Abstract
The generation of neurons by neural stem cells is a highly choreographed process that requires extensive and dynamic remodelling of the cytoskeleton at each step of the process. The atypical RhoGTPase Rnd3 is expressed by progenitors in the embryonic brain but its role in early steps of neurogenesis has not been addressed. Here we show that silencing Rnd3 in the embryonic cerebral cortex interferes with the interkinetic nuclear migration of radial glial stem cells, disrupts their apical attachment and modifies the orientation of their cleavage plane. These defects are rescued by co-expression of a constitutively active form of cofilin, demonstrating that Rnd3-mediated disassembly of actin filaments coordinates the cellular behaviour of radial glia. Rnd3 also limits the divisions of basal progenitors via a distinct mechanism involving the suppression of cyclin D1 translation. Interestingly, although Rnd3 expression is controlled transcriptionally by Ascl1, this proneural factor is itself required in radial glial progenitors only for proper orientation of cell divisions.
INTRODUCTION
The generation of neurons by stem cells involves cellular movements, cell divisions and dynamic changes in cell-cell interactions. How these different steps of neurogenesis, which all require extensive cytoskeleton rearrangement, are coordinately regulated and coupled to the activation of a neurogenic programme of gene expression is not well understood.
The neural stem cells of the embryonic brain or radial glial cells are highly polarized cells that are attached to one another in the ventricular zone (VZ) of the brain by apically-located adherens junctions. Their nuclei migrate during the cell cycle from a basal position during S-phase to an apical position during mitosis, and the nuclei of the daughter cells migrate back to enter S-phase on the basal side of the VZ, in a process of interkinetic nuclear migration (INM). During the peak of neurogenesis, most radial glial cells divide asymmetrically with vertical cleavage planes. In these divisions, one daughter remains a radial glial cell and continues to divide at the ventricular surface while the other loses its apical attachment and exits the cell cycle either immediately or after one or a few divisions as a basal progenitor 1-3. Fewer radial glial cells undergo oblique or horizontal divisions, and these divisions have been proposed to generate outer radial glia (oRG) 2,4.
The actin cytoskeleton has an important role in both the positioning of the mitotic spindles 5 and the formation and remodelling of adherens junctions 6,7. Actin has also been implicated in the regulation of INM 8-11 as well as other cortical progenitor behaviours 12-14. Members of the small RhoGTPase superfamily, which have important roles in the regulation of the actin cytoskeleton, are involved in the coordination of the different steps of neurogenesis. Cdc42 activity is essential for the maintenance of apical adherens junction and for INM 15,16. RhoA signaling has also a role in the maintenance of adherens junctions and in apico-basal polarity 17-19, in the orientation of mitotic cleavage planes 20,21 and in progenitor proliferation 22. Rac1 is required in neural progenitors for proper interkinetic nuclear movement, cytokinesis and for cell cycle re-entry 23-25.
We have previously shown that the small GTP-binding protein Rnd3 regulates the migration of cortical neuron downstream of the proneural protein Ascl1 by promoting the disassembly of actin filaments 26. Since Rnd3 is also expressed in cortical progenitors 26, we have asked whether it is also involved in earlier steps of neurogenesis that also require regulation of the actin cytoskeleton. We show here that Rnd3 regulates the movements of the nuclei of radial glia cells during INM, that it exerts a control over the orientation of mitotic spindles and that it is required for the maintenance of adherens junctions. As for its function in migrating neurons 26, Rnd3 regulates these early steps of neurogenesis by inhibiting actin filament polymerization. Rnd3 also limits the divisions of basal progenitors but interestingly, this involves a distinct mechanism of suppression of cyclin D1 translation. Our results thus reveal that Rnd3 exerts its pleiotropic functions in early steps of cortical neurogenesis by employing distinct mechanisms in radial glial cells and basal progenitors.
RESULTS
Rnd3 is required for interkinetic nuclear migration
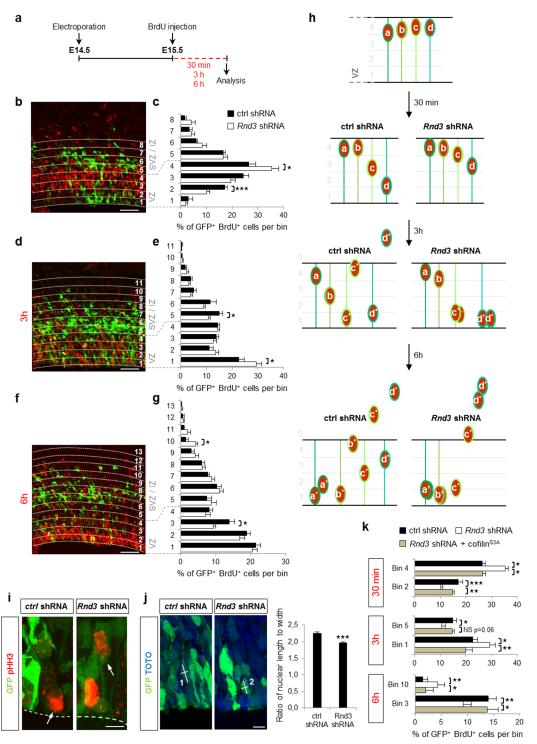
To assess the function of Rnd3 in cortical progenitors, we silenced the gene in the cortex of E14.5 embryos by in utero electroporation of a construct co-expressing Rnd3 shRNA, or scrambled shRNA in control experiments, and GFP to identify electroporated cells. Twenty-four hours later, we traced a cohort of cortical progenitors by labeling them in S-phase with BrdU and examining their positions after 30 min, 3 h or 6 h (Fig. 1a). 30 min after the pulse, nuclei that had incorporated BrdU had undergone INM and had migrated from the basal to the apical side of the ventricular zone (VZ) (Fig. 1b, h). Quantification of the distribution of VZ progenitors co-labeled for GFP and BrdU revealed that the nuclei of progenitors electroporated with the Rnd3 shRNA had migrated less than the nuclei of progenitors electroporated with the control shRNA and a larger fraction of them remained on the basal side of the VZ (Fig. 1c, h). 3h after the pulse, some control BrdU+, GFP+ cells had divided and one of the daughters had already moved out of the VZ whereas the nucleus of the radial glial daughter had moved back towards the basal side of the VZ (Fig. 1d, h). In contrast, Rnd3 silenced cells which were delayed in their basal-apical movement had just divided at the ventricular surface leading to an accumulation of cells at the ventricular surface (Fig. 1e, h). Altogether, these results indicate that silencing Rnd3 in cortical progenitors impairs INM.
Figure 1.
Rnd3 regulates INM of cortical neuron progenitors by depolymerizing F-actin. (a) Protocol used to study INM. Cerebral cortices from E14.5 embryos were electroporated in utero with Rnd3 shRNA or ctrl (control) shRNA, and subjected 24 h later to 30 min, 3 h or 6 h BrdU pulse labeling, followed by fixation and coimmunostaining for GFP (green) and BrdU (red). (b, d, f) Distribution of GFP+ BrdU+ cells after 30 min (b), 3 h (d) or 6 h BrdU pulse (f) across the VZ (ventricular zone), SVZ (subventricular zone) and IZ (intermediate zone), divided into bins as indicated, with bin 1 being at the ventricle surface and bin 4 at the VZ–SVZ boundary. (c, e, g) GFP+ BrdU+ cells in a given bin are expressed as percentage of the total number of GFP+ BrdU+-labeled cells in all bins. Data are presented as the mean ± s.e.m. from seven sections prepared from four embryos obtained from two or three litters. Student’s t-test; *p<0.05, ***p<0.001. (h) Schematic representation of the results obtained in (b-g). (i) Illustration of a mitotic radial glia cell dividing along or away from the ventricular surface (dashed line) 24 h after ctrl or Rnd3 shRNA electroporation respectively at E14.5. GFP identifies electroporated cells and pHH3 in red (phosphohistone-H3) marks mitotic cells. (j) Quantification of the ratio of nuclear length to width (indicated by white lines on the pictures; cell 1 ratio > cell 2 ratio) in GFP+ cells 24 h after electroporation (E14.5) of ctrl or Rnd3 shRNA. n = 163 cells for ctrl shRNA, n=162 cells for Rnd3 shRNA from ten sections prepared from four embryos obtained from two or three litters; mean ± s.e.m; Student’s t-test; ***p<0.001. (k) Coelectroporation of cofilinS3A, a non-phosphorylatable form of cofilin that depolymerizes F-actin, fully rescues the defects observed after Rnd3 silencing. Quantification is similar to c, e and g. mean ± s.e.m; one way ANOVA followed by a Fisher PLSD post hoc test; *p<0.05, **p<0.01, ***p<0.001. Scale bars represent 50 μm (b, d, f) and 5 μm (i, j).
See also Supplementary Fig. S1, S2 and S4.
Defects in the cell cycle progression of progenitors have been shown to interfere with INM 27,28. However, the duration of the different phases of the cycle was unaffected by Rnd3 silencing (Supplementary Fig. S1a-d), indicating that Rnd3 directly regulates the INM of cortical progenitors. Conversely, blocking INM during apical nuclear migration in G2 does not prevent cell cycle progression and results in ectopic mitoses 27,29. Accordingly, we observed progenitor cells dividing a short distance away from the ventricular surface one day after Rnd3 shRNA electroporation but not after control shRNA electroporation (Fig. 1i). Nuclei undergoing INM normally have an elongated morphology and INM impairment results in nuclei adopting a more rounded shape 30-32. We measured the length-to-width ratio of nuclei of transfected progenitors and observed that Rnd3-silenced progenitors have significantly less elongated nuclei (Fig. 1j). Together, these results indicate that Rnd3 is required for normal INM of cortical neuron progenitors.
To determine whether Rnd3 acts by regulating actin depolymerization in VZ progenitors as in migrating neurons 26, we co-electroporated Rnd3 shRNA with cofilinS3A, a non-phosphorylatable form of cofilin that constitutively depolymerizes F-actin. CofilinS3A fully rescued the INM defects of Rnd3-silenced progenitors (Fig. 1k and Supplementary Fig. S2), thus demonstrating that Rnd3 regulates INM of cortical progenitors by promoting F-actin depolymerization.
BrdU-pulsed cortices were also examined 6 h later, at which time some BrdU+ cells had left the VZ and migrated in the SVZ and IZ in control experiments (Fig. 1f, h and Supplementary Fig. S1e). Rnd3 silencing increased the fraction of BrdU+ cells that reached the SVZ and IZ and reduced the fraction that remained in the VZ (Fig. 1g, h and Supplementary Fig. S1e), suggesting that Rnd3 activity regulates not only INM in the VZ but also the movement of cells out of the VZ. This distinct function is examined in the next section.
Rnd3 controls cleavage plane orientation
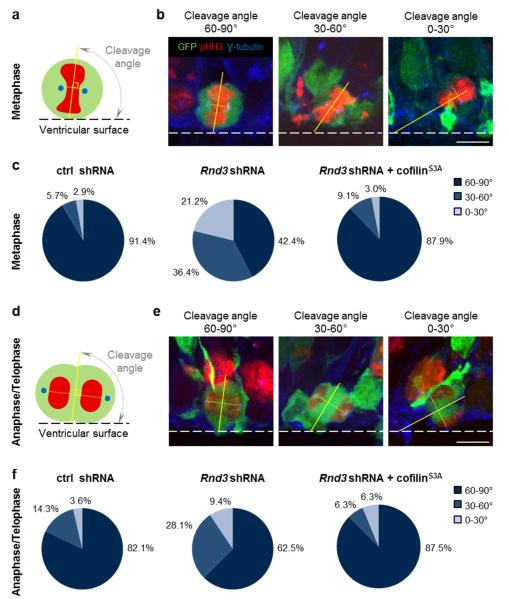
Radial glial cells in the cortical VZ divide mostly with a vertical cleavage plane and their two daughter cells remain attached to the ventricular surface immediately after mitosis 1,2. When a progenitor divides with an oblique or horizontal cleavage plane, one of its daughters lacks an apical process attached to the ventricular surface and migrates away from the VZ. The increase in number of cells located outside of the VZ after Rnd3 knockdown (Fig. 1g) therefore suggested that cleavage planes might be abnormal oriented, resulting in cells detaching prematurely from the ventricular surface. In control experiments, cleavage planes at metaphase and anaphase-telophase were mainly vertical, and horizontal or oblique cleavage planes were relatively rare (8.6% and 17.9%, respectively; Fig. 2c, f) as previously observed 1,33,34. When Rnd3 was knocked down, the fraction of cells dividing with an oblique or horizontal cleavage plane increased significantly (to 57.7% and 37.5%, respectively; Fig. 2c, f). The increase in divisions with non-vertical cleavage planes might account for the increased fraction of Rnd3-silenced, BrdU-labeled cells that migrate out of the VZ 6 h after the BrdU pulse (Fig. 1g). Like INM, orientation of the cleavage plane is an actomyosin-dependent process 5. Co-electroporation of cofilinS3A restored vertical cleavage plane orientation in Rnd3-silenced progenitors (Fig. 2c, f), thus indicating that Rnd3 maintains the vertical orientation of apical divisions by remodelling the actin cytoskeleton.
Figure 2.
Rnd3 is required for vertical divisions of cortical progenitors. (a, d) The cleavage angles of electroporated (green) apical progenitors were measured during metaphase and anatelophase as shown schematically in (a) and (d) respectively. DNA is in red and centrosomes are in blue. (b, e) Cells in metaphase (b) and in anaphase/telophase (e) with a vertical (60-90°), oblique (30-60°) or horizontal (0-30°) cleavage angles. Electroporated cells (GFP+) were labeled for pHH3 (mitotic cell, red) and γ-tubulin (centrosome, blue). The dashed line shows the ventricular surface. (c, f) Pie charts show the percentage of cells with colour coded cleavage angles during metaphase (c) and ana- and telophase (f) 24 h after the electroporation (E14.5) of ctrl shRNA, Rnd3 shRNA or Rnd3 shRNA + cofilinS3A. Metaphase: ctrl shRNA n=35 cells, Rnd3 shRNA n=33 cells, Rnd3 shRNA + cofilinS3A n=33 cells. Anaphase/Telophase: ctrl shRNA n=20 cells, Rnd3 shRNA n=22 cells, Rnd3 shRNA + cofilinS3A n=32 cells. Cells were analyzed from at least six embryos obtained from three litters. Scale bars represent 5 μm.
See also Supplementary Fig. S4.
Rnd3 is required for the maintenance of adherens junctions
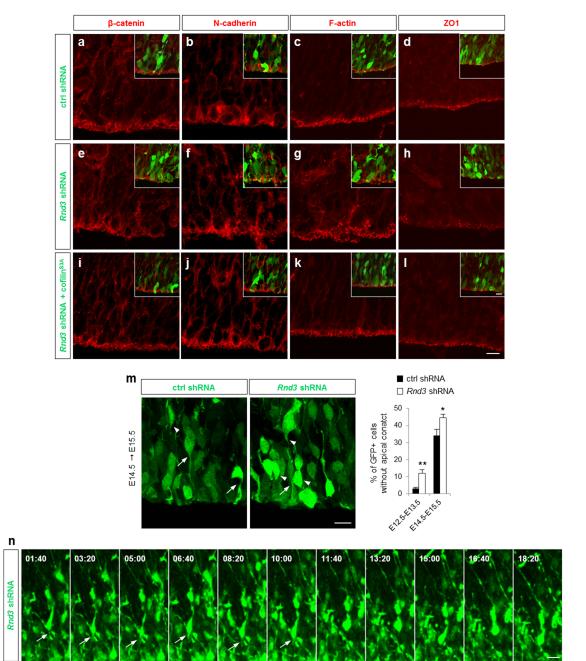
Another mechanism that could account for the premature movement of Rnd3-silenced cells out of the VZ is the loss of their junctions to neighbouring cells. Rnd3 expression in cortical progenitors is enriched at the ventricular surface (Fig. 6j) and Rnd3 has been implicated in the formation of tight junctions between epithelial tumor cells 35. Moreover, Rnd3 regulates RhoA signaling in migrating neurons 26 and RhoA is involved in the regulation of adherens junctions between progenitors in the cortical VZ and the spinal cord 17-19. To investigate whether Rnd3 is required to maintain adherens junctions between radial glia cells, we examined the expression of components of adherens junctions in electroporated cortices. In Rnd3 silenced cortices, expression of ZO1 was reduced and the ring-like distribution of β-catenin, N-cadherin and F-actin observed in control cortices, was disrupted (compare Fig. 3ad with Fig. 3e-h and Supplementary Fig. S3). As with the other VZ progenitor defects described above, the expression and localization of junction markers was restored by coelectroporation of Rnd3 shRNA and cofilinS3A but not by co-electroporation of a mutant form of Rnd3 (Rnd3T55V) that cannot interact with p190RhoGAP and therefore does not inhibit RhoA signaling (Fig. 3i-l and Supplementary Fig. S4). These results indicate that Rnd3 maintains the integrity of the junctions between radial glia cells through regulation of RhoA and the actin cytoskeleton.
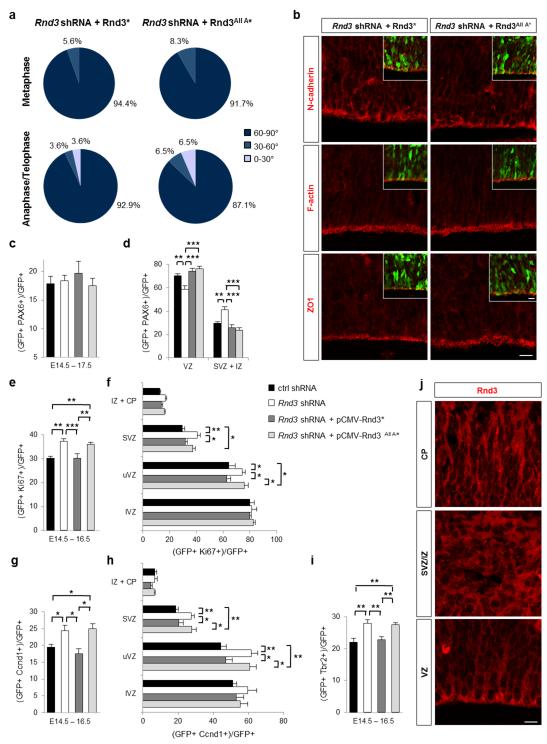
Figure 6.
Rnd3 activity in basal progenitor proliferation does not require its localization to the plasma membrane. (a) Rnd3* and Rnd3All A* restore cleavage plane orientation in Rnd3-deleted apical progenitors (the star indicates silent point mutations in the sequence recognized by Rnd3 shRNA). Pie charts show the percentage of cells with the color-coded cleavage angle during metaphase and ana-telophase in Rnd3 shRNA + pCMV-Rnd3* (metaphase n=36 cells, ana/telophase n= 28 cells) and Rnd3 shRNA + pCMV-Rnd3All A* cortices (metaphase n=36 cells, ana/telophase n=31 cells). (b) Immunostainings for N-cadherin, F-actin and ZO1 (in red) in Rnd3 shRNA + pCMV-Rnd3* and Rnd3 shRNA + pCMV-Rnd3All A* electroporated cortices show no disruption of adherens junctions, similar to the pattern of expression observed in control sections. (c) Quantification of the percentage of electroporated cells that are PAX6+. (d) The quantification of the proportion of GFP+ PAX6+ cells inside and outside the VZ shows that Rnd3All A* restores the position of PAX6+ progenitors after Rnd3 knockdown. (e-i) However Rnd3All A* does not rescue the increase of proliferation and basal progenitor population observed after Rnd3 silencing suggesting that Rnd3 is not localized at the plasma membrane to mediate these effects. (e-i) Quantification 2 days after the electroporation (E14.5) of the percentage of electroporated cells that are Ki67+ (e), Ccnd1 (g) or Tbr2+ (i) in the entire electroporated cortex or in the different zones of the cortex (f, h). Data in c-i are presented as the mean ± s.e.m; one way ANOVA followed by a Fisher PLSD post hoc test; *p<0.05, **p<0.01, ***p<0.001 (j) Expression of Rnd3 in the different domains of the cerebral cortex at E15.5. In the VZ and CP, Rnd3 is essentially localized at the plasma membrane whereas it is expressed throughout the cell in SVZ/IZ. Scale bars represent 10 μm (b and inset, j).
Figure 3.
Rnd3 is required for the maintenance of adherens junctions. (a-l) Cortices immunostained for β-catenin (red in a, e, i), N-cadherin (red in b, f, j), F-actin (red in c, g, k) and ZO1 (red in d, h, l), 24 h after the co-electroporation at E14.5 of GFP (green) and ctrl shRNA (a-d), Rnd3 shRNA (e-h) or Rnd3 shRNA + cofilinS3A (i-l). (m) Detachment of the apical process from the ventricular surface after Rnd3 knockdown. White arrows and arrowheads indicate electroporated cells with and without an apical contact respectively. The graph presents the quantification of the proportion of GFP+ cells without an apical contact within 60 μm from the ventricle surface. E12.5-E13.5: ctrl shRNA n=239 cells, Rnd3 shRNA n= 201 cells. E14.5-E15.5: ctrl and Rnd3 shRNA n =163 cells. Data represents the mean ± s.e.m. from at least eight sections prepared from four embryos obtained from two or three litters. Student’s t-test; *p<0.05, **p<0.01. (n) Time-lapse series of a GFP+ cell located into the upper part of the VZ analyzed 24 h after electroporation (E14.5) of Rnd3 shRNA, showing the retraction of the apical process after 11 h 40 min of live imaging. White arrows indicate the apical process. Scale bars represent 10 μm (l and inset, m, n).
See also Supplementary Fig. S3, S4, S5 and Supplementary Movie 1.
We then asked whether the disruption of adherens junctions between endfeets of radial glial progenitors was accompanied by a detachment of the apical process and premature delamination of the progenitors. We examined whether apical processes of GFP+ electroporated cells in the VZ contact the ventricular surface. We found that Rnd3 knockdown resulted in a higher proportion of VZ cells having an apical process detached from the ventricular surface (Fig. 3m), a defect also rescued by cofilinS3A coelectroporation (Supplementary Fig. S5a). VZ cells were also electroporated at E12.5 and examined at E13.5, a stage when delamination of radial glial cells is still infrequent. In control experiments, 3.0±0.8 % of GFP+ cells had an apical process that failed to reach the apical surface and Rnd3 silencing increased this percentage to 12.1±2.0 % (Fig. 3m). The premature detachment of the apical process was also observed by time-lapse imaging following Rnd3 knockdown but not in control sections (Fig. 3n, Supplementary Movie 1). Therefore Rnd3 activity is required to maintain adherens junctions between radial glial cells. The premature delamination of Rnd3-silenced progenitors from the ventricular surface, revealed by the increased fraction of BrdU-labeled cells that move out of the VZ 6 h after the BrdU pulse (Fig. 1g), might thus be the result of the disruption of adherens junctions (Fig. 3) and/or of the abnormal orientation of the cleavage planes of dividing VZ progenitors (Fig. 2).
Rnd3 regulates apical progenitor localization
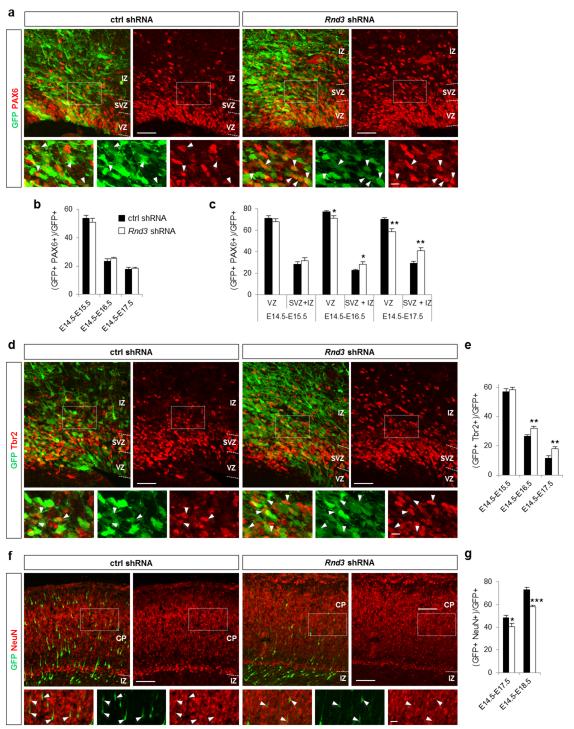
The premature detachment of radial glial cells from the ventricular surface could result in the displacement of apical progenitors (identified by Pax6 expression; 36) and/or in their conversion to basal progenitors (identified by Tbr2 expression; 37). Examination of Pax6 and Tbr2 expression in electroporated cells showed that the overall number of Pax6+ GFP+ cells was not affected by Rnd3 knockdown (Fig. 4b). However their distribution was altered, with more Pax6, GFP double labeled cells present in the SVZ and IZ and fewer in the VZ than in control brains (Fig. 4a, c). This suggests that radial glial cells with impaired Rnd3 expression prematurely leave the VZ and enter the SVZ while transiently maintaining their radial glial molecular phenotype and thus show similarities with oRG 38-40. Interestingly, this Rnd3 shRNA-induced displacement was prevented when cofilinS3A was coelectroporated (Supplementary Fig. S5b, c), indicating that it is actin-dependent. Rnd3 knockdown also increased the fraction of GFP+ cells expressing Tbr2 (Fig. 4d, e) and decreased the fraction expressing NeuN (Fig. 4f, g). Since the size of the Pax6+ population was not changed (Fig. 4b), the change in number of Tbr2+ cells might reflect an increased proliferation of basal progenitors rather than an effect of Rnd3 knockdown on the maturation of apical progenitors (see below).
Figure 4.
Rnd3 silencing results in translocation of apical progenitors to basal positions. (a, d, f) E17.5 cortices electroporated at E14.5 with ctrl shRNA or Rnd3 shRNA and immunostained for GFP (green) and the radial glial cell marker Pax6 (red in a) or the basal progenitor marker Tbr2 (red in d) or the neuronal marker NeuN (red in f). White rectangles show the areas enlarged in the insets and white arrowheads indicate double positive cells. (b, e, g) Percentage of electroporated cells that are PAX6+ (b), or Tbr2+ (e), or NeuN+ (g). (c) The change in the proportion of GFP+ PAX6+ cells inside and outside the VZ shows that Rnd3 silencing affects the location of apical progenitors. Data in b, c, e and g are presented as the mean ± s.e.m. from at least six sections prepared from four embryos obtained from two or three litters. Student’s t-test; *p<0.05, **p<0.01. Scale bars represents 10 μm (insets a, d), 20 μm (inset f), 50 μm (a, d) and 100 μm (f).
See also Supplementary Fig. S4 and S5.
Rnd3 controls basal progenitor proliferation via cyclin D1
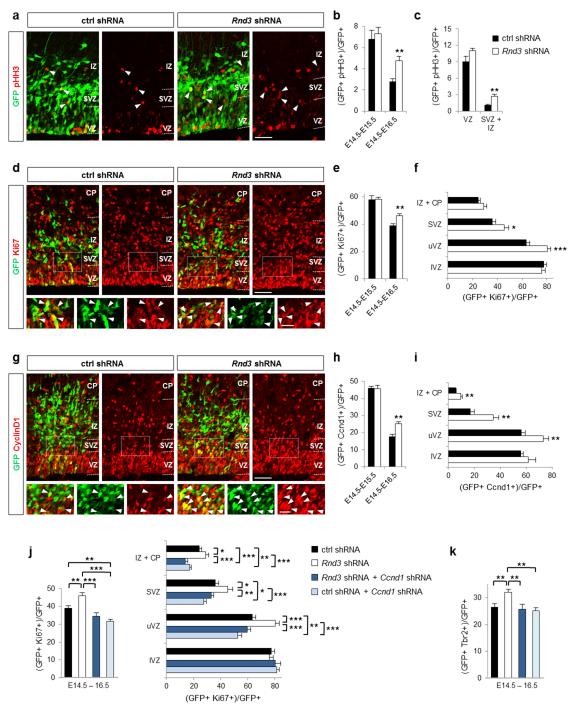
To determine whether Rnd3 regulates the proliferation of basal progenitors, we examined the expression of cell division markers. Rnd3 silencing significantly increased the number of cells expressing the mitotic marker phosphohistone H3 (pHH3) in the SVZ (where basal progenitors divide) but not in the VZ (where apical progenitors divide) (Fig. 5a-c). The fraction of cells expressing the proliferation marker Ki67 was also increased in the upper part of the VZ (uVZ) and the SVZ but not in the lower VZ (lVZ) (Fig. 5d-f). Moreover, time-lapse imaging showed that Rnd3-silenced progenitors can undergo more than one round of division in the SVZ (Supplementary Movie 2), which is uncommon in control brains 3,41. Therefore Rnd3 inhibits specifically the proliferation of basal progenitors.
Figure 5.
Rnd3 inhibits the proliferation of basal progenitors. (a, d, g) E16.5 cortices electroporated at E14.5 with ctrl shRNA or Rnd3 shRNA and immunostained for GFP (green) and pHH3 (red in a), GFP and Ki67 (red in d) or GFP and Ccnd1 (cyclin D1, red in g). White rectangles show the areas enlarged in the insets and white arrowheads indicate double positive cells. (b, e, h) Percentage of electroporated cells that are pHH3+ (b), Ki67+ (e) or Ccnd1+ (h). (c, f, i) Quantification 2 days after electroporation of the percentage of electroporated cells that are pHH3+ (c), Ki67+ (f) or Ccnd1+ (i) in the different zones of the cortex: lVZ (lower ventricular zone), uVZ (upper ventricular zone), SVZ, IZ and CP (cortical plate). Data in b, c, e, f, h, i are presented as the mean ± s.e.m. from at least six sections prepared from four embryos obtained from two or three litters. Student’s t-test; *p<0.05, **p<0.01. (j, k) Coelectroporation of Ccnd1 shRNA fully rescues the defects in proliferation and Tbr2 expression observed after Rnd3 silencing. Data in j and k are presented as the mean ± s.e.m; one way ANOVA followed by a Fisher PLSD post hoc test; *p<0.05, **p<0.01, ***p<0.001. Scale bars represent 20 μm (insets d, g) and 50 μm (a, d, g).
See also Supplementary Fig. S4, S5 and Supplementary Movie 2.
Rnd3 has previously been shown to inhibit cell cycle progression in fibroblasts and glioblastoma cells 42. Interestingly, this activity does not involve the regulation of RhoA/ROCK signaling as other functions of Rnd3, but the inhibition of cyclin D1 (Ccnd1) translation 43-45. In agreement with these studies, we found that RhoA signaling and the actin cytoskeleton were not involved in the increase of basal progenitor proliferation observed after Rnd3 knockdown (Supplementary Fig. S4f-h and Supplementary Fig. S5d-h). We then examined Ccnd1 expression and found that Rnd3 silencing increased the number of Ccnd1+ cells in the uVZ, SVZ and IZ of the cortex (Fig. 5g-i). Co-electroporation of a Ccnd1 shRNA construct 46 reduced the proliferation of Rnd3-silenced progenitors to control levels, indicating that Rnd3 suppresses the proliferation of basal progenitors by inhibiting Ccnd1 expression (Fig. 5j). The increased number of Tbr2+ cells in Rnd3-silenced cortices was also corrected by co-electroporation of Ccnd1 shRNA (Fig. 5k), indicating that this defect is due to an excessive proliferation of basal progenitors.
The function and localization of Rnd3 are regulated by phosphorylation 47,48. However, we showed previously that a non-phosphorylatable version of Rnd3 which is only expressed at the plasma membrane (Rnd3All A) could fully perform Rnd3 functions in migrating neurons 26. Similarly, co-electroporation of Rnd3All A with Rnd3 shRNA rescued the defects induced by Rnd3 knockdown in radial glial cells, including the non-vertical cleavage planes, the disruption of apical junctions and the increase in non-VZ Pax6+ cells (Fig. 6a-d). These results thus demonstrate that Rnd3 activities in radial glial cells do not require phosphorylation and suggest that Rnd3 is localized at the plasma membrane to mediate these functions (Fig. 6j). However, Rnd3All A did not revert the excessive proliferation of SVZ progenitors or the increased number of Tbr2+ cells (Fig. 6e-i). This suggests that phosphorylation of Rnd3 is required for its function in basal progenitor proliferation. This result also confirms that Rnd3 uses distinct downstream mechanisms in radial glial cells (RhoA inhibition and depolymerisation of actin filaments) and in basal progenitors (inhibition of cyclin D1 expression).
Ascl1 promotes vertical progenitor divisions via Rnd3
The expression of Rnd3 in the embryonic cerebral cortex is controlled transcriptionally by the proneural protein Ascl1. Moreover, Rnd3 mediates the function of Ascl1 in cortical neuron migration 26. We thus asked whether Rnd3 also acts downstream of Ascl1 in cortical progenitors. We first determined whether Ascl1 inactivation results in similar progenitor defects as Rnd3 knockdown. Ascl1 silencing by electroporation of a specific shRNA or Ascl1 deletion in Emx1Cre, Ascl1flox/flox embryos resulted in an increase in the fraction of radial glia progenitors that divide with an oblique or horizontal cleavage plane (Supplementary Fig. S6a, b), a phenotype reminiscent of that of Rnd3 silenced progenitors, although less pronounced (Fig. 2c, f). We then asked whether loss of Rnd3 contributes to this phenotype. Co-expressing Rnd3 with the Ascl1 shRNA rescued the division plane defect of Ascl1 silenced progenitors (Supplementary Fig. S6b), demonstrating that Ascl1 maintains the vertical angle of radial glial cell divisions by inducing Rnd3 expression.
In contrast with the increase in non-vertical divisions, Ascl1 knockdown or deletion did not produce other cellular defects observed in Rnd3 silenced progenitors, including delayed INM and disruption of adherens junctions (Supplementary Fig. S7). Acute deletion of Ascl1 by electroporation of Cre in the cortical VZ of Ascl1flox/flox embryos resulted in a reduction in the number of Ki67+ progenitors in the IZ (Supplementary Fig. S6c, d). This proliferation defect is similar to that observed in Ascl1 deficient ventral telencephalon progenitors 49, but it is opposite to that produced by Rnd3 knockdown (Fig. 5d-f). This is likely due to the regulation by Ascl1 of additional targets that regulate cell division in an opposite way to Rnd3 49.
DISCUSSION
Extensive and dynamic remodelling of the cytoskeleton underlies many of the steps that dividing progenitors and newborn neurons go through during neurogenesis. We have previously shown that Rnd3, downstream of the proneural factor Ascl1, is a key player in the regulation of neuronal migration in the embryonic cerebral cortex via its action on the actin cytoskeleton. In this study, we have asked i) whether Rnd3 also regulates earlier steps of neurogenesis in the cerebral cortex, ii) whether its activities in cortical progenitors involve actin remodelling and iii) whether it acts downstream of Ascl1.
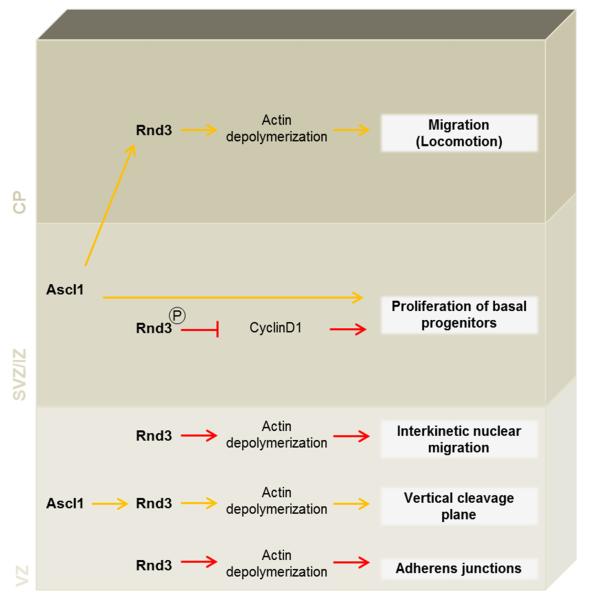
Our results indicate that Rnd3 uses distinct downstream mechanisms in radial glial cells and in basal progenitors to mediate its pleiotropic functions (Fig.7). In radial glia cells, Rnd3 regulates the movement of the nuclei occurring during INM, exerts a control on the orientation of mitotic spindles and is required for the maintenance of adherens junctions. As for its function in migrating neurons 26, Rnd3 regulates these different steps of neurogenesis by inhibiting actin filament polymerization. Indeed, the defects observed after Rnd3 knockdown were rescued when a constitutively active form of cofilin (cofilinS3A) that disrupts actin filaments was co-electroporated. Reorganization of the actin cytoskeleton is known to drive cell rounding during mitosis, but the actin cortex has been shown to also guide mitotic spindle orientation through interactions with astral microtubules 5,50. Our results suggest that the polymerisation of actin filaments must be tightly regulated to maintain mitotic spindles aligned to the ventricular surface and that excessive polymerisation perturbs the orientation of divisions. Interestingly, similar defects were observed in mice when LGN (the homologue of Partner of Inscuetable) was inactivated 1. LGN is enriched in a ring located at the lateral cell cortex where it concentrates pulling forces on astral microtubules 51. Rnd3 knockdown, by increasing cortical filamentous actin, might thus prevent the anchoring or the correct distribution of LGN complex and consequently proper spindle positioning. In migrating neurons, Rnd3 promotes actin cytoskeleton remodelling by inhibiting RhoA signaling at the plasma membrane through stimulation of the Rho GTPase activating protein p190RhoGAP 26. We used an in vivo rescue assay with mutated forms of Rnd3 that cannot interact with p190RhoGAP (Rnd3T55V) or that is only expressed at the plasma membrane (Rnd3All A), to show that Rnd3 uses a similar mechanism in apical progenitors to regulate actin-dependent processes (Fig. 6 and S4a-e).
Figure 7.
Roles of Rnd3 in cortical neurogenesis. In radial glia cells, Rnd3 regulates interkinetic nuclear migration, exerts a control on the orientation of the mitotic spindles and maintain the adherens junctions by inhibiting actin filament polymerization. In basal progenitors, Rnd3 controls proliferation through cyclin D1. Rnd3 also regulates migration of postmitotic neurons via its action on the actin cytoskeleton.
See also Supplementary Fig. S6 and S7.
Adherens junctions interact with circumferential actin filaments bundles, via the α-catenin component of cadherin-catenin complexes and other actin-binding proteins, and these interactions are essential for the development, remodelling and function of the junctions 7,52,53. Small GTPases are recruited and activated at adherens junctions and have a pivotal role in the dynamic remodelling of the junctions 18,53,54. In particular RhoA and its downstream effector mDia1 control the localisation and stabilisation of cadherin-catenin complexes at the junctions by mediating their association to the cortical actin cytoskeleton 6,17,19. Interestingly, excessive RhoA activity in colon carcinoma and non-tumorigenic epithelial cells results in contraction of the actomyosin cytoskeleton and disruption of adherens junctions, an effect mediated by another downstream RhoA effector, ROCK 6. If RhoA has similar activities in the cortical neuroepithelium, then Rnd3 may maintain the integrity of adherens junctions by moderating the level of RhoA-ROCK signaling, thus preventing excessive contraction of the actomyosin cytoskeleton.
Rnd3 knockdown also increased the number of Tbr2+ cells. This effect did not occur at the expense of Pax6+ progenitor cells, suggesting that it is the result of excessive proliferation rather than of a change of progenitor fate. Accordingly, Rnd3 silencing resulted in an increase in the number of cells expressing cyclin D1 and the mitotic marker Ki67+, particularly in the upper VZ and SVZ where basal progenitors are located. Rnd3 inhibits cell cycle progression in fibroblasts and glioblastoma cells independently of the inhibition of RhoA/ROCK signaling, by interfering with the release of the eukaryotic initiation factor eIF4E from the translation repressor 4E-BP1, resulting in the suppression of cyclin D1 translation 43-45. Similarly, we found that the silencing of cyclin D1 together with Rnd3 eliminated the supernumerary Ki67+ and Tbr2+ cells. Therefore, our results demonstrate that Rnd3 limits the proliferation of Tbr2+ basal progenitors independently of the RhoA-actin pathway but through regulation of cyclin D1 expression, in agreement with earlier work demonstrating the importance of cyclin D1 regulation in controlling the rate of generation and expansion of basal progenitors 46.
Rnd3 expression in cortical progenitors and postmitotic neurons is controlled transcriptionally by the proneural protein Ascl1, and Rnd3 acts downstream of Ascl1 in the regulation of cortical neuron migration 26. Our finding that Ascl1 inactivation in cortical progenitors has more limited effects than Rnd3 knockdown was therefore unexpected. Elimination of Ascl1 altered the orientation of the mitotic spindles in dividing radial glia, a defect corrected by expression of Rnd3. Thus Ascl1 promotes vertical divisions in the cortical VZ mainly by modulating the actin cytoskeleton via Rnd3 regulation. However, we did not observe defects in INM or in adherens junction integrity when Ascl1 was inactivated as when Rnd3 is silenced. One hypothesis is that loss of Ascl1 does not completely abolish Rnd3 expression in the ventricular zone 26 and the different Rnd3-dependent processes could be differently sensitive to reduced Rnd3 levels. A moderate decrease of Rnd3 expression after Ascl1 inactivation may not impact INM and junctions but may affect the orientation of the cleavage plane. Another possible explanation for the discrepancy between Rnd3 knockdown and Ascl1 knockdown/knockout results is that Ascl1 inactivation modifies the expression of other targets of Ascl1 that act in an opposite direction from Rnd3 and thus compensate the effect of Rnd3 silencing on INM and adherens junctions. These targets might not regulate cleavage plane orientation. Moreover, Ascl1 inactivation resulted in a decrease in proliferation in the IZ and CP, i.e. an opposite phenotype from that of Rnd3 knockdown. Ascl1 has been shown to induce the expression of cell cycle promoting genes such as E2f1 and Cdk1/2 in the ventral telencephalon and in cultivated neural stem cells 49. If these genes are also regulated by Ascl1 in the cortex, the net effect of their down regulation and that of Rnd3 when Ascl1 is inactivated might be a slowdown of divisions. The identification and analysis of additional targets of Ascl1 in cortical progenitors will allow us to test this hypothesis.
METHODS
Animals
Mice were housed, bred and treated according to the guidelines approved by the Home Office under the Animal (Scientific procedures) Act 1986. The following mouse lines were used and genotyped as described previously: Ascl1flox 26 and Emx1Cre 55.
In utero electroporation and tissue processing
In utero electroporation of the cerebral cortex was performed as described previously 56. A concentration of 1 μg/μl was used for each construct. The different expression plasmids and shRNA constructs used have been described previously 26. cyclinD1 shRNA was a kind gift from Dr Federico Calegari 46. At the desired time point after electroporation, pregnant mice were sacrificed by neck dislocation and embryos were processed for tissue analyses. Embryonic brains were fixed in 4% PFA overnight and then placed in 20% sucrose/PBS overnight. Embryonic brains were then embedded in OCT Compound and frozen before sectioning using a cryostat.
Immunostaining
After washing in PBS, sections were treated with PBS - 0.01% TritonX100 - 10% serum for 30 minutes. They were then incubated overnight at 4°C with the following primary antibodies diluted in the blocking buffer: rat anti-BrdU (1/1000, AbD Serotec), rabbit anti-cyclinD1 (1/25, Thermo Fisher Scientific), chicken anti-GFP (1/700, Millipore), mouse anti-Ki67 (1/50, BD PharmingenTM), mouse anti-NeuN (1/1000, Millipore), rabbit anti-PAX6 (1/200, Covance), rabbit anti-pHH3 (1/200, Upstate), mouse anti-Rnd3 (1/500, Abcam), rabbit anti-Tbr2 (1/500, Abcam) and mouse anti γ-tubulin (1/100, Abcam). Sections were then incubated with appropriate fluorescent secondary antibodies. Pretreatment with 2N HCl for 30 min at 37°C prior to pre-incubation with primary antibody was performed to detect BrdU. To detect NeuN, a step of antigen retrieval with citrate was performed before the blocking (sodium citrate pH=6 for 15 min at 90°C).
The junctional markers were immunostained as described previously 57 with minor modifications. After 5 min in PBS - 1% TritonX100, sections were treated with a blocking buffer containing PBS - 1% TritonX100 and bovine serum albumin (3%) for 1h. Primary antibodies were incubated overnight at room temperature in the blocking buffer without detergent: rabbit anti β-catenin (1/800, Sigma), mouse anti N-cadherin (1/100, BD Transduction Laboratories), rabbit anti ZO1 (1/200, Zymed). Appropriate fluorescent secondary antibodies diluted in PBS were incubated for 1h.
F-actin filaments were visualized using rhodamine-labeled phalloidin (Sigma). Sections were permeabilized 10 min with 0.1% Triton X-100-PBS, incubated with Rhodamine-labeled phalloidin diluted in PBS (0.2 μg/ml) for 40 minutes and washed several times in PBS.
All images were acquired with a laser scanning confocal microscope (Radiance 2100, BioRad). Cell counts were performed using MetaMorph software (Molecular Devices).
BrdU labeling
Interkinetic nuclear migration
Cerebral cortices were electroporated in utero at E14.5 and subjected to 30 min, 3 h or 6 h BrdU (50 mg/kg) pulse labeling 24 h after the electroporation. After the pulse, coimmunostaining for GFP and BrdU was performed. The cortex was then divided into bins of 20 μm in height and the proportion of GFP+/BrdU+ cells was determined in each bin with bin 1 at the ventricle surface.
Labeling index
Cerebral cortices were electroporated ex vivo at E14.5 as previously described 26. After 2 DIV, BrdU (final concentration of 20 μg/ml) was added every 10 h. Sections were fixed 1 h, 3.5 h, 5.5 h, 8 h and 11.5 h after the first addition of BrdU.
Measure of G1 duration
To measure G1 phase duration in the electroporated population in vivo, BrdU pulse injection (30 min, 50 mg/kg) was performed 8, 10, 12, 14 and 16 h after in utero electroporation at E14.5 as previously described 58. The percentage of electroporated (GFP+) cells that had incorporated BrdU was then determined at the different time points.
Live imaging
One day after ex vivo electroporation, GFP was imaged in live brain slices using 900nm multiphoton excitation (Spectraphysics DeepSee) with a Leica SP5 confocal scanner on a DM6000 CFS upright microscope. A 10x, 0.4NA (dry) lens was used and reflected excitation collected with a non descanned PMT through a 525/50 filter (Chroma).
Statistical analysis
A statistical analysis was performed using either unpaired two-tailed Student’s t-test between control and experimental condition, or one-way ANOVA followed by a PLSD Fisher post hoc test for multiple comparisons (StatView software, version 5). Details for each experiment are described in figure legends.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Guillemot laboratory for suggestions and comments on the manuscript, Dr. Federico Calegari for cyclin D1 shRNA plasmid. E.P. was supported by fellowships from the Federation of European Biochemical Societies (FEBS) and the Medical Research Council (MRC) and R.A. by an MRC studentship. This work was supported by a project grant from the Wellcome Trust (086947/Z/08/Z) and by a Grant-in-Aid from the Medical Research Council to F.G. (U117570528).
REFERENCES
- 1.Konno D, et al. Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat Cell Biol. 2008;10:93–101. doi: 10.1038/ncb1673. [DOI] [PubMed] [Google Scholar]
- 2.Morin X, Bellaiche Y. Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev Cell. 2011;21:102–119. doi: 10.1016/j.devcel.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 3.Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 4.Shitamukai A, Matsuzaki F. Control of asymmetric cell division of mammalian neural progenitors. Dev Growth Differ. 2012;54:277–286. doi: 10.1111/j.1440-169X.2012.01345.x. [DOI] [PubMed] [Google Scholar]
- 5.Kunda P, Baum B. The actin cytoskeleton in spindle assembly and positioning. Trends in cell biology. 2009;19:174–179. doi: 10.1016/j.tcb.2009.01.006. doi:10.1016/j.tcb.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Sahai E, Marshall CJ. ROCK and Dia have opposing effects on adherens junctions downstream of Rho. Nat Cell Biol. 2002;4:408–415. doi: 10.1038/ncb796. doi:10.1038/ncb796. [DOI] [PubMed] [Google Scholar]
- 7.Yonemura S. Cadherin-actin interactions at adherens junctions. Current opinion in cell biology. 2011;23:515–522. doi: 10.1016/j.ceb.2011.07.001. doi:10.1016/j.ceb.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Messier PE, Auclair C. Effect of cytochalasin B on interkinetic nuclear migration in the chick embryo. Dev Biol. 1974;36:218–223. doi: 10.1016/0012-1606(74)90206-1. [DOI] [PubMed] [Google Scholar]
- 9.Murciano A, Zamora J, Lopez-Sanchez J, Frade JM. Interkinetic nuclear movement may provide spatial clues to the regulation of neurogenesis. Mol Cell Neurosci. 2002;21:285–300. doi: 10.1006/mcne.2002.1174. [DOI] [PubMed] [Google Scholar]
- 10.Norden C, Young S, Link BA, Harris WA. Actomyosin is the main driver of interkinetic nuclear migration in the retina. Cell. 2009;138:1195–1208. doi: 10.1016/j.cell.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schenk J, Wilsch-Brauninger M, Calegari F, Huttner WB. Myosin II is required for interkinetic nuclear migration of neural progenitors. Proc Natl Acad Sci U S A. 2009;106:16487–16492. doi: 10.1073/pnas.0908928106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferland RJ, et al. Disruption of neural progenitors along the ventricular and subventricular zones in periventricular heterotopia. Human molecular genetics. 2009;18:497–516. doi: 10.1093/hmg/ddn377. doi:10.1093/hmg/ddn377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lian G, et al. Filamin a regulates neural progenitor proliferation and cortical size through Wee1-dependent Cdk1 phosphorylation. J Neurosci. 2012;32:7672–7684. doi: 10.1523/JNEUROSCI.0894-12.2012. doi:10.1523/jneurosci.0894-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagano T, et al. Filamin A-interacting protein (FILIP) regulates cortical cell migration out of the ventricular zone. Nat Cell Biol. 2002;4:495–501. doi: 10.1038/ncb808. doi:10.1038/ncb808. [DOI] [PubMed] [Google Scholar]
- 15.Cappello S, et al. The Rho-GTPase cdc42 regulates neural progenitor fate at the apical surface. Nat Neurosci. 2006;9:1099–1107. doi: 10.1038/nn1744. doi:10.1038/nn1744. [DOI] [PubMed] [Google Scholar]
- 16.Chen L, et al. Cdc42 deficiency causes Sonic hedgehog-independent holoprosencephaly. Proc Natl Acad Sci U S A. 2006;103:16520–16525. doi: 10.1073/pnas.0603533103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herzog D, et al. The small GTPase RhoA is required to maintain spinal cord neuroepithelium organization and the neural stem cell pool. J Neurosci. 2011;31:5120–5130. doi: 10.1523/JNEUROSCI.4807-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katayama K, et al. Loss of RhoA in neural progenitor cells causes the disruption of adherens junctions and hyperproliferation. Proc Natl Acad Sci U S A. 2011;108:7607–7612. doi: 10.1073/pnas.1101347108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thumkeo D, et al. Deficiency of mDia, an actin nucleator, disrupts integrity of neuroepithelium and causes periventricular dysplasia. PLoS One. 2011;6:e25465. doi: 10.1371/journal.pone.0025465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gauthier-Fisher A, et al. Lfc and Tctex-1 regulate the genesis of neurons from cortical precursor cells. Nat Neurosci. 2009;12:735–744. doi: 10.1038/nn.2339. doi:10.1038/nn.2339. [DOI] [PubMed] [Google Scholar]
- 21.Roszko I, Afonso C, Henrique D, Mathis L. Key role played by RhoA in the balance between planar and apico-basal cell divisions in the chick neuroepithelium. Dev Biol. 2006;298:212–224. doi: 10.1016/j.ydbio.2006.06.031. doi:10.1016/j.ydbio.2006.06.031. [DOI] [PubMed] [Google Scholar]
- 22.Cappello S, et al. A radial glia-specific role of RhoA in double cortex formation. Neuron. 2012;73:911–924. doi: 10.1016/j.neuron.2011.12.030. [DOI] [PubMed] [Google Scholar]
- 23.Chen L, Melendez J, Campbell K, Kuan CY, Zheng Y. Rac1 deficiency in the forebrain results in neural progenitor reduction and microcephaly. Dev Biol. 2009;325:162–170. doi: 10.1016/j.ydbio.2008.10.023. doi:10.1016/j.ydbio.2008.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leone DP, Srinivasan K, Brakebusch C, McConnell SK. The rho GTPase Rac1 is required for proliferation and survival of progenitors in the developing forebrain. Developmental neurobiology. 2010;70:659–678. doi: 10.1002/dneu.20804. doi:10.1002/dneu.20804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minobe S, et al. Rac is involved in the interkinetic nuclear migration of cortical progenitor cells. Neuroscience research. 2009;63:294–301. doi: 10.1016/j.neures.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Pacary E, et al. Proneural transcription factors regulate different steps of cortical neuron migration through Rnd-mediated inhibition of RhoA signaling. Neuron. 2011;69:1069–1084. doi: 10.1016/j.neuron.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taverna E, Huttner WB. Neural progenitor nuclei IN motion. Neuron. 2010;67:906–914. doi: 10.1016/j.neuron.2010.08.027. [DOI] [PubMed] [Google Scholar]
- 28.Ueno M, Katayama K, Yamauchi H, Nakayama H, Doi K. Cell cycle progression is required for nuclear migration of neural progenitor cells. Brain Res. 2006;1088:57–67. doi: 10.1016/j.brainres.2006.03.042. [DOI] [PubMed] [Google Scholar]
- 29.Spear PC, Erickson CA. Interkinetic nuclear migration: a mysterious process in search of a function. Dev Growth Differ. 2012;54:306–316. doi: 10.1111/j.1440-169X.2012.01342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chenn A, McConnell SK. Cleavage orientation and the asymmetric inheritance of Notch1 immunoreactivity in mammalian neurogenesis. Cell. 1995;82:631–641. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- 31.Ge X, Frank CL, Calderon de Anda F, Tsai LH. Hook3 interacts with PCM1 to regulate pericentriolar material assembly and the timing of neurogenesis. Neuron. 2010;65:191–203. doi: 10.1016/j.neuron.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sauer F. Mitosis in the neural tube. J Comp Neurol. 1935;62:337–420. [Google Scholar]
- 33.Asami M, et al. The role of Pax6 in regulating the orientation and mode of cell division of progenitors in the mouse cerebral cortex. Development. 2011;138:5067–5078. doi: 10.1242/dev.074591. [DOI] [PubMed] [Google Scholar]
- 34.Godin JD, et al. Huntingtin is required for mitotic spindle orientation and mammalian neurogenesis. Neuron. 2010;67:392–406. doi: 10.1016/j.neuron.2010.06.027. [DOI] [PubMed] [Google Scholar]
- 35.Rubenstein NM, Chan JF, Kim JY, Hansen SH, Firestone GL. Rnd3/RhoE induces tight junction formation in mammary epithelial tumor cells. Exp Cell Res. 2005;305:74–82. doi: 10.1016/j.yexcr.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 36.Gotz M, Stoykova A, Gruss P. Pax6 controls radial glia differentiation in the cerebral cortex. Neuron. 1998;21:1031–1044. doi: 10.1016/s0896-6273(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 37.Englund C, et al. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fietz SA, Huttner WB. Cortical progenitor expansion, self-renewal and neurogenesis-a polarized perspective. Current opinion in neurobiology. 2011;21:23–35. doi: 10.1016/j.conb.2010.10.002. doi:10.1016/j.conb.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 39.Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. doi:10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shitamukai A, Konno D, Matsuzaki F. Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. J Neurosci. 2011;31:3683–3695. doi: 10.1523/JNEUROSCI.4773-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci U S A. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. doi:10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riou P, Villalonga P, Ridley AJ. Rnd proteins: multifunctional regulators of the cytoskeleton and cell cycle progression. BioEssays : news and reviews in molecular, cellular and developmental biology. 2010;32:986–992. doi: 10.1002/bies.201000060. doi:10.1002/bies.201000060. [DOI] [PubMed] [Google Scholar]
- 43.Poch E, et al. RhoE interferes with Rb inactivation and regulates the proliferation and survival of the U87 human glioblastoma cell line. Exp Cell Res. 2007;313:719–731. doi: 10.1016/j.yexcr.2006.11.006. doi:10.1016/j.yexcr.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 44.Villalonga P, Fernandez de Mattos S, Ridley AJ. RhoE inhibits 4E-BP1 phosphorylation and eIF4E function impairing cap-dependent translation. J Biol Chem. 2009;284:35287–35296. doi: 10.1074/jbc.M109.050120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villalonga P, Guasch RM, Riento K, Ridley AJ. RhoE inhibits cell cycle progression and Ras-induced transformation. Mol Cell Biol. 2004;24:7829–7840. doi: 10.1128/MCB.24.18.7829-7840.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lange C, Huttner WB, Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell. 2009;5:320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 47.Madigan JP, et al. Regulation of Rnd3 localization and function by protein kinase C alpha-mediated phosphorylation. Biochem J. 2009;424:153–161. doi: 10.1042/BJ20082377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riento K, et al. RhoE function is regulated by ROCK I-mediated phosphorylation. Embo J. 2005;24:1170–1180. doi: 10.1038/sj.emboj.7600612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Castro DS, et al. A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev. 2011;25:930–945. doi: 10.1101/gad.627811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thery M, et al. The extracellular matrix guides the orientation of the cell division axis. Nat Cell Biol. 2005;7:947–953. doi: 10.1038/ncb1307. doi:10.1038/ncb1307. [DOI] [PubMed] [Google Scholar]
- 51.Peyre E, Morin X. An oblique view on the role of spindle orientation in vertebrate neurogenesis. Dev Growth Differ. 2012;54:287–305. doi: 10.1111/j.1440-169X.2012.01350.x. doi:10.1111/j.1440-169X.2012.01350.x. [DOI] [PubMed] [Google Scholar]
- 52.Harris TJ, Tepass U. Adherens junctions: from molecules to morphogenesis. Nature reviews. Molecular cell biology. 2010;11:502–514. doi: 10.1038/nrm2927. doi:10.1038/nrm2927. [DOI] [PubMed] [Google Scholar]
- 53.Samarin S, Nusrat A. Regulation of epithelial apical junctional complex by Rho family GTPases. Frontiers in bioscience : a journal and virtual library. 2009;14:1129–1142. doi: 10.2741/3298. [DOI] [PubMed] [Google Scholar]
- 54.Watanabe T, Sato K, Kaibuchi K. Cadherin-mediated intercellular adhesion and signaling cascades involving small GTPases. Cold Spring Harbor perspectives in biology. 2009;1:a003020. doi: 10.1101/cshperspect.a003020. doi:10.1101/cshperspect.a003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iwasato T, et al. Dorsal telencephalon-specific expression of Cre recombinase in PAC transgenic mice. Genesis. 2004;38:130–138. doi: 10.1002/gene.20009. [DOI] [PubMed] [Google Scholar]
- 56.Pacary E, et al. Visualization and genetic manipulation of dendrites and spines in the mouse cerebral cortex and hippocampus using in utero electroporation. Journal of visualized experiments : JoVE. 2012 doi: 10.3791/4163. doi:10.3791/4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marthiens V, ffrench-Constant C. Adherens junction domains are split by asymmetric division of embryonic neural stem cells. EMBO Rep. 2009;10:515–520. doi: 10.1038/embor.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pilaz LJ, et al. Forced G1-phase reduction alters mode of division, neuron number, and laminar phenotype in the cerebral cortex. Proc Natl Acad Sci U S A. 2009;106:21924–21929. doi: 10.1073/pnas.0909894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.