Abstract
Antibody arrays provide a valuable method of obtaining multiple protein measurements from low volumes of biological samples. Antibody arrays can be designed to target not only core protein abundances (relative or absolute abundances, depending on the availability of standards for calibration), but also protein post-translational modifications, provided antibodies or other affinity reagents are available to detect the modifications. Glycosylation is a common modification that has important and diverse functions, both in normal and disease biology. The methods for measuring glycan levels on multiple, specific proteins using antibody arrays and glycan-binding reagents have made significant progress. Here we describe practical approaches to develop, optimize and use antibody array assays to determine both protein abundances and glycosylation states. We cover the use of low-volume arrays to reduce sample consumption and a new way to improve the binding strength of particular glycan-binding reagents through multimerization. These methods could be useful for a wide range of biological studies in which glycosylation may be changing or affect protein function.
INTRODUCTION
Immunoassays have a central place in biological research as the primary method to obtain specific measurements of a particular protein in a complex, biological sample. This central spot in the toolkit owes to the features of sensitive, reproducible detection while maintaining high specificity for a particular analyte. The versatility of immunoassays also is valuable, as they have been implemented in microtiter plates, in situ sensors, microfluidic devices, and microarrays, coupled to various detection methods including enzymatic assays, electrochemiluminescence, fluorescence, Raman spectroscopy, electrochemical detection, and others. The miniaturization of the assays in microarrays and microfluidic devices has been important for research using clinical samples, which can be limited by low sample availability. For example biopsy material, or human-derived specimens that are divided between many different research projects, could be available in only small quantities. In addition, miniaturization allows the analysis of multiple, different proteins with low sample consumption, which is especially relevant to biomarker research and systems biology.
Another feature of immunoassays is that they can be designed not just to measure core protein levels, but also protein modifications, provided antibodies or other affinity reagents are available to measure those modifications. Post translational modifications to proteins are fundamentally important to their function, so the ability to measure changes in the modified states is important for protein studies. One of the most common modifications, occurring on nearly all secreted and membrane-bound proteins and on many intracellular proteins, is glycosylation—the covalent addition of carbohydrate structures onto proteins. Protein glycosylation is involved in central functions such as folding, degradation, trafficking, modulation of protein-protein interactions, signal transduction, and many others (Varki et al., 2009). Immunoassays and affinity-based assays to measure glycan modifications on proteins have made significant progress, and it is increasingly possible to measure specific glycan changes on specific proteins in biological samples (Chen et al., 2007; Li et al., 2009; Li et al., 2011). This ability has applications in biomarker research and in a wide variety of biological research (Haab, 2010; Hirabayashi, 2004).
The topic of this article is low volume, multiplexed immunoassays for protein abundance and glycosylation. We cover practical approaches for implementing these approaches in an academic lab setting. The article first compares some of the major formats, to provide the reader with an understanding of the important experimental considerations (see Strategic Planning). Second, we provide detailed instructions for using antibody arrays to reliably measure the levels of multiple proteins out of low sample volumes (see Basic Protocol 1). In the third section, we cover methods for assessing the glycosylation state of the proteins (see Basic Protocol 2).
BASIC PROTOCOL 1: Array-based sandwich assays for protein detection and quantification
BASIC PROTOCOL 2: Detecting glycans on proteins captured by antibody arrays
STRATEGIC PLANNING
Comparison of approaches
Several options of assay format, substrates, and detection methods are available for antibody arrays. The selections may depend on what is practical and available to the researcher. For example, some researchers have microarrayers readily available, which would facilitate the production of planar arrays on microscope slides, while other institutions have invested in suspension bead arrays (for example using the Luminex technology). Planar arrays refer to the immobilization of distinct antibody in ordered locations on the surface of a planar substrate (Haab, 2006), like a microscope slide, and bead arrays refer to suspensions of beads, with each bead coated with a particular antibody (Schwenk et al., 2008). The performance characteristics, including sensitivity, specificity, linear range, and throughput, can be very comparable between these two approaches.
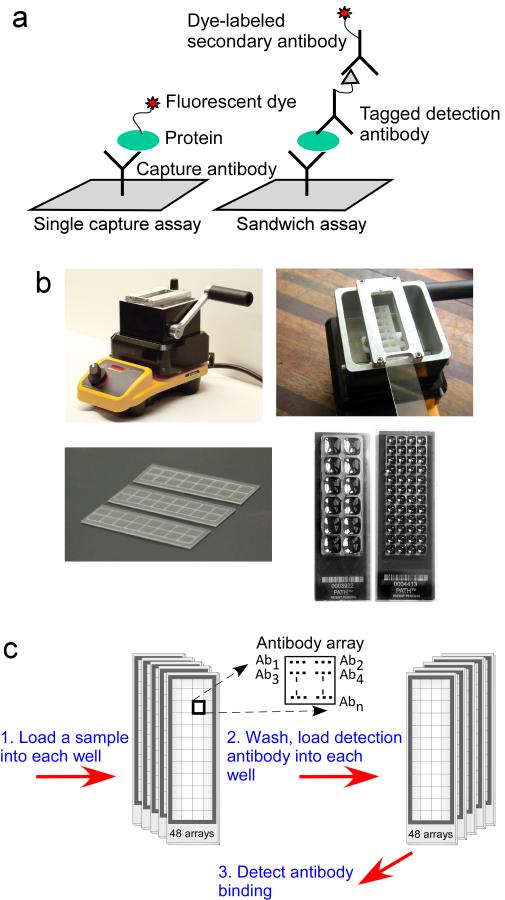
Researchers also can choose between sandwich assays and single capture assays for the recognition of each protein (Fig. 1a). Sandwich assays use two antibodies for each analyte, one to capture the protein (Kingsmore, 2006; Schweitzer et al., 2000), and a second to detect the captured protein, and single capture assays use just the capture antibody (Haab et al., 2001). To detect analyte binding in the single capture assay, usually the entire sample is labeled with a fluorescent dye or another tag like biotin to allow subsequent detection. Sandwich assays have greater specificity of detection, owing to the use of two antibodies, but they usually require more up-front development and optimization and can be difficult to multiplex. Single capture assays are simple to multiplex but are more susceptible to non-specific binding.
Figure 1. Low volume antibody arrays.
a) Schematic representation of single capture and sandwich assays. b) Hydrophobic borders to define wells. A hydrophobic, wax-based composition is melted in the Slide Imprinter (upper left), and a slide is inserted upside-down above the wax bath (upper right). Pulling the lever lifts a stamp out of the bath to contact the slide and imprint a pattern of hydrophobic boundaries (lower left). Stamps of various patterns can be used to define large or small wells, each containing a microarray (lower right). c) Running multiple samples on a single microscope slide. An example is shown of processing 48 samples on one slide.
Finally, two complementary approaches are available in how to achieve multiple protein measurements from low sample volumes. One approach is to multiplex—to detect several, different proteins in the same sample—thus reducing the amount of sample used per protein. The microarray is especially amenable to multiplexing, because each element on the array can be used to measure a different analyte. Multiplexing is less important when only a handful of analytes are targeted or when cross reactivity is a problem. Cross reactivity can be a problem when multiplexing sandwich assays by pooling detection antibodies, as off-target binding by the detection antibodies is increasing likely with increasing analyte numbers (Perlee et al., 2004). Another approach which avoids the problem of cross reactivity is to use only one detection antibody per array, but reduce the volume of each array (Forrester et al., 2007) (Fig. 1b). Separate arrays could be used for distinct assays, thus removing the possibility of cross reactivity, yet total volume used could still be low due to the low volume of each array.
We previously introduced a convenient method for achieving low-volume arrays (Forrester et al., 2007; Haab and Yue, 2011) (Fig. 1b). Hydrophobic borders are imprinted onto the surface of a coated microscope slide, which define the boundaries of each array. The boundaries of each array can be made very small (for example just a few millimeters per array), so that many arrays can be fit onto each slide, with a very low volume requirement for each array. An array defined by hydrophobic boundaries of 4×4 mm requires only 6 μL of sample to fully cover the array, and a 2×2 mm array requires only 1.5 μL. The ability to print many, identical arrays on a microscope slide also facilitates high-throughput sample processing, because many different samples could be processed on each slide in a manner compatible with multi-channel pipetting (Fig. 1c).
This article focuses on low-volume sandwich assays using antibody arrays, designed to detect either protein abundances or protein glycosylation states. We cover approaches for optimizing the assays and for setting proper dilution factors according to linear ranges. In addition, a novel approach for improving the binding strength of lectins for detecting glycans is described.
Preparation of antibody arrays on coated microscope slides
Several types of coated microscope slides can be used for the production of antibody arrays; the optimal type depends on many factors such as the type of microarrayer, the detection method, and the sample type. The methods described above for optimizing the assay can be used to compare the performance of the various available substrates.
The antibodies to be spotted on the array should be as pure as possible, since protein or other contaminants could cause non-specific signals. Small molecule contaminants can be removed by dialysis, and larger contaminants and aggregates can be removed by ultracentrifugation. The addition of a small concentration of detergent, such as 0.005% Tween-20, can improve the consistency of spot morphology for certain antibodies. It can be valuable to prepare each antibody in two or more different buffer or detergent conditions to see which is best on the particular substrate being used. Each antibody should be spotted three or more times in each array to allow an assessment of variability within each array. If the printed slides are not to be used right away, they should be vacuum sealed with desiccant and refrigerated to prevent degradation of the antibodies.
If using hydrophobic borders to segregate replicate arrays on each slide (Fig. 1b), the borders can be imprinted using the Slide Imprinter (Gel Company, San Francisco, CA) after printing is complete. Follow the manufacturer protocol for imprinting the hydrophobic borders. If using gaskets to segregate the arrays, the gaskets can be applied to the slides immediately prior to sample loading.
BASIC PROTOCOL 1 Array-based sandwich assays for protein detection and quantification
Optimization of the antibodies and control proteins for sandwich assays
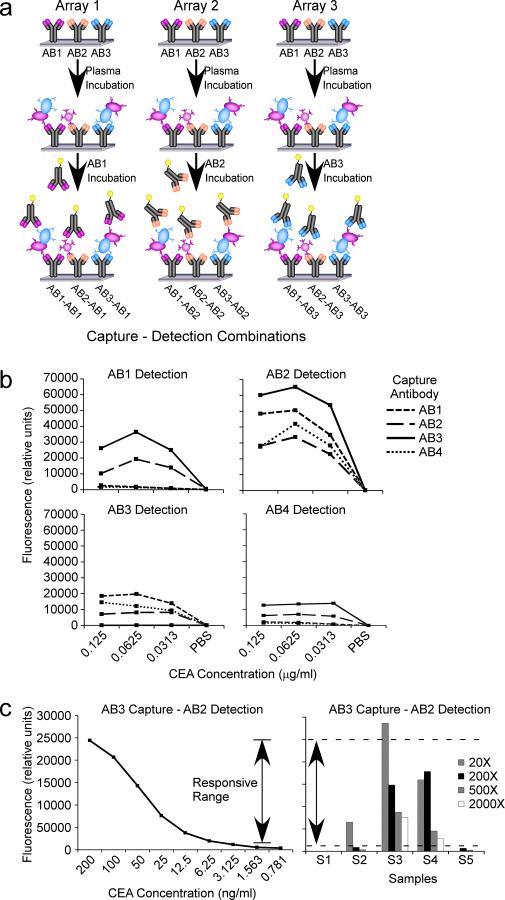
In the development of a new sandwich assay, optimization is required to select the combination of capture and detection antibodies that work best together. Some antibodies work well as capture antibodies (the antibody immobilized to the surface) but not as detection antibodies (the antibody that detects the captured protein), and vice versa, and some antibodies do not work together well as a capture-detection pair because their epitopes are too close together or interfere in some other way. Therefore, it is important to test all combinations of capture-detection pairs to see which work together best. Antibody arrays provide an efficient way to do that testing (Fig. 2a). Antibody arrays can be generated with all available antibodies, the samples incubated on the arrays in replicate, and each array probed with one of the antibodies. Each array gives a measure of one of the detection antibodies with each capture antibody.
Figure 2. Optimizing capture-detection pairs.
a) Examining all capture-detection combinations using antibody arrays. Three different antibodies are spotted in three identical arrays. Each array is incubated with the same sample and detected with one of the antibodies, yielding nine different capture-detection combinations. b) Comparing dilution curves of capture-detection combinations. The protein CEA was prepared in various dilutions, and the dilution series was examined among four different antibodies and 16 different capture-detection combinations. The best combination was AB3 capture and AB2 detection. c) Setting dilution factors. A dilution curve was run to define the boundaries of the responsive range (left), and representative experimental samples were run at four different dilutions each. Sample S3 was out of the linear range at the 20X dilution, and sample S2 was out of the linear range (at the low end) at the 200X and higher dilutions.
To assess which capture-detection pair is best, the purified protein, or a sample containing a high level of the protein, should be incubated on the arrays in a series of dilutions, covering the range of concentrations that should be detected by the assay, ending in a blank solution. One can examine the dilution curve for each capture-detection pair to determine which is best (Fig. 2b), based on smoothness of the curve (which is an indicator of the reproducibility of the assay) and the limit of detection (determine by the lowest concentration at which changes in signal are still observed). Sometimes a few options are available for control proteins or samples, and this method of comparison is useful for deciding which control sample to use for the calibration of future experiments.
Setting the dilution factor and using calibrator and control samples
When running the experimental samples, it is important to know if the experimental measurements are in the responsive range of the assay, which is the range of analyte concentrations in which changes in concentration can be observed. The responsive range can be assessed by running a dilution series of the control sample with each set of experiments and then determining the lowest and highest signals at which changes are observed (Fig. 2c). That determination can be imprecisely done by visual inspection of the dilution series, or it can be more precisely done by a technique called recovery analysis. In recovery analysis, a curve is fitted to the dilution series (usually a four-parameter fit), and the data used to make the curve are plugged back into the resulting equation to solve for concentration (or dilution factor). The concentrations calculated by the equation are compared to the known concentrations, and a percent change (or percent recovery) between the two is determined. The regions of the curve that are more flat will return an less accurate concentrations, because the signal does not change in response to concentration changes. The range of the curve that returns an acceptable percent recovery (determined by the user according to the requirements of the assay) is the responsive range. The experimental samples should be diluted to a concentration that will give a signal in the responsive range. Since the starting concentrations of the experimental samples are not known beforehand, before running a large experiment set it is useful to run representative samples at a few dilutions each, to see which dilution best places the samples in the responsive range (Fig. 2c).
Another factor to consider when running experimental samples is the stability of the assay between runs and over long periods of time. The use of control samples with each experiment set can help assess the consistency of the assay over time. A small set (5–10 samples) of representative samples with a range of analyte concentrations can be run with each experiment, randomly interspersed with the experimental samples and treated identically, and the measurements of those samples can be compared to previous experiment sets.
Materials
Nitrocellulose or NHS-hydrophilic polymer coated slides(several commercial varieties available).
Capture and detection antibodies (see below for buffer and concentration recommendations).
Slide Imprinter (The Gel Company, San Francisco, CA) or hybridization gaskets (ArrayIt, Sunnyvale, CA) for defining wells on the slides.
Biotinylation reagent (EZ-link Sulfo-NHS-LC-Biotin, Pierce Biotechnology, Rockford, IL).
Streptavidin-β-phycoerythrin (Invitrogen, Carlsbad, CA).
1X phosphate buffered saline (PBS), pH 7.4: 137 mM sodium chloride, 2.7 mM potassium chloride, 4.3 mM sodium phosphate dibasic, 1.4 mM potassium phosphate monobasic.
PBST0.1: 1X PBS + 0.1% Tween-20.
PBST0.5: 1X PBS + 0.5% Tween-20.
PBST0.5 + 1% bovine serum albumin (BSA).
PBST0.1 + 0.1% BSA.
10X sample buffer: 1X PBS + 1% Tween-20 + 1% Brij-35.
4X IgG blocking cocktail (400 μg/mL each of mouse, sheep, and goat IgG, and 800 μg/mL rabbit IgG in 1X PBS, antibodies from Jackson Immunoresearch). The IgG cocktail is included to prevent non-specific binding to the capture antibodies. Certain individuals have blood antibodies that are reactive with IgG from species such as mouse, goat, sheep and rabbit, and the binding of these antibodies to the capture antibodies on the arrays can cause noise within the assay. The addition of IgG into the human serum sample will block the binding of the human antibodies to the immobilized capture antibodies. Prepare the IgG Cocktail to include antibodies from animals representative of the capture antibodies on the arrays.
10X protease inhibitor (Complete Tablet, Roche Applied Science, Indianapolis, IN). Dissolve one protease inhibitor tablet in 1 mL 1X PBS to make the 10X stock.
2X sample dilution buffer: 2X sample buffer + 2X protease inhibitor + 2X IgG cocktail in 1X PBS.
Protocol
The protocol consists of preparation of the samples, incubation of the samples, incubation of detection reagents, and detecting signals. It is presented as a three-day protocol but can be significantly shortened at certain steps, as indicated.
Day 1
-
1.
Prepare the samples. The final dilution factor of the biological sample should be determined beforehand as described above (Fig. 2c). Each sample should be diluted to two-fold the final concentration and then mixed 1:1 with the 2X sample dilution buffer (see Materials). For example, for a final 4X dilution, mix the sample 1:1 with 1X PBS and then mix the diluted sample 1:1 with the 2X sample dilution buffer. [Authors; so any initial dilutions are made with PBS and only the final dilution uses 2X dilution buffer?]
-
2.Mix thoroughly and place at 4°C overnight with gentle shaking.The overnight mixing allows the IgG blockers to fully interact with components in the samples. The protocol may be shorted to a two day protocol by prepping samples at least two hours before beginning of day two of the protocol.
-
3.Prepare the biotinylated detection antibodies. Follow the manufacturer's protocol for protein biotinylation and sample cleanup. A high-concentration (~1 mg/mL) stock solution in 1X PBS can be aliquoted and frozen for long-term use.A modification to the biotinylation procedure is to limit the reaction to prevent the labeling of all available amine groups, because the complete labeling of the antibody might negatively affect activity. The reaction can be slowed by performing it at pH 7 and at 4 °C (instead of pH 9 at room temperature) and by quenching after one hour.
Day 2
-
4.Briefly rinse slides in PBST0.5 once and wash the slides in PBST 0.5 for 10–15 minutes with gentle shaking to remove unbound antibodies from the surface. Dry the slides by centrifuging in a swinging-bucket clinical centrifuge for 2 min at ~200 × g.A convenient method to wash and dry the slides is to load the slides into a slide rack, load the entire slide rack into a staining chamber for washing, and then transfer the slide rack to a swinging-bucket centrifuge for spin-drying. Place a folded paper towel under the slide rack to capture the liquid when spin-drying. Make sure that the transfer to the centrifuge and spinning takes place as rapidly as possible to prevent evaporative drying on the slide. Slides should be completely dry before moving to the next step. To decrease possible buildup of residue from the drying, alternate the direction of the slides in the slide carrier for subsequent spins.
-
5.Block the slides. The composition of the blocking solution depends on the slide coating. A solution that works well for nitrocellulose and amine coatings is PBST 0.5% containing 1% BSA. For slides containing amine-reactive groups such as epoxy or N-hydroxysuccinimide, use 25 mM ethanolamine in sodium borate buffer (pH 9.0). Pipette the blocking solution onto each array and incubate for 1 hour at room temperature in a humidified chamber with gentle shaking. The volume depends on the size of the array. For 4.5 mm square arrays, 7 μL easily covers each array. The subsequent descriptions will refer to the 4.5 mm size array.The slides should be incubated in a humidified box to prevent evaporation. A standard slide box provides a good holder for slides, and humidity can be achieved by placing a damp paper towel in the bottom of the box. The use of a paper towel prevents splashing if the box is jarred.
-
6.
Quickly rinse the slides in PBST 0.5 twice to remove the samples, and then wash 3 times in PBST 0.5 for 3 minutes eachThe quick rinses serve to immediately remove the samples and prevent a high protein concentration in the wash baths. Change to new buffer for each wash. Centrifuge at ~200 × g for 2 min to dry slides.
-
7.Load the slides into a humidified slide box and pipette 6 μL of each sample prepped the day before onto an array. Repeat until all arrays from each microscope slide are loaded and place at 4 °C overnight.Alternatively, the samples can be incubated on the slides for two hours at room temperature.
Day 3
-
8.
Rinse slides in PBST 0.1 twice and then wash 3 times in PBST 0.1 for 3 minutes eachChange to new buffer for each wash. Centrifuge at ~200 × g for 2 min to dry slides.
-
9.
Prepare the primary detection antibodies. The primary detection antibodies should be diluted into PBST0.1 with 0.1% BSA at a final concentration optimized for each detection antibody. Optimal concentrations are usually 1–3 μg/ml.
-
10.
Pipette 6 μL of each detection antibody solution onto each array and place the slides in a humidified box for 1 hour at room temperature with gentle shaking.
-
11.
Wash and dry the slides (repeat Step 8).
-
12.
Prepare the fluorescence-labeled secondary detection reagents. Use streptavidin-phycoerythrin or dye-labeled anti-biotin to detect the biotinylated primary detection antibodies. Prepare in 0.1% BSA with PBST0.1 at a final concentration of 2 μg/mL.
-
13.
Pipette 6 μL of the detection solution onto each array and incubate in a humidified box for 1 hour at room temperature with gentle shaking.
-
14.
Wash and dry the slides (repeat Step 8
-
15.
Scan the slides. If planning to scan the slides at a later time, vacuum seal the slides with desiccant and store in 4°C for up to 3 months. An example of raw scan data is given in Figure 3c. Image and data analysis depends on the systems used. A typical strategy is to use image analysis software (such as the GenePix Pro software) to locate the spots on the array and quantify the fluorescence signal at each spot. The next steps are to match the fluorescence data with the known identities of the spots, average the data between replicate spots, and then perform the analyses relevant to the goals of the experiment. Normalization of microarray data may be required if experimental bias is present. Methods to detect and normalize for bias in antibody microarray data are discussed elsewhere (Hamelinck et al., 2005).
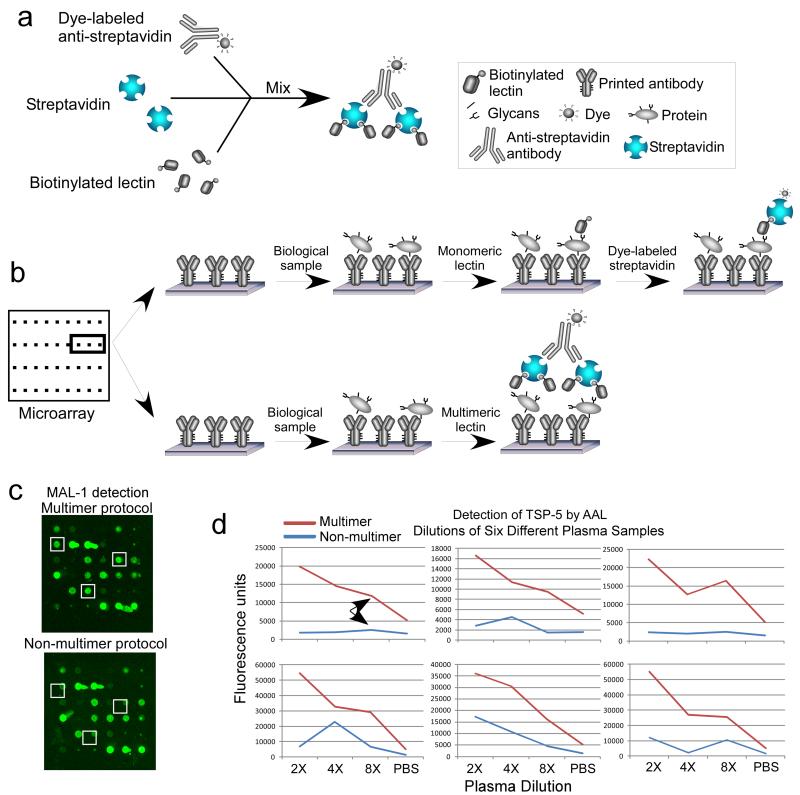
Figure 3. Improving lectin binding using multimerization.
a) Lectins are pre-incubated with dye-labeled streptavidin to form multi-lectin units. b) After samples are incubated on antibody arrays, the capture proteins are probed with the multimers (top), in contrast to the standard protocol in which the lectins are not linked (bottom). c) Signals can be improved for certain assays, resulting in d) lower detection limits as shown by dilution curves for the detection of the protein TSP-5 by the lectin AAL.
BASIC PROTOCOL 2 Detecting glycans on proteins captured by antibody arrays
Assay optimization and choosing glycan-binding affinity reagents
The principles and methods for development and optimization are the same as Basic Protocol 1. For the detection of specific glycans, the options are glycan-binding antibodies and lectins. A variety of glycan-binding antibodies have been generated with a range of binding strengths and specificities (Manimala et al., 2007). Some are named after a particular hybridoma clone, cancer antigen, or blood group, so that their status as a glycan-binding antibody is not immediately clear, such as the CA 19–9 antibody, which binds a specific tetra-saccharide that is overexpressed in some cancers. Lectins are naturally-occurring proteins with glycan-binding activity, and those with high specificity and binding affinity, such as concanavalin A and wheat germ agglutinin have been useful as analytical reagents to detect their target glycans. Lectins should be processed and biotinylated according to the guidelines given for antibodies Basic Protocol 1
Various sources in the literature can be consulted to find lectins with defined specificities or to find more information about particular lectins, such as a studies of lectin specificities using frontal affinity chromatography (Nakamura-Tsuruta et al., 2006) or glycan arrays (Manimala et al., 2006; Porter et al., 2010) and experiments using lectin arrays (Pilobello et al., 2007). Another source for lectin specificity information is publicly-available glycan array data provided by the Consortium for Functional Glycomics (CFG) (Liu et al., 2009). Glycan arrays enable the parallel comparison of lectin binding to hundreds of different glycans, from which we can derive much information about specificity. Investigators can send a protein of interest to the CFG for analysis on their glycan array (Blixt et al., 2004), and the Consortium provides back a report of the binding to each of the glycans. Since the introduction of the CFG glycan array in 2004, several thousand experiments covering hundreds of different glycan binding proteins have been performed. These data are publicly available via a web portal. Efforts are underway to automate the analysis of these data (Maupin et al., 2011; Porter et al., 2010) so they can be provided in a comprehensive, searchable format.
Improving detection sensitivity through multimerization
Some glycan-binding proteins have a weak affinity for their target glycan, at least when used as a reagent outside the biological context. Weak affinity leads to poor sensitivity in an analytical assay; this is a serious limitation, because rarely is any glycan found at a high abundance in a biological sample. Thus, certain lectins have not had broad use in biomarker studies.
In biology, lectins frequently rely upon multivalent interactions to affect their functions. For example, the glycan ligands of a lectin may be tethered to a cell surface and clustered together in multiple interactions when bound by lectins. The galectin family of lectins uses a well-described system of forming lattices in order to induce downstream effects (Brewer et al., 2002). In such systems, the strength of any one protein-glycan interaction is not great, but linking several such interactions together increases the overall interaction strength.
Given the biological interactions of certain lectins, it follows that the in vitro binding strength of such lectins to their target glycans may be improved by inducing multivalent interactions. Such an improvement might be possible if multiple target glycans are present on a particular protein (or on closely packed proteins) and if the lectins used as detection probes can be linked together. This strategy was used to improve the in vitro binding of the influenza hemaglutinin (HA) protein to its sialic acid ligand in a glycan array assay (Stevens et al., 2006). The HA proteins were recombinantly produced with a His-6 tag, and the spontaneously formed trimers of the His-labeled HA proteins were linked together with a goat antibody against the His tag. Two of the antibody-bound clusters were further joined through an anti-goat IgG antibody to form a final cluster of 12 HA proteins.
The same type of strategy could be pursued using any type of lectin. One way to cluster lectins would use the multiple binding sites of streptavidin, which has four subunits that each contain a binding site for biotin. Therefore, by mixing biotinylated lectins with streptavidin, clusters of one to four lectins should be formed (Fig. 3a). To induce further multimerization, anti-streptavidin could be added, which would link two streptavidin molecules to form clusters of two to eight lectins. The pre-formed complex of multiple lectins then can be applied to probed closely-spaced glycans on captured proteins (Fig. 3b). We recently demonstrated that this strategy indeed can dramatically increase signal strength when used in the ALSA assay (Fig. 3c), which led to increased sensitivity for for certain lectins (Fig. 3d).
The level of improvement gained by multimerization would depend on the geometry and organization of the target glycans and the structural ability of the lectin to properly bind adjacent glycans. Therefore, no protocol will be optimal for every lectin; rather, each lectin will require individual testing and optimization using a variety of multimer and non-multimer protocols. But by individual optimization of each lectin, it may be possible to greatly enhance the effectiveness of a wide range of lectins that previously were not useful as analytical reagents. The protocol below includes the steps for forming and using multimer complexes.
Protocol
The antibody array, sample preparation, and sample incubation steps are identical to the sandwich array protocol above, so this protocol begins with the day 3 description. If not using the multimer protocol, the steps are identical to Basic Protocol 1 except that glycan-binding detection reagents are used instead of protein-binding antibodies.
Another consideration when detecting antibody arrays with lectins is that the capture antibodies are glycosylated, resulting in possible non-specific binding to the capture antibodies by the lectins. Such an interaction can be probed by incubating the detection lectins on the arrays in the absence of a sample. If high levels of lectin-capture antibody interactions are observed, the glycans on the capture antibodies can be chemically modified using mild oxidation and reaction with a hydrazide reagent, thus reducing lectin binding. This procedure is described in detail elsewhere (Chen et al., 2007; Li et al., 2009; Li et al., 2011).
Follow steps 1–7 from Basic Protocol 1.
- Prepare the multimer complex. Pre-incubate the primary biotinylated lectin and the secondary streptavidin-phycoerythrin detection reagent by mixing them at a molar ratio of 4:1 (final concentration for streptavidin-phycoerythrin should be about 3 μg/mL) on ice for 1 hour.Another option for forming complexes is to incubate the biotinlylated detection reagents with anti-biotin. In that case, label the anti-biotin with a fluorescent dye such as Cy3 (using a Cy3-NHS ester reagent from GE Healthcare, for example) and mix the lectin with the anti-biotin at a 2:1 molar ratio.
Rinse the slides (after sample incubation) in PBST 0.1 twice and then wash 3 times in PBST 0.1 for 3 minutes each. Change to new buffer for each wash. Centrifuge at 200 × g for 2 min to dry slides.
Pipette 6 μL of each detection solution onto each array and place the slides in a humidified box for 1 hour at room temperature with gentle shaking.
Wash and dry the slides (repeat step 2).
Scan the slides. If planning to scan the slides at a later time, vacuum seal the slides with desiccant and store in 4°C for up to 3 months. Follow the indications given in Basic Protocol 1, Step 15.
COMMENTARY
Background Information
Antibody microarray methods were introduced in 2001 (Haab et al., 2001) and have been in continuous development since then. The ability to obtain parallel measurements of multiple proteins in small sample volumes (<10 μL) gave clear advantages over conventional protein analysis methods such as ELISA, in which 100–200 μL of sample would be required for a single assays and a corresponding high consumption of reagents. These capabilities have been especially useful in studies of protein profiles in clinical samples with limited amounts of material available, such as in studies of mouse tissue (Hung et al., 2009), wound fluid (Trostrup et al., 2011), cyst fluid (Haab et al., 2010), and sera from cancer patients (Orchekowski et al., 2005; Yue et al., 2009; Yue et al., 2011). The antibody microarray may provide a useful link between the discovery of candidate biomarkers and the validation and clinical implementation of biomarkers (Haab, 2010). Discovery technologies such as mass spectrometry can provide a wealth of protein information but with low reproducibility and low sample throughput. Antibody arrays can be used to perform the initial testing and validation of some of the best candidates coming out of the mass-spectrometry discovery studies, since several candidates could be efficiently profiled with good precision, sensitivity, and sample throughput. The addition of glycan detection further expands the application of antibody array methods, as the glycan moiety of a protein may contain disease-related information. Therefore, by detection both protein abundance and glycan variation, improved accuracy for disease may be possible in certain cases.
The antibody array method described here provides similar performance to the main alternate method, the bead-based arrays described above, but may have advantages in some respects. For example, the ability to visually inspect and evaluate the raw primary image data (Fig. 3c) is an added level of quality control, and users of planar antibody arrays have the flexibility of choosing between a wide variety of available substrates. Planar arrays also offer more flexibility in detection methods, as enzymatic methods using color change or chemiluminescence could be used (Huang et al., 2001). Such enzymatic detection methods have the advantage of not requiring expensive scanning instrumentation, and they can be very sensitive, although precision and sensitivity typically are not as good as fluorescence detection. For those with access to scanning instrumentation, fluorescence offers a reliable and sensitive option.
Critical Parameters and Troubleshooting
The successful use of this technology depends upon many individual factors being optimized and functioning properly. These factors include the quality and purity of the antibodies, the quality of the printing substrate (the coated microscope slide) and the production of the arrays, the purity of the reagents, and the accuracy of the analysis. Above we addressed some approaches to optimizing the individual components of the technology. An important tool for assessing the quality of each component and for guiding the resolution of problems is the proper use of positive and negative controls, which are discussed in more detail below.
Primary data quality inspection
The first step in assessing the quality of the data is to visually inspect the primary image. The array should show consistent, low signal from the substrate surrounding the spots, and the spots should have consistent, even morphology with signals higher than the background substrate, at least in some of the samples. Low-quality data sometimes have high or uneven signal from the background substrate, uneven spot morphology, or very low signals from the antibodies. A number of potential solutions exist for each of these problems.
Poor quality spots indicate a potential problem with the microarrayer, which usually involves dirty print tips. Rigorous cleaning of the print tips can often solve that problem. Another potential cause of poor quality spots is contamination of the antibodies, which can be solved by more rigorous purification (for example using dialysis or ultracentrifugation). In addition, the antibodies could be tested in alternate buffers or detergent additives such as Brij-35, Triton X, or CHAPS.. High signals from the substrate itself (the slide on which the antibodies are spotted) could be due to poor blocking of the slides or the presence of substances or precipitates in the samples that are particularly sticky on the surface. A solution in certain cases is to centrifuge the samples for at least 10 minutes at >10,000 × g in order to remove precipitates. Some researchers remove high-abundance components from serum/plasma preparations using affinity columns designed to capture albumin, immunoglobulin, and other proteins. Other approaches are simply to dilute the sample more or to test detergent additives to the sample. Finally, various substrates can be tested (as described above) to see which gives the most consistent and lowest backgrounds.
Another basic quality test is reproducibility between spots on the same array and between replicate arrays. It is a good idea to print replicate spots of each antibody—ideally six or more to allow good assessment of the coefficient of variation (CV) between the replicate spots. Sometimes it is not possible to print so many replicate spots due to space limitations, but at least duplicate or triplicate spots should be used. The CV between replicate spots ideally is no more than 5–10%. Arrays with internal CVs greater than 10% should be inspected for clear visual problems, as the high CV may be due to just one or two clearly faulty spots that can be removed from the analysis. An automated removal of “outlier” spots is possible using a outlier test such as the Grubb's test, if six or more replicate spots are present. A general, high variability within an array could indicate dirty print tips or a poor quality surface. These problems can be addressed as described above.
Reproducibility between replicate arrays (the same sample repeated on separate arrays) ideally is less than 25% in this manual protocol, but slightly higher levels (<40%) can be acceptable in early-stage, discovery research. Inter-array reproducibility in a manual protocol is highly dependent on user technique and the steps taken to reduce variability between arrays in every step of the protocol. Variability also can be affected by the surface and print quality, as discussed above. Replicate arrays (at least three per sample) should be used for critical applications in which the precision of the measurements needs to be confirmed. Replicate arrays that show CVs above the thresholds cited above should be visually inspected for obvious problem areas that could be removed from the analysis. If no clear cause of the high variability is apparent, the experiment should be repeated after accounting for any potential print tip, substrate, or procedural contributions to variability.
Checking the controls
Properly designed controls can provide extremely valuable information for assessing the quality of the data and determining how to improve the data. Several positive and negative controls should be included and checked in each experiment. Each array should include antibodies that should not capture anything (negative control antibodies) and antibodies that capture proteins known to be present in certain samples (positive control antibodies). The negative control antibodies could be mouse monoclonal antibodies against something never present in the type of samples to be analyzed. For example, an antibody against the HA fusion tag would be expected to never capture anything from human serum, and the signals from this antibody should extremely low. As a positive control example, an antibody against human IgM should show predictable signal in all human serum samples. A positive control for the secondary detection reagent is a biotinylated antibody, which should show strong signal from streptavidin-based detection.
An observation of high signal at the negative control antibody indicates that something from the sample is non-specifically binding the antibody. Above we described the addition of blocking IgGs to serum samples to prevent such non-specific binding, but if the non-specific signals persist, higher concentrations of blocking IgGs could be used, or the removal of high-abundance serum components.
Negative control arrays, which should be included in each experiment set, are incubated with buffer instead of sample. These arrays should show no signal except at the biotinylated antibody spots (the positive control spots). If measurable signals are observed at some of the capture antibody spots, the cause could be non-specific binding of the detection reagents to the spots or contamination of the antibody preparations. Other negative control arrays can sort out these alternatives, namely by detecting with just the secondary reagent (the dye-labeled streptavidin) or nothing at all. The solution depends on the identified source of non-specific signal. Contaminants in the antibodies can be removed as described above, and non-specific binding of lectins to the capture antibodies can be reduced using chemical derivatization of the antibodies (Chen et al., 2007).
Positive control arrays are incubated with samples known to contain particular proteins that are targeted by particular antibodies on the arrays. For example, an array could be incubated with a normal serum sample in order to observe expected signal at an antibody targeting fibronectin, a protein present in high abundance in all sera. The signals from these spots can be compared to previous results to be sure that the detection reagents are working properly. In addition, samples containing the targeted proteins can be incubated in a series of dilutions on a set of arrays. The signal at each of the antibodies should decrease with increasing dilution, finally reaching a minimum in the array incubated with just buffer. Ideally, the shape of the dilution curve is sigmoidal, with a plateau at high concentrations and a tapering off at low concentrations. A lack of consistent data from the positive control arrays could indicate a need to replace the capture or detection reagents. These controls will help to determine the quality of the data and identify routes for solving problems.
Anticipated Results
Figures 2 and 3 provide examples of the type of data that can be expected from properly functioning antibody array experiments. Experiments that pass the quality control checks described above should provide interpretable information in the form of dilution curves with a characteristic sigmoidal shape and differences in signals between individual samples. When properly designed, the experiments can provide valuable information about the associations between the glycosylation states of particular proteins and various biological conditions. Comparing the results to findings from other types of experiments, such as Western blots of the protein levels or mass spectrometry analysis of the glycans, can provide further insights into the data and the biology.
Time Considerations
We have presented these protocols as requiring two or three days, depending on how they are structured. The preparation for the experiment, including acquisition of samples and antibodies and the printing of the arrays, could be a lengthy process. The time required for the production of the arrays depends on the equipment used (some are capable of extremely rapid printing), the number of elements to be printed, and the number of slides to be printed. With a relatively fast microarrayer and a medium sized batch of slides (for example enough for two sets of experiments), the arrays could be produced concurrently while the samples are prepared. Arrays can be printed in larger batches and saved for later use (stored under vacuum), but after a few months the quality of the arrays can drop due to the gradual breakdown of antibodies.
Another consideration not addressed in the protocol is the time required for data analysis. The analysis could require significantly more time than the experiments themselves, depending on the complexity of the experimental design and the software available. The first step in the analysis is to quantify the image data. This step typically involves software, such as GenePix Pro and Array-Pro, for locating the antibody spots, quantifying the signals in and around the spots, and outputting the processed data into a spreadsheet for further analysis. The time required for this primary analysis depends on the software and the size of the arrays; a day or two should be sufficient for most experiments. The next levels of analysis can address the statistical qualities of the data, the testing of hypotheses regarding differences between the samples, and other types of data mining.
ACKNOWLEDGEMENTS
We thank the National Cancer Institute (R33 CA122890 and 1U01CA152653 to B.B.H.) and the Van Andel Research Institute for support of this work.
Footnotes
CONFLICT OF INTEREST STATEMENT B.B.H. is an inventor on the Slide Imprinter technology licensed to The Gel Company.
LITERATURE CITED
- Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N, Stevens DJ, Skehel JJ, van Die I, Burton DR, Wilson IA, Cummings R, Bovin N, Wong CH, Paulson JC. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer CF, Miceli MC, Baum LG. Clusters, bundles, arrays and lattices: novel mechanisms for lectin-saccharide-mediated cellular interactions. Current opinion in structural biology. 2002;12:616–623. doi: 10.1016/s0959-440x(02)00364-0. [DOI] [PubMed] [Google Scholar]
- Chen S, LaRoche T, Hamelinck D, Bergsma D, Brenner D, Simeone D, Brand RE, Haab BB. Multiplexed analysis of glycan variation on native proteins captured by antibody microarrays. Nature methods. 2007;4:437–444. doi: 10.1038/nmeth1035. [DOI] [PubMed] [Google Scholar]
- Forrester S, Kuick R, Hung KE, Kucherlapati R, Haab BB. Low-volume, high-throughput sandwich immunoassays for profiling plasma proteins in mice: identification of early-stage systemic inflammation in a mouse model of intestinal cancer. Molecular Oncology. 2007;1:216–225. doi: 10.1016/j.molonc.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haab BB. Applications of antibody array platforms. Current opinion in biotechnology. 2006;17:415–421. doi: 10.1016/j.copbio.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Haab BB. Antibody-lectin sandwich arrays for biomarker and glycobiology studies. Expert review of proteomics. 2010;7:9–11. doi: 10.1586/epr.09.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haab BB, Dunham MJ, Brown PO. Protein microarrays for highly parallel detection and quantitation of specific proteins and antibodies in complex solutions. Genome biology. 2001;2:RESEARCH0004. doi: 10.1186/gb-2001-2-2-research0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haab BB, Porter A, Yue T, Li L, Scheiman J, Anderson MA, Barnes D, Schmidt CM, Feng Z, Simeone D. Glycosylation Variants of Mucins and CEACAMs as Candidate Biomarkers for the Diagnosis of Pancreatic Cystic Neoplasms. Annals of surgery. 2010;251:937–945. doi: 10.1097/SLA.0b013e3181d7738d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haab BB, Yue T. High-throughput studies of protein glycoforms using antibody-lectin sandwich arrays. Methods in molecular biology (Clifton, N.J. 2011;785:223–236. doi: 10.1007/978-1-61779-286-1_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamelinck D, Zhou H, Li L, Verweij C, Dillon D, Feng Z, Costa J, Haab BB. Optimized normalization for antibody microarrays and application to serum-protein profiling. Mol Cell Proteomics. 2005;4:773–784. doi: 10.1074/mcp.M400180-MCP200. [DOI] [PubMed] [Google Scholar]
- Hirabayashi J. Lectin-based structural glycomics: glycoproteomics and glycan profiling. Glycoconjugate journal. 2004;21:35–40. doi: 10.1023/B:GLYC.0000043745.18988.a1. [DOI] [PubMed] [Google Scholar]
- Huang R-P, Huang R, Fan Y, Lin Y. Simultaneous detection of multiple cytokines from conditioned media and patient's sera by an antibody-based protein array system. Anal. Biochem. 2001;294:55–62. doi: 10.1006/abio.2001.5156. [DOI] [PubMed] [Google Scholar]
- Hung KE, Faca V, Song K, Sarracino DA, Richard LG, Krastins B, Forrester S, Porter A, Kunin A, Mahmood U, Haab BB, Hanash SM, Kucherlapati R. Comprehensive proteome analysis of an Apc mouse model uncovers proteins associated with intestinal tumorigenesis. Cancer Prev Res (Phila Pa) 2009;2:224–233. doi: 10.1158/1940-6207.CAPR-08-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsmore SF. Multiplexed protein measurement: technologies and applications of protein and antibody arrays. Nat Rev Drug Discov. 2006;5:310–320. doi: 10.1038/nrd2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Simeone D, Brenner D, Anderson MA, Shedden K, Ruffin MT, Lubman DM. Pancreatic Cancer Serum Detection Using a Lectin/Glyco-Antibody Array Method. Journal of proteome research. 2009;8:483–492. doi: 10.1021/pr8007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tao SC, Bova GS, Liu AY, Chan DW, Zhu H, Zhang H. Detection and verification of glycosylation patterns of glycoproteins from clinical specimens using lectin microarrays and lectin-based immunosorbent assays. Analytical chemistry. 2011;83:8509–8516. doi: 10.1021/ac201452f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Palma AS, Feizi T. Carbohydrate microarrays: key developments in glycobiology. Biol Chem. 2009;390:647–656. doi: 10.1515/BC.2009.071. [DOI] [PubMed] [Google Scholar]
- Manimala JC, Roach TA, Li Z, Gildersleeve JC. High-throughput carbohydrate microarray analysis of 24 lectins. Angewandte Chemie (International ed. 2006;45:3607–3610. doi: 10.1002/anie.200600591. [DOI] [PubMed] [Google Scholar]
- Manimala JC, Roach TA, Li Z, Gildersleeve JC. High-throughput carbohydrate microarray profiling of 27 antibodies demonstrates widespread specificity problems. Glycobiology. 2007;17:17C–23C. doi: 10.1093/glycob/cwm047. [DOI] [PubMed] [Google Scholar]
- Maupin KA, Liden D, Haab BB. The fine specificity of mannose-binding and galactose-binding lectins revealed using outlier-motif analysis of glycan array data. Glycobiology. 2011;22:160–169. doi: 10.1093/glycob/cwr128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura-Tsuruta S, Uchiyama N, Hirabayashi J. High-throughput analysis of lectin oligosaccharide interactions by automated frontal affinity chromatography. Methods in enzymology. 2006;415:311–325. doi: 10.1016/S0076-6879(06)15019-3. [DOI] [PubMed] [Google Scholar]
- Orchekowski R, Hamelinck D, Li L, Gliwa E, vanBrocklin M, Marrero JA, Vande Woude GF, Feng Z, Brand R, Haab BB. Antibody microarray profiling reveals individual and combined serum proteins associated with pancreatic cancer. Cancer research. 2005;65:11193–11202. doi: 10.1158/0008-5472.CAN-05-1436. [DOI] [PubMed] [Google Scholar]
- Perlee L, Christiansen J, Dondero R, Grimwade B, Lejnine S, Mullenix M, Shao W, Sorette M, Tchernev V, Patel D, Kingsmore S. Development and standardization of multiplexed antibody microarrays for use in quantitative proteomics. Proteome science. 2004;2:9. doi: 10.1186/1477-5956-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilobello KT, Slawek DE, Mahal LK. A ratiometric lectin microarray approach to analysis of the dynamic mammalian glycome. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:11534–11539. doi: 10.1073/pnas.0704954104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter A, Yue T, Heeringa L, Day S, Suh E, Haab BB. A motif-based analysis of glycan array data to determine the specificities of glycan-binding proteins. Glycobiology. 2010;20:369–380. doi: 10.1093/glycob/cwp187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer B, Wiltshire S, Lambert J, O'Malley S, Kukanskis K, Zhu Z, Kingsmore SF, Lizardi PM, Ward DC. Imunoassays with rolling circle DNA amplification: a versatile platform for ultrasensitive antigen detection. Proc. Natl. Acad. Sci. USA. 2000;97:10113–10119. doi: 10.1073/pnas.170237197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk JM, Gry M, Rimini R, Uhlen M, Nilsson P. Antibody suspension bead arrays within serum proteomics. Journal of proteome research. 2008;7:3168–3179. doi: 10.1021/pr700890b. [DOI] [PubMed] [Google Scholar]
- Stevens J, Blixt O, Glaser L, Taubenberger JK, Palese P, Paulson JC, Wilson IA. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. Journal of molecular biology. 2006;355:1143–1155. doi: 10.1016/j.jmb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Trostrup H, Lundquist R, Christensen LH, Jorgensen LN, Karlsmark T, Haab BB, Agren MS. S100A8/A9 deficiency in nonhealing venous leg ulcers uncovered by multiplexed antibody microarray profiling. The British journal of dermatology. 2011;165:292–301. doi: 10.1111/j.1365-2133.2011.10384.x. [DOI] [PubMed] [Google Scholar]
- Varki A, Cummings R, Esko J, Freeze H, Stanley P, Bertozzi CR, Hart G, Etzler ME. Essentials of Glycobiology. 2nd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2009. [PubMed] [Google Scholar]
- Yue T, Goldstein IJ, Hollingsworth MA, Kaul K, Brand RE, Haab BB. The prevalence and nature of glycan alterations on specific proteins in pancreatic cancer patients revealed using antibody-lectin sandwich arrays. Mol Cell Proteomics. 2009;8:1697–1707. doi: 10.1074/mcp.M900135-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue T, Maupin KA, Fallon B, Li L, Partyka K, Anderson MA, Brenner DE, Kaul K, Zeh H, Moser AJ, Simeone DM, Feng Z, Brand RE, Haab BB. Enhanced discrimination of malignant from benign pancreatic disease by measuring the CA 19-9 antigen on specific protein carriers. PLoS ONE. 2011;6:e29180. doi: 10.1371/journal.pone.0029180. [DOI] [PMC free article] [PubMed] [Google Scholar]