Abstract
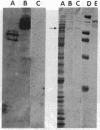
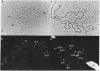
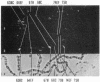
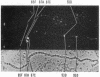
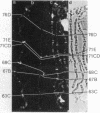
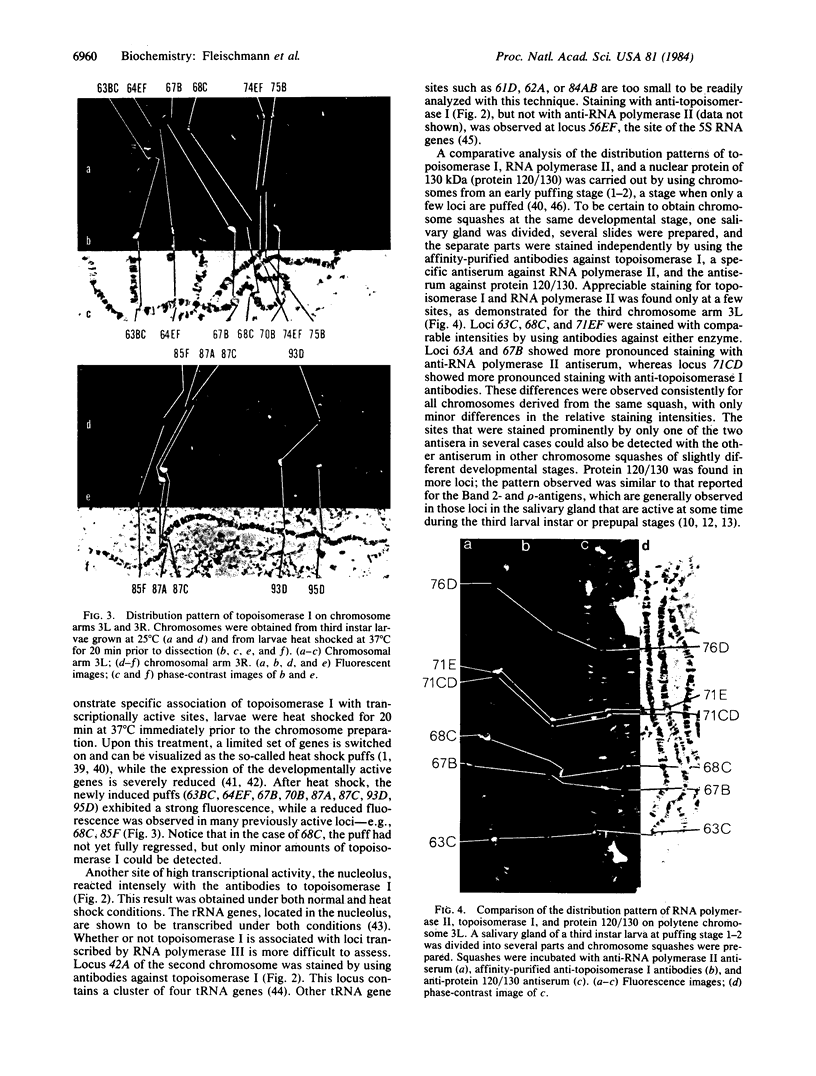
The distribution of DNA topoisomerase I within Drosophila polytene chromosomes was observed by immunofluorescent staining with affinity-purified antibodies. The enzyme is preferentially associated with active loci, as shown by prominent staining of puffs. The heat shock loci 87A-87C are stained after, but not before, heat shock induction. A detailed comparison of the distribution of topoisomerase I with that of RNA polymerase II reveals a similar, although not identical, pattern of association. Topoisomerase I is also found in association with the nucleolus, the site of transcription by RNA polymerase I.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alfageme C. R., Rudkin G. T., Cohen L. H. Locations of chromosomal proteins in polytene chromosomes. Proc Natl Acad Sci U S A. 1976 Jun;73(6):2038–2042. doi: 10.1073/pnas.73.6.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BECKER H. J. [The puffs of salivary gland chromosomes of Drosophilia melanogaster. Part 1. Observations on the behavior of a typical puff in the normal strain and in two mutants, giant and lethal giant larvae]. Chromosoma. 1959;10:654–678. doi: 10.1007/BF00396591. [DOI] [PubMed] [Google Scholar]
- Bloom K. S., Anderson J. N. Conformation of ovalbumin and globin genes in chromatin during differential gene expression. J Biol Chem. 1979 Oct 25;254(20):10532–10539. [PubMed] [Google Scholar]
- Bloom K. S., Anderson J. N. Fractionation of hen oviduct chromatin into transcriptionally active and inactive regions after selective micrococcal nuclease digestion. Cell. 1978 Sep;15(1):141–150. doi: 10.1016/0092-8674(78)90090-9. [DOI] [PubMed] [Google Scholar]
- Bonner J. J., Pardue M. L. The effect of heat shock on RNA synthesis in Drosophila tissues. Cell. 1976 May;8(1):43–50. doi: 10.1016/0092-8674(76)90183-5. [DOI] [PubMed] [Google Scholar]
- Champoux J. J. Proteins that affect DNA conformation. Annu Rev Biochem. 1978;47:449–479. doi: 10.1146/annurev.bi.47.070178.002313. [DOI] [PubMed] [Google Scholar]
- Christensen M. E., LeStourgeon W. M., Jamrich M., Howard G. C., Serunian L. A., Silver L. M., Elgin S. C. Distribution studies on polytene chromosomes using antibodies directed against hnRNP. J Cell Biol. 1981 Jul;90(1):18–24. doi: 10.1083/jcb.90.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen L. H., Gotchel B. V. Histones of polytene and nonpolytene nuclei of Drosophila melanogaster. J Biol Chem. 1971 Mar 25;246(6):1841–1848. [PubMed] [Google Scholar]
- Cozzarelli N. R. DNA gyrase and the supercoiling of DNA. Science. 1980 Feb 29;207(4434):953–960. doi: 10.1126/science.6243420. [DOI] [PubMed] [Google Scholar]
- Elgin S. C., Boyd J. B. The proteins of polytene chromosomes of Drosophila hydei. Chromosoma. 1975;51(2):135–145. doi: 10.1007/BF00319831. [DOI] [PubMed] [Google Scholar]
- Elgin S. C., Serunian L. A., Silver L. M. Distribution patterns of Drosophila nonhistone chromosomal proteins. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):839–850. doi: 10.1101/sqb.1978.042.01.085. [DOI] [PubMed] [Google Scholar]
- Ellgaard E. G., Clever U. RNA metabolism during puff induction in Drosophila melanogaster. Chromosoma. 1971;36(1):60–78. doi: 10.1007/BF00326422. [DOI] [PubMed] [Google Scholar]
- Garel A., Axel R. Selective digestion of transcriptionally active ovalbumin genes from oviduct nuclei. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3966–3970. doi: 10.1073/pnas.73.11.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M. DNA topoisomerases. Annu Rev Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- Gocke E., Bonven B. J., Westergaard O. A site and strand specific nuclease activity with analogies to topoisomerase I frames the rRNA gene of Tetrahymena. Nucleic Acids Res. 1983 Nov 25;11(22):7661–7678. doi: 10.1093/nar/11.22.7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashinakagawa T., Wahn H., Reeder R. H. Isolation of ribosomal gene chromatin. Dev Biol. 1977 Feb;55(2):375–386. doi: 10.1016/0012-1606(77)90180-4. [DOI] [PubMed] [Google Scholar]
- Hill R. J., Mott M. R., Burnett E. J., Abmayr S. M., Lowenhaupt K., Elgin S. C. Nucleosome repeat structure is present in native salivary chromosomes of Drosophila melanogaster. J Cell Biol. 1982 Oct;95(1):262–266. doi: 10.1083/jcb.95.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard G. C., Abmayr S. M., Shinefeld L. A., Sato V. L., Elgin S. C. Monoclonal antibodies against a specific nonhistone chromosomal protein of Drosophila associated with active genes. J Cell Biol. 1981 Jan;88(1):219–225. doi: 10.1083/jcb.88.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T., Brutlag D. ATP-dependent DNA topoisonmerase from D. melanogaster reversibly catenates duplex DNA rings. Cell. 1980 Aug;21(1):115–125. doi: 10.1016/0092-8674(80)90119-1. [DOI] [PubMed] [Google Scholar]
- Jamrich M., Greenleaf A. L., Bautz E. K. Localization of RNA polymerase in polytene chromosomes of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1977 May;74(5):2079–2083. doi: 10.1073/pnas.74.5.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamrich M., Haars R., Wulf E., Bautz F. A. Correlation of RNA polymerase B and transcriptional activity in the chromosomes of Drosophila melanogaster. Chromosoma. 1977 Dec 6;64(4):319–326. doi: 10.1007/BF00294939. [DOI] [PubMed] [Google Scholar]
- Javaherian K., Tse Y. C., Vega J. Drosophila topoisomerase I: isolation, purification and characterization. Nucleic Acids Res. 1982 Nov 11;10(21):6945–6955. doi: 10.1093/nar/10.21.6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kister K. P., Müller B., Eckert W. A. Complex endonucleolytic cleavage pattern during early events in the processing of pre-rRNA in the lower eukaryote, Tetrahymena thermophila. Nucleic Acids Res. 1983 Jun 11;11(11):3487–3502. doi: 10.1093/nar/11.11.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korge G. Direct correlation between a chromosome puff and the synthesis of a larval saliva protein in Drosophila melanogaster. Chromosoma. 1977 Jul 5;62(2):155–174. doi: 10.1007/BF00292637. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Lawson G. M., Knoll B. J., March C. J., Woo S. L., Tsai M. J., O'Malley B. W. Definition of 5' and 3' structural boundaries of the chromatin domain containing the ovalbumin multigene family. J Biol Chem. 1982 Feb 10;257(3):1501–1507. [PubMed] [Google Scholar]
- Liu L. F. DNA topoisomerases--enzymes that catalyse the breaking and rejoining of DNA. CRC Crit Rev Biochem. 1983;15(1):1–24. doi: 10.3109/10409238309102799. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Miller K. G. Eukaryotic DNA topoisomerases: two forms of type I DNA topoisomerases from HeLa cell nuclei. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3487–3491. doi: 10.1073/pnas.78.6.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield J. E., Serunian L. A., Silver L. M., Elgin S. C. A protein released by DNAase I digestion of drosophila nuclei is preferentially associated with puffs. Cell. 1978 Jul;14(3):539–544. doi: 10.1016/0092-8674(78)90240-4. [DOI] [PubMed] [Google Scholar]
- McGinnis W., Farrell J., Jr, Beckendorf S. K. Molecular limits on the size of a genetic locus in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7367–7371. doi: 10.1073/pnas.77.12.7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie S. L., Henikoff S., Meselson M. Localization of RNA from heat-induced polysomes at puff sites in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1117–1121. doi: 10.1073/pnas.72.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muskavitch M. A., Hogness D. S. An expandable gene that encodes a Drosophila glue protein is not expressed in variants lacking remote upstream sequences. Cell. 1982 Jul;29(3):1041–1051. doi: 10.1016/0092-8674(82)90467-6. [DOI] [PubMed] [Google Scholar]
- Muskavitch M. A., Hogness D. S. Molecular analysis of a gene in a developmentally regulated puff of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7362–7366. doi: 10.1073/pnas.77.12.7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. A. The regulation of yeast mating-type chromatin structure by SIR: an action at a distance affecting both transcription and transposition. Cell. 1982 Sep;30(2):567–578. doi: 10.1016/0092-8674(82)90253-7. [DOI] [PubMed] [Google Scholar]
- Olmsted J. B. Affinity purification of antibodies from diazotized paper blots of heterogeneous protein samples. J Biol Chem. 1981 Dec 10;256(23):11955–11957. [PubMed] [Google Scholar]
- Plagens U., Greenleaf A. L., Bautz E. K. Distribution of RNA polymerase on Drosophila polytene chromosomes as studied by indirect immunofluorescence. Chromosoma. 1976 Dec 16;59(2):157–165. doi: 10.1007/BF00328484. [DOI] [PubMed] [Google Scholar]
- Rodriguez Alfageme C., Rudkin G. T., Cohen L. H. Isolation, properties and cellular distribution of D1, a chromosomal protein of Drosophila. Chromosoma. 1980;78(1):1–31. doi: 10.1007/BF00291907. [DOI] [PubMed] [Google Scholar]
- Rubin G. M., Spradling A. C. Genetic transformation of Drosophila with transposable element vectors. Science. 1982 Oct 22;218(4570):348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Saumweber H., Symmons P., Kabisch R., Will H., Bonhoeffer F. Monoclonal antibodies against chromosomal proteins of Drosophila melanogaster: establishment of antibody producing cell lines and partial characterization of corresponding antigens. Chromosoma. 1980;80(3):253–275. doi: 10.1007/BF00292684. [DOI] [PubMed] [Google Scholar]
- Seed B. Diazotizable arylamine cellulose papers for the coupling and hybridization of nucleic acids. Nucleic Acids Res. 1982 Mar 11;10(5):1799–1810. doi: 10.1093/nar/10.5.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shermoen A. W., Beckendorf S. K. A complex of interacting DNAase I-hypersensitive sites near the Drosophila glue protein gene, Sgs4. Cell. 1982 Jun;29(2):601–607. doi: 10.1016/0092-8674(82)90176-3. [DOI] [PubMed] [Google Scholar]
- Silver L. M., Elgin S. C. A method for determination of the in situ distribution of chromosomal proteins. Proc Natl Acad Sci U S A. 1976 Feb;73(2):423–427. doi: 10.1073/pnas.73.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver L. M., Elgin S. C. Distribution patterns of three subfractions of drosophila nonhistone chromosomal proteins: possible correlations with gene activity. Cell. 1977 Aug;11(4):971–983. doi: 10.1016/0092-8674(77)90308-7. [DOI] [PubMed] [Google Scholar]
- Spradling A., Penman S., Pardue M. L. Analysis of drosophila mRNA by in situ hybridization: sequences transcribed in normal and heat shocked cultured cells. Cell. 1975 Apr;4(4):395–404. doi: 10.1016/0092-8674(75)90160-9. [DOI] [PubMed] [Google Scholar]
- Steiner E. K., Eissenberg J. C., Elgin S. C. A cytological approach to the ordering of events in gene activation using the Sgs-4 locus of Drosophila melanogaster. J Cell Biol. 1984 Jul;99(1 Pt 1):233–238. doi: 10.1083/jcb.99.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissières A., Mitchell H. K., Tracy U. M. Protein synthesis in salivary glands of Drosophila melanogaster: relation to chromosome puffs. J Mol Biol. 1974 Apr 15;84(3):389–398. doi: 10.1016/0022-2836(74)90447-1. [DOI] [PubMed] [Google Scholar]
- Tjian R., Stinchcomb D., Losick R. Antibody directed against Bacillus subtilis rho factor purified by sodium dodecyl sulfate slab gel electrophoresis. Effect on transcription by RNA polymerase in crude extracts of vegetative and sporulating cells. J Biol Chem. 1975 Nov 25;250(22):8824–8828. [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks J. R., Coulter D. E., Greenleaf A. L. Immunological studies of RNA polymerase II using antibodies to subunits of Drosophila and wheat germ enzyme. J Biol Chem. 1982 May 25;257(10):5884–5892. [PubMed] [Google Scholar]
- Weintraub H. A dominant role for DNA secondary structure in forming hypersensitive structures in chromatin. Cell. 1983 Apr;32(4):1191–1203. doi: 10.1016/0092-8674(83)90302-1. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weisbrod S. T. Properties of active nucleosomes as revealed by HMG 14 and 17 chromatography. Nucleic Acids Res. 1982 Mar 25;10(6):2017–2042. doi: 10.1093/nar/10.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimber D. E., Steffensen D. M. Localization of 5S RNA genes on Drosophila chromosomes by RNA-DNA hybridization. Science. 1970 Nov 6;170(3958):639–641. doi: 10.1126/science.170.3958.639. [DOI] [PubMed] [Google Scholar]
- Woodcock C. L., Safer J. P., Stanchfield J. E. Structural repeating units in chromatin. I. Evidence for their general occurrence. Exp Cell Res. 1976 Jan;97:101–110. doi: 10.1016/0014-4827(76)90659-5. [DOI] [PubMed] [Google Scholar]
- Wu C., Wong Y. C., Elgin S. C. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell. 1979 Apr;16(4):807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]
- Yen P. H., Sodja A., Cohen M., Jr, Conrad S. E., Wu M., Davidson N. Sequence arrangement of tRNA genes on a fragment of Drosophila melanogaster DNA cloned in E. coli. Cell. 1977 Aug;11(4):763–777. doi: 10.1016/0092-8674(77)90290-2. [DOI] [PubMed] [Google Scholar]