 1990;18:6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baneyx F, Mujacic M. Recombinant protein folding and misfolding in Escherichia coli. Nat Biotechnol. 2004;22:1399–1408. doi: 10.1038/nbt1029. [DOI] [PubMed] [Google Scholar]
- Bernard HU, Helinski DR. Use of the lambda phage promoter PL to promote gene expression in hybrid plasmid cloning vehicles. Methods Enzymol. 1979;68:482–492. doi: 10.1016/0076-6879(79)68037-0. [DOI] [PubMed] [Google Scholar]
- Blommel PG, Fox BG. A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expr Purif. 2007;55:53–68. doi: 10.1016/j.pep.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J, Holy A. Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc Natl Acad Sci U S A. 1984;81:6929–6933. doi: 10.1073/pnas.81.22.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Jin X, Huang H, Derebe MG, Levin EJ, Kabaleeswaran V, Pan Y, Punta M, Love J, Weng J, Quick M, Ye S, Kloss B, Bruni R, Martinez-Hackert E, Hendrickson WA, Rost B, Javitch JA, Rajashankar KR, Jiang Y, Zhou M. Crystal structure of a potassium ion transporter, TrkH. Nature. 2011a;471:336–340. doi: 10.1038/nature09731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Jin X, Levin EJ, Huang H, Zong Y, Quick M, Weng J, Pan Y, Love J, Punta M, Rost B, Hendrickson WA, Javitch JA, Rajashankar KR, Zhou M. Crystal structure of a phosphorylation-coupled saccharide transporter. Nature. 2011b;473:50–54. doi: 10.1038/nature09939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae PS, Rasmussen SG, Rana RR, Gotfryd K, Chandra R, Goren MA, Kruse AC, Nurva S, Loland CJ, Pierre Y, Drew D, Popot JL, Picot D, Fox BG, Guan L, Gether U, Byrne B, Kobilka B, Gellman SH. Maltose-neopentyl glycol (MNG) amphiphiles for solubilization, stabilization and crystallization of membrane proteins. Nat Methods. 2010;7:1003–1008. doi: 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Hu L, Punta M, Bruni R, Hillerich B, Kloss B, Rost B, Love J, Siegelbaum SA, Hendrickson WA. Homologue structure of the SLAC1 anion channel for closing stomata in leaves. Nature. 2010;467:1074–1080. doi: 10.1038/nature09487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czyzewski BK, Wang DN. Identification and characterization of a bacterial hydrosulphide ion channel. Nature. 2012;483:494–497. doi: 10.1038/nature10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessailly BH, Nair R, Jaroszewski L, Fajardo JE, Kouranov A, Lee D, Fiser A, Godzik A, Rost B, Orengo C. PSI-2: structural genomics to cover protein domain family space. Structure. 2009;17:869–881. doi: 10.1016/j.str.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubendorff JW, Studier FW. Creation of a T7 autogene. Cloning and expression of the gene for bacteriophage T7 RNA polymerase under control of its cognate promoter. J Mol Biol. 1991;219:61–68. doi: 10.1016/0022-2836(91)90857-3. [DOI] [PubMed] [Google Scholar]
- Eschenfeldt WH, Lucy S, Millard CS, Joachimiak A, Mark ID. A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol Biol. 2009;498:105–115. doi: 10.1007/978-1-59745-196-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshaghi S. High-throughput expression and detergent screening of integral membrane proteins. Methods Mol Biol. 2009;498:265–271. doi: 10.1007/978-1-59745-196-3_17. [DOI] [PubMed] [Google Scholar]
- Everett JK, Acton TB, Montelione GT. Primer Prim'er: a web based server for automated primer design. J Struct Funct Genomics. 2004;5:13–21. doi: 10.1023/B:JSFG.0000029238.86387.90. [DOI] [PubMed] [Google Scholar]
- Graslund S, Nordlund P, Weigelt J, Hallberg BM, Bray J, Gileadi O, Knapp S, Oppermann U, Arrowsmith C, Hui R, Ming J, dhe-Paganon S, Park HW, Savchenko A, Yee A, Edwards A, Vincentelli R, Cambillau C, Kim R, Kim SH, Rao Z, Shi Y, Terwilliger TC, Kim CY, Hung LW, Waldo GS, Peleg Y, Albeck S, Unger T, Dym O, Prilusky J, Sussman JL, Stevens RC, Lesley SA, Wilson IA, Joachimiak A, Collart F, Dementieva I, Donnelly MI, Eschenfeldt WH, Kim Y, Stols L, Wu R, Zhou M, Burley SK, Emtage JS, Sauder JM, Thompson D, Bain K, Luz J, Gheyi T, Zhang F, Atwell S, Almo SC, Bonanno JB, Fiser A, Swaminathan S, Studier FW, Chance MR, Sali A, Acton TB, Xiao R, Zhao L, Ma LC, Hunt JF, Tong L, Cunningham K, Inouye M, Anderson S, Janjua H, Shastry R, Ho CK, Wang D, Wang H, Jiang M, Montelione GT, Stuart DI, Owens RJ, Daenke S, Schutz A, Heinemann U, Yokoyama S, Bussow K, Gunsalus KC. Protein production and purification. Nat Methods. 2008;5:135–146. doi: 10.1038/nmeth.f.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L, Drogh A, Sonnhammer E. An HMM posterior decoder for sequence feature prediction that includes homology information. Bioinformatics. 2005;21(Suppl 1):i251–i257. doi: 10.1093/bioinformatics/bti1014. [DOI] [PubMed] [Google Scholar]
- Käll L, Drogh A, Sonnhammer E. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res. 2007;35:W429–432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloppmann E, Punta M, Rost B. Structural genomics plucks high-hanging membrane proteins. Curr Opin Struct Biol. 2012;22:326–332. doi: 10.1016/j.sbi.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Levin EJ, Cao Y, Enkavi G, Quick M, Pan Y, Tajkhorshid E, Zhou M. Structure and permeation mechanism of a mammalian urea transporter. Proc Natl Acad Sci U S A. 2012;109:11194–11199. doi: 10.1073/pnas.1207362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin EJ, Quick M, Zhou M. Crystal structure of a bacterial homologue of the kidney urea transporter. Nature. 2009;462:757–761. doi: 10.1038/nature08558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundstrom K. Structural genomics: the ultimate approach for rational drug design. Mol Biotechnol. 2006a;34:205–212. doi: 10.1385/MB:34:2:205. [DOI] [PubMed] [Google Scholar]
- Love J, Mancia F, Shapiro L, Punta M, Rost B, Girvin M, Wang DN, Zhou M, Hunt JF, Szyperski T, Gouaux E, MacKinnon R, McDermott A, Honig B, Inouye M, Montelione G, Hendrickson WA. The New York Consortium on Membrane Protein Structure (NYCOMPS): a high-throughput platform for structural genomics of integral membrane proteins. J Struct Funct Genomics. 2010;11:191–199. doi: 10.1007/s10969-010-9094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundstrom K. Structural genomics for membrane proteins. Cell Mol Life Sci. 2006b;63:2597–2607. doi: 10.1007/s00018-006-6252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundstrom K. Structural genomics and drug discovery. J Cell Mol Med. 2007;11:224–238. doi: 10.1111/j.1582-4934.2007.00028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L, Vaiphei ST, Shimazu T, Schneider WM, Tang Y, Mani R, Roth MJ, Montelione GT, Inouye M. The E. coli single protein production system for production and structural analysis of membrane proteins. J Struct Funct Genomics. 2009;11:81–84. doi: 10.1007/s10969-009-9070-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancia F, Love J. High-throughput expression and purification of membrane proteins. J Struct Biol. 2010;172:85–93. doi: 10.1016/j.jsb.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancusso R, Gregorio GG, Liu Q, Wang DN. Structure and mechanism of a bacterial sodium-dependent dicarboxylate transporter. Nature. 2012;491:622–626. doi: 10.1038/nature11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsson MK, Kronberg B, Claesson PM. Enhanced adsorption of alkyl glucosides on the silica/water interface by addition of amine oxides. Langmuir. 2005;21:2766–2772. doi: 10.1021/la049173i. [DOI] [PubMed] [Google Scholar]
- Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- Moffatt BA, Studier FW. T7 lysozyme inhibits transcription by T7 RNA polymerase. Cell. 1987;49:221–227. doi: 10.1016/0092-8674(87)90563-0. [DOI] [PubMed] [Google Scholar]
- Moller JV, le Maire M. Detergent binding as a measure of hydrophobic surface area of integral membrane proteins. J Biol Chem. 1993;268:18659–18672. [PubMed] [Google Scholar]
- Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- Parks TD, Leuther KK, Howard ED, Johnston SA, Dougherty WG. Release of proteins and peptides from fusion proteins using a recombinant plant virus proteinase. Anal Biochem. 1994;216:413–417. doi: 10.1006/abio.1994.1060. [DOI] [PubMed] [Google Scholar]
- Punta M, Love J, Handelman S, Hunt JF, Shapiro L, Hendrickson WA, Rost B. Structural genomics target selection for the New York consortium on membrane protein structure. J Struct Funct Genomics. 2009;10:255–268. doi: 10.1007/s10969-009-9071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg AH, Lade BN, Chui DS, Lin SW, Dunn JJ, Studier FW. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene. 1987;56:125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- Sahdev S, Khattar SK, Saini KS. Production of active eukaryotic proteins through bacterial expression systems: a review of the existing biotechnology strategies. Mol Cell Biochem. 2008;307:249–264. doi: 10.1007/s11010-007-9603-6. [DOI] [PubMed] [Google Scholar]
- Shaffer PL, Goehring A, Shankaranarayanan A, Gouaux E. Structure and mechanism of a Na+-independent amino acid transporter. Science. 2009;325:1010–1014. doi: 10.1126/science.1176088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skolnick J, Fetrow JS, Kolinski A. Structural genomics and its importance for gene function analysis. Nat Biotechnol. 2000;18:283–287. doi: 10.1038/73723. [DOI] [PubMed] [Google Scholar]
- Sonnhammer ELL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology. 1998:175–182. [PubMed] [Google Scholar]
- Speers AE, Wu CC. Proteomics of integral membrane proteins--theory and application. Chem Rev. 2007;107:3687–3714. doi: 10.1021/cr068286z. [DOI] [PubMed] [Google Scholar]
- Stevens RC. Design of high-throughput methods of protein production for structural biology. Structure. 2000;8:R177–185. doi: 10.1016/s0969-2126(00)00193-3. [DOI] [PubMed] [Google Scholar]
- Stroud RM, Choe S, Holton J, Kaback HR, Kwiatkowski W, Minor DL, Riek R, Sali A, Stahlberg H, Harries W. 2007 annual progress report synopsis of the Center for Structures of Membrane Proteins. J Struct Funct Genomics. 2009;10:193–208. doi: 10.1007/s10969-008-9058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J Mol Biol. 1991;219:37–44. doi: 10.1016/0022-2836(91)90855-z. [DOI] [PubMed] [Google Scholar]
- Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Tan S, Tan HT, Chung MC. Membrane proteins and membrane proteomics. Proteomics. 2008;8:3924–3932. doi: 10.1002/pmic.200800597. [DOI] [PubMed] [Google Scholar]
- Thain A, Gaston K, Jenkins O, Clarke AR. A method for the separation of GST fusion proteins from co-purifying GroEL. Trends Genet. 1996;12:209–210. doi: 10.1016/s0168-9525(96)90022-0. [DOI] [PubMed] [Google Scholar]
- von Heijne G. The membrane protein universe: what's out there and why bother? J Intern Med. 2007;261:543–557. doi: 10.1111/j.1365-2796.2007.01792.x. [DOI] [PubMed] [Google Scholar]
- Wagner S, Baars L, Ytterberg AJ, Klussmeier A, Wagner CS, Nord O, Nygren PA, van Wijk KJ, de Gier JW. Consequences of membrane protein overexpression in Escherichia coli. Mol Cell Proteomics. 2007;6:1527–1550. doi: 10.1074/mcp.M600431-MCP200. [DOI] [PubMed] [Google Scholar]
- Wagner S, Bader ML, Drew D, de Gier JW. Rationalizing membrane protein overexpression. Trends Biotechnol. 2006;24:364–371. doi: 10.1016/j.tibtech.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Wagner S, Klepsch MM, Schlegel S, Appel A, Draheim R, Tarry M, Hogbom M, van Wijk KJ, Slotboom DJ, Persson JO, de Gier JW. Tuning Escherichia coli for membrane protein overexpression. Proc Natl Acad Sci U S A. 2008;105:14371–14376. doi: 10.1073/pnas.0804090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waight AB, Love J, Wang DN. Structure and mechanism of a pentameric formate channel. Nat Struct Mol Biol. 2010;17:31–37. doi: 10.1038/nsmb.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt J. Structural genomics-impact on biomedicine and drug discovery. Exp Cell Res. 2010;316:1332–1338. doi: 10.1016/j.yexcr.2010.02.041. [DOI] [PubMed] [Google Scholar]
- Xie H, Guo XM, Chen H. Making the most of fusion tags technology in structural characterization of membrane proteins. Mol Biotechnol. 2009;42:135–145. doi: 10.1007/s12033-009-9148-x. [DOI] [PubMed] [Google Scholar]
Key References
- Punta M, Love J, Handelman S, Hunt JF, Shapiro L, Hendrickson WA, Rost B. Structural genomics target selection for the New York consortium on membrane protein structure. J Struct Funct Genomics. 2009;10:255–268. doi: 10.1007/s10969-009-9071-1. Describes target selection of bacterial αIMPs for the NYCOMPS high through-put expression pipeline and discusses the potential impact of structure determination of these targets on a larger uncharacterized pool of proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloppmann E, Punta M, Rost B. Structural genomics plucks high-hanging membrane proteins. Curr Opin Struct Biol. 2012;22:326–332. doi: 10.1016/j.sbi.2012.05.002. Reviews recent progress of IMP structure determination by structural genomics (SG) initiatives and discusses the progress of SG centers in covering important protein families. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graslund S, Nordlund P, Weigelt J, Hallberg BM, Bray J, Gileadi O, Knapp S, Oppermann U, Arrowsmith C, Hui R, Ming J, dhe-Paganon S, Park HW, Savchenko A, Yee A, Edwards A, Vincentelli R, Cambillau C, Kim R, Kim SH, Rao Z, Shi Y, Terwilliger TC, Kim CY, Hung LW, Waldo GS, Peleg Y, Albeck S, Unger T, Dym O, Prilusky J, Sussman JL, Stevens RC, Lesley SA, Wilson IA, Joachimiak A, Collart F, Dementieva I, Donnelly MI, Eschenfeldt WH, Kim Y, Stols L, Wu R, Zhou M, Burley SK, Emtage JS, Sauder JM, Thompson D, Bain K, Luz J, Gheyi T, Zhang F, Atwell S, Almo SC, Bonanno JB, Fiser A, Swaminathan S, Studier FW, Chance MR, Sali A, Acton TB, Xiao R, Zhao L, Ma LC, Hunt JF, Tong L, Cunningham K, Inouye M, Anderson S, Janjua H, Shastry R, Ho CK, Wang D, Wang H, Jiang M, Montelione GT, Stuart DI, Owens RJ, Daenke S, Schutz A, Heinemann U, Yokoyama S, Bussow K, Gunsalus KC. Protein production and purification. Nat Methods. 2008;5:135–146. doi: 10.1038/nmeth.f.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens RC. Design of high-throughput methods of protein production for structural biology. Structure. 2000;8:R177–185. doi: 10.1016/s0969-2126(00)00193-3. These two reviews describe the different cloning, expression and purification methods employed for high throughput protein production. The former also summarizes the different approaches used by structural genomics centers. [DOI] [PubMed] [Google Scholar]