Monovalent engagement can trigger BCR signal transduction, and fine-tuning of BCR-ligand recognition can lead to B cell nonresponsiveness, activation, or inhibition.
Abstract
Valency requirements for B cell activation upon antigen encounter are poorly understood. OB1 transnuclear B cells express an IgG1 B cell receptor (BCR) specific for ovalbumin (OVA), the epitope of which can be mimicked using short synthetic peptides to allow antigen-specific engagement of the BCR. By altering length and valency of epitope-bearing synthetic peptides, we examined the properties of ligands required for optimal OB1 B cell activation. Monovalent engagement of the BCR with an epitope-bearing 17-mer synthetic peptide readily activated OB1 B cells. Dimers of the minimal peptide epitope oriented in an N to N configuration were more stimulatory than their C to C counterparts. Although shorter length correlated with less activation, a monomeric 8-mer peptide epitope behaved as a weak agonist that blocked responses to cell-bound peptide antigen, a blockade which could not be reversed by CD40 ligation. The 8-mer not only delivered a suboptimal signal, which blocked subsequent responses to OVA, anti-IgG, and anti-kappa, but also competed for binding with OVA. Our results show that fine-tuning of BCR-ligand recognition can lead to B cell nonresponsiveness, activation, or inhibition.
The B cell receptor (BCR) displays the dual function of sensing tonic signals for B cell survival at rest and of triggering B cell activation and differentiation into antibody-producing cells upon ligation with the appropriate antigen. The valency requirements for each of these functions remain incompletely understood. To achieve full B cell activation, the prevailing view holds that the BCR remains monomeric in resting B cells and clusters upon cross-linking only by a multivalent antigen (Woodruff et al., 1967). High-resolution live cell imaging has clarified our view of the BCR distribution in resting and activated B cells. Total internal reflection fluorescence microscopy suggests that most BCRs are apparently monomeric on the cell surface and diffuse freely, with a lesser percentage comprised of dimers and immobile oligomers; BCR engagement leads to BCR clustering (Tolar et al., 2005). Studies on the BCR complex reconstituted in insect cells provide an alternative view and indicate that BCRs are present as autoinhibited oligomeric complexes at rest; ligand binding then improves accessibility of immunoreceptor tyrosine-based activation motifs (ITAMs) and “opening” of these oligomers, culminating in B cell activation (Yang and Reth, 2010). Consistent with this theory, stochastic optical reconstruction microscopy (STORM) allowed identification of IgM and IgD clusters on resting B cells (Mattila et al., 2013). Diffusion of the BCR and signaling depend on the actin cytoskeleton; the actin-depolymerizing agents latrunculin A and cytochalasin D promoted BCR activation and diffusion even in the absence of antigen (Treanor et al., 2010). Thus, at rest, BCR diffusion is restricted, whereas upon antigen binding the BCR diffuses more rapidly, likely disaggregates, and disperses to help capture more antigen (Fleire et al., 2006). BCRs may then form caps, which lead to internalization and, ultimately, presentation of captured antigen on MHC class II molecules. Antigens that favor such BCR movement may indeed be best at attaining full B cell activation.
The aforementioned studies evaluated BCR dynamics but did not address the valency of the BCR-stimulating ligand. Indeed, the valency requirements for efficient BCR activation continue to be an underexplored aspect of B cell biology. Polyclonal activation of B cells is usually achieved using the F(ab)′2 portion of anti–mouse IgM, which targets constant regions of BCR rather than its antigen-binding site. Existing transgenic mice are directed to protein antigens such as hen egg lysozyme (HEL; Goodnow et al., 1988), DNA (Erikson et al., 1991), or hapten (Shih et al., 2002). Existing transgenic BCR models are ill-suited for valency studies because of the tendency of proteins to form aggregates in serum-containing medium and thus yield ligands of unknown valency. The repetitive nature of DNA and the requirement for a carrier protein or other polymer in the case of hapten-specific BCR complicate the use of correspondingly specific transgenic BCR models to address valency. Still, using anti-HEL BCR transgenic mice, monomeric HEL triggered BCR responses but was inefficient at inducing antigen presentation (Kim et al., 2006). Varying the number of 3-nitro-4-hydroxy-5-iodo-phenylacetate (NIP) hapten molecules in peptides demonstrated that low valency antigen could still activate B cell responses (Minguet et al., 2010). Thus, cross-linking of the BCR by multivalent antigen may not be strictly required to activate B cells.
To explore BCR activation of an antigen-specific B cell population, we generated mice by somatic cell nuclear transfer, using the nucleus of an OVA-specific B cell as donor. The resulting OB1 mice produce an OVA-specific IgG1 that recognizes OVA in both native and denatured forms. Short synthetic peptides mimic the epitope recognized by OB1 (Dougan et al., 2012). By controlling the size of the peptide epitope and its display as a monovalent, bivalent, or cell surface entity, we here examine the activation requirements of the IgG1 OB1 BCR. Monovalent engagement of this BCR suffices to activate OB1 B cells. Dimerization further enhances activation, with N-N orientation of the epitope being somewhat more favorable when compared with C-C. Reduction of peptide length is accompanied by a decrease in B cell activation. A suboptimal 8-mer peptide species prevents further responses to cell-bound antigen. Our data are also the first report of an antigen-specific response of primary B cells carrying a complete IgG1 BCR.
RESULTS
OVA-specific B cells are activated by OB1 peptide containing the epitope FGD
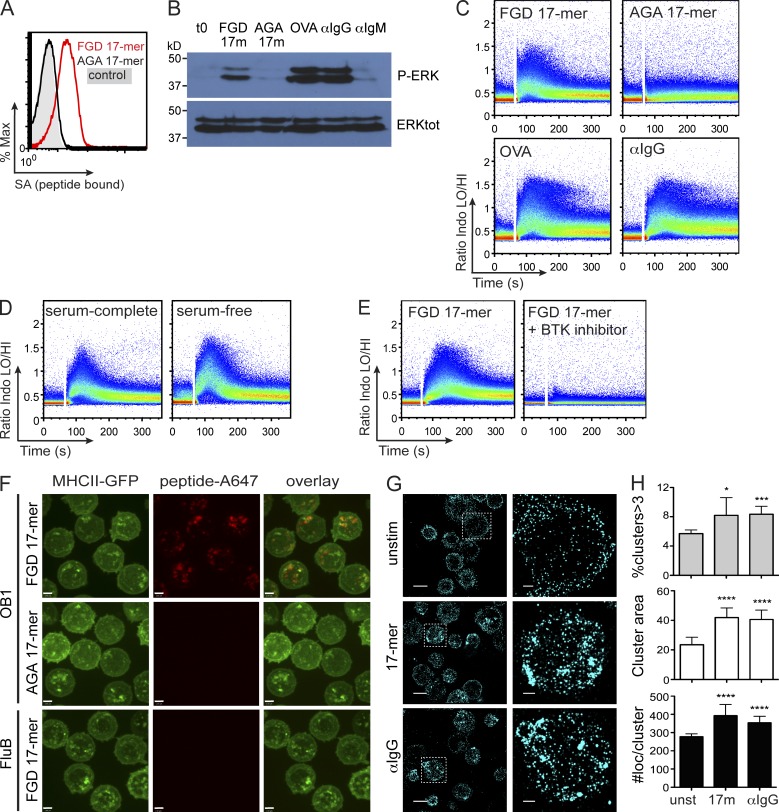
The binding of OB1 serum to a series of peptides that span the OVA sequence allowed identification of the epitope recognized by the OB1 BCR, centered on the peptide DKLPGFGDSI, where the FGD sequence is essential (Dougan et al., 2012). To further explore the requirement of this sequence for binding of OVA to the BCR, we generated a 17-residue biotinylated peptide that contains the core FGD sequence, FDKLPGFGDSIEAQGGK (FGD 17-mer) and tested binding to OB1 B cells from OB1 mice. We compared its binding with that of a 17-mer in which AGA replaced the FGD segment, FDKLPGAGASIEAQGGK (AGA 17-mer). OB1 B cells bound the FGD 17-mer but not the AGA 17-mer (Fig. 1 A). The FGD 17-mer but not the AGA 17-mer elicited robust phosphorylation of the MAP kinase ERK1/2, a downstream effector of BCR engagement (Fig. 1 B; Kurosaki et al., 2010). Accordingly, the FGD 17-mer but not the AGA 17-mer elicited a calcium (Ca2+) flux (Kurosaki et al., 2010) comparable with that elicited by OVA (Fig. 1 C). The 17-mer peptide is water-soluble and shows no signs of aggregation. Given its size and hydrophilicity, we considered oligomerization of the FGD 17-mer unlikely. Still, we tested whether inclusion of serum could lead to peptide aggregation. B cells stimulated with FGD 17-mer in serum-free medium responded comparably with those cultured in serum-containing medium (Fig. 1 D). Ca2+ flux evoked by the FGD 17-mer was blocked by an inhibitor of Bruton’s tyrosine kinase (Btk), required for the initial steps of BCR signaling (Fig. 1 E; Mohamed et al., 2009). To determine the path followed by FGD 17-mer upon binding to the BCR, we prepared Alexa Fluor 647–conjugated versions of FGD and AGA 17-mer and added them to OB1-MHC class II GFP B cells for 10 min. We removed unbound material by washing and determined the extent of antigen internalization by confocal microscopy. FGD 17-mer was internalized and localized to MHC II GFP+ compartments; as expected, we did not detect any AGA 17-mer inside cells (Fig. 1 F), arguing against pinocytosis as the means of peptide acquisition. To further ensure that peptide internalization required the BCR and was not attributable to pinocytosis, we incubated FGD 17-mer with FluB1 MHC II GFP transnuclear B cells, which are specific for hemagglutinin (HA; Dougan et al., 2013) and thus should not internalize FGD 17-mer, unrelated in sequence to flu HA. The FGD 17-mer did not accumulate in FluB1 cells (Fig. 1 F), thus confirming the requirement of BCR specificity for internalization.
Figure 1.
OVA-specific OB1 B cells are activated by peptide containing the epitope FGD. (A) OB1 B cells were incubated on ice in the absence (control) or presence of biotinylated versions of 0.5 µM FGD 17-mer and AGA 17-mer peptides. Binding was detected by flow cytometry using SA-APC. (B) OB1 B cells were stimulated with equimolar (0.5 µM) FGD 17-mer, AGA 17-mer, or OVA or 5 µg/ml anti-IgG1 or anti-IgM for 3 min, and then lysates were run in SDS-PAGE and immunoblotted with anti–phospho-ERK (top) or anti–total ERK (bottom). (C) OB1 B cells were loaded with Ca2+-sensitive Indo-1 AM dye and collected for 1 min (unstimulated baseline). After that, 5 µM FGD 17-mer, AGA 17-mer, or OVA or 5 µg/ml anti-IgG1 was added and collected for five additional minutes. (D) OB1 B cells were resuspended in serum-free or serum-complete RPMI, and Ca2+ trace was collected as in C upon incubation with 0.5 µM FGD 17-mer. (E) OB1 B cells loaded with Indo-1 AM dye were preincubated with 50 nM Btk inhibitor for 30 min at 37°C, and Ca2+ trace was collected upon stimulation with 0.5 µM FGD 17-mer. (F) Render of z-stacks of confocal microscopy of OB1 MHC II GFP B cells that were incubated for 10 min with Alexa Fluor 647–labeled FGD or AGA 17-mer or FluB1 MHC II B cells stimulated with Alexa Fluor 647–labeled FGD 17-mer. (G) OB1 B cells were either left unstimulated or stimulated with 17-mer or anti-IgG for 15 min in the presence of 100 µM dynasore at 37°C and stained with anti–mouse IgG antibody conjugated with Alexa Fluor 405 and Alexa Fluor 647 for STORM imaging. Representative fields (left) and single-cell images from selected box regions (right) are shown. Bars: (F) 2 µm; (G, left) 5 µm; (G, right) 1 µm. (H) The percentage of clusters that have more than three BCRs (top), the cluster size (middle), and the number of localizations within clusters (bottom) between unstimulated, 17-mer, and anti-IgG conditions were quantified, and means and SDs are plotted. Unpaired Student’s t test was performed comparing the total number of clusters contained in unstimulated versus the 17-mer or the anti-IgG condition. The number of cells analyzed was 67 (unstimulated), 102 (17-mer), and 118 (anti-IgG). P values of resulting analysis are shown: *, P = 0.024; ***, P = 0.0004; ****, P < 0.0001. Results shown are representative of two (A, G, and H) and at least three independent experiments (B–F).
Only multivalent antigens can cross-link the BCR molecules. Aggregation of the BCR subsequent to initial engagement may require the involvement of cytoskeletal elements, but not necessarily cross-linking of the BCR by exposure to monovalent antigen. Based on its size, amino acid composition, and behavior as a monomer in solution, we did not expect the 17-mer to elicit BCR cross-linking in the manner of a multivalent ligand. To determine whether 17-mer could promote BCR aggregation, we performed STORM experiments, with a lateral resolution of ∼20–30 nm (Rust et al., 2006; Huang et al., 2010) to directly examine the distribution of the BCR. To ensure that the BCR would remain confined to the cell surface, we incubated OB1 B cells with dynasore to prevent BCR internalization, a rapid process (Chaturvedi et al., 2011). OB1 B cells were either left unstimulated or treated with 17-mer or anti-IgG (positive control to evoke BCR aggregation) in the presence of dynasore for 15 min, fixed, and stained with Alexa Fluor 405– and Alexa Fluor 647–conjugated anti–mouse IgG to visualize the BCR and imaged by STORM. Anti-IgG treatment readily evoked a strong aggregation of BCR at the cell surface (Fig. 1 G), as reflected by a significant increase of the percentage of clusters with more than three BCRs (as defined by more than three antibody localizations used to detect the BCR), a larger cluster size, and with higher number of localizations per cluster (Fig. 1 H). The 17-mer promoted BCR clustering to a level comparable with that elicited by anti-IgG (Fig. 1, G and H). The low initial percentage of clusters with more than three BCRs (∼5%) suggests that in OB1 B cells most surface BCR behaves as a monomer. Thus, a monomeric 17-mer peptide can signal and trigger clustering of the BCR.
Shorter peptides containing the FGD sequence bind to OB1 B cells but fail to elicit a response
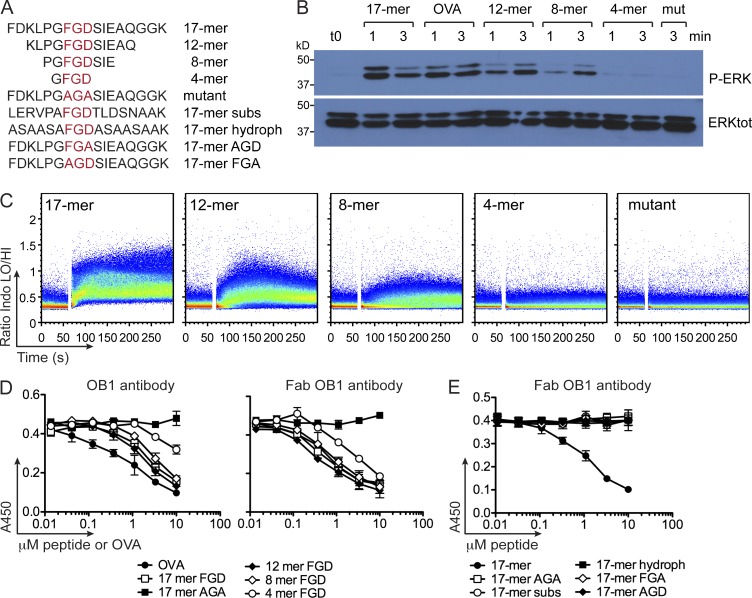
To determine the extent to which a response required the FGD-flanking residues, we synthesized a nested set of peptides centered on the FGD sequence, from the benchmark 17-mer down to a 4-residue peptide (Fig. 2 A). We then recorded phosphorylation of ERK1/2. Shorter peptides caused delayed ERK1/2 phosphorylation, the 4-mer being inactive (Fig. 2 B). As expected, the AGA 17-mer did not stimulate ERK1/2 phosphorylation. We observed a similar correlation between peptide length and the ability to elicit a Ca2+ flux (Fig. 2 C). Thus, amino acids flanking the FGD motif must be present to efficiently trigger a response.
Figure 2.
Shorter peptides containing the FGD sequence bind to OB1 B cells but fail to elicit a response. (A) Sequence of peptides nested on the FGD epitope bearing 17, 12, 8, and 4 amino acids, the mutant (mut) AGA 17-mer, and 17-mer peptides where the flanking residues were either substituted by same-polarity amino acids (17-mer subs), by hydrophobic amino acids with the exception of four serines (17-mer hydroph), or 17-mer peptides in which the core FGD was mutated to AGD or FGA. (B) OB1 B cells were stimulated with 0.5 µM of each FGD 17-mer, OVA, 12-, 8-, and 4-mer peptides for 1 and 3 min, and AGA 17-mer (mut) for 3 min, and then lysates were run in SDS-PAGE and immunoblotted with anti–phospho-ERK (top) or anti–total ERK (bottom). (C) OB1 B cells were loaded with Indo-1 AM, stimulated with 0.5 µM FGD 17-, 12-, 8-, and 4-mer and 17-mer AGA (mut), and Ca2+ flux traces were collected. (D) ELISA plates were coated with 0.1 µM OVA, and then 50 ng/ml of whole OB1 antibody (left) or 500 ng/ml OB1 antibody Fab (right) plus increasing concentrations of FGD 17-, 12-, 8-, and 4-mer, AGA 17-mer mutant peptide, or OVA were added. Plates were developed with anti-IgG1 HRP (left) or anti-Fab HRP (right). (E) Competitive ELISA was performed as in D but adding increasing concentrations of 17-mer subs, hydroph, AGD, or FGA, according to the sequences in A. Results are representative of three (B and C) independent experiments. In D and E, mean + SEM of two (whole antibody) and three (Fab) independent experiments is shown.
To examine whether the diminishing responses to progressively shorter peptides were caused by deficiencies in binding to OB1 IgG1, we used OB1 B cells to generate hybridomas and purified the OB1 antibody from hybridoma supernatant. To compare binding of nested peptides to that of OVA, we constructed a competitive ELISA assay using plate-bound OVA and added a nonsaturating amount of OB1 antibody, together with a range of concentrations of OVA and peptides as competitors. OVA and the longer peptides competed efficiently for binding of OB1 to the OVA coat, but as the peptide was shortened, its ability to compete for binding was hindered (Fig. 2 D, left). We prepared Fab fragments from OB1 antibody by digestion with immobilized papain. All FGD-containing peptides competed for binding, with only the 4-mer peptide somewhat less efficient at doing so (Fig. 2 D, right). Thus, the residues flanking the FGD sequence appear more important for binding to the whole antibody and less so for binding to Fab.
We next explored the possible contribution of the residues flanking the FGD sequence to binding and activation. We generated peptides that retained the FGD core sequence but either with the adjacent residues altered to amino acids of similar polarity (17-mer substituted) or exchanged for alanine for all but four residues (17-mer hydrophobic). We also made single amino acid replacements in the FGD motif (17-mer FGA and AGD mutants; Fig. 2 A). None of these peptides bound to the OB1 Fab fragment (Fig. 2 E) nor did they activate B cells (not depicted). Thus, optimal binding and activation of OB1 B cells by 17-mer requires the presence of both the FGD core and its adjacent residues.
Peptide dimerization and N-N orientation favor enhanced B cell stimulation
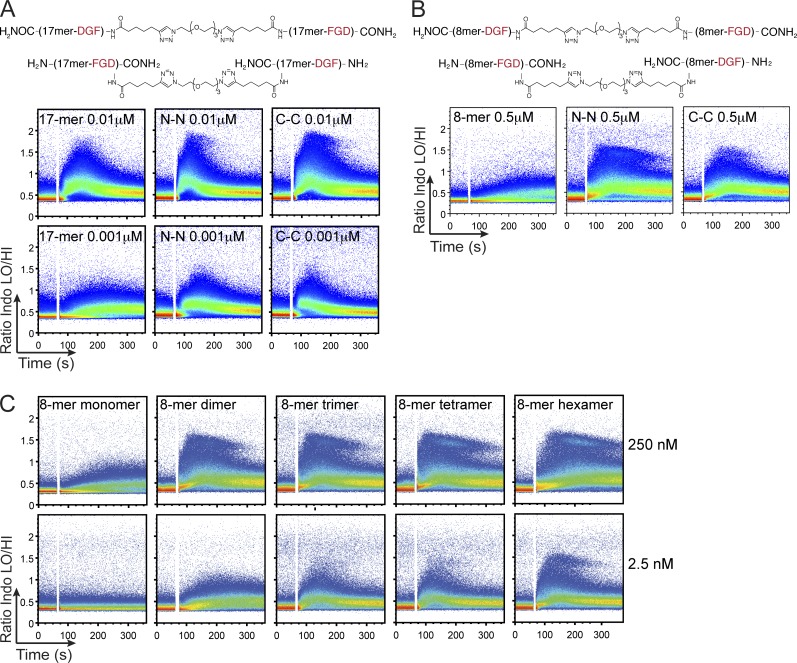
To bring BCR molecules in close proximity, the epitope in dimeric versions of antigen may interact with BCR in a preferred orientation (head to head, tail to tail, or head to tail). To define which epitope orientation best favors BCR activation, we created dimeric ligands of defined topology by synthetizing peptides connected in head to head (N-N) or tail to tail (C-C) fashion (Fig. 3, A and B, top) by means of a click reaction. The two peptides remain separated by a flexible linker, which in fully extended form is calculated to span a distance of ∼3 nm (∼30 Å). Regardless of the FGD orientation in these dimers, their stimulatory capacity as recorded by Ca2+ flux was comparable with, if not higher, than that recorded for 17-mer peptide alone at a concentration of 0.01 µM (Fig. 3 A, middle). When the concentration of 17-mer was brought down to suboptimal levels (0.001 µM), we could see the enhancement in stimulation by 17-mer dimerization with no apparent difference between the N-N and C-C orientation (Fig. 3 A, bottom). Because the FGD 17-mer peptide was already strongly stimulatory, we instead focused our attention on the FGD 8-mer peptide to more rigorously test the most favorable epitope orientation required for stimulation. The FGD 8-mer at 0.5 µM as monomer was poor in its ability to stimulate ERK1/2 phosphorylation and Ca2+ flux (Fig. 2, B and C). We generated N-N and C-C dimers for the FGD 8-mer peptides, which induced responses at a concentration of 0.5 µM, at which FGD 8-mer monomer was unable to do so (Fig. 3 B, bottom). Dimerization thus improves BCR responses, and the N-N orientation appears more favorable when compared with the C-C orientation for the FGD 8-mer. We next synthesized progressively longer peptides containing two, three, four, or six repeats of the FGD 8-mer in N-C configuration. As the number of epitopes increased, so did the ability to flux Ca2+ at a faster rate and to higher levels (Fig. 3 C). Thus, increasing the number of epitopes correlates with better responses, and this must be caused by enhancement of BCR cross-linking. Our results also show a subpopulation of stronger responders that becomes more prominent as the number of epitopes increases. This population was enriched in marginal zone B cells (not depicted), but the identity of these cells remains to be elucidated.
Figure 3.
Increased valency and N-N orientation favor enhanced B cell stimulation. (A and B) FGD 17-mer (A) or FGD 8-mer (B) peptides were linked on an N-N (top structure) or C-C (bottom structure) orientation by click chemistry. Monomers or N-N or C-C dimers for FGD 17-mer at 0.01 and 0.001 µM (A) or FGD 8-mer at 0.5 µM (B) were added to OB1 B cells loaded with Indo-1 AM, and Ca2+ flux traces were collected. (C) Peptides carrying one, two, three, four, and six copies of FGD 8-mer (monomer, dimer, trimer, tetramer, and hexamer, respectively) at 250 nM (top) or 2.5 nM (bottom) were added to OB1 B cells that were loaded with Indo-1 AM, and Ca2+ flux traces were collected. Results are representative of two (A and C) and three (B) independent experiments.
Suboptimal stimulation of BCR leads to suppression of response to antigen
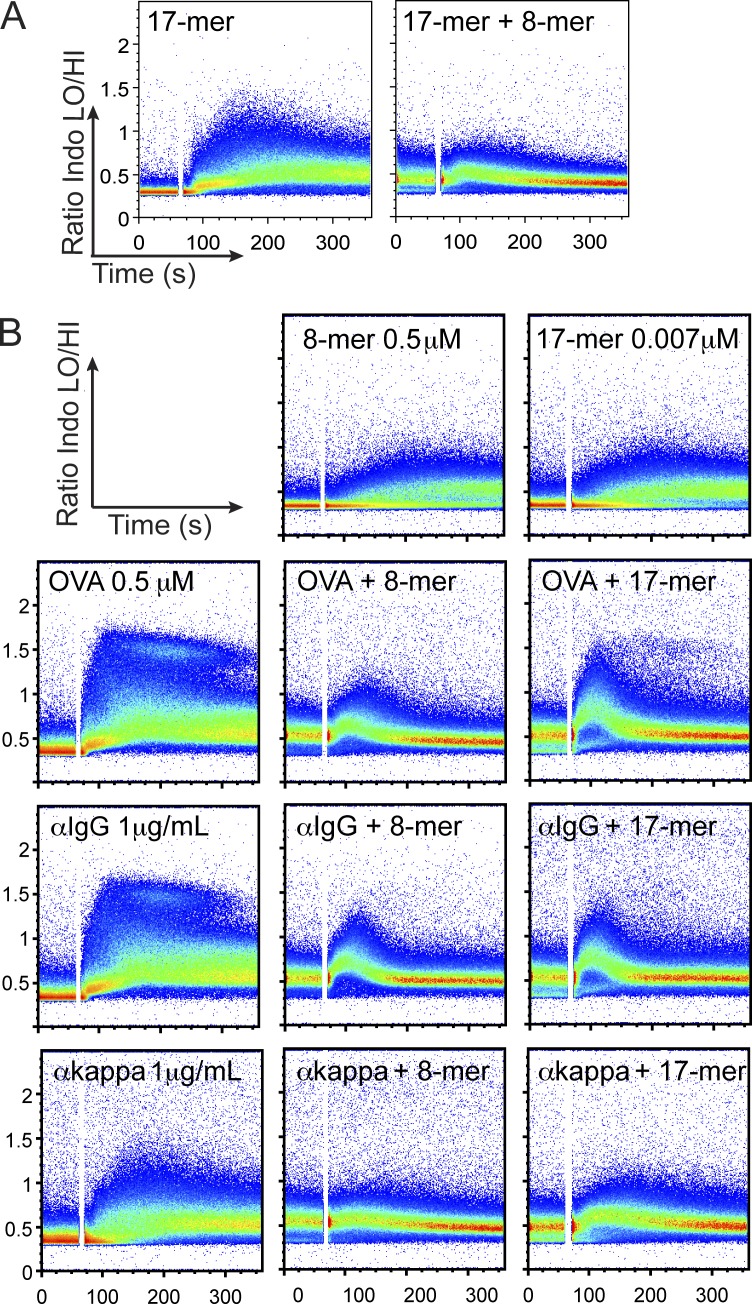
The OB1 antibody and Fab fragment bound the FGD 8-mer (8-mer from now on) and FGD 17-mer (17-mer from now on) peptide equally well, yet the 8-mer triggered a suboptimal Ca2+ response and failed to elicit optimal ERK phosphorylation (Fig. 2). To explore the consequences of this mode of signaling, we asked whether preincubation with 8-mer would affect subsequent responses to 17-mer. Upon incubation with 0.5 µM 8-mer, OB1 B cells generated a sustained low-grade Ca2+ flux that prevented a subsequent Ca2+ spike in response to equimolar concentrations of 17-mer (Fig. 4 A). The 8-mer interfered with stimulation by OVA at equimolar concentrations and also in response to anti-IgG and anti-kappa stimulation, albeit to a lesser extent (Fig. 2 B). The suppression of responses to anti-IgG1 and anti-kappa were surprising because these antibodies are directed to conserved portions of the BCR and not to the antigen-binding site. We asked whether suppression was caused by the suboptimal signal evoked by 8-mer. If this were the case, then presumably any ligand that triggers a comparable signal should inhibit subsequent responses to antigen. Thus, we lowered the concentration of 17-mer to a concentration that elicited a comparable signal with that of the 8-mer at 0.5 µM; this concentration was 70-times less at 0.007 µM for 17-mer. Low concentrations (0.007 µM) of 17-mer still blocked responses to OVA, anti-IgG, or anti-kappa, almost as effectively as did the 8-mer (Fig. 4 B). Preincubation with AGA 17-mer did not affect subsequent responses to equimolar FGD 17-mer (not depicted). Thus, when the BCR is stimulated suboptimally, it cannot then be efficiently triggered by optimal ligand.
Figure 4.
Peptides that bind but do not elicit a response block activation by antigen or cross-linking agents. (A) OB1 B cells were loaded with Indo-1 AM and incubated for 5 min with 0.5 µM FGD 8-mer or media before addition of equimolar FGD 17-mer. (B, top two panels) OB1 B cells were incubated with 8-mer at 0.5 µM or 17-mer at 0.007 µM. (bottom nine panels) OB1 B cells were preincubated with 8-mer as stated in A (middle) or with 17-mer at 0.007 µM (right) for 6 min before addition of 0.5 µM OVA (top), anti-IgG at 1 µg/ml (middle), or anti-kappa at 1 µg/ml (bottom), and Ca2+ flux traces were collected. Results are representative of three independent experiments, except for anti-kappa (two).
Computational modeling reveals different behavior of 17-mer, 8-mer, and OVA despite comparable Kd
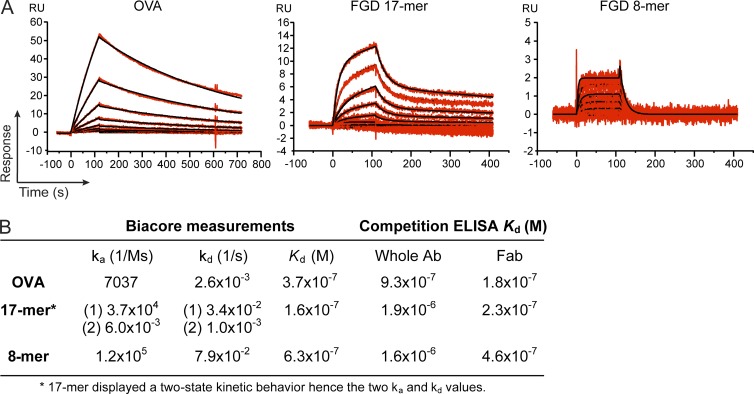
Our ELISA results showed that the 8-mer and 17-mer competed equally for OVA binding to the OB1 antibody, yet the 8-mer blocked responses to equimolar 17-mer, while being incapable of inducing a robust Ca2+ response when added on its own. These observations stressed the need for a more accurate measurement of the kinetic parameters of antigen binding. To arrive at estimates for the parameters that characterize the kinetics of association and dissociation (ka and kd), we immobilized OB1 antibody on a sensor chip for surface plasmon resonance (SPR) measurements, using intact OVA, 17-mer, and 8-mer as analytes over a range of concentrations. The behavior of these ligands differed markedly in their on and off rates, despite having rather similar values for affinity, all in the order of 10−7 M (Fig. 5, A and B): OVA showed a slow on and slow off rate, and 8-mer displayed a very fast on and very fast off rate. 17-mer shows clearly biphasic binding kinetics with an initial rapid on rate, followed by a slower on rate and a rapid off rate, followed by slower dissociation. The behavior of both OVA and 8-mer as analytes could be fitted using a 1:1 mathematical model (one class of homogeneous binding sites) to yield the values for ka and kd (Fig. 5 B). However, the behavior of 17-mer is inconsistent with simple binding to a homogeneous class of binding sites. The biphasic association and dissociation kinetics of 17-mer is consistent with a model in which two-state binding related to a conformational change occurs. The values obtained by SPR were comparable with the apparent Kd calculated by our competition ELISA based on the Orosz and Ovádi method (Fig. 5 B; Orosz and Ovádi, 2002).
Figure 5.
FGD 17-mer, FGD 8-mer, and OVA share similar Kd but different binding behavior. (A) CM5 chips coated with OB1 antibody were subject to Biacore measurements using OVA (left), FGD 17-mer (middle), and FGD 8-mer (right) peptides as analytes, and binding was recorded as relative units (RU) at different concentrations of analyte. Data recorded are shown in red, and the model prediction is shown in black. (B) Kinetic parameters calculated by Biacore experiments and competitive ELISAs.
It was puzzling that the 8-mer was unable to fully activate, yet blocked responses to the 17-mer and OVA despite comparable Kd values. We therefore calculated BCR occupancy by these three ligands using a monovalent model for SPR and Fab ELISA data (Stone et al., 2011) and a bivalent model for intact antibody ELISA data. At 0.5 µM (concentration of 8-mer used for Ca2+ flux inhibition experiments), 8-mer is estimated to occupy 44% of BCR molecules, and for 17-mer and OVA the values are 76% and 57%, respectively, with a similar trend for Fab ELISA (8-mer = 52%, 17-mer = 68%, and OVA = 73%). Using a bivalent model, results were also comparable (8-mer occupies 38%, 17-mer 34%, an OVA 52% of BCRs). Thus, BCR occupancy does not explain the suboptimal stimulation mediated by 8-mer.
To explain the inability of 8-mer to activate the BCR, we examined the kinetic behavior of 8-mer, 17-mer, and OVA in more detail using computational modeling. 17-mer and OVA lead to enhanced Ca2+ flux compared with 8-mer, indicating that the off rate of 8-mer is such that the Src family kinase Lyn phosphorylated the ITAMs in Igα and Igβ suboptimally (Fig. 6 A, inset). This would be the case if the time scale for Lyn engagement, as set by the concentration of Lyn times the on rate for Lyn/BCR engagement, is greater than the dwell time of the 8-mer but shorter than that of 17-mer and OVA. To further explore this hypothesis, we performed stochastic computer simulations of the earliest events in B cell signaling using the Gillespie algorithm (Gillespie, 1977). Our simulation included only the earliest downstream events involving the Src family kinase Lyn and the phosphatase SHP-1. Our read-out of signaling was ITAM phosphorylation. We performed 1,000 simulation runs with the same initial configurations for systems in which the B cell is exposed to OVA, 17-mer, and 8-mer (represented by the kinetics of their binding to BCR) and recorded the mean fraction of doubly phosphorylated BCR ITAMs at various time points.
Figure 6.
Computational modeling of OVA, 17-mer, and 8-mer binding and activation. (A) The fraction of doubly phosphorylated ITAM versus time for FGD 8-mer, 17-mer, and OVA from stochastic simulations: (left) for the case when and all peptide concentrations are the same; inset shows the level of phosphorylated ITAMs in Igα in response to 17-mer, 8-mer, or OVA; (middle) for the case when and all peptide concentrations are the same; (right) for the case when and 17-mer is 8% of the OVA and 8-mer concentration. (B) Contour plots showing the probability of 8-mer rebinding as a function of 8-mer diffusion coefficient (y axis) and 17-mer concentration (x axis).
When the value of (the rate for Lyn engagement at a single BCR) is greater than the off rate for FGD 8-mer, all three ligands elicit similar signaling patterns of doubly phosphorylated BCR (Fig. 6 A, left). However, when we decrease the value of such that is smaller than the off rate of 8-mer but larger than the off rates of 17-mer or OVA, the results for the three ligands separate (Fig. 6 A, middle). The ligand that yields the most doubly phosphorylated ITAM is the 17-mer because it has the slowest off rate. The ligand that elicits the least doubly phosphorylated ITAM is the 8-mer because it has the fastest off rate. If a threshold level of doubly phosphorylated ITAMs is required to propagate downstream signals, the 17-mer and OVA but not the 8-mer will induce signaling. This result is simply a reflection of kinetic proofreading, which applies only when Because we used the same ligand concentration in all of our simulations, 17-mer appears to dominate OVA in terms of generating doubly phosphorylated ITAMs. However, the initial fast off rate of 17-mer during the biphasic dissociation recorded by SPR suggests that there may be two populations of 17-mer bound to BCR, one that is bound more strongly than the other. This would result in the effective concentration of signaling-competent 17-mers to be less than that for OVA. When we use a lower effective ligand concentration for 17-mer, it has a simulated phosphorylation curve comparable with that elicited by OVA (Fig. 6 A, right). In light of these calculations, our experimental data suggests that indeed and that BCRs may be exposed to lower effective concentration of 17-mer.
Because we observed that 8-mer was better at blocking responses to OVA when compared with a low concentration of 17-mer, we sought to model whether the kinetics of binding of 8-mer could explain this behavior. Several studies (Bell, 1978; Aleksic et al., 2010; Govern et al., 2010; Abel et al., 2012; Tubo et al., 2013) have noted the importance of ligand rebinding when the on rate of receptor-ligand binding is high. We hypothesized that, upon preincubation with 8-mer, its high on rate may lead to many rebinding events before OVA or 17-mer could bind to the BCR. To estimate the probability of rebinding of 8-mer upon disengaging from the receptor, we took into account two mechanisms for rebinding. One possibility is that an 8-mer that has just dissociated from a BCR can rebind immediately before it diffuses over a distance commensurate with a characteristic protein molecular length scale of 100 Å. The second possibility is that, after dissociation, the ligand can make one or many diffusive excursions away from the BCR but returns and ultimately rebinds, with none of the diffusive excursions exceeding a threshold distance. To examine whether this process can occur before a 17-mer can bind, the threshold distance is taken to be the expected distance to the nearest 17-mer, which we compute assuming a uniform concentration of 17-mers (corresponding to experimental conditions). Once the 8-mer has diffused as far away as the nearest 17-mer, we can treat the two as effectively well mixed, with both now being on equal footing vis-à-vis binding. We computed the probability of 8-mer rebinding as a function of both its diffusion coefficient and 17-mer concentration in one (1D), two (2D), and three dimensions (3D). We carry out calculations as a function of dimensionality because the B cell surface is not a perfectly smooth sphere, but rather has protrusions and indentations that yield closely apposed surfaces that may restrict antigen diffusion. Our calculations show that decreasing the dimensionality of the system from 3D to 2D to 1D results in a significantly enhanced probability of rebinding (Fig. 6 B). Because 8-mer rebinding out-competes the 17-mer, it is likely that rebinding of 8-mers interferes with binding of 17-mers. The effective dimensionality of the B cell surface thus is lower than 3D because the distance between opposing membrane surfaces can be shorter than the expected distance to the nearest 17-mer. Thus, rebinding effects can be significant, given the fast on rate of the 8-mer and may explain why 8-mer blocks OVA stimulation better than a low concentration of 17-mer.
Our calculations, along with the results of these three experiments (8-mer inhibition of 17-mer, 8-mer vs. 17-mer inhibition of OVA, and 8-mer vs. 17-mer inhibition of anti-IgG and anti-kappa), suggest that a combination of effects are at play when preincubation is inhibitory. First, preincubation with a ligand that leads to suboptimal signaling results in a refractory effect or exhaustion, thus limiting signaling upon restimulation with another antigen. This effect is clearly reflected in the experimental results using anti-IgG and anti-kappa, which show that suboptimal binding alone is enough to inhibit subsequent signals (Fig. 4 B), perhaps as the result of local depletion of ATP in the vicinity of the BCR. Second, competition for ligand binding to the antigen-binding site constitutes an additional form of inhibition. 8-mer at 0.5 µM shows a better inhibitory effect when compared with a low concentration (0.007 µM) of 17-mer upon addition of OVA (Fig. 4 B), and this is possibly caused by the 8-mer’s fast rebinding kinetics as our calculations show (Fig. 6 B). Finally, preincubation of 8-mer before addition of 17-mer shows that despite the off rate of 8-mer being much higher than that of 17-mer, the very rapid binding kinetics allow for multiple rebinding events that can inhibit a 17-mer despite its longer dwell time. This is a result of both exhaustive effects and direct competition at the binding site.
Binding by 8-mer renders B cells nonresponsive to cell-bound 17-mer in the presence of CD40 ligation
Because 17-mer and OVA promoted comparable early BCR signaling events, we then asked whether later events in B cell activation by 17-mer and OVA could be similar also. The 17-mer induced up-regulation of the co-stimulatory molecule CD86, but to a lesser extent than that elicited by OVA and anti-IgG1 (Fig. 7 A, left). To test whether 8-mer could block CD86 up-regulation, we preincubated OB1 B cells with 8-mer before addition of equimolar concentrations of 17-mer or OVA or a saturating amount of anti-IgG1. Preincubation with 8-mer (Fig. 7 A, middle) but not mutant (AGA) 17-mer (Fig. 7 A, right) modestly decreased CD86 up-regulation by 17-mer but not by OVA or anti-IgG1. In vivo, antigen presentation on cell surface by macrophages and follicular dendritic cells appears as a more efficient means of inducing BCR responses when compared with soluble antigen (Batista et al., 2001; Batista and Harwood, 2009). Because OB1 B cells showed modest CD86 responses to soluble 17-mer, we explored whether presentation of 17-mer in a cell-bound form would be more efficient and also whether 8-mer could prevent responses to 17-mer presented on the cell surface. In the process, we sought to establish whether co-stimulatory signals such as CD40 ligation on B cells could reverse the blockade produced by incubation with the 8-mer. We took advantage of a sortase-catalyzed transacylation reaction, which allows ligation in flexible linkage of a substrate that contains N-terminal (Gly)n, such as a suitably (Gly)n-modified FGD 17-mer peptide, to a target protein that carries the sequence LPXTG at its C terminus. This method has been used for the modification of type II membrane proteins such as human CD40L (Popp et al., 2007). To determine whether the 8-mer could block both stimulation by cell-bound 17-mer and the contribution of CD40 ligation in a cell-based system, we created a Madin-Darby canine kidney (MDCK) cell line that stably expresses mouse CD40L modified with the sortase recognition sequence LPETG, followed by a HA tag. HA-tagged CD40L is present in transduced MDCK cells but not in control cells (Fig. 7 B). We synthesized a 17-mer extended at its N terminus with GGG and performed a sortase reaction using the MDCK CD40L-LPETG line. A successful reaction covalently ligates the GGG-extended 17-mer to surface-exposed CD40L (CD40L-peptide) with concomitant loss of the HA tag located downstream of the LPETG sequence. Upon incubation with sortase and peptide substrate, levels of surface CD40L were unchanged, whereas the percentage of HA-labeled cells decreased (Fig. 7 C, left and middle), indicative of a successful transacylation reaction. Peptide linkage to CD40L was monitored by using a biotinylated version of the GGG-17-mer, detected upon completion of the reaction by streptavidin (SA)-HRP (Fig. 7 C, right) and by Western blot (Fig. 7 D). Upon co-culture of OB1 B cells with such modified MDCK cells, which carry both CD40L and the 17-mer, ligation of CD40 on B cells and 17-mer peptide engagement of BCR should up-regulate CD86 on B cells (Fig. 7 E). Untransduced cells failed to up-regulate CD86 in the absence or presence of 17-mer alone or with sortase (Fig. 7 F, left). As expected, MDCK cells transduced with CD40L were able to up-regulate CD86 on B cells in the absence of peptide, indicating that CD40L was engaging CD40 on B cells (Fig. 7, F [right, blue histogram] and G). Upon covalent conjugation of peptide to CD40L on MDCK cells, OB1 B cells expressed higher levels of CD86 than OB1 B cells co-cultured with MDCK cells in the absence of covalently conjugated FGD 17-mer (Fig. 7, F [right, black and red histograms] and G). In this setting, preincubation of OB1 B cells with 8-mer for 2 h precluded responses to CD40L-linked 17-mer (Fig. 7, F [right, gray histogram] and G). Thus, preincubation with 8-mer blocks responses to cell-bound FGD 17-mer, even in the presence of the CD40L co-stimulatory signal.
Figure 7.
FGD 8-mer renders B cells unresponsive to cell-bound antigen with engaged CD40L-CD40. (A) OB1 B cells were preincubated with 5 µM FGD 8-mer or mutant AGA 17-mer for 2 h before addition of 5 µM FGD 17-mer, OVA, or 1 µg/ml anti-IgG1 and incubated for 18 h. Up-regulation of CD86 was detected by flow cytometry using anti–CD86-APC. (B) MDCK cells were stably transduced or not with CD40L-LPETG-HA and stained with anti-CD40L and anti-HA. Cells expressing both CD40L and HA were FACS-sorted and cultured in DMEM in the presence of 250 mg/ml hygromycin B. (C) Untransduced or CD40L-LPETG-HA–expressing MDCK cells were incubated with 200 µM FGD 17-mer peptide coupled to GGG and biotin in the absence or presence of 100 µM sortase A for 30 min on ice. After that, cells were washed three times with PBS, detached with 8 mM EDTA, and FACS-stained. (left) Total CD40L levels. (middle) HA levels. (right) Incorporated biotinylated peptide detected with SA. (D) Untransduced (UT) and CD40L-transduced MDCK cells (CD40L-tag) were incubated with peptide alone or in the presence of sortase A as in C, and cells were lysed and analyzed by immunoblotting for covalent incorporation of biotinylated peptide to CD40L by SA conjugated to HRP. (E) Representative scheme showing stimulation of B cells by cross-linked FGD 17-mer peptide on CD40L. (F) Sortase reaction on untransduced or CD40L-carrying MDCK cells was performed as in C. After washing, untransduced or CD40L-LPETG-HA–expressing MDCK cells were co-cultured for 18–20 h with OB1 B cells that were untreated or previously incubated with 10 µM FGD 8-mer for 2 h. Histograms show CD86 levels on B cells at the indicated conditions. Dashed lines on histograms delimit CD86 negative/low and positive/high populations. (G) Graph shows the percentage of B cells expressing high levels of CD86 under different treatments. Results shown are representative of three (A, B, and F), two (C and D), and the mean + SD of two (G) independent experiments.
DISCUSSION
OVA has a long history as a model antigen to study adaptive immune responses, especially for CD4 and CD8 T cells, but comparatively little is known about OVA-specific B cells. The OVA-specific OB1 mouse is currently the only animal model with monoclonal primary IgG1 B cells that recognize a sequence-defined epitope on an intact protein, OVA. Equally important, this epitope can be displayed functionally as a short synthetic peptide. Using peptides of defined composition as ligands, we explored valency requirements for BCR engagement. Our data support the ability of monovalent antigen to elicit a response in two ways. First, we showed that 17-mer was stimulatory in the absence of serum. Second, suboptimal stimulation by the 8-mer was vastly improved by dimerization, suggesting indeed that the 8-mer was present as a monomer in solution. Dimerization and multimerization of antigen increase the population of B cells able to flux Ca2+, with N-N dimers somewhat more favorable than C-C dimers. By estimating the dimensions of Fab as 65 × 35 Å (Green, 1969), the volume of the 8-mer at ∼1,000 Å3 and the linker between dimers at ∼30 Å when fully extended, we can now model how peptide orientation may favor more or less extensive cross-linking of the BCR. Thus, epitope dimerization with an N-N orientation contributes to better activation, as more BCR molecules are brought in close proximity, a feature further enhanced by increasing copy number of the epitope through linear extension.
By changing the length of the peptide, we varied the quality of the antigenic stimulus and discerned three qualitatively different responses, depending on signal strength and binding site of ligand. Both intact OVA and the FGD 17-mer elicit a robust response through engagement via the BCR’s antigen-binding site, which leads to BCR aggregation. The FGD 8-mer triggers a blunted, yet sustained response that fails to recruit active kinases to the threshold concentration required to achieve an optimal response (Fig. 6 A, inset). Cross-linking of the BCR by an anti-Ig reagent, which binds sites on the constant regions of IgG, produces a strong response by bringing BCRs in close proximity through cross-linking. Although the qualities of early signals evoked by the 17-mer and OVA were comparable, the 17-mer caused less robust up-regulation of CD86 when compared with OVA, a likely result of the allowed overall conformations of these ligands and their binding kinetics. Thus, ligands that can achieve B cell activation must therefore either be monomeric antigens that engage the antigen-binding site and satisfy a defined signaling threshold or ligands that can bring together (cross-link) BCR molecules, either through their constant or variable regions. We believe this model explains the results obtained with types of antigens that activate B cells but that do not fit the accepted multivalent antigen structure (Kim et al., 2006; Minguet et al., 2010) and the inability of monomeric Fab anti-IgM to activate B cells (Woodruff et al., 1967).
Our findings that the 8-mer could prevent early responses to 17-mer or OVA suggested that the 8-mer serves as a weak agonist that renders the B cell temporarily nonresponsive. The quality of the signal delivered through the BCR determined this suppression. Thus, antigens that can bind and signal may temporarily incapacitate the BCR to respond to further antigen challenge until a B cell fate decision is made. This blockade extended not only to bonafide antigens, but also to treatment with anti-IgG and anti-kappa as cross-linking agents, suggesting that the reduction in the response may be caused by a conformational change in BCR induced by 8-mer or by a feedback inhibitory mechanism perhaps under the agency of phosphatases that act downstream of the BCR. The low-grade yet persistent Ca2+ flux elicited by 8-mer, still observed after 1 h of stimulation, is consistent with observations of increased basal intracellular Ca2+ in anergic B cells from the HEL and Ars/A1 models (Cambier et al., 2007). In the Ars model, blockade required constant receptor occupancy (Gauld et al., 2005). In our hands, the inhibitory effect elicited by the 8-mer also required its continuous presence; B cells preincubated with 8-mer regained responsiveness to the 17-mer after washing cells in a timeframe consistent with dissociation of the 8-mer (unpublished data). Moreover, the 8-mer could still block responses to B cells receiving antigen presented in a cell-bound high-avidity configuration (signal 1), in conjunction with provision of the CD40-CD40L signal (signal 2). Thus, the 8-mer may induce a state of anergy that can still persist in the presence of co-stimulation.
An important aspect of our model is the presence of IgG1 BCR on the surface. The OB1 IgG1 displays moderate affinity for OVA and peptides (10−7) even having undergone somatic hypermutation (Dougan et al., 2012). It has been reported that class-switched BCRs such as IgG1 respond better to antigen because of their higher affinity for antigen, the presence of a cytoplasmic tail absent from IgM and IgD (Kaisho et al., 1997) that enhances antibody production and clonal expansion (Martin and Goodnow, 2002), and the ability to better cluster and activate in response to membrane-bound antigen (Liu et al., 2010). The identity of the constant region of Ig may also influence the variable regions; antibodies of different isotypes but with identical variable regions can exhibit differences in specificity, affinity, and effector function (Casadevall and Janda, 2012; Tudor et al., 2012). Thus, class-switched BCRs may exhibit antigen requirements that differ from their IgM counterparts to ensure prompt responses to antigen reencounter in the course of a memory response. Further studies comparing antigen-specific BCRs with different isotypes should help clarify whether this is indeed the case, and somatic cell nuclear transfer may prove to be an efficient means to generate such models.
MATERIALS AND METHODS
Mice and reagents.
Transnuclear OB1 RAG1−/− (mixed C57BL/6 and 129 background), OB1 RAG-proficient (C57BL/6 background), and FluB1 mice have been described previously (Dougan et al., 2012, 2013). OB1/MHC II GFP and FluB1/MHC II GFP were generated by crossing animals in the C57BL/6 background. All animals were maintained under specific pathogen–free conditions, and experiments were performed with approval of the Committee for Animal Care and the Association for the Assessment and Accreditation of Laboratory Animal Care. Monomeric peptides bearing biotin at the N-terminal end (N-terminal biotin; FGD 12-mer, 8-mer, 4-mer, 8-mer dimer, trimer, tetramer, and hexamer) or at the epsilon amine of lysine (C-terminal biotin; FGD 17-mer, AGA 17-mer, 17-mer subs, 17-mer hydroph, 17-mer AGD, and 17-mer FGA), (Gly)3 at the N terminus, or FGD 8-mer, 17-mer, and AGA 17-mer bearing cysteine at the C terminus for conjugation of maleimide–Alexa Fluor 647 were synthetized at the Biopolymers Laboratory at MIT. OVA was purchased from Sigma-Aldrich. Antibodies and SA used for flow cytometry were obtained from BD. Indo-1 AM was purchased from Invitrogen. Anti-IgG1 (unlabeled and conjugated to HRP) was obtained from SouthernBiotech. Anti–Fab-HRP was obtained from Sigma-Aldrich. O-benzotriazole-N,N,N′,N′-tetramethyl-uronium hexafluorophosphate (HBTU) was purchased from EMD Millipore. DriSolv anhydrous CH2Cl2, DriSolv anhydrous MeOH, and DriSolv anhydrous DMF were purchased from EMD Millipore. Redistilled, anhydrous N,N′-diisopropylethylamine (DiPEA), TFA, triisopropylsilane (TIS), N-methylpyrrolidone (NMP), heptynoic acid, and ethanedithiol were obtained from Sigma-Aldrich. 1,5-diazido-3-oxopentane was purchased from Santa Cruz Biotechnology, Inc. 1,11-diazido-3,6,9-trioxaundecane was synthesized as described in the literature (Lüning et al., 1990). Btk inhibitor was synthetized in house and was previously reported (Strijbis et al., 2013).
B cell purification.
Untouched B cells from OB1 RAG1−/− mice were purified by negative selection using CD43 Dynabeads (Life Technologies) according to the manufacturer’s recommendations. Purity was always >90–95%.
Synthesis of dimeric peptides.
The protected peptides were synthesized on an automated solid phase peptide synthesizer and subsequently the C or N terminus was modified with heptynoic acid. For N-terminal modification, the resin was swollen in NMP, deprotected with 20% piperidine/NMP, washed, coupled with heptynoic acid (3 equiv) using HBTU (3 equiv) and DiPEA (6 equiv) in NMP, and finally washed again. C-terminally modified peptides were obtained by removing the Mtt group of the C-terminal lysine residue with 1% TFA, 1% TIS in CH2Cl2, washing the resin, coupling of heptynoic acid, washing, deprotecting with 20% piperidine/NMP, and washing again. After the final washing step, the modified peptides were cleaved off the resin with 94% TFA, 2% H2O, 2% TIS, and 2% ethanedithiol, precipitated in ice-cold diethyl ether, collected by centrifugation, dried, and purified using reverse-phase HPLC. Homodimeric peptides were prepared by incubating the monomers (2.0–2.15 equiv) with a mixture of diazido linker (1 equiv), CuSO4 (0.1 equiv), and sodium ascorbate (0.15 equiv) in tBuOH/H2O. The resulting azido-linker-peptide products were purified by size exclusion column chromatography before being conjugated to the second peptide. All dimeric peptides were purified to homogeneity by reverse-phase HPLC followed by size-exclusion chromatography.
Labeling of peptides with Alexa Fluor 647.
FGD 17-mer and AGA 17-mer peptides carrying a cysteine at the C terminus were dissolved in 200 µl PBS/DMSO (1/1 vol/vol) and added to 1 mg Alexa Fluor 647–maleimide. The reaction was incubated at room temperature for 16 h before being quenched with 0.1% aqueous TFA. The labeled peptides were purified by reverse-phase HPLC, and the product fractions were lyophilized to a powder.
Confocal microscopy.
OB1 MHC II GFP or FluB1 MHC II GFP B cells were purified and attached to poly-l-lysine–coated chamber slides in phenol red–free RPMI medium supplemented with 10% heat-inactivated FBS. The temperature was at 22°C. Alexa Fluor 647 versions of FGD 17-mer and AGA 17-mer were added to B cells at 1 µM for 10 min and washed three times with RPMI medium and imaged immediately in an Ultraview spinning disk confocal microscope (PerkinElmer) equipped with an ORCA-ER CCD camera (Hamamatsu Photonics). Image acquisition and analysis were performed using the Volocity software (PerkinElmer), and the intensity of display signal was normalized between all images.
STORM imaging.
OB1 B cells in serum-free RPMI were attached to poly-l-lysine–coated chamber slides, incubated with 100 µM dynasore for 1 h, and placed on ice for 2 min, stimuli were added (17-mer at 2.5 µM and anti-IgG at 5 µg/ml) for 10 min on ice, and then chamber slides were moved to 37°C for 15 min. Samples were fixed in 4% paraformaldehyde for 10 min and stained with donkey anti–mouse IgG (Jackson ImmunoResearch Laboratories, Inc.) conjugated with Alexa Fluor 405 and Alexa Fluor 647 for 3D STORM imaging in MEA imaging buffer (He et al., 2013). STORM experiments were performed as previously described on a homemade STORM microscope constructed around an Olympus IX71 body (Jones et al., 2011) at room temperature. Near total internal reflection angle was used during imaging acquisition to reduce background fluorescence from the rest of the cell. The depth of illumination was several hundred nanometers axially, so that only a section of the cell is imaged. Two lasers (657 nm, RCL-300-656 [CrystaLaser]; and 405 nm, CUBE 405-50C [Coherent]) were combined and focused to the back focal plane of a high numerical aperture (NA) oil-immersion objective (100× UPlanSapo, NA1.4; Olympus), which is imaged onto a back-illuminated electron-multiplying charge-coupled device (EMCCD) camera (iXON DU-897; Andor). A dichroic long-pass mirror (T660LPXR; Chroma) was used to reflect the excitation beams, and the fluorescence emission was further filtered with a band-pass filter (ET705/72m; Chroma). A cylindrical lens (focal length = 10 m) was inserted into the imaging path to create astigmatism for 3D imaging (Huang et al., 2008). STORM imaging was performed at 60 Hz and imaged onto 256 × 256 pixels in the EMCCD camera. The images were drift-corrected and generated using similar methods as previously described (Huang et al., 2008). The spatial resolution measured was ∼20–30 nm laterally and ∼50–60 nm axially for Alexa Fluor 647. We randomly selected different regions of the sample to image for each treatment, which usually contained several cells per image, and there was no bias on the choice of imaging focal plane. After imaging acquisition, for cluster analysis, we used a density-based clustering of application with noise (DBSCAN) cluster algorithm with an ε value of 53 and n value of 25, as previously described for STORM image cluster analysis (Kim et al., 2013). Single antibody signal scattered on the chamber slide was used to calculate the mean number of localizations for an antibody. Since binding of antibody to its target does not alter the switching property of STORM dye pairs, the number of localizations coming from a single antibody bound to monomeric BCR should be the same as that of antibody bound to the chamber slide. The parameters were chosen to make sure the algorithm could distinguish single antibody and background signal. Using the mean number of localizations from a single antibody as a reference for all experimental conditions, we then quantified the clusters having more than three antibodies.
We systematically quantified the number of localizations for all the identified clusters, the size area of each cluster, and the molecule density (number of localization within the cluster/size area). The percentage of clusters that have more than three single antibody binding (presumably more than three BCRs), the mean size area, and the number of localizations per cluster were calculated. At least 60 cells were analyzed for each treatment condition (unstimulated 67, 17-mer 102, and anti-IgG 118 cells). Unpaired Student’s t test was performed using the total numbers of localizations/clusters and size of clusters contained in the cells (unstimulated 498, 17-mer 1,159, and anti-IgG 899), and the mean percentage of clusters above three BCRs in five (unstimulated) and six (17-mer and anti-IgG) independent fields was calculated.
Generation of hybridomas and antibody purification.
OB1 B cells were cultured for 5 d in the presence of 20 ng/ml TGF-β and 40 µg/ml LPS. Cells were then fused with NSObcl2 myeloma partner (B. Diamond, The Feinstein Institute for Medical Research, Manhasset, NY), and positive colonies were recloned by limiting dilution and screened by ELISA using plates coated with OVA and developed using anti–IgG1-HRP. OB1 IgG1 antibody was purified from hybridoma supernatants using Protein G, eluted using 0.1 M glycine, pH 2.7, on 1:20 volume of Tris-HCL, pH 8.5, to neutralize the pH and then dialyzed against PBS. Fab fragments were generated by digestion with immobilized papain activated with Cysteine HCl (Thermo Fisher Scientific) overnight at 37°C with shaking. Resulting Fab and Fc fragments were dialyzed against PBS and separated by size-exclusion using Sephadex S75.
Measurement of Ca2+ flux.
OB1 B cells were resuspended in 10% heat-inactivated FCS or serum-free phenol red–free RPMI and labeled with Indo-1 AM for 30 min at 37°C. Cells were washed and left at room temperature to enable desterification (and thus impermeabilization) of the Indo dye. Cells were then equilibrated at 37°C, and signal at 485 nm (HI) and 410 nm (LO) was collected for 1 min to record baseline Ca2+ levels as the ratio of Indo LO/HI. Cells were then stimulated and Ca2+ levels recorded for additional 5 min. For inhibition experiments, B cells were preincubated at 37°C for 30 min (Btk inhibitor) or 5–8 min (8-mer) before data collection. Data were collected in a FACS LSR instrument (BD) and analyzed using FlowJo software (Tree Star).
ELISA.
96-well plates were coated with 0.1 µM OVA in PBS overnight at 4°C, blocked in 10% FBS/PBS, and 25 µl OB1 antibody at 50 ng/ml or Fab at 500 ng/ml was added in the presence of 25 µl of increasing concentrations of peptides diluted in 10% FBS/PBS as described in the Results and incubated for 2 h at room temperature. Plates were washed three times with PBS-T and then incubated with anti–IgG1-HRP (SouthernBiotech) or anti–Fab-HRP (Sigma-Aldrich) at a 1:1,000 dilution in 10% FBS/PBS for 1 h and developed with an OptEIA TMB substrate reagent kit (BD), and the reaction was stopped with 1M sulfuric acid and plate read at 450-nm absorbance.
Biacore.
The binding kinetics between the OB1 mAb and antigens (FGD 8-mer, FGD 17-mer, and OVA) was determined using SPR on a Biacore T100 with Series S CM5 sensor chip and standard running buffer 1× HSB-EP+ (GE Healthcare). The OB1 antibody was immobilized onto the sensor chip using direct amine coupling or via anti–mouse Ig capture kit (GE Healthcare) according to the manufacturer’s specifications. The mAb was captured as 1,500–2,500 RU, and antigens were then injected at various concentrations (9.75–2,500 nM for FGD 8-mer and FDG 17-mer, 7.8–1,000 nM for OVA) at 100 µl/min for 110 s and allowed to dissociate for 300–600 s. To regenerate the chip surface, 10 mM glycine, pH 2.2, was injected for 60 s at 30 µl/min. The association, dissociation, and equilibrium dissociation constants were determined from double referenced sensorgram with Biacore T100 Evaluation Software version 1.1.1, using either 1:1 binding or conformational change model.
Calculation of fraction of occupied BCR.
The BCR occupancy of FGD 8-mer, FGD 17-mer, and OVA was calculated using a monovalent model for SPR and Fab ELISA data (Stone et al., 2011); a bivalent model was used for intact antibody ELISA data. The method is detailed in the Supplemental materials and methods.
Stochastic simulations of earliest events of B cell signaling.
Stochastic simulations were performed inside a simulation box with an area of 4 µm2 and a total volume of V = 0.08 µm3. The volume was chosen in such a way that diffusion within the box is much faster than the time scales of the chemical reactions. Under these conditions, the simulation box is well mixed. The ligand concentration was held constant throughout the simulation. The fraction of doubly phosphorylated ITAMs was averaged over 1,000 simulation runs. Parameters used in calculations are described in the Supplemental materials and methods. Rate constants used in the stochastic simulations are shown in Table S1. Concentrations used in the stochastic simulations are shown in Table S2.
Calculation of the probability of rebinding of 8-mers.
To compute the probability for 8-mer rebinding in 1D, 2D, and 3D cases, we consider both immediate rebindings and also rebindings that occur after the FGD 8-mer has made one or many diffusive excursions >100 Å away (a molecular distance), but closer than the expected distance to the nearest FGD 17-mer. The procedure for calculating the FGD 8-mer rebinding probability is detailed in the Supplemental materials and methods.
Stable transduction of MDCK cells with CD40L-LPETG construct.
Mouse CD40L bearing an LPETG-HA tag was cloned into pLHCX vector and used to make retrovirus by transfecting HEK 293T cells with pLHCX-CD40L, VSVG, and Gag-Pol constructs. Retrovirus was used to infect MDCK cells as described previously (Popp et al., 2012), and cells were selected and maintained in 250 µg/ml hygromycin B.
Immunoblotting.
OB1 B cells were equilibrated at 37°C before addition of stimuli. Lysates were analyzed by SDS-PAGE and immunoblotting using anti–P-ERK or anti–P-CD79A (phosphor-Igα) and anti–total ERK or anti–β-actin for equal loading (CST). For detection of peptide linkage to CD40L on MDCK cells by sortase, lysates of MDCK cells that were treated as described above were lysed in RIPA buffer and analyzed by immunoblotting using SA-HRP.
Peptide binding and CD86 up-regulation in response to soluble antigen.
For binding, OB1 B cells were incubated with biotinylated peptides on ice and stained with SA-APC. For CD86 up-regulation, untouched OB1 B cells were incubated with peptides, OVA, or anti-IgG1 for 18–20 h and stained with anti–CD86-APC. To detect inhibition by FGD 8-mer, B cells were preincubated with FGD 8-mer for 2 h before stimulation with soluble peptide, OVA, or anti-IgG1. Flow cytometry data were plotted using FlowJo software.
Sortase-catalyzed covalent linkage of peptide to CD40L on MDCK cells and incubation with B cells.
Untransduced or CD40L-transduced MDCK cells were plated at a density of 50,000 in 24-well plates and allowed to attach overnight. Cells were washed twice with PBS and 200 µM biotinylated GGG FGD 17-mer peptide alone or peptide + sortase A (Chen et al., 2011) at 100 µM was added in DME/10% FBS and incubated on ice for 30 min. Cells were washed three times in PBS, and untouched OB1 B cells were added at a density of 106. To detect inhibition by FGD 8-mer, OB1 B cells were preincubated with FGD 8-mer for 2 h before seeding on top of untransduced or CD40L-transduced MDCK cells that were reacted with sortase and GGG 17-mer peptide. MDCK–B cell co-cultures were incubated for 18–20 h, and CD86 up-regulation on B cells was detected by flow cytometry.
Online supplemental material.
Table S1 shows rate constants used in the stochastic simulations. Table S2 shows concentrations used in the stochastic simulations. The supplemental materials and methods includes detailed equations used for stochastic simulations of earliest events of B cell signaling, calculation of the probability of rebinding events for 8-mer, and the model description and calculation of fraction of occupied BCRs. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20131603/DC1.
Supplementary Material
Acknowledgments
We wish to thank Dr. Herman Eisen for helpful discussion and Dr. Gabriel Victora for critical reading of the manuscript, the Flow Cytometry Core Facility at Whitehead Institute for analyzer and sorter use, the Biopolymers and Proteomics Core Facility at MIT for peptide synthesis, John Jackson for mouse husbandry, and Yiling Chen for technical assistance.
This work was supported by the National Institutes of Health (grants R01 AI087879 and R01 GM100518 to H.L. Ploegh and P01 AI091580 to A.K. Chakraborty). M.D. Witte was supported by the Netherlands Organization for Scientific Research (NWO).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- BCR
- B cell receptor
- Btk
- Bruton’s tyrosine kinase
- HA
- hemagglutinin
- HEL
- hen egg lysozyme
- ITAM
- immunoreceptor tyrosine-based activation motif
- MDCK
- Madin-Darby canine kidney
- NMP
- N-methylpyrrolidone
- SA
- streptavidin
- SPR
- surface plasmon resonance
- STORM
- stochastic optical reconstruction microscopy
- TIS
- triisopropylsilane
References
- Abel S.M., Roose J.P., Groves J.T., Weiss A., Chakraborty A.K. 2012. The membrane environment can promote or suppress bistability in cell signaling networks. J. Phys. Chem. B. 116:3630–3640 10.1021/jp2102385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleksic M., Dushek O., Zhang H., Shenderov E., Chen J.L., Cerundolo V., Coombs D., van der Merwe P.A. 2010. Dependence of T cell antigen recognition on T cell receptor-peptide MHC confinement time. Immunity. 32:163–174 10.1016/j.immuni.2009.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista F.D., Harwood N.E. 2009. The who, how and where of antigen presentation to B cells. Nat. Rev. Immunol. 9:15–27 10.1038/nri2454 [DOI] [PubMed] [Google Scholar]
- Batista F.D., Iber D., Neuberger M.S. 2001. B cells acquire antigen from target cells after synapse formation. Nature. 411:489–494 10.1038/35078099 [DOI] [PubMed] [Google Scholar]
- Bell G.I. 1978. Models for the specific adhesion of cells to cells. Science. 200:618–627 10.1126/science.347575 [DOI] [PubMed] [Google Scholar]
- Cambier J.C., Gauld S.B., Merrell K.T., Vilen B.J. 2007. B-cell anergy: from transgenic models to naturally occurring anergic B cells? Nat. Rev. Immunol. 7:633–643 10.1038/nri2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A., Janda A. 2012. Immunoglobulin isotype influences affinity and specificity. Proc. Natl. Acad. Sci. USA. 109:12272–12273 10.1073/pnas.1209750109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi A., Martz R., Dorward D., Waisberg M., Pierce S.K. 2011. Endocytosed BCRs sequentially regulate MAPK and Akt signaling pathways from intracellular compartments. Nat. Immunol. 12:1119–1126 10.1038/ni.2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I., Dorr B.M., Liu D.R. 2011. A general strategy for the evolution of bond-forming enzymes using yeast display. Proc. Natl. Acad. Sci. USA. 108:11399–11404 10.1073/pnas.1101046108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan S.K., Ogata S., Hu C.C., Grotenbreg G.M., Guillen E., Jaenisch R., Ploegh H.L. 2012. IgG1+ ovalbumin-specific B-cell transnuclear mice show class switch recombination in rare allelically included B cells. Proc. Natl. Acad. Sci. USA. 109:13739–13744 10.1073/pnas.1210273109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan S.K., Ashour J., Karssemeijer R.A., Popp M.W., Avalos A.M., Barisa M., Altenburg A.F., Ingram J.R., Cragnolini J.J., Guo C., et al. 2013. Antigen-specific B-cell receptor sensitizes B cells to infection by influenza virus. Nature. 503:406–409 10.1038/nature12637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erikson J., Radic M.Z., Camper S.A., Hardy R.R., Carmack C., Weigert M. 1991. Expression of anti-DNA immunoglobulin transgenes in non-autoimmune mice. Nature. 349:331–334 10.1038/349331a0 [DOI] [PubMed] [Google Scholar]
- Fleire S.J., Goldman J.P., Carrasco Y.R., Weber M., Bray D., Batista F.D. 2006. B cell ligand discrimination through a spreading and contraction response. Science. 312:738–741 10.1126/science.1123940 [DOI] [PubMed] [Google Scholar]
- Gauld S.B., Benschop R.J., Merrell K.T., Cambier J.C. 2005. Maintenance of B cell anergy requires constant antigen receptor occupancy and signaling. Nat. Immunol. 6:1160–1167 10.1038/ni1256 [DOI] [PubMed] [Google Scholar]
- Gillespie D.T. 1977. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 81:2340–2361 10.1021/j100540a008 [DOI] [Google Scholar]
- Goodnow C.C., Crosbie J., Adelstein S., Lavoie T.B., Smith-Gill S.J., Brink R.A., Pritchard-Briscoe H., Wotherspoon J.S., Loblay R.H., Raphael K., et al. 1988. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature. 334:676–682 10.1038/334676a0 [DOI] [PubMed] [Google Scholar]
- Govern C.C., Paczosa M.K., Chakraborty A.K., Huseby E.S. 2010. Fast on-rates allow short dwell time ligands to activate T cells. Proc. Natl. Acad. Sci. USA. 107:8724–8729 10.1073/pnas.1000966107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green N.M. 1969. Electron microscopy of the immunoglobulins. Adv. Immunol. 11:1–30 10.1016/S0065-2776(08)60476-9 [DOI] [PubMed] [Google Scholar]
- He J., Sun E., Bujny M.V., Kim D., Davidson M.W., Zhuang X. 2013. Dual function of CD81 in influenza virus uncoating and budding. PLoS Pathog. 9:e1003701 10.1371/journal.ppat.1003701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Wang W., Bates M., Zhuang X. 2008. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 319:810–813 10.1126/science.1153529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Babcock H., Zhuang X. 2010. Breaking the diffraction barrier: super-resolution imaging of cells. Cell. 143:1047–1058 10.1016/j.cell.2010.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S.A., Shim S.H., He J., Zhuang X. 2011. Fast, three-dimensional super-resolution imaging of live cells. Nat. Methods. 8:499–505 10.1038/nmeth.1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaisho T., Schwenk F., Rajewsky K. 1997. The roles of gamma 1 heavy chain membrane expression and cytoplasmic tail in IgG1 responses. Science. 276:412–415 10.1126/science.276.5311.412 [DOI] [PubMed] [Google Scholar]
- Kim I., Pan W., Jones S.A., Zhang Y., Zhuang X., Wu D. 2013. Clathrin and AP2 are required for PtdIns(4,5)P2-mediated formation of LRP6 signalosomes. J. Cell Biol. 200:419–428 10.1083/jcb.201206096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.M., Pan J.Y., Korbel G.A., Peperzak V., Boes M., Ploegh H.L. 2006. Monovalent ligation of the B cell receptor induces receptor activation but fails to promote antigen presentation. Proc. Natl. Acad. Sci. USA. 103:3327–3332 10.1073/pnas.0511315103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T., Shinohara H., Baba Y. 2010. B cell signaling and fate decision. Annu. Rev. Immunol. 28:21–55 10.1146/annurev.immunol.021908.132541 [DOI] [PubMed] [Google Scholar]
- Liu W., Meckel T., Tolar P., Sohn H.W., Pierce S.K. 2010. Intrinsic properties of immunoglobulin IgG1 isotype-switched B cell receptors promote microclustering and the initiation of signaling. Immunity. 32:778–789 10.1016/j.immuni.2010.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüning U., Baumstark R., Peters K., von Schnering H.G. 1990. Concave reagents. 3. Synthesis, basicity, and conformation of new concave pyridines. Liebigs Annalen der Chemie. 1990:129–143 10.1002/jlac.199019900124 [DOI] [Google Scholar]
- Martin S.W., Goodnow C.C. 2002. Burst-enhancing role of the IgG membrane tail as a molecular determinant of memory. Nat. Immunol. 3:182–188 10.1038/ni752 [DOI] [PubMed] [Google Scholar]
- Mattila P.K., Feest C., Depoil D., Treanor B., Montaner B., Otipoby K.L., Carter R., Justement L.B., Bruckbauer A., Batista F.D. 2013. The actin and tetraspanin networks organize receptor nanoclusters to regulate B cell receptor-mediated signaling. Immunity. 38:461–474 10.1016/j.immuni.2012.11.019 [DOI] [PubMed] [Google Scholar]
- Minguet S., Dopfer E.P., Schamel W.W. 2010. Low-valency, but not monovalent, antigens trigger the B-cell antigen receptor (BCR). Int. Immunol. 22:205–212 10.1093/intimm/dxp129 [DOI] [PubMed] [Google Scholar]
- Mohamed A.J., Yu L., Bäckesjö C.M., Vargas L., Faryal R., Aints A., Christensson B., Berglöf A., Vihinen M., Nore B.F., Smith C.I. 2009. Bruton’s tyrosine kinase (Btk): function, regulation, and transformation with special emphasis on the PH domain. Immunol. Rev. 228:58–73 10.1111/j.1600-065X.2008.00741.x [DOI] [PubMed] [Google Scholar]
- Orosz F., Ovádi J. 2002. A simple method for the determination of dissociation constants by displacement ELISA. J. Immunol. Methods. 270:155–162 10.1016/S0022-1759(02)00295-8 [DOI] [PubMed] [Google Scholar]
- Popp M.W., Antos J.M., Grotenbreg G.M., Spooner E., Ploegh H.L. 2007. Sortagging: a versatile method for protein labeling. Nat. Chem. Biol. 3:707–708 10.1038/nchembio.2007.31 [DOI] [PubMed] [Google Scholar]
- Popp M.W., Karssemeijer R.A., Ploegh H.L. 2012. Chemoenzymatic site-specific labeling of influenza glycoproteins as a tool to observe virus budding in real time. PLoS Pathog. 8:e1002604 10.1371/journal.ppat.1002604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust M.J., Bates M., Zhuang X. 2006. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods. 3:793–796 10.1038/nmeth929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih T.A., Roederer M., Nussenzweig M.C. 2002. Role of antigen receptor affinity in T cell-independent antibody responses in vivo. Nat. Immunol. 3:399–406 10.1038/ni776 [DOI] [PubMed] [Google Scholar]
- Stone J.D., Artyomov M.N., Chervin A.S., Chakraborty A.K., Eisen H.N., Kranz D.M. 2011. Interaction of streptavidin-based peptide-MHC oligomers (tetramers) with cell-surface TCRs. J. Immunol. 187:6281–6290 10.4049/jimmunol.1101734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strijbis K., Tafesse F.G., Fairn G.D., Witte M.D., Dougan S.K., Watson N., Spooner E., Esteban A., Vyas V.K., Fink G.R., et al. 2013. Bruton’s Tyrosine Kinase (BTK) and Vav1 contribute to Dectin1-dependent phagocytosis of Candida albicans in macrophages. PLoS Pathog. 9:e1003446 10.1371/journal.ppat.1003446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolar P., Sohn H.W., Pierce S.K. 2005. The initiation of antigen-induced B cell antigen receptor signaling viewed in living cells by fluorescence resonance energy transfer. Nat. Immunol. 6:1168–1176 10.1038/ni1262 [DOI] [PubMed] [Google Scholar]
- Treanor B., Depoil D., Gonzalez-Granja A., Barral P., Weber M., Dushek O., Bruckbauer A., Batista F.D. 2010. The membrane skeleton controls diffusion dynamics and signaling through the B cell receptor. Immunity. 32:187–199 10.1016/j.immuni.2009.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubo N.J., Pagán A.J., Taylor J.J., Nelson R.W., Linehan J.L., Ertelt J.M., Huseby E.S., Way S.S., Jenkins M.K. 2013. Single naive CD4+ T cells from a diverse repertoire produce different effector cell types during infection. Cell. 153:785–796 10.1016/j.cell.2013.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudor D., Yu H., Maupetit J., Drillet A.S., Bouceba T., Schwartz-Cornil I., Lopalco L., Tuffery P., Bomsel M. 2012. Isotype modulates epitope specificity, affinity, and antiviral activities of anti-HIV-1 human broadly neutralizing 2F5 antibody. Proc. Natl. Acad. Sci. USA. 109:12680–12685 10.1073/pnas.1200024109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff M.F., Reid B., James K. 1967. Effect of antilymphocytic antibody and antibody fragments on human lymphocytes in vitro. Nature. 215:591–594 10.1038/215591a0 [DOI] [PubMed] [Google Scholar]
- Yang J., Reth M. 2010. Oligomeric organization of the B-cell antigen receptor on resting cells. Nature. 467:465–469 10.1038/nature09357 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.