Abstract
MicroRNAs (miRNAs) represent a class of small non-coding RNA molecules that regulate gene expression at the post-transcriptional level. They play a crucial role in diverse cellular pathways. Ionizing radiation (IR) is one of the most important treatment protocols for patients that suffer from cancer and affects directly or indirectly cellular integration. Recently it has been discovered that microRNA-mediated gene regulation interferes with radio-related pathways in ionizing radiation. Here, we review the recent discoveries about miRNAs in cellular response to IR. Thoroughly understanding the mechanism of miRNAs in radiation response, it will be possible to design new strategies for improving radiotherapy efficiency and ultimately cancer treatment.
Key Words: Cellular response, ionizing radiation, microRNA
Ionizing radiation (IR) is widely used for the treatment of cancer. IR directly damage cellular components or generate reactive oxygen (ROS) and nitrogen (RNS) species that can disrupt atomic structure of macromolecules (1). DNA is the primary target for cell damage from IR (2). There are two major types of IR including photons (x-rays and γ-rays), which are most widely used in radiotherapy and particle radiation (electrons, protons, neutrons, carbon ions, α-particles, and β-particles). In X and γ-rays, most of the radiation effects are through free radicals whereas in particle radiation, direct damage of DNA is more important. In this review, the word "ionizing radiation" refers to all these radiations and their effects. A series of signaling pathways begin in cells following exposure to IR that may arrest the cell cycle and repair the damage or induce apoptosis if damage is extensive or in some cases enhance cell division and resistance to cytotoxic stresses (3).
MicroRNAs (miRNAs) are a newly discovered class of non-coding and endogenous RNA molecules that regulate gene expression at the post-transcriptional level (4). These single-strand RNA molecules are about 18-22 nucleotides long that incorporate and act in a nuclear protein complex named RNA induced silencing complex (RISC). In this complex miRNAs interact with 3′ untranslated region (3′ UTR) of target mRNAs at specific sites (5, 6). If complete complementation occurs between the miRNA and target mRNA, Argonaute 2 (Ago2) (an RNase enzyme), cleaves the mRNA, otherwise translation is repressed without degradation of target mRNA (7-9). So far, about 1500 miRNAs have been identified in human. A single miRNA can bind many mRNAs and multiple miRNAs can target a single mRNA. Therefore, there is a complex regulatory network between miRNAs and their target genes (10, 11).
IR triggers several biochemical and signaling events and alters expression of many genes involved in cell cycle regulation, checkpoints, apoptosis, and signal transduction pathways (12). Since miRNAs participate in control of an estimated >30% of all human genes, it is logical that miRNAs be also involved in the regulation of IR-induced gene expression. In this review, we illustrate the detailed regulatory mechanisms of miRNAs in modulating cellular response to ionizing radiation from diverse aspects.
IR-induced DNA damage response (DDR)
The main toxic biological effect of IR is DNA single (SSB) or double (DSB) strand break. Cells are very sensitive to DNA breaks especially at late G2, and during M phase (13, 14). Several genes upregulate after DNA break. The product of these genes could be categorized in three groups including sensors, transducers, and effectors. The final effect of all these processes is cell cycle arrest and DNA repair, or if the damage is extensive, apoptosis will occur (15). The first group of molecules that detect DNA breaks are sensors. The major known sensors are Mre11/Rad50/NBS1 (MRN)-complex and ataxia telangiectasia mutated (ATM). The main known transducers are Mouse double minute 2 homolog (MDM2), p53, Silent information regulator 1 (SIRT1), Checkpoint kinase 2 (Chk2), breast cancer type 1 susceptibility protein (BRCA1), and finally the major effectors are Bcl-2–associated X protein (BAX), cyclin-dependent kinase inhibitor 1 (p21), Cdk2/Cycline E, Rad51 and Rad50/Mre11 (-). Recently with discovery of miRNAs, a new layer of complexity has been added to IR signaling (19).
Role of miRNAs in IR-induced DDR
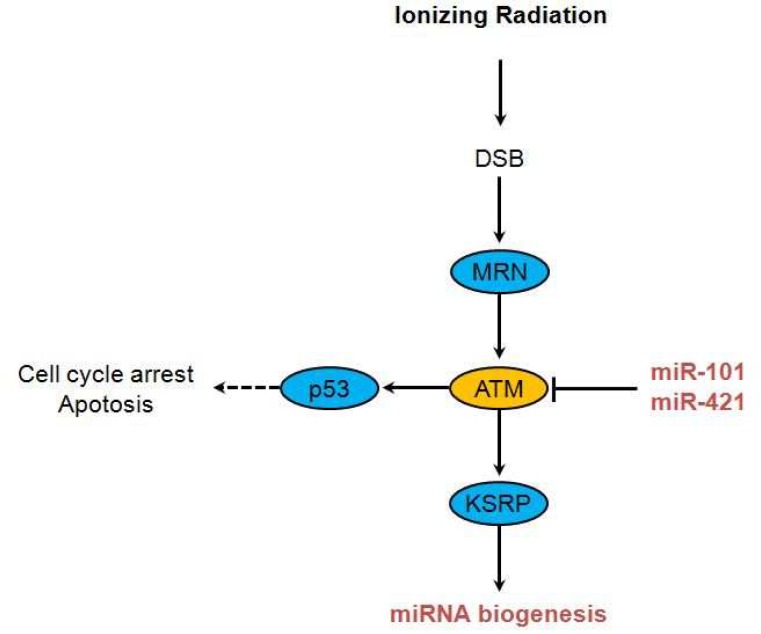
Following double-strand breaks by IR, the MRN complex binds to broken ends of DNA. MRN complex recruits ATM to broken DNA molecules and fully activates ATM (20). ATM, in its active form, phosphorylates several key proteins that initiate activation of the DNA damage checkpoint, leading to cell cycle arrest, DNA repair or apoptosis (21, 22). Hu et al. showed that miR-421, suppresses ATM expression by targeting the 3′ UTR of ATM transcripts. They also observed that ectopic expression of miR-421 results in S-phase cell cycle checkpoint changes and an increased sensitivity to ionizing radiation, creating a cellular phenotype similar to that of cells derived from ataxia-telangiectasia (A-T) patients (23). MiR-101 can efficiently target ATM via binding to the 3′ UTR of ATM mRNA and upregulated miR-101 decreases the protein levels of ATM. Upregulating miR-101, sensitizes the tumor cells to radiation in vitro and in vivo. MiRNA biogenesis is globally induced upon DNA damage in an ATM-dependent manner (24). About one fourth of miRNAs are significantly upregulated after DNA damage, while loss of ATM abolishes their induction. ATM phosphorylates KH-type splicing regulatory protein (KSRP), a key player of miRNA biogenesis, and increases its activity and thereby miRNA processing (Fig. 1) (25).
Fig 1.
ATM associated miRNAs. After ionizing radiation and DSB, MRN complex binds to broken ends of DNA, then recruits and activates ATM. MiR-101 and miR-421 are suppressors of ATM expression. This information is resulted from references (21-26)
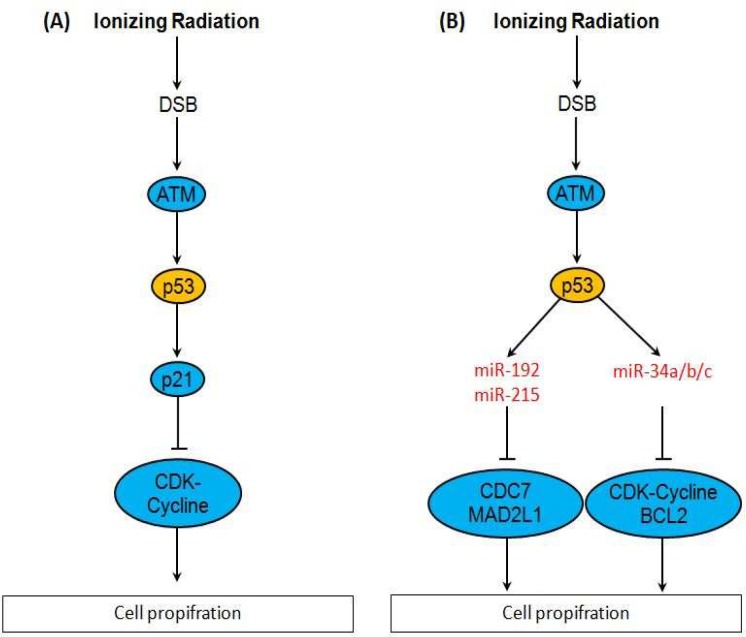
Active ATM in its monomeric form has about 700 substrates that one of them is p53. ATM phosphorylates p53 that leads to its dissociation from MDM-2, an inhibitor of p53, and activation of p53 (27). p53 in its active form phosporylates and activates p21. Activated p21 binds to and inactivates CDK-Cyclin complexes and thereby arrests cell cycle in G1→S, S phase and G2→M. p53 induces apoptosis through transcriptional activation of several pro-apoptotic factors (28). However, there are newly discovered pathways mediated by some miRNAs. Activated p53 induces the expression of miR-34a/b/c (-). miR-34a/b/c downregulate CDK4 and CDK6 to induce cell cycle arrest and downregulate BCL2 to promote apoptosis (34). p53 also enhances expression of miR-192 and miR-215. These miRNAs, downregulate transcription of CDC7, MAD2L1 and CUL5, that are involved in cell proliferation (Fig. 2) (35, 36).
Fig 2.
DSB activates p21 that inhibits cell proliferation by inactivating CDK-Cycline complexes (A). p53 induces miR-34a/b/c, miR-192 and miR-215 transcription. These miRNAs inhibit cell proliferation by downregulating CDC2, MAD2L1, CDK-Cycline and BCL2 (B) (27-36)
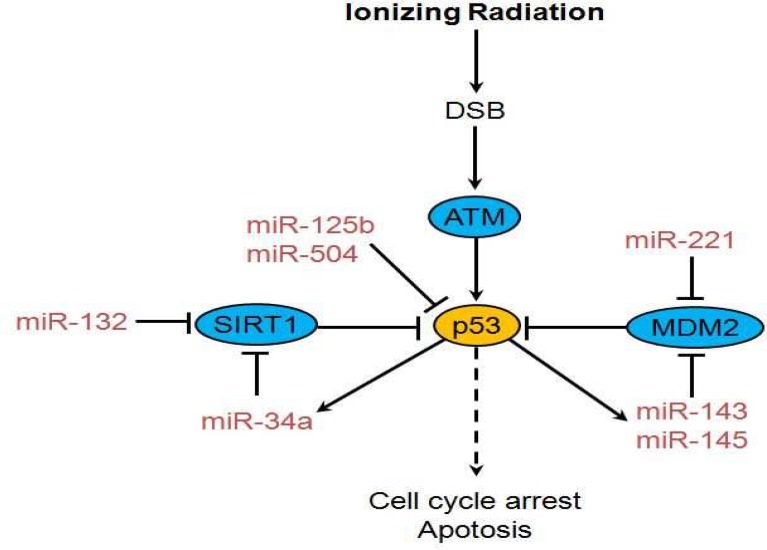
Several miRNAs directly or indirectly effect p53 expression. Hu et al. reported that miR-504 acts as a negative regulator of human p53 through its direct binding to two sites in p53 3′ UTR. They reported that overexpression of miR-504 decreases p53 protein levels and functions in cells, including p53 transcriptional activity, p53-mediated apoptosis and cell cycle arrest in response to stress (37). Le et al. demonstrated that miR-125b, is a negative regulator of p53 in both zebrafish and humans. They found that miR-125b-mediated downregulation ability of p53, is strictly dependent on the binding of miR-125b to a microRNA response element in the 3′ UTR of p53 mRNA. They also observed that knockdown of miR-125b elevates the level of p53 protein and induces apoptosis in human lung fibroblasts and in the zebrafish brain. Interestingly, miR-125b was downregulated when zebrafish embryos are treated with γ-irradiation, corresponding to the rapid increase in p53 protein in response to DNA damage. Ectopic expression of miR-125b suppressed the increase of p53 and stress-induced apoptosis (38).
MDM2 is a direct inhibitor of p53. There is an inverse correlation between miR-221 and Mdm2 protein levels. MiR-221 directly regulates Mdm2 expression and increases p53 protein levels and stability (39). Both miR-143 and miR-145, which belong to the same miRNA cluster, target the miRNA regulatory elements (MREs) in the 3′ UTR of MDM2. These molecules are post-transcriptionally activated by p53, thereby generating a short miRNAs-MDM2-p53 feedback loop (Fig 3) (40). Silent information regulator 1 (SIRT1) is a negative regulator of p53. MiR-132 was shown to inhibit SIRT1 expression through a miR-132 binding site in the 3′ UTR of SIRT1 (41). Yamakuchi et al. showed that MiR-34a inhibits SIRT1 expression through a miR-34a-binding site within the 3′ UTR of SIRT1 (42).
Fig 3.
A simplified working model for p53-associated miRNAs. See text for more information (37-42)
MiRNAs and radiation induced signal trans-duction pathways
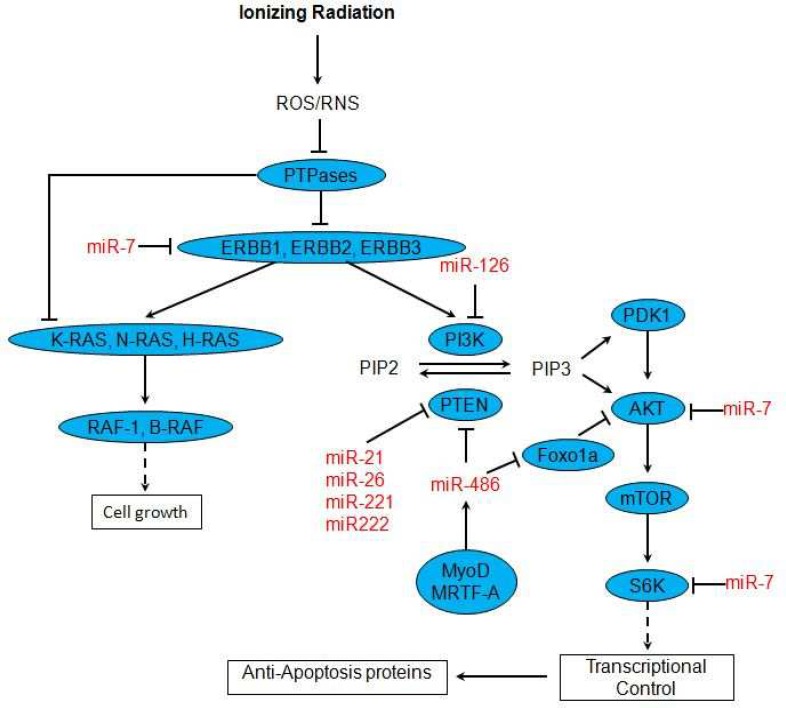
IR not only causes cell death, but also has potential of enhancing proliferation in the surviving fraction of cells (43, 44). IR damages DNA and generates ROS that the later products in addition to activation of p53 and ATM, are able to activate growth factor receptors in the plasma membrane (44). Protein tyrosine phosphatases (PTPases) are key regulatory components in signal transduction pathways that regulate the phosphorylation state of many important signaling molecules. PTPases dephosphorylate and activate several intermediates of signal transduction pathways. ROS is an inhibitor of PTPases. Therefore, radiation decreases PTPases activities by generating ROS and consequently increased phosphoryation and activity of several proteins especially receptor tyrosin kinases will occur (45, 46).
Recent evidences suggested that microRNA-mediated gene regulation interconnects with the radio-related signal transduction pathways. Webster et al. in a study on several cancer cell lines (lung, breast, and glioblastoma) found that miR-7 downregulates EGFR mRNA and protein expression via two sites on 3' UTR of EGFR mRNA, inducing cell cycle arrest and cell death (47). Furthermore in another study, researchers showed that ectopic over expression of miR-7, attenuated EGFR and Protein Kinase B (Akt) expression and radio-sensitized MDA-MB-468 breast cancer cells (48). p85β is a regulatory subunit involved in stabilizing and propagating the PI3K signal. p85β 3' UTR is directly targeted by miR-126 (49). Phosphoinositide 3-kinase catalytic subunit delta (PIK3CD) and p70S6K are also miR-7 targets (50).
The seed sequence of miR-221 and miR-222 matched the 3' UTR of Phosphatase and tensin homolog (PTEN) mRNA, and PTEN-3' UTR luciferase reporter assay confirmed PTEN as a direct target of miR-221 and miR-222 (51). MiR-26a and miR-21 are direct regulators of PTEN expression (52, 53). Transcription of miR-486 is directly controlled by Serum Response Factor (SRF), Myocardin-related transcription factor A (MRTF-A) and MyoD. PTEN and Forkhead box protein O1A (Foxo1a) are targets of miR-486, which negatively affect phosphoinositide-3-kinase (PI3K)/Akt signaling. MRTF-A promotes PI3K/Akt signaling by upregulating miR-486 expression, (Fig4) (54).
Fig 4.
A simplified working model for role of miRNAs in radiation induced signal transduction pathways. See text for more explanation (43-54).
Conclusion
Although it is not long time from the discovery of miRNAs, it have been cleared that these molecules have some roles in several cellular processes such as stress response, apoptosis, cellular differentiation and proliferation, cellular response to ionizing radiation and also these molecules have aberrant expression in a variety of diseases (55, 56). The main effect of IR on cells is DNA damage that results to cell cycle arrest or cell death especially in dividing cells. IR can also induce proliferation in the surviving fraction of cells. It has been discovered that miRNAs have fundamental role in several IR induced cell signaling events. In this review some miRNAs involved in cellular response to ionizing radiation have been discussed briefly; However, there are still many questions to be addressed. With thorough knowledge about the role of miRNAs in radiation response, it will be possible to design new strategies for improving radiotherapy efficiency and ultimately cancer treatment.
Conflict of interest: None declared.
References
- 1.Azzam EI, Jay-Gerin JP, Pain D. Ionizing radiation-induced metabolic oxidative stress and prolonged cell injury. Cancer Lett. 2012;327:48–60. doi: 10.1016/j.canlet.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warters RL, Hofer KG. Radionuclide toxicity in cultured mammalian cells. Elucidation of the primary site for radiation-induced division delay. Radiat Res. 1977;69:348–58. [PubMed] [Google Scholar]
- 3.Spitz DR, Azzam EI, Li JJ, et al. Metabolic oxidation/reduction reactions and cellular responses to ionizing radiation: a unifying concept in stress response biology. Cancer Metastasis Rev. 2004;23:311–22. doi: 10.1023/B:CANC.0000031769.14728.bc. [DOI] [PubMed] [Google Scholar]
- 4.Wiemer EAC. The role of microRNAs in cancer: No small matter. Eur J Cancer. 2007;43:1529–44. doi: 10.1016/j.ejca.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Rigoutsos I. New Tricks for Animal MicroRNAs: Targeting of Amino Acid Coding Regions at Conserved and Nonconserved Sites. Cancer Res. 2009;69:3245–8. doi: 10.1158/0008-5472.CAN-09-0352. [DOI] [PubMed] [Google Scholar]
- 6.Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Bio. 2009;10:141–8. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 7.He L, Hannon GJ. Micrornas: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 8.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–11. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brennecke J, Stark A, Russell RB, et al. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomson DW, Bracken CP, Goodall GJ. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011;39:6845–53. doi: 10.1093/nar/gkr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lhakhang TW, Chaudhry MA. Interactome of Radiation-Induced microRNA-Predicted Target Genes. Comp Funct Genomics. 2012 doi: 10.1155/2012/569731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li L, Story M, Legerski RJ. Cellular responses to ionizing radiation damage. Int J Radiat Oncol Biol Phys. 2001;49:1157–62. doi: 10.1016/s0360-3016(00)01524-8. [DOI] [PubMed] [Google Scholar]
- 13.Quiet CA, Weichselbaum RR, Grdina DJ. Variation in radiation sensitivity during the cell cycle of two human squamous cell carcinomas. Int J Radiat Oncol Biol Phys. 1991;20:733–8. doi: 10.1016/0360-3016(91)90016-w. [DOI] [PubMed] [Google Scholar]
- 14.Tell R, Heiden T, Granath F, et al. Comparison between radiation-induced cell cycle delay in lymphocytes and radiotherapy response in head and neck cancer. Brit J Cancer. 1998;77:643–9. doi: 10.1038/bjc.1998.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson SP. Sensing and repairing DNA double-strand breaks - Commentary. Carcinogenesis. 2002;23:687–96. doi: 10.1093/carcin/23.5.687. [DOI] [PubMed] [Google Scholar]
- 16.Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Gene Dev. 2011;25:409–33. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wouters MD, van Gent DC, Hoeijmakers JHJ, et al. MicroRNAs, the DNA damage response and cancer. Mutat Res-Fund Mol M. 2011;717:54–66. doi: 10.1016/j.mrfmmm.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 18.Hu HL, Gatti RA. MicroRNAs: new players in the DNA damage response. J Mol Cell Biol. 2011;3:151–8. doi: 10.1093/jmcb/mjq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joly-Tonetti N, Lamartine J. The Role of MicroRNAs in the Cellular Response to Ionizing Radiations. In: Nenoi M, editor. Current Topics in Ionizing Radiation Research. Shanghai: InTech InC; 2012. pp. 149–74. [Google Scholar]
- 20.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–4. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 21.Zhou JQ, Lim CUK, Li JJ, et al. The role of NBS1 in the modulation of PIKK family proteins ATM and ATR in the cellular response to DNA damage. Cancer Letters. 2006;243:9–15. doi: 10.1016/j.canlet.2006.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khoronenkova SV, Dianov GL. Regulation of USP7/HAUSP in response to DNA damage Yet another role for ATM. Cell Cycle. 2012;11:2409–10. doi: 10.4161/cc.20800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu HL, Du LT, Nagabayashi G, et al. ATM is down-regulated by N-Myc-regulated microRNA-421. P Natl Acad Sci. 2010;107:1506–11. doi: 10.1073/pnas.0907763107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan D, Ng WL, Zhang XM, et al. Targeting DNA-PKcs and ATM with miR-101 Sensitizes Tumors to Radiation. Plos One. 2010;5:e11397. doi: 10.1371/journal.pone.0011397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang XN, Wan GH, Berger FG, et al. The ATM Kinase Induces MicroRNA Biogenesis in the DNA Damage Response. Mol Cell. 2011;41:371–83. doi: 10.1016/j.molcel.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JH, Paull TT. Activating ATM with Broken DNA. Sci. STKE. 2005:tw161. [Google Scholar]
- 27.Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–8. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- 28.Levine AJ, Finlay CA, Hinds PW. P53 is a tumor suppressor gene. Cell. 2004;116:S67–9. doi: 10.1016/s0092-8674(04)00036-4. 1 p following S9. [DOI] [PubMed] [Google Scholar]
- 29.Rokhlin OW, Scheinker VS, Taghiyev AF, et al. MicroRNA-34 mediates AR-dependent p53-induced apoptosis in prostate cancer. Cancer Biol Ther. 2008;7:1288–96. doi: 10.4161/cbt.7.8.6284. [DOI] [PubMed] [Google Scholar]
- 30.Corney DC, Flesken-Nikitin A, Godwin AK, et al. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 31.Tarasov V, Jung P, Verdoodt B, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–93. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 32.Raver-Shapira N, Marciano E, Meiri E, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–43. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 33.Chang TC, Wentzel EA, Kent OA, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–52. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cole KA, Attiyeh EF, Mosse YP, et al. A functional screen identifies miR-34a as a candidate neuroblastoma tumor suppressor gene. Mol Cancer Res. 2008;6:735–42. doi: 10.1158/1541-7786.MCR-07-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song B, Wang Y, Kudo K, et al. miR-192 Regulates Dihydrofolate Reductase and Cellular Proliferation through the p53-microRNA Circuit. Clin Cancer Res. 2008;14:8080–6. doi: 10.1158/1078-0432.CCR-08-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Georges SA, Biery MC, Kim SY, et al. Coordinated Regulation of Cell Cycle Transcripts by p53-inducible microRNAs, miR-192 and miR-215. Cancer Res. 2008;68:10105–12. doi: 10.1158/0008-5472.CAN-08-1846. [DOI] [PubMed] [Google Scholar]
- 37.Hu WW, Chan CS, Wu R, et al. Negative Regulation of Tumor Suppressor p53 by MicroRNA miR-504. Mol Cell. 2010;38:689–99. doi: 10.1016/j.molcel.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le MTN, Teh C, Shyh-Chang N, et al. MicroRNA-125b is a novel negative regulator of p53. Gene Dev. 2009;23:862–76. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fornari F, Milazzo M, Giovannini C, et al. Mir-221/Mdm2/P53 Control Loop Sensitizes Hepatocellular Carcinoma Cells to Anti-Cancer Treatments. J Hepatol. 2012;56:S113–S. [Google Scholar]
- 40.Zhang J, Sun Q, Zhang Z, et al. Loss of microRNA-143/145 disturbs cellular growth and apoptosis of human epithelial cancers by impairing the MDM2-p53 feedback loop. Oncogene. 2013;32:61–9. doi: 10.1038/onc.2012.28. [DOI] [PubMed] [Google Scholar]
- 41.Strum JC, Johnson JH, Ward J, et al. MicroRNA 132 Regulates Nutritional Stress-Induced Chemokine Production through Repression of SirT1. Mol Endocrinol. 2009;23:1876–84. doi: 10.1210/me.2009-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci. 2008;105:13421–6. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bentzen SM, Atasoy BM, Daley FM, et al. Epidermal growth factor receptor expression in pretreatment biopsies from head and neck squamous cell carcinoma as a predictive factor for a benefit from accelerated radiation therapy in a randomized controlled trial. J Clin Oncol. 2005;23:5560–7. doi: 10.1200/JCO.2005.06.411. [DOI] [PubMed] [Google Scholar]
- 44.Valerie K, Yacoub A, Hagan MP, et al. Radiation-induced cell signaling: inside-out and outside-in. Mol Cancer Ther. 2007;6:789–801. doi: 10.1158/1535-7163.MCT-06-0596. [DOI] [PubMed] [Google Scholar]
- 45.Stoker AW. Protein tyrosine phosphatases and signalling. J Endocrinol. 2005;185:19–33. doi: 10.1677/joe.1.06069. [DOI] [PubMed] [Google Scholar]
- 46.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–46. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 47.Webster RJ, Giles KM, Price KJ, et al. Regulation of Epidermal Growth Factor Receptor Signaling in Human Cancer Cells by MicroRNA-7. J Biol Chem. 2009;284:5731–41. doi: 10.1074/jbc.M804280200. [DOI] [PubMed] [Google Scholar]
- 48.Lee KM, Choi EJ, Kim IA. microRNA-7 increases radiosensitivity of human cancer cells with activated EGFR-associated signaling. Radiother Oncol. 2011;101:171–6. doi: 10.1016/j.radonc.2011.05.050. [DOI] [PubMed] [Google Scholar]
- 49.Guo C, Sah JF, Beard L, et al. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer. 2008;47:939–46. doi: 10.1002/gcc.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fang Y, Xue JL, Shen Q, et al. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology. 2012;55:1852–62. doi: 10.1002/hep.25576. [DOI] [PubMed] [Google Scholar]
- 51.Chun-Zhi Z, Lei H, An-Ling Z, et al. MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer. 2010;10 doi: 10.1186/1471-2407-10-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huse JT, Brennan C, Hambardzumyan D, et al. The PTEN-regulating microRNA miR-26a is amplified in high-gradeglioma and facilitates gliomagenesis in vivo. Gene Dev. 2009;23:1327–37. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang JG, Wang JJ, Zhao F, et al. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC) Clin Chim Acta. 2010;411:846–52. doi: 10.1016/j.cca.2010.02.074. [DOI] [PubMed] [Google Scholar]
- 54.Small EM, O'Rourke JR, Moresi V, et al. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. P Natl Acad Sci. 2010;107:4218–23. doi: 10.1073/pnas.1000300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 56.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–24. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]