CD73 expression is induced in response to TCR ligation and identifies a population of thymocytes that are committed to the γδ T cell fate.
Abstract
Numerous studies indicate that γδ T cell receptor (γδTCR) expression alone does not reliably mark commitment of early thymic progenitors to the γδ fate. This raises the possibility that the γδTCR is unable to intrinsically specify fate and instead requires additional environmental factors, including TCR–ligand engagement. We use single cell progenitor assays to reveal that ligand acts instructionally to direct adoption of the γδ fate. Moreover, we identify CD73 as a TCR ligand-induced cell surface protein that distinguishes γδTCR-expressing CD4−CD8− progenitors that have committed to the γδ fate from those that have not yet done so. Indeed, unlike CD73− γδTCR+ progenitors, which largely adopt the αβ fate upon separation from the intrathymic selecting environment, those that express CD73 remain CD4−CD8− and committed to the γδ fate. CD73 is expressed by >90% of peripheral γδ cells, suggesting this is a common occurrence during development. Moreover, CD73 induction appears to mark a metastable intermediate stage before acquisition of effector function, suggesting that γδ lineage and effector fate are specified sequentially. These findings have important implications for the role of ligand in γδ lineage commitment and its relationship to the specification of effector fate.
T lymphocytes comprise two distinct lineages that express either αβ or γδ TCR complexes and perform nonoverlapping roles in immune responses. Although αβ T cells generally respond to peptide ligands in the context of MHC class I and II, the types of antigens recognized by γδTCRs are more diverse and include nonclassical MHC molecules, heat shock proteins, and lipids. γδ T cells make up a small proportion of T cells in the peripheral lymphoid organs but predominate in the epithelial tissues that form the inner and outer surfaces of the body (Hayday, 2000; Carding and Egan, 2002; Vantourout and Hayday, 2013). Furthermore, γδ T cells are thought be an important link between the innate and adaptive immune systems because they recognize pathogen-derived and host stress-induced ligands at epithelial barriers (Born et al., 2006; Witherden and Havran, 2011). Although γδ T cells have been shown to play a vital role in certain types of responses, it has been difficult to identify the factors that govern their divergence from the αβ lineage during development. Accordingly, the molecular mechanisms that control lineage commitment, shape the γδ T cell repertoire, and specify effector fate during development are not well understood.
Both αβ and γδ lineage T cells arise from immature CD4−CD8− (double negative [DN]) precursors in the thymus (Petri et al., 1992; Dudley et al., 1995). γδ lineage T cells largely remain DN and develop in response to signals from the γδTCR complex, whereas signals transduced through the preTCR complex are required for adoption of the αβ fate and differentiation of αβ progenitors to the CD4+CD8+ (double positive [DP]) stage (Kreslavsky et al., 2010; Lee et al., 2010). Therefore, αβ and γδ lineage cells are usually identified by their expression of either TCR isotype in combination with whether they progress to the DP stage (αβ) or remain DN (γδ). However, it has become apparent that fate decisions are not always matched by TCR expression and that expression of the TCR type alone is not sufficient to direct lineage commitment. Studies using gene-targeted (Tcrb−/− or Ptcra−/−) mice, which allow expression of only the γδTCR, have revealed the presence of progenitors that were diverted to the DP stage (Bruno et al., 1996; Passoni et al., 1997). Likewise, studies using Rag2−/− KN6 γδTCR transgenic (Tg) mice have also demonstrated that TCR type does not exclusively determine lineage fate (Haks et al., 2005). The KN6 model provides a unique system for studying lineage fate because, unlike most γδTCRs, the ligand for the KN6 TCR is known. KN6 Tg thymocytes recognize an endogenous nonclassical MHC class I molecule (T-10/22) whose surface expression is β2M-dependent (Bonneville et al., 1989). In the presence of ligand, most KN6 thymocytes remain DN, adopt the γδ fate, down-modulate CD24 expression, and acquire effector function (Pereira et al., 1992). However, when surface expression of ligand is attenuated in β2M-deficient mice, adoption of the γδ fate by KN6 Tg thymocytes is abrogated and they are instead diverted to the DP stage of the αβ lineage (Haks et al., 2005). These studies and others have demonstrated that γδTCR+ DN T cell progenitors retain the ability to adopt either the αβ or γδ lineage, regardless of the TCR isotype they express (Terrence et al., 2000; Lacorazza et al., 2001).
Attempts to explain the role of the TCR in αβ/γδ lineage commitment have been distilled into two basic models: stochastic and instructional. The stochastic model predicts that lineage fate is determined independently of TCR expression and that TCR signaling serves only to reinforce the previously established fate decision, provided the TCR isotype matches the preordained lineage fate (Narayan and Kang, 2010). Conversely, the instructional model proposes that TCR signaling “instructs” an uncommitted precursor to adopt either the αβ or γδ fate (Wong and Zúñiga-Pflücker, 2010). That is, the signals transduced through the preTCR or γδTCR actively specify the αβ and γδ fates, respectively. These models share the basic tenet that the preTCR or γδTCR complexes transduce unique signals inextricably linked to specification of the αβ and γδ fates, respectively. However, these models are not adequate to explain the status of TCR gene rearrangements in αβ and γδ lineage cells, nor do they appropriately explain the lineage infidelity observed in TCR Tg and gene-targeted mice (Lee et al., 2010). To address these inconsistencies, a signal strength model was proposed which posits that strong signaling through a TCR promotes adoption of the γδ lineage, whereas weaker signals lead to adoption of the αβ lineage, irrespective of the isotype of the TCR complex from which those signals originate (Hayes and Love, 2006). Compelling support for the signal strength model was provided by the demonstration that thymocytes expressing a single γδTCR transgene could adopt either the αβ or γδ fate upon manipulating the γδTCR to transduce weak or strong TCR signals, respectively (Haks et al., 2005; Hayes et al., 2005). Evidence using Ab ligation of the TCR suggests that the strong TCR signals linked to adoption of the γδ fate are functioning in an instructional manner (Kreslavsky et al., 2008).
Although the signal strength model is now widely regarded as providing the best explanation for the role of the TCR complex in lineage commitment, it remains unclear how the γδTCR complex transduces the stronger signals required for adoption of the γδ fate. The role of ligand in regulating the γδTCR signal remains controversial (Kreslavsky and von Boehmer, 2010; Meyer et al., 2010). Based on the restriction of the chain usage and CDR3 sequences of the γδTCR complex that characterizes the dendritic epidermal T cell (DETC) subset of γδ T cells, it is likely that their development is ligand-dependent (Havran and Allison, 1988). DETC development requires expression of Skint1, although whether Skint1 functions as a selecting ligand remains to be fully established (Boyden et al., 2008). Analysis of the only other γδ population for which a selecting ligand has been identified, T-10/22 binding γδ T cells, has produced conflicting results. Data from γδTCR Tg models have provided very clear evidence in support of a role for ligand in their selection (Haks et al., 2005; Lauritsen et al., 2009), but this did not appear to be true for polyclonal T-10/22 reactive γδ progenitors identified by tetramer binding (Jensen et al., 2008).
Efforts to gain clear insights into the molecular processes involved in αβ/γδ fate specification have been hampered by the absence of markers that distinguish DN γδTCR-expressing progenitors that have committed to the γδ fate from those yet to do so. Sox13 was initially advanced as a marker of developing γδ T cells and mediator of their development, but more recent analysis now reveals that its expression marks only a subset of γδ T cells (Melichar et al., 2007; Kreslavsky et al., 2008). Moreover, analysis of mice with a spontaneous mutation in Sox13 has revealed that this transcription factor is not required for lineage commitment and likely has a role in maturation or specification of effector fate (Gray et al., 2013). The Hayday laboratory previously identified a γδ-biased expression profile consisting of genes highly expressed in mature γδ T cells; however, this signature corresponds with acquisition of effector function and not necessarily lineage commitment (Pennington et al., 2003; Silva-Santos et al., 2005). Recent expression profiling of γδ subsets has identified new candidates, in addition to confirming a previous study linking particular transcription factors to effector fate (e.g., Sox13 and RORγt with IL-17 production and Egr3 with IFN-γ production; Turchinovich and Hayday, 2011; Narayan et al., 2012). Furthermore, analysis of gene expression in γδ subsets has led to the identification of a network of high-mobility group transcription factors, including Sox4, Sox13, Tcf1, and Lef1, responsible for directing the programming of IL-17 production in certain γδ T cells subsets (Malhotra et al., 2013). Nonetheless, a molecular marker for γδ lineage commitment remains to be identified.
In this study, we investigate the role of TCR–ligand engagement in γδ T cell commitment and use single cell progenitor studies to show that ligand instructs the fate of individual progenitors. We also identified Nt5e (CD73), which is up-regulated upon ligand engagement of the γδTCR, is highly expressed in 25–30% of γδTCR+ DN progenitors, and marks their commitment to the γδ fate. Indeed, CD73+ but not CD73− γδTCR-expressing progenitors remain DN and adopt the γδ fate, even after removal from intrathymic selecting conditions in vivo or from ligand in vitro. Expression of CD73 on immature CD24+ DN progenitors also defines an intermediate developmental stage after commitment but before acquisition of effector fate, where the transcription factor networks involved in specifying effector fate are scrambled. These networks are then resolved upon subsequent maturation into CD73+CD24low cells that are functionally competent, providing the first indication that γδ lineage commitment and specification of effector fate may occur sequentially. Altogether, these findings describe a role for CD73 as a novel, ligand-dependent marker of γδ T cell commitment and provide important insights into its relationship to acquisition of effector fate.
RESULTS
Ligand instructs the lineage fate of T cell progenitors expressing the KN6 γδTCR
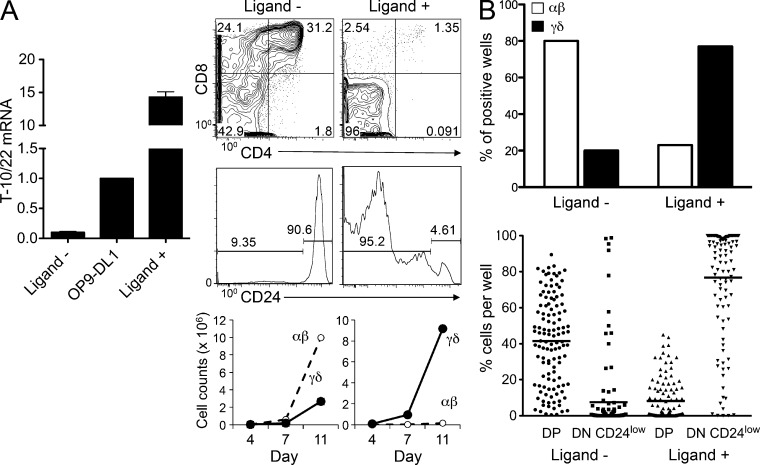
We have previously shown that KN6 γδTCR Tg thymocytes adopt the γδ fate in the presence of T-10/22 ligand in vivo, as indicated by their remaining DN and down-regulating the CD24 maturation marker (Pereira et al., 1992; Haks et al., 2005). KN6 thymocytes are diverted to the αβ fate and differentiate to the DP stage when ligand expression is indirectly attenuated in β2M-deficient hosts because T-10/22 ligands depend upon β2M for surface expression (Haks et al., 2005). Because β2M deficiency indirectly attenuates surface expression of T-10/22, along with all other β2M-dependent molecules, we evaluated the role of KN6 TCR–ligand interactions in determining lineage fate using an in vitro culture system where T-10/22 expression could be directly and specifically attenuated by shRNA knockdown (Fig. 1). We generated OP9-DL1 cells (Schmitt and Zúñiga-Pflücker, 2002) that expressed very low levels of T-10/22 (Ligand−) or elevated levels of the low-affinity T-10d (Ligand+) ligand and then monitored the development of KN6-expressing T cell progenitors on these monolayers (Fig. 1 A). KN6 γδTCR-transduced Rag2−/− progenitors that were cultured on ligand-expressing monolayers adopted the γδ fate, as indicated by remaining DN and down-regulating CD24 (Fig. 1 A). Consistent with our finding that KN6 Tg thymocytes are diverted to the αβ lineage in β2M-deficient mice, KN6-expressing progenitors cultured on OP9-DL1 cells in which T-10/22 expression was knocked down (Ligand−) were diverted to the αβ lineage, as indicated by their development to the DP stage and their failure to down-regulate CD24 (Fig. 1 A). These findings demonstrate that adoption of the γδ fate by KN6-expressing progenitors is specifically dependent on T-10d ligand. Although our results demonstrate that TCR–ligand interactions are required for adoption of the γδ fate in this model, because our analysis was performed on bulk cultures, it remained unclear whether the strong TCR signals induced by ligand engagement were acting to instruct the lineage fate of uncommitted precursors (instructional) or, alternatively, were rescuing the survival of precommitted progenitors (stochastic). To distinguish these possibilities, we performed single-cell progenitor fate assays by plating individual KN6 TCR-expressing DN3 progenitors onto monolayers either lacking (Ligand−) or expressing (Ligand+) ligand (Fig. 1 B). Our analysis indicated that 80% of cells developing in the absence of ligand adopted the αβ lineage, as defined by the presence of >15% DP cells, the criterion used in our previous single-cell analysis (Ciofani et al., 2006). Conversely, 80% of cells developing on ligand-expressing monolayers adopted the γδ fate as indicated by their remaining DN (Fig. 1 B). The scatter plot illustrating the percentage of DP and mature DN cells in each well likewise indicates that most wells from cultures developing on ligand were γδ lineage because they comprised predominantly mature DN, whereas those cultured in the absence of ligand were αβ lineage because they comprised predominantly CD24hi DP (Fig. 1 B, bottom). KN6 γδTCR-expressing progenitors exhibited a similarly high cloning frequency on both Ligand− and Ligand+ monolayers, as did progenitors expressing the preTCR (i.e., TCR-β–transduced; not depicted; Ciofani et al., 2006). Altogether, these single cell progenitor studies reveal different lineage fates in the presence or absence of ligand and suggest that lineage commitment of KN6 γδTCR-expressing progenitors is regulated by ligand in an instructional manner. This is consistent with previous analysis using Ab ligation to mimic ligand engagement (Kreslavsky et al., 2008).
Figure 1.
TCR ligand dictates the fate of KN6 progenitors in an instructional manner. (A, Left) T-10/22 expression was determined by real-time PCR for OP9-DL1 cells that were retrovirally transduced with shRNA (Ligand−) targeting T-10/22 or with a T-10d–encoding construct (Ligand+). Expression was normalized to β-actin and is shown relative to nontransduced OP9-DL1. Mean ± SD for triplicates is shown. (A, Right) Rag2−/− DN3 fetal liver progenitors were transduced with a KN6 γδTCR-IRES-YFP retroviral construct and cultured on Ligand− or Ligand+ OP9-DL1 cells. Development was assessed at various time points by flow cytometry to evaluate CD4 and CD8 expression as well as CD24 levels on DN-gated cells. Flow cytometry plots shown are for analysis on day 11 and the gate frequencies for each population are indicated on the histograms. Graphs show cell counts of αβ (DP, CD24high) and γδ (DN, CD24low) lineage progeny present in cultures on days 4, 7, and 11. Data are representative of at least three independent experiments. (B) Single Rag2−/− DN3 fetal liver progenitors were transduced with KN6 γδTCR and placed into individual wells of 96-well plates containing either Ligand− or Ligand+ OP9-DL1 cells. On day 10, plates were analyzed by flow cytometry and lineage fate was determined in reconstituted wells (119 wells for Ligand− and 112 for Ligand+ out of 120 total plated for each). (Top) Positive wells containing >15% DP cells were recorded as αβ and those containing <15% DP are indicated as γδ. Data are expressed as a percentage of total reconstituted wells. (Bottom) The percentages of DP and DN-gated CD24low cells are shown for individual reconstituted wells (each symbol represents one well). Horizontal bars indicate means. Results are representative of two independent experiments.
CD73 is highly expressed among γδ T cells
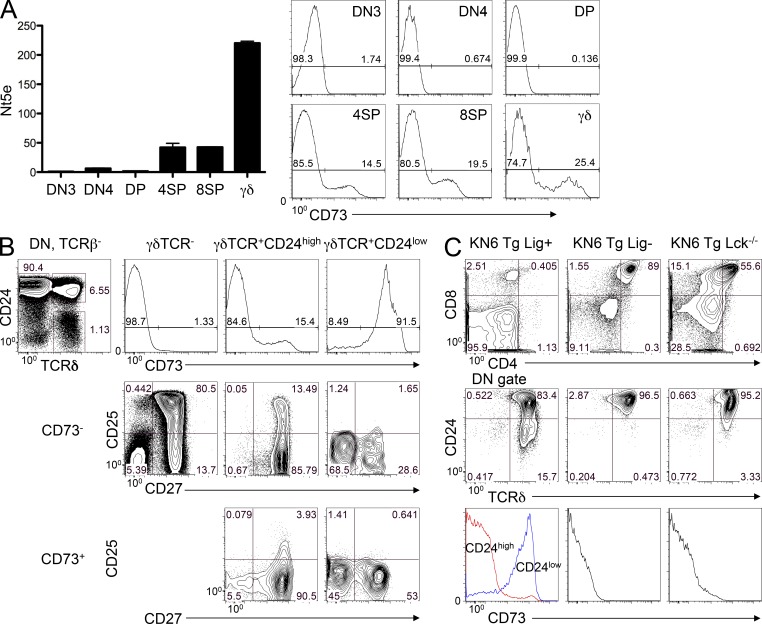
Our findings, and those of others, demonstrate that expression of γδTCR alone is not sufficient to dictate the lineage fate of progenitors (Lee et al., 2010); however, to date, aside from the absence of development to the DP stage, there is no other marker that distinguishes DN γδTCR-expressing cells that have adopted the γδ fate from those that have yet to do so. To identify such markers, we performed microarray analysis to reveal genes that are highly expressed, compared with αβ lineage DP cells, in both KN6 γδTCR Tg thymocytes that are adopting the γδ fate in response to ligand engagement and in polyclonal non-Tg γδTCR-expressing cells. Among the genes identified in this manner as being more highly expressed in γδ progenitors than in αβ lineage DP was Nt5e (ecto-5′-nucleotidase, CD73; Fig. 2 A). Analysis of Nt5e expression in populations of sorted adult C57BL/6 thymocytes confirmed that the gene is highly expressed in γδTCR+ cells and expressed at low levels in DN3, DN4, and DP thymocytes, with somewhat higher levels expressed in mature CD4 and CD8 single-positive thymocytes. Flow cytometry analysis revealed that CD73 surface expression was consistent with the mRNA levels, being highly expressed on 25% of γδTCR+ cells. The elevated expression in γδTCR+ DN thymocytes, and the fact that CD73 is a cell surface molecule, made it an appealing candidate for lineage commitment studies. Analysis of γδTCR-positive thymocytes revealed that 15% of CD24high immature γδTCR-expressing cells were CD73+, whereas CD24low cells uniformly expressed CD73 at high levels (Fig. 2 B). Therefore, we speculated that CD73 expression might be an indicator of initial γδ commitment that precedes CD24 down-modulation. Ribot et al. (2009) previously reported that CD27+CD25+ γδTCR-expressing DN thymocytes constitute a highly proliferative progenitor subset that gives rise to more mature CD25− populations. Interestingly, the CD73−CD24high population of γδTCR-expressing cells contains some (∼13%) CD25+CD27+ cells, but these are diminished upon induction of CD73, suggesting that CD73 induction occurs as the earliest CD25+CD27+ progenitors differentiate into CD25−CD27+ cells (Fig. 2 B). In the KN6 Tg γδTCR model where all thymocytes express the same TCR specificity, we found CD73 to be highly expressed among CD24low cells that have adopted the γδ fate. However, CD73 was not expressed on thymocytes that have been diverted to the αβ fate by attenuating TCR signaling, either through abrogation of ligand expression using β2M deficiency or impairing TCR signaling through elimination of the key TCR signaling kinase, p56lck (Fig. 2 C). Together, these data indicate that CD73 expression is bimodal in the earliest γδTCR-expressing progenitors and that weakening TCR signaling attenuates its induction.
Figure 2.
CD73 is inducible by ligand engagement and is highly expressed in γδ lineage T cells. (A) The indicated thymocyte populations were isolated by cell sorting: DN3, TCR-β−TCR-γδ−CD25+CD44−; DN4, TCR-β−TCR-γδ−CD25−CD44−; DP, CD4+CD8+; 4SP, CD4+; 8SP, CD8+; and γδ, CD4−CD8−γδTCR+. Expression of CD73 mRNA was quantified by real-time PCR and levels were normalized to β-actin. Graphed values indicate mean ± SD. Surface expression of CD73 on the sorted populations was measured by flow cytometry to determine the frequency of CD73− and CD73+ cells. Gate frequencies of the CD73− and CD73+ populations are listed on the histograms. (B) CD73 expression was measured by flow cytometry on the indicated γδTCR-expressing thymocyte populations from adult mice. The left panel shows the gating strategy and percentages for each population, with the gate frequencies indicated on the histograms. (C) KN6 γδTCR Tg thymocytes were exposed in vivo to strong TCR signals (T-10d; KN6 Tg Lig+, KN6 Tg Rag2−/−) or to diminished TCR signaling, produced either by eliminating ligand (KN6 Tg Lig−, KN6 Tg Rag2−/−B2m−/−) or by ablation of the p56lck tyrosine kinase (KN6 Tg Rag2−/−Lck−/−). The numbers shown indicate the percentage of cells within gated quadrants and data are representative of results from analysis of at least five mice of each genotype from at least three independent experiments.
CD73 marks γδ lineage committed KN6 γδTCR-expressing progenitors
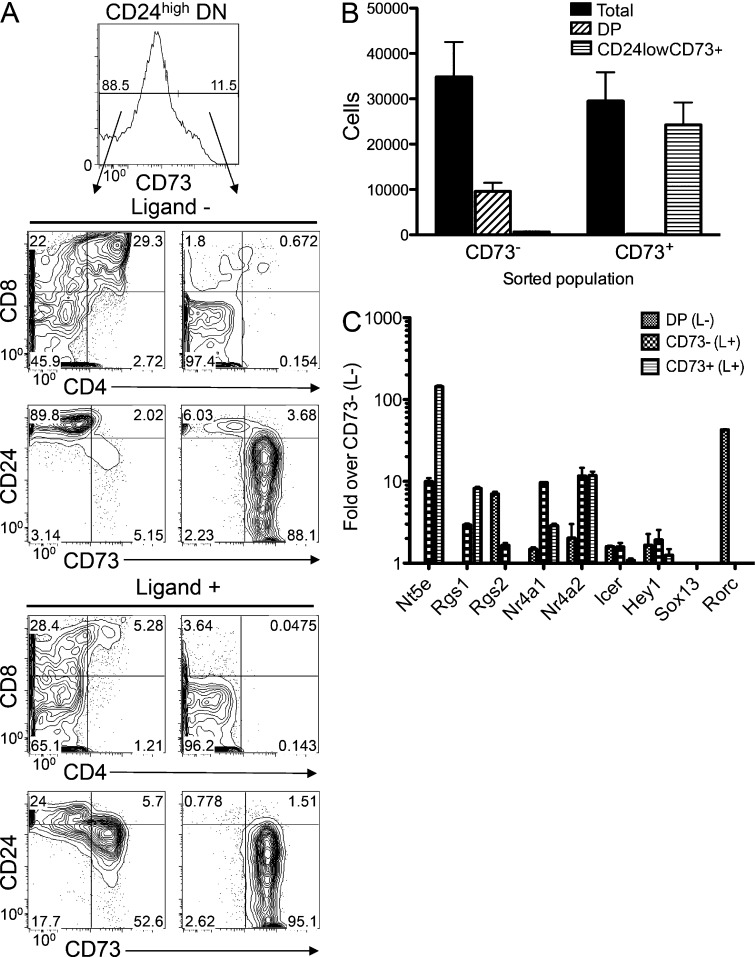
Because the expression of CD73 by KN6 Tg progenitors was induced by TCR ligand engagement and was correlated with adoption of the γδ fate, we asked whether the induction of CD73 actually marked γδ lineage commitment. To address this possibility, KN6-expressing DN3 progenitors were cultured on ligand-expressing OP9-DL1 cells until CD73 began to be induced (5 d), after which the CD24highCD73+ and CD24highCD73− progenitors were independently transferred to cultures lacking ligand (Fig. 3 A). After additional culture on OP9-DL1 cells lacking ligand, the progenitors that had not yet induced CD73 adopted the αβ fate, as indicated by differentiation to the DP stage and retention of the CD73−CD24high phenotype (Fig. 3 A). When these CD73− cells were transferred to OP9 cultures expressing ligand, they induced CD73 and down-modulated CD24 (Fig. 3 A, bottom), indicating that they remained capable of committing to the γδ fate in response to prolonged exposure to ligand. Importantly, when CD73+ progenitors were transferred to cultures lacking ligand, they remained DN and down-regulated CD24, indicating that they had committed to the γδ fate (Fig. 3, A and B). Notably, although the vast majority of CD73+ progenitors retained CD73 expression, a small CD73− population emerged in these cultures, perhaps because of incomplete separation of CD73+ cells from the CD73− population during cell sorting. Altogether, these data suggest that CD73 expression identifies KN6 progenitors that have engaged ligand and committed to the γδ lineage, whereas cells that have not induced CD73 appear to remain bipotential and capable of adopting either the αβ or γδ lineage upon receipt of appropriate stimulation. Based on these findings, we conclude that CD73 induction precedes CD24 down-modulation and distinguishes γδTCR+ DNs that have committed to the γδ fate from those that retain αβ fate potential.
Figure 3.
CD73 identifies γδ lineage–committed KN6 progenitors. (A and B) Rag2−/− fetal liver DN3 progenitors were transduced with the KN6 γδTCR as in Fig. 1 and cultured on ligand-expressing OP9-DL1 cells for 5 d. On day 5, CD24highCD73− and CD73+ populations were isolated by cell sorting and transferred to Ligand− or Ligand+ OP9-DL1 cultures. After an additional 4 d of culture, development was assessed by flow cytometry using the indicated Ab to determine the frequency of gated populations, which are indicated on the histograms. (B) The graph shows absolute cell numbers of the indicated phenotypes of cells transferred to Ligand− OP9-DL1 cultures (mean ± SD) for triplicate wells seeded with 5,000 progenitors on Ligand− OP9-DL1. Data shown are representative of results from three independent experiments. (C) KN6 γδTCR-expressing DN3 progenitors were cultured for 6 d on Ligand− (L−) OP9 DL1 cells to generate αβ lineage DP or Ligand+ (L+) OP9-DL1 cells to generate CD73− and CD73+ DN thymocytes. Expression of the indicated genes was quantified by real-time PCR. Values were normalized to β-actin and the mean ± SD depicted graphically as fold change in expression over that in CD73− DN isolated from L− cultures. Results are representative of 3 experiments performed.
To determine whether previously identified genes that are enriched in γδ lineage cells might also exhibit expression patterns linked to γδ lineage commitment, we analyzed their expression using real-time PCR in ligand-exposed progenitors (Fig. 3 C). We found that ligand exposure increased CD73 mRNA levels even in those KN6 γδTCR-expressing cells that were CD73− by flow cytometry; however, expression of CD73 mRNA increased a further 20-fold in the cells that were CD73+ by flow cytometry and had committed to the γδ fate (Fig. 3 C). Interestingly, with the exception of Rgs1, which we previously confirmed to be closely linked to adoption of the γδ fate (Lauritsen et al., 2009), none of the genes in this profile were further enriched in the CD73+ (Nt5ehigh) population, which had committed to the γδ fate (Fig. 3 C). Rgs2, Nr4a1, Nr4a2, Crem (Icer), and Hey1 were expressed to varying degrees but, unlike CD73 (Nt5e), none of these other genes appeared to be up-regulated in the γδ lineage-committed CD73+ population. Notably, although Sox13 had previously been considered a marker of γδ T cells, it was not expressed by γδ-committed KN6-expressing thymocytes. Based on these data, we propose that CD73 expression is an early identifier of γδ T cell lineage commitment that precedes expression of other, previously identified γδ-biased genes. Therefore, CD73 can be used to identify other genes and molecular pathways that are pertinent to lineage fate decisions.
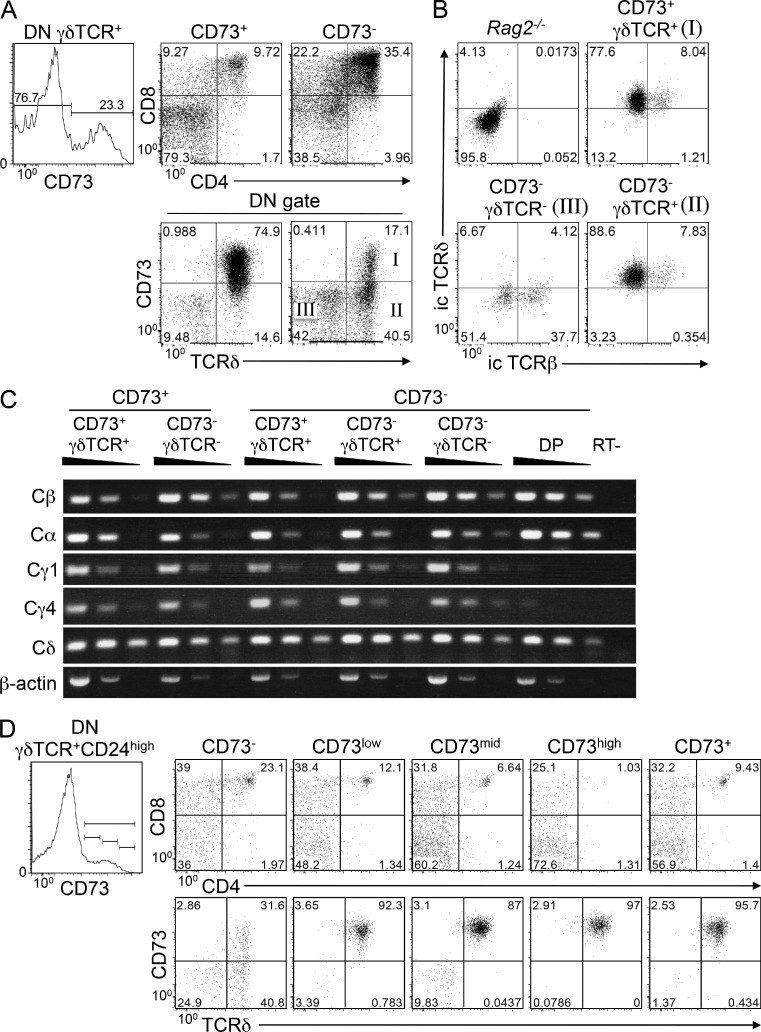
To determine whether CD73 expression also marked the γδ lineage-committed cells among polyclonal γδTCR+ DN from adult mice, we isolated the CD73+ and CD73− subpopulations and cultured them on OP9-DL1 monolayers for 4 d (Fig. 4). The vast majority of CD73+ cells remained DN and γδTCR+, indicating that they had committed to the γδ fate (Fig. 4 A, left). For the CD73− population, ∼35% were diverted to the αβ fate and differentiated to the DP stage (Fig. 4 A; right). This appears to be an underestimate of the propensity of CD73−γδTCR+ cells to fate switch, as 17% of these cells up-regulated CD73 upon culture on OP9-DL1 cells, presumably in response to encounter with ligands expressed by OP9 cells. Interestingly, although all of the CD73− cells that up-regulated CD73 during culture on OP9 cells retained expression of the γδTCR, about half of those that remained CD73-negative silenced surface expression of the γδTCR (Fig. 4 A, bottom right). The silencing of γδTCR expression by cultured CD73− cells was accompanied by loss of intracellular TCR-δ protein (Fig. 4 B, bottom left), but not mRNA encoding the constant domains of TCR-γ and TCR-δ (Fig. 4 C). As expected, Cγ expression was lost in DP thymocytes as the TCR-γ silencing element is activated at this stage (Fig. 4 C; Ferrero et al., 2006). Accordingly, γδTCR expression appears to be silenced at a posttranscriptional level. It is important to note that many of the CD73−γδTCR− cells also expressed intracellular TCR-β, consistent with the notion that the switch to the αβ fate by the CD73− progenitors that silence γδTCR expression, is likely promoted by up-regulation of the preTCR. Altogether, these data indicate that CD73 induction marks commitment to the γδ fate and retention of γδTCR expression; however, when γδ progenitors are removed from the intrathymic milieu before CD73 induction, many of these DN thymocytes silence γδTCR expression, express TCR-β protein, switch to the αβ fate, and develop to the DP stage. Among the heterogeneous γδTCR-expressing DN thymocytes used in this experiment was a subpopulation that were stimulated by ligands expressed on OP9-DL1 cells, as indicated by the up-regulation of CD73 during culture. Accordingly, this suggests that the extent of fate-switching by the CD73− γδTCR-expressing DN cells is likely to be underestimated in this system.
Figure 4.
CD73+ polyclonal γδTCR+ DN cells are enriched for those that have committed to the γδ lineage. (A) CD73− and CD73+ γδTCR-expressing DN thymocytes were isolated by flow cytometry from adult C57BL/6 mice (gating shown in left plot) and cultured 4 d on OP9-DL1 cells. Development was assessed by flow cytometry based on expression of CD4, CD8, CD73, and TCR-δ and numerical values indicate the frequency of cells in each quadrant. Data shown are representative of analysis from three independent experiments. (B) As in A, CD73− cells were cultured for 4 d and stained to identify DN populations that were CD73+TCRδ+ (I), CD73−TCRδ+ (II), and CD73−TCRδ− (III). These populations were then sorted, fixed, permeabilized, and stained to determine the frequencies of cells (indicated on the histograms) that expressed intracellular (i.c.) TCR-β and TCR-δ protein. Rag2−/− thymocytes served as a negative control. Data shown are representative of analysis from three independent experiments. (C) Expression of mRNA encoding the indicated clonotypic TCR subunits was evaluated by reverse transcription PCR and ethidium bromide staining on the subsets identified in A. Data shown are representative of two independent experiments. (D) CD73−, CD73low, CD73mid, CD73high, and total CD73+ γδTCR-expressing CD24high DN thymocytes were isolated by flow cytometry from adult C57BL/6 mice and cultured for 4 d on OP9-DL1 cells, after which development was assessed by flow cytometry based on expression of CD4, CD8, CD73, and TCR-δ. Data shown in all panels are representative of two independent experiments, with gate frequencies indicated on the histograms.
Although the vast majority of CD73-expressing cells in this experiment remained DN and committed to the γδ fate, ∼10% of cells in these cultures differentiated to the DP stage. To determine the basis for this, we conducted further analysis to determine if this resulted from incomplete separation of the CD73+ and CD73− populations or, alternatively, reflected a threshold of CD73 expression that must be achieved for commitment. Interestingly, when CD73low, CD73mid, or CD73high γδTCR+CD24high DNs were cultured on OP9-DL1 cells, we found that the fraction of DP decreased with increasing CD73 levels, suggesting perhaps that a particular threshold of TCR-dependent CD73 induction must be met in order for lineage commitment to be achieved (Fig. 4 D).
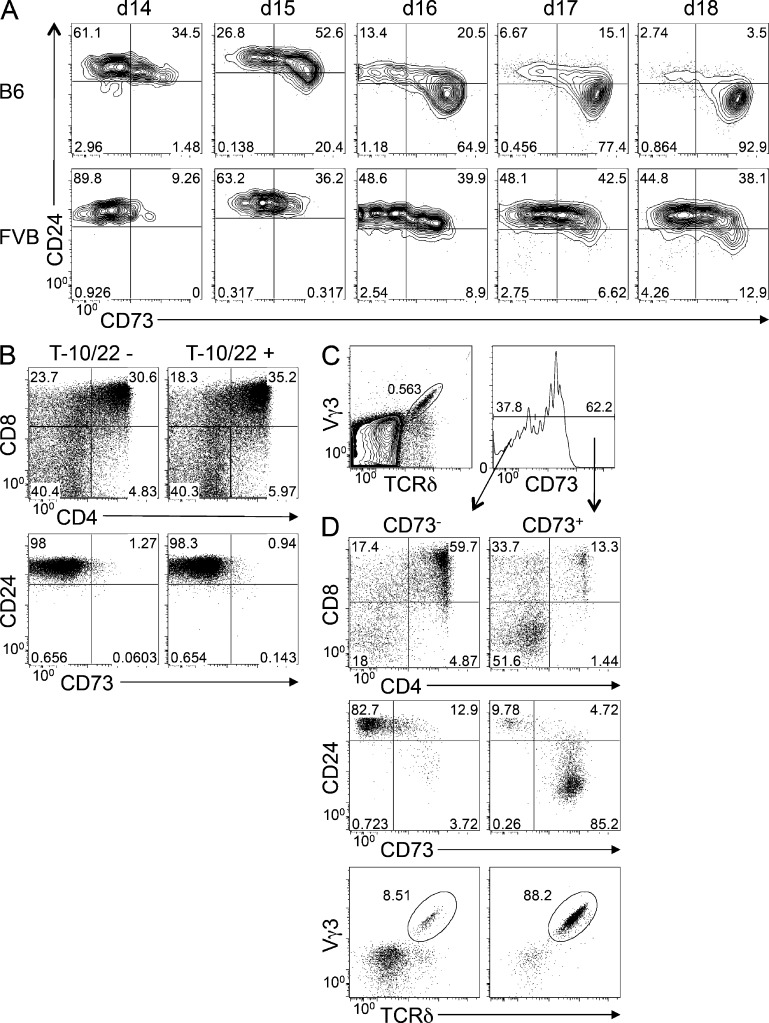
These data indicate that CD73 induction marks cells that have lost the ability to fate-switch to the αβ lineage. Thus, CD73 induction could mark the commitment process itself or be a maturation marker that precedes all other maturation markers identified to date. To address this possibility, we used FVB/N (FVB.Tac) mice in which a point mutation in the Skint1 gene blocks maturation of Vγ3+ (a.k.a., Vγ5+) DETC progenitors during development in fetal thymus, but not their commitment to the γδ lineage (Fig. 5 A; Lewis et al., 2006; Boyden et al., 2008). Daily analysis of Vγ3+ cells in fetal thymic organ culture (FTOC) revealed that CD73 induction in FVB.Tac Vγ3+ cells was delayed relative to that in Vγ3+ cells from C57BL/6 mice, but induction was observed in a substantial fraction of cells. The Vγ3+ cells from FVB.Tac mice fail to mature in the absence of Skint1 protein, as indicated by the failure to down-regulate CD24 (Fig. 5 A), consistent with previous studies (Lewis et al., 2006). Together, these findings suggest that CD73 is a marker of γδ lineage commitment, not a marker of maturation.
Figure 5.
CD73 expression identifies γδ lineage–committed Vγ3+ DETC progenitors. (A) The expression of CD73 and CD24 was evaluated by flow cytometry on Vγ3+ γδTCR-expressing progenitors from C57BL/6 and FVB.Tac (FVB) FTOCs on the indicated days. Gate frequencies of the indicated populations are indicated on the histograms. Data are representative of two experiments performed. (B) Rag2−/− DN3 fetal liver progenitors were retrovirally transduced with the DETC TCR (Vγ3Vδ1−IRES-YFP) and cultured on OP9-DL1 cells that lack or express T-10/22 for 7 d. Development was assessed by flow cytometry based on expression of CD4, CD8, CD24, and CD73. Frequencies of each of the indicated populations are listed on the histograms. Results are representative of 3 experiments performed. (C and D) CD73− and CD73+ Vγ3-expressing fetal thymic progenitors were isolated by flow cytometry (gating shown in top panels) from fetal thymi at embryonic day 16.5 and cultured on OP9-DL1 monolayers for 4 d. Developmental progression was evaluated by flow cytometry based on expression of CD4, CD8, CD24, and CD73, with gate frequencies listed on the histograms. In addition, gated DN cells were analyzed to determine the percentage of cells coexpressing Vγ3 and TCR-δ. Results represent at least 15 pooled embryos and 3 independent experiments.
Because a large fraction of the polyclonal adult γδTCR+ cells that we analyzed appeared to be stimulated by culture on OP9 cells (Fig. 4), we wished to assess the extent of fate switching among endogenous Vγ3+ DETC progenitors where agonist stimulation can be readily controlled. To determine whether OP9 cultures supported the development and maturation of DETC progenitors, we retrovirally transduced Rag2−/− fetal DN3 cells with the canonical DETC γδTCR and assessed development on OP9-DL1 monolayers either expressing or lacking the T-10/22 KN6 ligand. Unlike the progenitors transduced with KN6 γδTCR, which adopt the γδ fate and remain DN (Fig. 1 A), those expressing the DETC receptor did not induce CD73 or adopt the γδ fate; instead, they committed to the αβ fate and developed to the DP stage, irrespective of the presence of T-10/22 (Fig. 5 B). This confirms that OP9 cells do not express the DETC TCR agonist necessary for selection and adoption of the γδ fate, and that the OP9 model can be used to conduct studies with DETC progenitors under culture conditions devoid of additional agonist stimulation (Barbee et al., 2011). We sorted CD73− and CD73+ Vγ3-expressing thymocytes from day 16.5 C57BL/6 fetal thymuses and cultured them on OP9-DL1 cells (Fig. 5, C and D). Importantly, the majority of CD73+Vγ3+ progenitors remained DN, down-regulated CD24, and retained expression of the γδTCR, indicating that despite being separated from the intrathymic selecting environment, these cells remained committed to the γδ lineage (Fig. 5 D). Conversely, upon separation from the intrathymic selecting environment most CD73−Vγ3+ progenitors silenced the γδTCR, presumably expressed the preTCR complex, remained CD24high, and developed to the DP stage, indicating that they had not yet committed to the γδ fate and in fact switched fates to the αβ lineage (Fig. 5 D). Altogether, these data suggest that the induction of CD73 serves as a marker that distinguishes DN γδTCR+ progenitors that have committed to the γδ fate from those that have yet to do so. Moreover, these analyses suggest that a robust mechanism supporting fate switching exists in γδTCR-expressing progenitors that have not yet encountered conditions resulting in CD73 induction (i.e., ligand).
CD73 expression identifies functional γδ T cell subsets
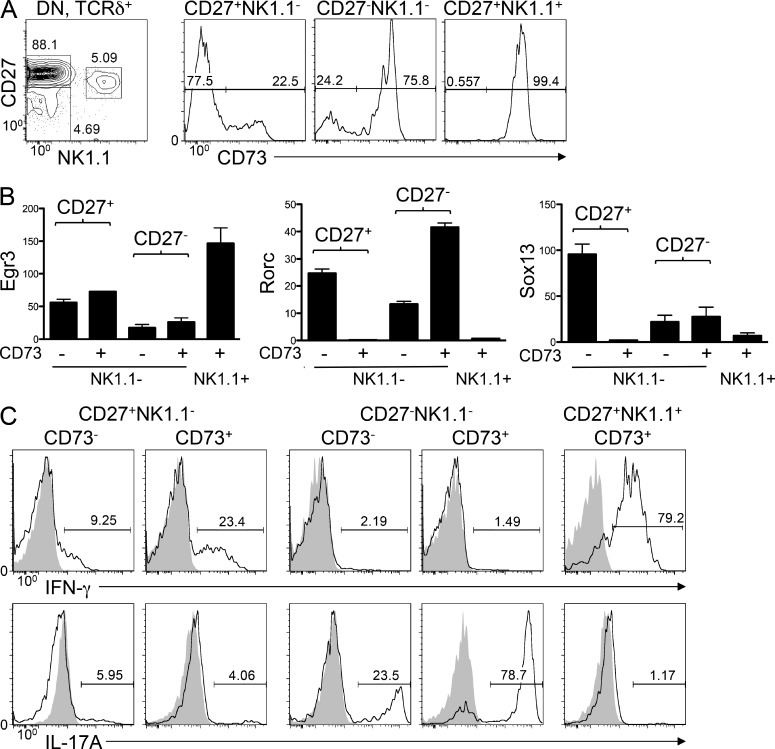
γδ T cell effector fate is largely programmed during development in the thymus (Bonneville et al., 2010; O’Brien and Born, 2010); however, the temporal relationship of specification of effector fate to lineage commitment remains unclear. Consequently, because CD73 appears to mark γδTCR-expressing cells that have adopted the γδ fate, we asked whether CD73 might also mark the acquisition of effector function. Ligand engagement has been reported to be responsible for the development of CD27+NK1.1− IFN-γ–producing γδ cells (Jensen et al., 2008; Ribot et al., 2009). Conversely, adoption of the IL-17–producing fate by CD27−NK1.1− progenitors has been suggested to occur in a ligand-independent manner (Jensen et al., 2008). Given that CD73 appears to be induced upon ligand engagement of the TCR, we used the gating schema proposed by the Hayday laboratory to determine whether CD73 expression distinguished IFN-γ–producing cells purported to undergo ligand-mediated selection (CD27+NK1.1− IFN-γ producers) from IL-17–producing γδ cells (CD27−NK1.1−) thought to develop in the absence of ligand engagement (Turchinovich and Hayday, 2011). Consistent with their purported dependence on ligand engagement, just over 20% of cells whose phenotype is associated with IFN-γ production (CD27+NK1.1−) expressed CD73 (Fig. 6 A). Surprisingly, despite reports that the IL-17–producing subset (CD27−NK1.1−) is thought to arise in the absence of ligand engagement, the vast majority of this subpopulation expressed CD73, a nominally TCR ligand-inducible molecule (Fig. 2 C; Chalmin et al., 2012). CD27+NK1.1+ innate-like γδ cells were uniformly CD73+ (Fig. 6 A), consistent with the observation that the PLZF transcription factor that characterizes their development can be induced by anti-TCR Ab stimulation (Kreslavsky et al., 2009).
Figure 6.
CD73-expressing γδ lineage cells are enriched for the ability to produce cytokines. (A) CD73 expression was measured by flow cytometry on the indicated DN subsets of adult γδTCR-expressing thymocytes. The left panel shows gating based on CD27 and NK1.1 expression and the frequencies of the gated populations. (B) The populations identified in A were isolated by flow cytometry, and the expression of Egr3, Rorc, and Sox13 was quantified by real-time PCR. Expression was normalized to β-actin and to the level observed in DN3 cells, and mean ± SD of triplicate measurements was depicted graphically. (C) Thymic γδ T cell populations identified in A were stimulated with 50 ng/ml PMA and 1 µg/ml ionomycin for 5 h, and the frequency of IL-17A and IFN-γ–producing cells was determined by flow cytometry using intracellular staining. Shaded histograms show analysis of unstimulated controls. The frequency of cytokine-producing cells is indicated on the histograms. Data shown are representative of three independent experiments.
A recent study indicated that Egr3 induction is linked to specification of the IFN-γ–producing effector fate, whereas Sox13 and RORγt were linked to IL-17 production (Turchinovich and Hayday, 2011). To determine whether expression of these genes segregated with CD73 expression, we measured their mRNA levels by real-time PCR (Fig. 6 B). Interestingly, upon induction of CD73 among CD27+NK1.1− IFN-γ–producing cells, expression of the genes linked to the alternate, IL-17–producing fate (Rorc and Sox13) was suppressed, whereas that of Egr3 was essentially unchanged (Fig. 6 B). Conversely, among IL-17–producing CD27−NK1.1− cells, Rorc was elevated in the CD73-expressing subset, whereas Egr3 and Sox13 remained low (Fig. 6 B). Because CD73 induction is associated with enrichment of transcription factors linked to their effector fate (e.g., Rorc in CD27− IL-17 producers) and a relative reduction of transcription factors required for the alternate fate, we reasoned that CD73 induction might also mark function. To investigate this possibility, we assessed the ability of CD73− and CD73+ subsets to produce cytokine upon stimulation with PMA and ionomycin (Fig. 6 C). Consistent with the changes in Rorc expression that accompany CD73 induction, roughly three times more CD73-expressing cells were competent to produce cytokine than cells in the CD73− fraction (Fig. 6 C). This was true for both IFN-γ and IL-17 production (Fig. 6 C). These data indicate that in addition to marking cells that have committed to the γδ fate, CD73 expression also seems to identify a cell population that is enriched for the ability to produce cytokine.
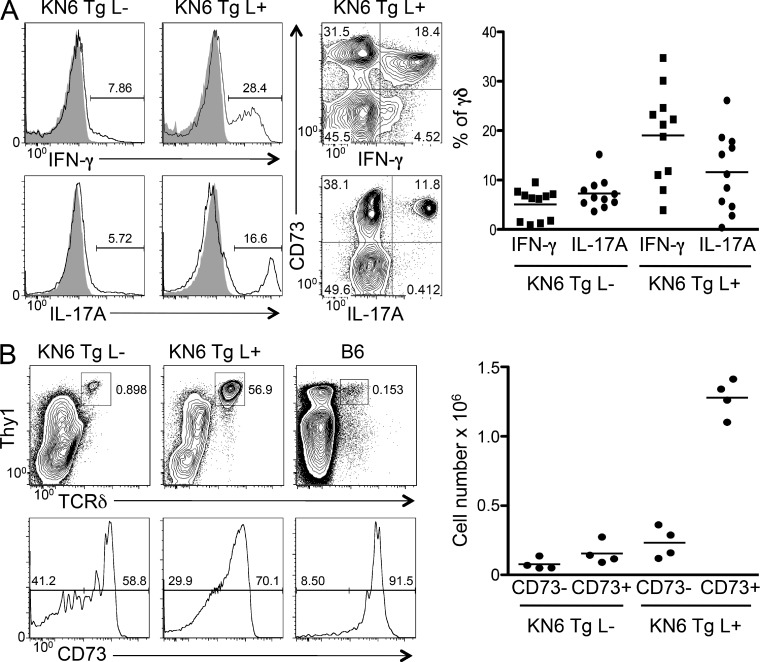
To further assess the role of TCR signaling in adoption of effector fate, we evaluated the production of cytokines by KN6 Tg thymocytes. We isolated DN thymocytes from ligand-sufficient (T-10d–expressing) and ligand-deficient (β2m−/−) KN6 Tg mice and stimulated these cells with PMA and ionomycin. Although few KN6 Tg thymocytes from β2m−/− mice were capable of producing either cytokine, ligand-sufficient mice were capable of producing both IFN-γ and IL-17, with IFN-γ production being more robust (Fig. 7 A). In addition, co-staining with CD73 revealed that the majority of cytokine-producing KN6 Tg thymocytes in ligand-expressing mice were also CD73+ (Fig. 7 A). Therefore, although previous studies have suggested that adoption of the IL-17–producing effector fate can occur in a ligand-independent manner, this did not appear to be true for KN6 γδTCR Tg thymocytes deprived of ligand. In addition, although the number of splenic γδ T cells in KN6 Tg β2m−/− was found to be greatly reduced, more than half of the cells that were able to mature and exit the thymus expressed high levels of CD73. The CD73high fraction was enriched for IFN-γ–producing cells, suggesting they may have been selected on the low levels of residual ligand present in β2m−/− mice, as previously reported (Haks et al., 2005). The requirement for ligand in determining both IFN-γ and IL-17 effector fates reveals that, in addition to TCR signals, other environmental factors likely influence specification of effector fate (Haas et al., 2012). Nevertheless, our finding that >90% of peripheral γδ lineage cells express CD73 (Fig. 7 B) suggests that ligand plays an extensive role in γδ lineage commitment and may also do so in the specification of effector fate.
Figure 7.
Ligand is required for acquisition of function by KN6 γδTCR Tg thymocytes. (A) Thymocytes from adult KN6 Tg L+ and KN6 Tg L− mice were stimulated with 50 ng/ml PMA and 1 mg/ml ionomycin for 5 h, and the production of IFN-γ and IL-17A was assessed by intracellular flow cytometry. Shaded histograms show analysis of unstimulated controls. Gate frequencies of the indicated populations are listed on the histograms. Middle panels show IFN-γ and IL-17A production by CD73+ and CD73− populations. The graph shows the percentage of cytokine-producing cells among total γδ T cells. Data from three independent experiments were combined, and each data point represents an individual mouse, with the horizontal bars indicating means. (B) Spleen cell suspensions from adult KN6 Tg L+, KN6 Tg L−, and C57BL/6 mice were analyzed by flow cytometry to assess the frequency of γδ T cells (top) and expression of CD73 on these cells (bottom). Gate frequencies of the indicated populations are listed on the histograms. Graph shows total numbers of CD73− and CD73+ γδ T cells in spleens from KN6 Tg L− and KN6 Tg L+ as calculated by flow cytometry. Points represent data from individual mice and horizontal bars indicate means.
CD73 expression identifies an intermediate stage of γδ effector specification
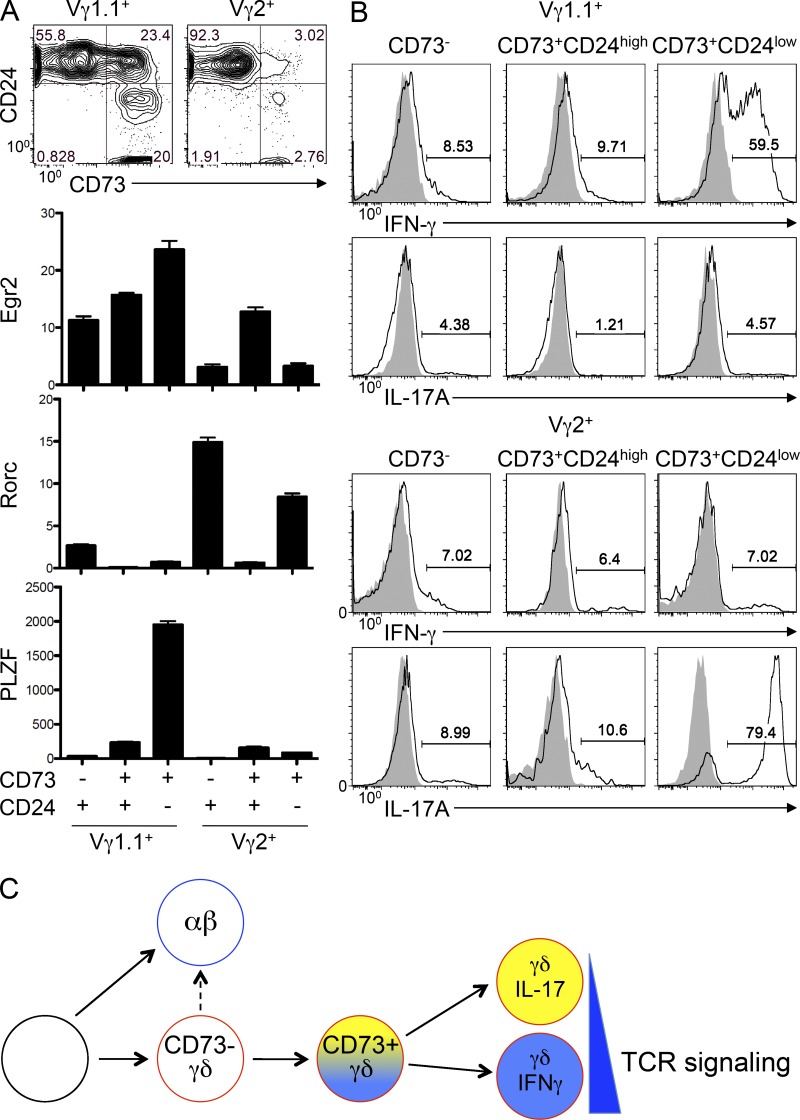
Because we found that CD73-expressing γδTCR+ cells had not only committed to the γδ fate but were also enriched for effector function, we wished to investigate the temporal relationship between lineage commitment and the specification of effector fate. γδ effector fate is often correlated with Vγ usage. Vγ2+ cells are often found to produce IL-17, whereas Vγ1+ cells produce either IFN-γ alone or adopt a PLZF-expressing innate fate characterized by simultaneous production of IFN-γ with IL-4 (O’Brien and Born, 2010). Moreover, recent analysis of gene expression in thymic γδ subsets demonstrated that gene signatures corresponding to the ultimate effector fates of Vγ subsets were already evident among immature CD24high populations, which the authors interpreted as evidence for predetermination of effector fate (Narayan et al., 2012). To address whether effector fate is predetermined and explore the relationship between lineage commitment and effector fate, we examined both the expression of effector fate-linked transcription factors and the functional competence of Vγ1.1+ and Vγ2+ progenitors subdivided based on CD73 and CD24 expression (Fig. 8 A, top). If an effector transcription signature were already evident in CD73−CD24high progenitors and preserved upon CD73 induction and lineage commitment, this would suggest that effector fate is preprogrammed. However, when we examined the expression of the genes linked to effector fate (Egr2 with IFN-γ, Rorc with IL-17, and PLZF with innate) in those Vγ subpopulations, we found they could be further subdivided based on CD73 expression (Fig. 8 A). We found that CD24+CD73− Vγ subpopulations were enriched in the transcription factors linked to their ultimate effector fate; however, the expression of those transcription factors was markedly altered upon CD73 induction, with Egr2 and PLZF being induced and Rorc being repressed in both Vγ1 and Vγ2 progenitors, despite linkage of these Vγ to distinct effector fates (Fig. 8 A). The discordant expression patterns of these transcription factors did resolve during maturation to the CD24low stage, where elevated levels of Egr2 and PLZF are expressed in Vγ1.1+ cells and Rorc is reexpressed in Vγ2+ cells. Importantly, the resolution of the transcription factor expression patterns is accompanied by the acquisition of effector function because the ability to produce IFN-γ or IL-17 was also largely restricted to CD24low Vγ1.1+ or Vγ2+ subsets, respectively (Fig. 8 B). The fact that the expression differences only became realigned with their effector fate upon maturation to the CD24low stage suggests that lineage commitment, as measured by CD73 induction, and effector fate may be specified sequentially. Collectively, these data suggest that in addition to serving as an indicator of commitment to the γδ fate, CD73 expression may also identify a novel, intermediate stage in the development of γδ thymocytes from which the ultimate effector fate is resolved (Fig. 8 C).
Figure 8.
CD73 expression marks an intermediate γδ population that has not yet acquired function and exhibits altered expression of effector fate genes. (A, Top) CD73 and CD24 expression was assessed by flow cytometry on Vγ1.1+ and Vγ2+ subsets of γδ T cells from adult thymus. Gate frequencies of the indicated populations are listed on the histograms. (A, Bottom) Subpopulations of Vγ1.1+ and Vγ2+ identified in A were isolated by cell sorting, and the expression of Egr2, Rorc, and PLZF were quantified by real-time PCR. Expression was normalized to β-actin and to the level observed in DN3 cells, and mean ± SD of triplicate measurements was depicted graphically. Results shown are representative of two independent experiments. (B) Vγ1.1+ and Vγ2+ populations identified in A were isolated by cell sorting and stimulated with 50 ng/ml PMA and 1 mg/ml ionomycin for 5 h, after which the percentage of cells that produced IL-17A or IFN-γ was determined by flow cytometry using intracellular staining. Shaded histograms show analysis of unstimulated controls. The frequencies of cytokine-producing cells are listed on the histograms. Results shown are representative of two independent experiments. (C) In the proposed model of sequential adoption of the γδ lineage and effector fate, CD73 expression distinguishes γδTCR+ progenitors that have committed to the γδ lineage from those that retain αβ lineage potential. CD73 induction on CD24high γδTCR+ cells also identifies an intermediate population in which effector fate specifying genes are remodeled (yellow/blue gradient). Expression of effector fate specifying genes resolves during maturation and loss of CD24 expression, which coincides with acquisition of effector function. These findings suggest that γδ lineage commitment and specification of effector fate are sequential, separable processes.
DISCUSSION
Although αβ and γδ lineage T cells are known to arise from a common progenitor in the thymus, the basis for specification of these lineages remains poorly understood, in part because of the inability to identify within the broader pool of γδTCR-expressing DN progenitors those that have committed to the γδ fate. Previously, the only means of determining whether progenitors had committed to the γδ fate was to culture them in vitro and assess their developmental potential. In this assay, γδ lineage–committed cells remain DN and retain γδTCR expression, whereas those not committed to the γδ lineage silence the γδTCR, switch to the αβ fate, and develop to the DP stage. We report here, in the KN6 γδ TCR Tg model where the selecting ligand is known, that TCR ligand engagement induces adoption of the γδ fate and does so in an instructional manner. This is accompanied by the TCR ligand-mediated induction of CD73, which is expressed by ∼25% of γδTCR+ DN progenitors, and is a marker of those progenitors that have committed to the γδ fate. Indeed, CD73 expression is induced under conditions that favor commitment to the γδ fate. Moreover, γδTCR+ progenitors that have induced CD73 can be separated from the selecting milieu and continue to be committed to the γδ fate, as indicated by remaining DN and retaining the γδTCR. Conversely, DN γδTCR+ progenitors that have not induced CD73 switch to the αβ fate upon separation from the selecting milieu, as indicated by silencing of the γδTCR and development to the DP stage. The use of CD73 as a marker of γδ lineage commitment has also enabled us to provide the first insights into the temporal relationship between γδ lineage commitment and specification of effector fate. Indeed, CD73 induction identifies a transitional CD24high stage after γδ lineage commitment but before acquisition of function, in which the linkage between fate-specifying transcription factors and effector fate is perturbed. This linkage is realigned upon maturation to the CD24low stage, which suggests that lineage commitment and the specification of effector fate are separable and occur sequentially.
The role of the γδTCR and preTCR complexes in αβ/γδ lineage commitment has long been debated. Reports from our laboratory, and others, demonstrated that neither strict instructional nor stochastic models provide an adequate explanation for the experimental data relating to αβ/γδ lineage commitment. Instead, these data provided strong support for a TCR signal strength model that now represents the prevailing view in the field (Haks et al., 2005; Hayes et al., 2005). Moreover, we demonstrated that ligand engagement was able to alter the fate of γδTCR-expressing DN progenitors, raising the possibility that ligand might be involved in regulating lineage commitment of polyclonal progenitors in vivo. Nevertheless, it remained unclear whether ligand was acting to instruct fate or to rescue viability of progenitors in which lineage fate had been predetermined. A recent study using Ab engagement as a surrogate for ligand indicated that a population of γδTCR-expressing cells that arose from a single cell adopted the αβ fate in the absence of Ab engagement and the γδ fate upon Ab stimulation (Kreslavsky et al., 2008), suggesting that Ab engagement was acting instructionally to dictate fate. Consistent with those findings, we used the KN6 γδTCR Tg model and single cell progenitor analysis to demonstrate that the T-10d ligand acted instructionally to promote adoption of the γδ fate. Nevertheless, in another study that used tetramer to identify T-10/22–reactive γδ cells, the indirect removal of ligand did not substantially reduce the number of T-10/22 binding γδ T cells in the periphery, leading the authors to conclude that ligand engagement is not involved in shaping the γδTCR repertoire, and thus is unlikely to play a role in selection (Jensen et al., 2008). We favor an alternative interpretation for three reasons. First, it is possible, if not likely, that γδ cells, like αβ T cells, are not selected on the same ligand responsible for their activation in the periphery (i.e., T-10/22). Specifically, selection of KN6 γδTCR Tg thymocytes is mediated by T-10d, which fails to activate in the periphery (Bonneville et al., 1989; Ito et al., 1990). Conversely, higher affinity T10b ligand activates KN6 cells in the periphery but causes deletion in the thymus (Pereira et al., 1992; Lauritsen et al., 2009). Second, T-10/22 tetramer binding γδ T cells have in common the utilization of the Dδ2 element, but their affinity for T-10/22 can vary by >10-fold (Adams et al., 2008), suggesting that members of the polyclonal T-10/22 binding progenitor pool are likely to exhibit a wide range of developmental outcomes upon encountering T-10/22 during development. Finally, our observation that CD73 is a γδTCR ligand-inducible molecule expressed by 25% of thymic progenitors and >90% of peripheral γδ T cells strongly suggests that ligand may be involved in development of a significant fraction of γδ progenitors.
CD73 is purportedly induced in response to antigen receptor signaling, in agreement with our findings; however, the molecular basis for control of CD73 expression remains poorly understood. Although CD73 is highly expressed in γδ lineage T cells, it has also been found on other murine lymphocyte populations, including αβ lineage CD4 and CD8 T cells, Th17 cells, and T reg T cells (Yamashita et al., 1998; Thompson et al., 2004; Deaglio et al., 2007; Chalmin et al., 2012). Recent analysis in Th17 cells suggests that the induction of CD73 in response to anti-TCR + CD28 stimulation is augmented by cytokines that signal through STAT3, including IL-6. TCR-induced CD73 expression was also augmented by TGF-β signaling, and this effect appeared to be mediated by the SMAD-dependent loss of the Gfi-1 transcriptional repressor (Chalmin et al., 2012). We have observed that induction of CD73 in developing γδ T cells is dependent on TCR–ligand interactions and requires signaling through the p56lck tyrosine kinase. Moreover, we have shown that KN6 γδTCR-expressing progenitors developing on Ligand− (T-10/22 shRNA-treated) OP9 cells failed to induce CD73, suggesting that the IL-7 signaling in these cultures is not sufficient to induce CD73 despite its ability to activate STAT3, providing additional support that CD73 induction requires TCR ligand engagement, and that cytokine signaling alone is insufficient to do so. Accordingly, if CD73 induction on γδ lineage progenitors always requires TCR–ligand interactions, then this has important implications for the control of γδ lineage commitment and the specification of effector fate.
Although CD73 is not exclusively expressed on γδ lineage cells, CD73 represents the first effective marker with which to distinguish immature DN γδTCR-expressing thymic progenitors that have committed to the γδ fate from those that have yet to do so. However, it remained unclear whether CD73 induction marked γδ lineage commitment itself or a subsequent, early stage in maturation. To address this question, we analyzed the development of Vγ3+ DETC progenitors from FVB.Tac mice, which are reported to commit to the γδ fate but fail to mature due to a mutation in Skint1 (Lewis et al., 2006; Boyden et al., 2008). We observed that CD73 was induced on ∼50% of FVB.Tac DETC progenitors, suggesting that CD73 does indeed mark lineage commitment and not maturation. Nevertheless, CD73 was induced on fewer DETCs from FVB.Tac mice than from B6 mice (∼90%). Because the precise role of Skint1 in promoting DETC development has not been established, the basis for this difference remains to be established. However, the decreased frequency in CD73-expressing DETC progenitors in FVB.Tac mice may reflect a difference in the background strain (FVB/N vs. C57BL/6) because Skint1 mRNA levels are far higher in C57BL/6 thymic stroma than in stroma from FVB (Barbee et al., 2011). Alternatively, because the number of DETC in the fetal thymus of Skint1 mutant FVB.Tac mice is reduced to about the same extent as is CD73 induction, it is likely that the Skint1 mutation also modestly impairs γδ lineage commitment (Lewis et al., 2006). Skint1 expression on thymic stroma is required for DETC selection (Barbee et al., 2011). Accordingly, the Skint1 mutation might adversely affect lineage commitment and CD73 induction by reducing the ability of Skint1 to function as a selecting ligand or may indirectly impair lineage commitment, perhaps by reducing the availability of intrathymic niches where DETC selection can occur. Efforts are in progress to determine how the absence of Skint1 might interfere with lineage commitment as well as maturation.
Although CD73+γδTCR+ progenitors remain committed to the γδ fate even after separation from the selecting milieu, CD73−γδTCR+ progenitors silence the γδTCR and switch to the αβ fate. The fate switching observed among CD73−γδTCR+ progenitors is quite robust, as under circumstances where agonist stimulation can be limited during culture in vitro (e.g., for DETC progenitors), the vast majority of CD73− cells switch fate and develop to the DP stage. Before developing to the DP stage, CD73−γδTCR+ DN thymocytes silence their γδTCRs. Two mechanisms have been proposed previously to explain the silencing of the γδTCR in αβ lineage DP thymocytes: cessation of TCR-γ expression by the transcriptional silencing element near the TCR-γ constant region (Ishida et al., 1990; Ferrero et al., 2006; Tani-ichi et al., 2011) and elimination of the TCR-δ locus by TCR-α rearrangement (Pennington et al., 2005). Interestingly, neither of these mechanisms appears to be responsible for silencing the γδTCR complex in CD73− progenitors because despite the loss of surface expression of the γδTCR complex, mRNA encoding both TCR-Cγ and TCR-Cδ continues to be expressed, whereas protein expression is absent, suggesting a posttranscriptional mechanism (Fig. 4). Efforts are in progress to establish the basis for silencing of the γδTCR by CD73− γδ progenitors, as well as to use CD73 induction to gain insight into the regulation of the γδ lineage commitment process.
Based both on published evidence and our own investigation, CD73 induction appears to require TCR–ligand interactions (Fig. 2; Chalmin et al., 2012), which has important implications for the role of ligand in γδ lineage commitment. The induction of CD73 appears to begin quite early among γδTCR-expressing progenitors, as it is first observed during maturation of the most immature γδTCR+ progenitors (γδTCR+CD27+CD25+) to their immediate CD27+CD25− progeny. At steady state, ∼25% of γδTCR+ thymocytes express CD73, indicating that a substantial proportion of γδTCR+ cells commit to the γδ fate in response to ligand engagement. This is likely to be an underestimate of the fraction of γδTCR+ thymocytes that encounter ligand in the thymus, as >90% of peripheral γδ T cells express CD73. Nevertheless, some CD73−γδTCR+ progenitors remained CD73− and DN upon removal from the selecting milieu, consistent with the notion that ligand-independent γδ lineage commitment may also be possible (Mahtani-Patching et al., 2011). Because so few γδTCR ligands are known, the identification of CD73 as a surrogate for ligand stimulation should facilitate progress in understanding how extensive a role ligand may play in γδ T cell development.
Although we have focused primarily on CD73 as a marker of lineage commitment, it is important to note that CD73 also functions to hydrolyze adenosine monophosphate to adenosine, which can have immunosuppressive properties (Ohtsuka et al., 2010). In fact, CD73 is expressed on several cell types including regulatory T cells and plays a role in their ability to repress immune responses (Deaglio et al., 2007). However, CD73 function appears to be dispensable for γδ T cell development because CD73-deficient mice have no apparent abnormalities in γδ T cell development (not depicted; Thompson et al., 2004). The effect of CD73 deficiency on γδ T cell function remains unclear.
Thymic γδ populations expressing CD73 are also enriched for those capable of producing cytokine. Indeed, the CD73+ populations of CD27+NK1.1− cells and CD27−NK1.1− cells contain a larger fraction of cells capable of producing IFN-γ and IL-17, respectively, than the corresponding CD73− populations (Fig. 6; Turchinovich and Hayday, 2011). The functional capability is linked to suppression of Sox13 and Rorc expression in CD73+ IFN-γ–producing cells and the gain of Rorc by CD73+ IL-17–producing cells (Fig. 6). Because we have found that CD73 is induced in γδTCR+ progenitors after ligand engagement, this suggests that the specification of effector fate follows γδTCR–ligand engagement, even in the IL-17–producing subset that has been purported to develop in an antigen-naive manner (Jensen et al., 2008). In support, we found that the acquisition of IL-17 and, to a greater extent, IFN-γ–producing capability by KN6 γδ T lineage progenitors was dependent on ligand engagement (Fig. 7).
Recent expression profiling analysis revealed that immature CD24+ Vγ subsets exhibited expression patterns of transcription factors portending their ultimate effector fate and concluded that effector fate was both pre-programmed and independent of influence of TCR signaling, perhaps resulting from the developmental timing of Vγ rearrangement. Specifically, Egr2 expression was elevated in immature Vγ1.1+ cells that usually produce IFN-γ, whereas Rorc was elevated in Vγ2+ cells that usually become IL-17 producers (Narayan et al., 2012). Nevertheless, by further subdividing CD24high immature Vγ1.1+ and Vγ2+ subsets based on CD73 expression, we came to different conclusions (Fig. 8). Consistent with the findings of Narayan et al. (2012), we found that CD73−CD24high Vγ subsets did exhibit elevated levels of transcription factors linked to effector fate. However, we found that CD73 induction among CD24high progenitors was accompanied by remodeling of the expression pattern of these transcription factors. Expression of Rorc (linked to IL-17 production) was decreased and Egr2 expression (linked to IFN-γ production) was increased, irrespective of the effector fate usually adopted by the Vγ subset. Expression of these transcription factors was then realigned with Vγ usage upon maturation into CD24low functionally competent effectors (Fig. 8). Based on these observations, we propose that although some predisposition may exist among CD24highCD73− Vγ subsets, the γδTCR–ligand engagement that induces CD73 and γδ lineage commitment remodels expression of the transcription factors that specify effector fate, severing the link with Vγ usage. Consequently, the CD24highCD73+ population appears to represent a developmental intermediate that is γδ lineage committed but has not yet acquired effector function. Moreover, we propose that specification of effector fate then occurs under the influence of a variety of factors, including signaling by Notch, the γδTCR, and cytokines. Under the influence of these factors, a subset of cells in this developmental intermediate is resolved into mature CD24low effectors in which the expression of transcription factors linked to effector fate has been realigned with Vγ usage. Accordingly, because CD73 induction appears to mark a metastable intermediate from which the effector fate is ultimately resolved, we suggest that γδ lineage commitment and effector fate are specified sequentially in separable processes. Therefore, unraveling the processes that control CD73 expression may lead to insights into additional signaling cascades and molecular effectors that specify γδ fate and effector function.
We have presented evidence suggesting that CD73 induction serves to mark those DN γδTCR+ progenitors that have adopted the γδ fate and entered an intermediate state from which effector fate is resolved. Because so few selecting ligands for γδ T cells are known, the manipulation of one of those ligands could reveal the role of that ligand in γδ T lineage commitment or adoption of effector fate by a particular γδ subset, but the question of whether those principles could be applied more generally to other γδ cells remained unanswered. We now show that CD73 is a TCR ligand-induced molecule that can serve as a surrogate indicator for ligand experience, irrespective of the identity of the ligand. Consequently, our observation that nearly all peripheral γδ T cells express CD73 suggests that most γδ T cells are ligand experienced. Likewise, a substantial fraction of IFN-γ–producing thymic γδ cells express CD73, in agreement with recent studies suggesting that their development requires ligand (Jensen et al., 2008). Surprisingly, however, CD73 is expressed by an even greater proportion of CD27−NK1.1− thymic γδ lineage cells that produce IL-17, raising the possibility that even IL-17–producing γδ cells, which are thought to develop in a ligand-independent manner (Jensen et al., 2008), may have encountered ligand. Accordingly, CD73 induction promises to be a useful tool with which to address some of the longstanding, unresolved questions regarding the way that γδ lineage fate is specified and how this relates to adoption of effector fate during development in the thymus.
MATERIALS AND METHODS
Mice.
All mice were maintained in Fox Chase Cancer Center’s AALAC-accredited animal colony and all procedures were approved by the Institutional Animal Care and Use Committee. KN6 Tg Rag2−/−, B2m−/−, Lck−/− mice have been previously described (Lauritsen et al., 2009). C57BL/6 mice were bred and maintained at the Fox Chase Cancer Center Animal Facility. For FTOC, timed pregnant FVB carrying a point mutation in Skint1 (FVB/NTac) and C57BL/6 were obtained from Taconic (Boyden et al., 2008).
Retroviral transduction and OP9 culture.
Viral particles were produced by transient calcium phosphate transfection of Phoenix cells. shRNAs targeting T-10/22 were expressed in the MSCV-based vector LMS, which was described previously (Dickins et al., 2005). The KN6 γδTCR was cloned into pMiY as a Tescovrius 2A linked fusion protein as previously described (Lauritsen et al., 2009). The DETC γδTCR-pMiY construct was a gift of W. Havran (Scripps Research Institute, La Jolla, CA). TCR-β–pMIG was produced as previously described (Ciofani et al., 2006). T-10d was amplified by PCR from cDNA generated from BALB/c splenocytes and cloned into pMiCherry. OP9-DL1 cells expressing T-10d (Ligand+) or in which low level expression of endogenous T-10/22 was knocked down (Ligand−) were produced by retroviral transduction with pMiCherry–T-10d or MLS-T-10 shRNA vectors, respectively. After 48 h, transduced cells were isolated by cell sorting (pMiCherry+ or GFP+), and T10 expression was quantified by real-time PCR. The altered levels of T10 in these cells were found to be stable. Fetal livers were harvested from Rag2−/− mice at day 14.5 of gestation and cell suspensions were centrifuged over a cushion of Lympholyte-M (Cedarlane Labs). Isolated cells were expanded on OP9-DL1 for 7 d in the presence of 5 ng/ml IL-7 and 5 ng/ml Flt3L (R&D Systems) and then harvested for spin infection using retroviral supernatant treated with 8 µg/ml polybrene (Sigma-Aldrich). Infected cells were cultured overnight, and YFP+ DN3 (Thy1.2+CD25+CD44−) cells were sorted the next day and cultured on OP9-DL1 for the duration of the experiment. For bulk cultures, cells were transferred to fresh monolayers every 4 d. For single-cell assays, progenitors were sorted directly into individual wells of 96-well plates containing 4,000 γ-irradiated OP9-DL1 cells (15 Gy). On day 5, half of the medium was replaced with fresh medium containing 10 ng/ml IL-7 and 10 ng/ml Flt3L.
Flow cytometry and intracellular TCR staining.
Abs against Thy1.2 (53–2.1), CD24 (M1/69), CD25 (PC61), CD44 (IM7), TCR-δ (GL3), TCR-β (H57-597), CD4 (GK1.5), CD8α (53–6.7), CD27 (LG.7F9), CD45RB (C363-16A), CD73 (Ty/11.8), Vγ2 (UC3-10A6), Vγ3 (536), and NK1.1 (PK136) were purchased from eBioscience, BD, or BioLegend. Anti-Vγ1.1 antibody was provided by R. O’Brien (National Jewish Medical Center, Denver, CO). Propidium iodide gating was included for dead cell exclusion. Data were collected on either a LSRII or FACSVantage SE (BD) and analyzed using FlowJo Software (Tree Star). For isolation of thymic γδ T cell populations, thymocytes were harvested from 6–7-wk-old C57BL/6 mice, depleted with anti-CD4 and anti-CD8 magnetic beads (Miltenyi Biotec), and isolated by cell sorting. A dump gate (NK1.1, B220, Ter11, Gr-1, and CD11b) was included. Intracellular TCR chains were detected by blocking surface TCR using unlabeled Ab, fixing for 10 min with 1% paraformaldehyde at room temperature, permeabilization with saponin and NP-40 detergents, and then staining with fluorochrome-conjugated anti–TCR-β and TCR-δ Ab.
FTOC.
Fetuses were harvested from timed-pregnant FvBN.Tac or C57BL/6 mice at day 14 of gestation, after which thymic lobes were cultured on Isopore membrane filters (Millipore) resting on Helistat collagen sponges (Integra Life Sciences) in Iscove’s medium containing 20% FCS as described previously (Haks et al., 2005). At the indicated time points, lobes were harvested and single cell suspensions were prepared for analysis by flow cytometry.
Analysis of cytokine production.
Sorted cell populations were stimulated with 1 µg/ml ionomycin and 50 ng/ml PMA for 5 h at 37°C in the presence of Brefeldin A (eBioscience). Cells were labeled with Live/Dead fixable violet dead cell stain kit (Invitrogen) and then fixed with 4% paraformaldehyde according to the manufacturer’s protocol. Cells were permeabilized and stained in Perm/Wash buffer (BD) using Ab against IFN-γ (XMG1.2) and IL-17A (TC11-18H10.1; BioLegend) for analysis by flow cytometry.
Quantitative real-time PCR.
Cell populations were purified by flow cytometry. RNA was extracted using the RNeasy kit according to manufacturer’s specifications (QIAGEN) and converted to cDNA using the Superscript II kit (Invitrogen) with oligo dT(12–18) primers (Invitrogen). Expression of the indicated genes was measured by real-time PCR using stock TaqMan primer/probe sets on an ABI Prism 7700 Real Time PCR machine (Applied Biosystems). β-Actin was measured using a custom primer/probe set generated by the Fox Chase Cancer Center Genomics Core Facility. β-Actin expression was used to normalize cycle thresholds, which were then converted into fold difference. All samples were analyzed in triplicates, and RT and nontemplate controls were added for each PCR reaction.
Acknowledgments
We thank Drs. R. O’Brien and W. Havran for reagents and gratefully acknowledge the assistance of the following core facilities of the Fox Chase Cancer Center: Cell Culture, DNA Sequencing, Flow Cytometry, Genomics, and Laboratory Animal.
D.L. Wiest was supported by National Institutes of Health (NIH) grants AI081314, AI073920, NIH core grant P01CA06927, Center grant #P30-DK-50306, and an appropriation from the Commonwealth of Pennsylvania. L.F. Thompson was supported by NIH grant AI018220. F. Coffey was supported by T32CA903536 and F32AI098241. S.-Y. Lee was supported by the Greenwald and Plain & Fancy Fellowships. J.C. Zúñiga-Pflücker is a Canada Research Chair in Developmental Immunology and is supported by the Canadian Institutes of Health Research (MOP-42387) and Krembil Foundation. L.F. Thompson holds the Putnam City Schools Distinguished Chair in Cancer Research. J.-P.H. Lauritsen is supported by the Novo Nordisk Foundation.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- DETC
- dendritic epidermal T cell
- DN
- double negative
- DP
- double positive
- FTOC
- fetal thymic organ culture
- Tg
- transgenic
References
- Adams E.J., Strop P., Shin S., Chien Y.H., Garcia K.C. 2008. An autonomous CDR3delta is sufficient for recognition of the nonclassical MHC class I molecules T10 and T22 by gammadelta T cells. Nat. Immunol. 9:777–784 10.1038/ni.1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbee S.D., Woodward M.J., Turchinovich G., Mention J.J., Lewis J.M., Boyden L.M., Lifton R.P., Tigelaar R., Hayday A.C. 2011. Skint-1 is a highly specific, unique selecting component for epidermal T cells. Proc. Natl. Acad. Sci. USA. 108:3330–3335 10.1073/pnas.1010890108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneville M., Ito K., Krecko E.G., Itohara S., Kappes D., Ishida I., Kanagawa O., Janeway C.A., Murphy D.B., Tonegawa S. 1989. Recognition of a self major histocompatibility complex TL region product by gamma delta T-cell receptors. Proc. Natl. Acad. Sci. USA. 86:5928–5932 10.1073/pnas.86.15.5928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneville M., O’Brien R.L., Born W.K. 2010. Gammadelta T cell effector functions: a blend of innate programming and acquired plasticity. Nat. Rev. Immunol. 10:467–478 10.1038/nri2781 [DOI] [PubMed] [Google Scholar]
- Born W.K., Reardon C.L., O’Brien R.L. 2006. The function of gammadelta T cells in innate immunity. Curr. Opin. Immunol. 18:31–38 10.1016/j.coi.2005.11.007 [DOI] [PubMed] [Google Scholar]
- Boyden L.M., Lewis J.M., Barbee S.D., Bas A., Girardi M., Hayday A.C., Tigelaar R.E., Lifton R.P. 2008. Skint1, the prototype of a newly identified immunoglobulin superfamily gene cluster, positively selects epidermal gammadelta T cells. Nat. Genet. 40:656–662 10.1038/ng.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno L., Fehling H.J., von Boehmer H. 1996. The alpha beta T cell receptor can replace the gamma delta receptor in the development of gamma delta lineage cells. Immunity. 5:343–352 10.1016/S1074-7613(00)80260-5 [DOI] [PubMed] [Google Scholar]
- Carding S.R., Egan P.J. 2002. Gammadelta T cells: functional plasticity and heterogeneity. Nat. Rev. Immunol. 2:336–345 10.1038/nri797 [DOI] [PubMed] [Google Scholar]
- Chalmin F., Mignot G., Bruchard M., Chevriaux A., Végran F., Hichami A., Ladoire S., Derangère V., Vincent J., Masson D., et al. 2012. Stat3 and Gfi-1 transcription factors control Th17 cell immunosuppressive activity via the regulation of ectonucleotidase expression. Immunity. 36:362–373 10.1016/j.immuni.2011.12.019 [DOI] [PubMed] [Google Scholar]
- Ciofani M., Knowles G.C., Wiest D.L., von Boehmer H., Zúñiga-Pflücker J.C. 2006. Stage-specific and differential notch dependency at the alphabeta and gammadelta T lineage bifurcation. Immunity. 25:105–116 10.1016/j.immuni.2006.05.010 [DOI] [PubMed] [Google Scholar]
- Deaglio S., Dwyer K.M., Gao W., Friedman D., Usheva A., Erat A., Chen J.F., Enjyoji K., Linden J., Oukka M., et al. 2007. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J. Exp. Med. 204:1257–1265 10.1084/jem.20062512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickins R.A., Hemann M.T., Zilfou J.T., Simpson D.R., Ibarra I., Hannon G.J., Lowe S.W. 2005. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat. Genet. 37:1289–1295 [DOI] [PubMed] [Google Scholar]
- Dudley E.C., Girardi M., Owen M.J., Hayday A.C. 1995. Alpha beta and gamma delta T cells can share a late common precursor. Curr. Biol. 5:659–669 10.1016/S0960-9822(95)00131-X [DOI] [PubMed] [Google Scholar]
- Ferrero I., Mancini S.J., Grosjean F., Wilson A., Otten L., MacDonald H.R. 2006. TCRgamma silencing during alphabeta T cell development depends upon pre-TCR-induced proliferation. J. Immunol. 177:6038–6043 [DOI] [PubMed] [Google Scholar]
- Gray E.E., Ramírez-Valle F., Xu Y., Wu S., Wu Z., Karjalainen K.E., Cyster J.G. 2013. Deficiency in IL-17-committed Vγ4(+) γδ T cells in a spontaneous Sox13-mutant CD45.1(+) congenic mouse substrain provides protection from dermatitis. Nat. Immunol. 14:584–592 10.1038/ni.2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas J.D., Ravens S., Düber S., Sandrock I., Oberdörfer L., Kashani E., Chennupati V., Föhse L., Naumann R., Weiss S., et al. 2012. Development of interleukin-17-producing γδ T cells is restricted to a functional embryonic wave. Immunity. 37:48–59 10.1016/j.immuni.2012.06.003 [DOI] [PubMed] [Google Scholar]
- Haks M.C., Lefebvre J.M., Lauritsen J.P., Carleton M., Rhodes M., Miyazaki T., Kappes D.J., Wiest D.L. 2005. Attenuation of gammadeltaTCR signaling efficiently diverts thymocytes to the alphabeta lineage. Immunity. 22:595–606 10.1016/j.immuni.2005.04.003 [DOI] [PubMed] [Google Scholar]
- Havran W.L., Allison J.P. 1988. Developmentally ordered appearance of thymocytes expressing different T-cell antigen receptors. Nature. 335:443–445 10.1038/335443a0 [DOI] [PubMed] [Google Scholar]
- Hayday A.C. 2000. [gamma][delta] cells: a right time and a right place for a conserved third way of protection. Annu. Rev. Immunol. 18:975–1026 10.1146/annurev.immunol.18.1.975 [DOI] [PubMed] [Google Scholar]
- Hayes S.M., Love P.E. 2006. Strength of signal: a fundamental mechanism for cell fate specification. Immunol. Rev. 209:170–175 10.1111/j.0105-2896.2006.00356.x [DOI] [PubMed] [Google Scholar]
- Hayes S.M., Li L., Love P.E. 2005. TCR signal strength influences alphabeta/gammadelta lineage fate. Immunity. 22:583–593 10.1016/j.immuni.2005.03.014 [DOI] [PubMed] [Google Scholar]
- Ishida I., Verbeek S., Bonneville M., Itohara S., Berns A., Tonegawa S. 1990. T-cell receptor gamma delta and gamma transgenic mice suggest a role of a gamma gene silencer in the generation of alpha beta T cells. Proc. Natl. Acad. Sci. USA. 87:3067–3071 10.1073/pnas.87.8.3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Van Kaer L., Bonneville M., Hsu S., Murphy D.B., Tonegawa S. 1990. Recognition of the product of a novel MHC TL region gene (27b) by a mouse gamma delta T cell receptor. Cell. 62:549–561 10.1016/0092-8674(90)90019-B [DOI] [PubMed] [Google Scholar]
- Jensen K.D., Su X., Shin S., Li L., Youssef S., Yamasaki S., Steinman L., Saito T., Locksley R.M., Davis M.M., et al. 2008. Thymic selection determines gammadelta T cell effector fate: antigen-naive cells make interleukin-17 and antigen-experienced cells make interferon gamma. Immunity. 29:90–100 10.1016/j.immuni.2008.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreslavsky T., von Boehmer H. 2010. gammadeltaTCR ligands and lineage commitment. Semin. Immunol. 22:214–221 10.1016/j.smim.2010.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreslavsky T., Garbe A.I., Krueger A., von Boehmer H. 2008. T cell receptor–instructed αβ versus γδ lineage commitment revealed by single-cell analysis. J. Exp. Med. 205:1173–1186 10.1084/jem.20072425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreslavsky T., Savage A.K., Hobbs R., Gounari F., Bronson R., Pereira P., Pandolfi P.P., Bendelac A., von Boehmer H. 2009. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of gammadelta T cells with restricted TCR diversity. Proc. Natl. Acad. Sci. USA. 106:12453–12458 10.1073/pnas.0903895106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreslavsky T., Gleimer M., von Boehmer H. 2010. Alphabeta versus gammadelta lineage choice at the first TCR-controlled checkpoint. Curr. Opin. Immunol. 22:185–192 10.1016/j.coi.2009.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacorazza H.D., Tucek-Szabo C., Vasović L.V., Remus K., Nikolich-Zugich J. 2001. Premature TCR alpha beta expression and signaling in early thymocytes impair thymocyte expansion and partially block their development. J. Immunol. 166:3184–3193 [DOI] [PubMed] [Google Scholar]
- Lauritsen J.P., Wong G.W., Lee S.Y., Lefebvre J.M., Ciofani M., Rhodes M., Kappes D.J., Zúñiga-Pflücker J.C., Wiest D.L. 2009. Marked induction of the helix-loop-helix protein Id3 promotes the gammadelta T cell fate and renders their functional maturation Notch independent. Immunity. 31:565–575 10.1016/j.immuni.2009.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.Y., Stadanlick J., Kappes D.J., Wiest D.L. 2010. Towards a molecular understanding of the differential signals regulating alphabeta/gammadelta T lineage choice. Semin. Immunol. 22:237–246 10.1016/j.smim.2010.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J.M., Girardi M., Roberts S.J., Barbee S.D., Hayday A.C., Tigelaar R.E. 2006. Selection of the cutaneous intraepithelial gammadelta+ T cell repertoire by a thymic stromal determinant. Nat. Immunol. 7:843–850 10.1038/ni1363 [DOI] [PubMed] [Google Scholar]
- Mahtani-Patching J., Neves J.F., Pang D.J., Stoenchev K.V., Aguirre-Blanco A.M., Silva-Santos B., Pennington D.J. 2011. PreTCR and TCRγδ signal initiation in thymocyte progenitors does not require domains implicated in receptor oligomerization. Sci. Signal. 4:ra47 10.1126/scisignal.2001765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra N., Narayan K., Cho O.H., Sylvia K.E., Yin C., Melichar H., Rashighi M., Lefebvre V., Harris J.E., Berg L.J., Kang J.; Immunological Genome Project Consortium 2013. A network of high-mobility group box transcription factors programs innate interleukin-17 production. Immunity. 38:681–693 10.1016/j.immuni.2013.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melichar H.J., Narayan K., Der S.D., Hiraoka Y., Gardiol N., Jeannet G., Held W., Chambers C.A., Kang J. 2007. Regulation of gammadelta versus alphabeta T lymphocyte differentiation by the transcription factor SOX13. Science. 315:230–233 10.1126/science.1135344 [DOI] [PubMed] [Google Scholar]
- Meyer C., Zeng X., Chien Y.H. 2010. Ligand recognition during thymic development and gammadelta T cell function specification. Semin. Immunol. 22:207–213 10.1016/j.smim.2010.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan K., Kang J. 2010. Disorderly conduct in gammadelta versus alphabeta T cell lineage commitment. Semin. Immunol. 22:222–227 10.1016/j.smim.2010.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan K., Sylvia K.E., Malhotra N., Yin C.C., Martens G., Vallerskog T., Kornfeld H., Xiong N., Cohen N.R., Brenner M.B., et al. Immunological Genome Project Consortium 2012. Intrathymic programming of effector fates in three molecularly distinct γδ T cell subtypes. Nat. Immunol. 13:511–518 10.1038/ni.2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien R.L., Born W.K. 2010. gammadelta T cell subsets: a link between TCR and function? Semin. Immunol. 22:193–198 10.1016/j.smim.2010.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka T., Changelian P.S., Bouïs D., Noon K., Harada H., Lama V.N., Pinsky D.J. 2010. Ecto-5′-nucleotidase (CD73) attenuates allograft airway rejection through adenosine 2A receptor stimulation. J. Immunol. 185:1321–1329 10.4049/jimmunol.0901847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passoni L., Hoffman E.S., Kim S., Crompton T., Pao W., Dong M.Q., Owen M.J., Hayday A.C. 1997. Intrathymic delta selection events in gammadelta cell development. Immunity. 7:83–95 10.1016/S1074-7613(00)80512-9 [DOI] [PubMed] [Google Scholar]
- Pennington D.J., Silva-Santos B., Shires J., Theodoridis E., Pollitt C., Wise E.L., Tigelaar R.E., Owen M.J., Hayday A.C. 2003. The inter-relatedness and interdependence of mouse T cell receptor gammadelta+ and alphabeta+ cells. Nat. Immunol. 4:991–998 10.1038/ni979 [DOI] [PubMed] [Google Scholar]
- Pennington D.J., Silva-Santos B., Hayday A.C. 2005. Gammadelta T cell development—having the strength to get there. Curr. Opin. Immunol. 17:108–115 10.1016/j.coi.2005.01.009 [DOI] [PubMed] [Google Scholar]
- Pereira P., Zijlstra M., McMaster J., Loring J.M., Jaenisch R., Tonegawa S. 1992. Blockade of transgenic gamma delta T cell development in beta 2-microglobulin deficient mice. EMBO J. 11:25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri H.T., Scollay R., Shortman K. 1992. Commitment to the T cell receptor-alpha beta or -gamma delta lineages can occur just prior to the onset of CD4 and CD8 expression among immature thymocytes. Eur. J. Immunol. 22:2185–2188 10.1002/eji.1830220836 [DOI] [PubMed] [Google Scholar]
- Ribot J.C., deBarros A., Pang D.J., Neves J.F., Peperzak V., Roberts S.J., Girardi M., Borst J., Hayday A.C., Pennington D.J., Silva-Santos B. 2009. CD27 is a thymic determinant of the balance between interferon-gamma- and interleukin 17-producing gammadelta T cell subsets. Nat. Immunol. 10:427–436 10.1038/ni.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt T.M., Zúñiga-Pflücker J.C. 2002. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 17:749–756 10.1016/S1074-7613(02)00474-0 [DOI] [PubMed] [Google Scholar]
- Silva-Santos B., Pennington D.J., Hayday A.C. 2005. Lymphotoxin-mediated regulation of gammadelta cell differentiation by alphabeta T cell progenitors. Science. 307:925–928 10.1126/science.1103978 [DOI] [PubMed] [Google Scholar]
- Tani-ichi S., Satake M., Ikuta K. 2011. The pre-TCR signal induces transcriptional silencing of the TCRγ locus by reducing the recruitment of STAT5 and Runx to transcriptional enhancers. Int. Immunol. 23:553–563 10.1093/intimm/dxr055 [DOI] [PubMed] [Google Scholar]
- Terrence K., Pavlovich C.P., Matechak E.O., Fowlkes B.J. 2000. Premature expression of T cell receptor (TCR)αβ suppresses TCRγδ gene rearrangement but permits development of γδ lineage T cells. J. Exp. Med. 192:537–548 10.1084/jem.192.4.537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson L.F., Eltzschig H.K., Ibla J.C., Van De Wiele C.J., Resta R., Morote-Garcia J.C., Colgan S.P. 2004. Crucial role for ecto-5′-nucleotidase (CD73) in vascular leakage during hypoxia. J. Exp. Med. 200:1395–1405 10.1084/jem.20040915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchinovich G., Hayday A.C. 2011. Skint-1 identifies a common molecular mechanism for the development of interferon-γ-secreting versus interleukin-17-secreting γδ T cells. Immunity. 35:59–68 10.1016/j.immuni.2011.04.018 [DOI] [PubMed] [Google Scholar]
- Vantourout P., Hayday A. 2013. Six-of-the-best: unique contributions of γδ T cells to immunology. Nat. Rev. Immunol. 13:88–100 10.1038/nri3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witherden D.A., Havran W.L. 2011. Molecular aspects of epithelial γδ T cell regulation. Trends Immunol. 32:265–271 10.1016/j.it.2011.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong G.W., Zúñiga-Pflücker J.C. 2010. gammadelta and alphabeta T cell lineage choice: resolution by a stronger sense of being. Semin. Immunol. 22:228–236 10.1016/j.smim.2010.04.005 [DOI] [PubMed] [Google Scholar]
- Yamashita Y., Hooker S.W., Jiang H., Laurent A.B., Resta R., Khare K., Coe A., Kincade P.W., Thompson L.F. 1998. CD73 expression and fyn-dependent signaling on murine lymphocytes. Eur. J. Immunol. 28:2981–2990 [DOI] [PubMed] [Google Scholar]