Abstract
Cholera is an infection of the small intestines caused by the bacterium V. cholerae. It is a major cause of health threat and also a major cause of death worldwide and especially in developing countries. The major virulence factor produced by V. cholerae during infection is the cholera toxin. Total mRNA extraction and reverse transcription was performed for making ctxAB cDNA. Relative Real-Time PCR analysis showed unequal enterotoxin production in V. cholerae strains. The results showed that, classical strain produces cholera toxin more than El Tor strain.
Key Words: Vibrio cholerae, RT-qPCR, ctxAB expression
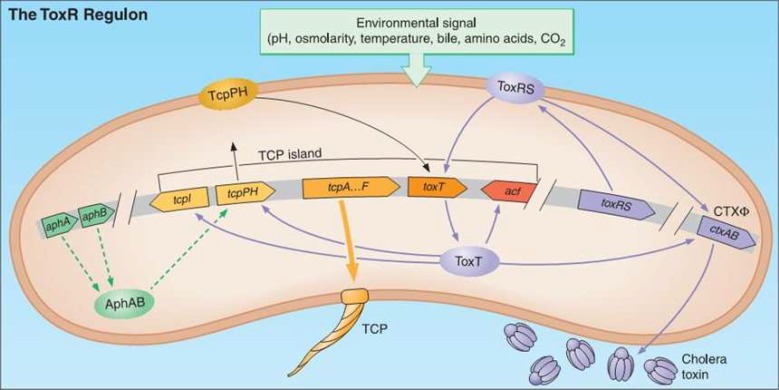
Cholera is one of the infectious diseases that still happens in developing countries. The 8th pandemic of cholera spreads from Southeast Asia across the Middle East and into Central America and Africa (1, 2). The important pathogenesis factor in Vibrio cholerae is a potent enterotoxin, cholera toxin, which causes the severe diarrhea of cholera (3, 4). The cholera toxin is produced by V. cholerae and CTXΦ phage corporation. Control of enterotoxin gene expression seems to be complex, so that environmental factors are very important in its expression (5, 6). The environmental signals affect TcpPH gene and cause its activation and finally affect ToxT gene. The ToxT protein is the most important agent for ctxAB toxin expression, because ToxT protein attaches to toxbox region at upstream of ctxAB gene and induces ctxAB toxin expression (7, 8) (Fig 1). Beside, other signaling systems such as ToxR, RS1, AphAB and quorum sensing have positive or negative effects on ToxT protein (9, 10). Moreover, H-NS protein has negative effect on TcpPH and ToxT genes that finally would decrease the ctxAB toxin production (11). The RS1 region contains rstA, rstB and rstC fragments which have positive effect on ToxR gene and therefore increase ctxAB toxin production (12). Bakhshi et al. reported several V. cholerae attacks in Southwest and Southeast of Iran between 2005-2009 (13, 14). As the level of production of a protein is somehow related to its mRNA quantity, we therefore aimed to determine V. cholerae strains that can produce more ctxAB toxin.
Fig 1.
Diagram of the vibrio cholerae ToxR regulon and ctxAB expression, with permission from ASM
Material and Method
Bacterial strains and growth conditions
We used two standard strains named V.cholerae O1 Classic ATCC 14035 & V. cholerae O1 El Tor N16961. The isolates were confirmed by biochemical and immunological tests. Serotyping was performed using monoclonal O1 antiserum and mono-specific Inaba and Ogawa antisera (Pasteur institute, Paris, France). All selected strains were cultured according to the AKI-SW method and standard growth curve were drawn (15).
Isolation of RNA and RT-PCR
Approximately, 2×108 cfu/ml from each strain, was used for total RNA extraction. Total RNA was isolated from the strains isolated randomly from each V. cholerae grown in AKI medium using the RNeasy® Protect Bacteria Mini Kit (Qiagen Inc, GMBH, Germany) and the integrity and purity was checked. Equivalent concentrations of total RNA from each strain were selected as template for RT-PCR. cDNA synthesis and PCR amplification were performed using QuantiTec Reverse Transcription Qiagen kit (Qiagen Inc, GMBH, Germany). RT-PCR was performed in the presence of random primer at 42°C for 10 min. After cDNA synthesis, the ctxAB and recA genes were PCR amplified for checking. PCR amplification was performed for 35 cycles as follows: initial denaturation at 94°C for 5 min, then denaturation at 94°C for 30 sec, annealing at 60°C for 30 sec, extension at 72°C for 30 sec. At the end of the 35th cycle, reaction mixtures were left at 72°C for another 3 min. Five microliters of each reaction mixture was loaded on a 1% agarose gel and subjected to electrophoresis to confirm that the unique amplified fragment correspond to the expected ctxAB gene fragment and recA as housekeeping gene (16).
Real-Time PCR
Prepared cDNA was quantified using SYBR green I dye. Four primers were designed by AlleleID 6 software, 5’-CAGTCAGGTGGTCT-TATGC-3’ (ctxAB-F) and 5’-ATCGTGCCTAAC-AAATCCC-3’ (ctxAB-R) for gene of interest and 5’ –ATTGAAGGCGAAATGGGCGATAG- 3’ (recA-F) and 5’ –TACACATACAGTTGGATTGCTTGAGG- 3’ (recA-R) for housekeeping gene. Those primers were specific to ctxAB and recA and amplified a 115 & 106 bp respectively. SYBR green Real-Time PCR assay was performed with a 20 µl PCR mixture volume containing 2x QuantiTec SYBR Green PCR Master Mix (Qiagen Inc, GMBH, Germany), 0.25 µM specific primer sets, and 2 µl of cDNA sample. Amplification of the primers, data acquisition, and relative analysis were carried out in Chromo4 BioRad Real-Time PCR. PCR reactions were performed as followings: one cycle of 95 °C for 5 min, then 40 cycles of 95 °C for 15 sec, 60 °C for 30 sec. Following the amplification, melting temperature analysis of the PCR products was performed to determine the specificity of the PCR. The standard curve was established by using genomic DNA for each studied gene to confirm that the primers amplified at the same rate and to validate the experiment (55-95°C with warming of 0.2°C per sec). Reverse transcription and PCR positive controls (RNA and DNA, respectively) and negative controls (distilled water) were included in each run. The Real-Time PCR reaction was performed twice assayed in triplicate. Classical V. cholerae O1 ATCC 14035 was used as a standard control.
Results
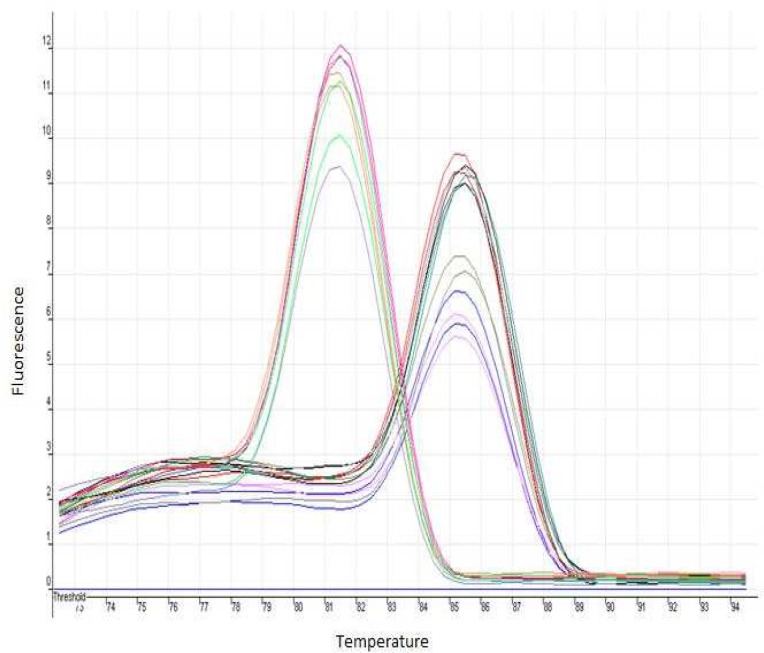
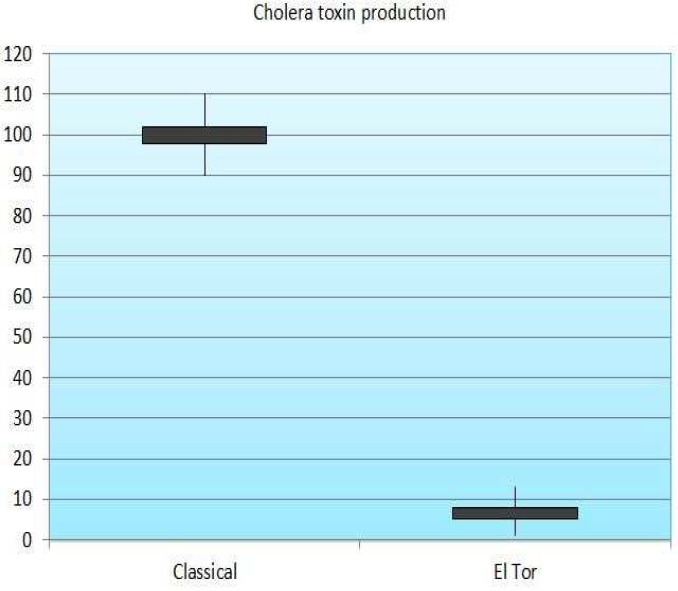
The specificity of each primer set for V. cholerae was tested by PCR with genomic DNA extracted via boiling. Only one size of amplicon was obtained by PCR reaction for ctxAB and recA genes when DNA from V. cholerae strains was used. The amplicons obtained for each gene were verified by sequencing. The presence of a single PCR product was confirmed by Real-Time PCR for each set of the primers using melting curve analysis that resulted in a single product-specific melting curve (Fig 2). The PCR efficiencies varied between 1.90 and 1.94. The relative expression ratio was calculated for each gene of interest by a mathematical model described by ΔCT method. The Cycle Threshold (CT ) results are showed in table 1. Histogram and samples CT values are indicated in Fig 3.
Fig 2.
Melting curves of ctxAB and recA genes
Table 1.
Cycle threshold (Ct) results for the V.cholerae O1 Classic ATCC 14035 & V. cholerae O1 El Tor 62013
| Strain | Mean Ct recA±SD | Mean Ct ctxAB±SD | Mean ΔΔCt | Mean Ratio* |
|---|---|---|---|---|
| Classic | 25.58±0.14 | 24.35±0.39 | 1.23 | 1.0 |
| El Tor | 22.85±0.13 | 26.02±0.15 | 4.35 | 0.06 |
*ΔΔCt was calculated as: ΔCt (test) - ΔCt (calibrator).Ratio=efficiency -ΔΔCt.
Fig 3.
Level of cholera toxin production in classical and EL Tor strains
Discussion
In our study, the results are derived by using “relative” method and ΔCT formula. By considering that ctxAB primers have been carefully designed, the amount of standard deviation results are close to zero, the primer-dimer bands were not seen, because the concentration of participating primers in the reaction had been set up. Also, for more accuracy and sensitivity, the PCR efficiency in Real-Time PCR reactions were calculated and replaced with ratio 2 in computational equations.
Dirita et al. showed that the level of cholera toxin was higher in the classical strain compared to El Tor strain because of the influence of toxR on cholera toxin production in the classical strain (17). In our study, in addition to Dirita et al. results, we determined quantitative cholera toxin production between classic and El Tor strains. Both classical and El Tor strains have been shown to express equivalent levels of ToxR. In contrast, the classical strain expresses more ToxT, which has a higher binding affinity to toxbox region, resulting in higher expression of cholera toxin (18).
The comparison of the CT of the El Tor strain with classical strain shows that toxin production in El Tor strain is approximately 16-17 times lower than in the classical strain (P Value<0.05 for each). This result is consistent with other reports because the amount of pathogenicity in classic strain is more than El Tor strains (18). Furthermore, histogram and samples CT results indicated that toxin production in classical strain is higher than El Tor strain (Fig 3). In conclusion, the results of our study suggest that other factors modulate the production of cholera toxin by regulating the CTX cassette, supporting the idea that cholera toxin production in V. cholerae classical and El Tor strains is a multi-factorial phenomenon.
Acknowledgment
We are grateful to Dr. Bita Bakhshi for shipment of the V. cholerae O1 classic ATCC 14035 and V. cholerae O1 El Tor ATCC N16961 strains.
References
- 1.Amin Marashi SM, Nasr Esfahani B, Tavakoli A, et al. Simultaneous detection of integrase and antibiotic resistance genes within SXT Constin in Vibrio cholerae O1 El Tor strains isolated from Iran using multiplex-PCR. Iran J Basic Med Sci. 2012;15:885–9. [PMC free article] [PubMed] [Google Scholar]
- 2.Taneja N, Mishra A, Sangar G, et al. Outbreaks caused by new variants of Vibrio cholerae O1 El Tor, India. Emerg Infect Dis. 2009;15:352–4. doi: 10.3201/eid1502.080943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baptista MA, Andrade JR, Vicente AC, et al. The Amazonia variant of Vibrio cholerae: molecular identification and study of virulence genes. Mem Inst Oswaldo Cruz. 1998;93:601–7. doi: 10.1590/s0074-02761998000500008. [DOI] [PubMed] [Google Scholar]
- 4.Bravo L, Monte RJ, Ramirez M, et al. Detection of toxigenic Vibrio cholerae 01 using polymerase chain reaction. Mem Inst Oswaldo Cruz. 1992;87:443–4. doi: 10.1590/s0074-02761992000300018. [DOI] [PubMed] [Google Scholar]
- 5.Bakhshi B, Pourshafie MR, Navabakbar F, et al. Genomic organisation of the CTX element among toxigenic Vibrio cholerae isolates. Clin Microbiol Infect. 2008;14:562–8. doi: 10.1111/j.1469-0691.2008.01976.x. [DOI] [PubMed] [Google Scholar]
- 6.Sa LL, Fonseca EL, Pellegrini M, et al. Occurrence and composition of class 1 and class 2 integrons in clinical and environmental O1 and non-O1/non-O139 Vibrio cholerae strains from the Brazilian Amazon. Mem Inst Oswaldo Cruz. 105:229–32. doi: 10.1590/s0074-02762010000200021. [DOI] [PubMed] [Google Scholar]
- 7.Withey JH, Dirita VJ. Vibrio cholerae ToxT independently activates the divergently transcribed aldA and tagA genes. J Bacteriol. 2005;187:7890–900. doi: 10.1128/JB.187.23.7890-7900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Withey JH, DiRita VJ. The toxbox: specific DNA sequence requirements for activation of Vibrio cholerae virulence genes by ToxT. Mol Microbiol. 2006;59:1779–89. doi: 10.1111/j.1365-2958.2006.05053.x. [DOI] [PubMed] [Google Scholar]
- 9.Klose KE. Regulation of virulence in Vibrio cholerae. Int J Med Microbiol. 2001;291:81–8. doi: 10.1078/1438-4221-00104. [DOI] [PubMed] [Google Scholar]
- 10.Zhu J, Miller MB, Vance RE, et al. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci. 2002;99:3129–34. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matson JS, Withey JH, DiRita VJ. Regulatory networks controlling Vibrio cholerae virulence gene expression. Infect Immun. 2007;75:5542–9. doi: 10.1128/IAI.01094-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis BM, Waldor MK. Filamentous phages linked to virulence of Vibrio cholerae. Curr Opin Microbiol. 2003;6:35–42. doi: 10.1016/s1369-5274(02)00005-x. [DOI] [PubMed] [Google Scholar]
- 13.Bakhshi B, Pourshafie MR. Assessing clonality of Vibrio cholerae strains isolated during four consecutive years (2004 - 2007) in Iran. Scand J Infect Dis. 2009;41:256–62. doi: 10.1080/00365540902767049. [DOI] [PubMed] [Google Scholar]
- 14.Pourshafie MR, Bakhshi B, Ranjbar R, et al. Dissemination of a single Vibrio cholerae clone in cholera outbreaks during 2005 in Iran. J Med Microbiol. 2007;56:1615–9. doi: 10.1099/jmm.0.47218-0. [DOI] [PubMed] [Google Scholar]
- 15.Iwanaga M, Yamamoto K, Higa N, et al. Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol Immunol. 1986;30:1075–83. doi: 10.1111/j.1348-0421.1986.tb03037.x. [DOI] [PubMed] [Google Scholar]
- 16.Amin Marashi M, Bakhshi B, Imani Fooladi AA, et al. Quantitative expression of cholera toxin mRNA in Vibrio cholerae isolates with different CTX cassette arrangements. doi: 10.1099/jmm.0.038752-0. [DOI] [PubMed] [Google Scholar]
- 17.DiRita VJ, Neely M, Taylor RK, et al. Differential expression of the ToxR regulon in classical and E1 Tor biotypes of Vibrio cholerae is due to biotype-specific control over toxT expression. Proc Natl Acad Sci. 1996;93:7991–5. doi: 10.1073/pnas.93.15.7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez-Bonilla C, Gutierrez-Cogco L, Moguel-Pech L, et al. [Evaluation of the ELISA method for cholera toxin determination in Vibrio cholerae cultures] Rev Latinoam Microbiol. 1994;36:273–6. [PubMed] [Google Scholar]