Abstract
Recent reports have identified a phenomenon by which hypoxia shifts glutamine metabolism from oxidation to reductive carboxylation. We now identify the mechanism by which HIF-1 activation results in a dramatic reduction in the activity of the key mitochondrial enzyme complex alpha-ketoglutarate dehydrogenase (αKGDH). HIF-1 activation promotes SIAH2 targeted ubiquitination and proteolysis of the 48 kDa splice variant of the E1 subunit of the αKGDH complex (OGDH2). Knockdown of SIAH2, or mutation of the ubiquitinated lysine residue on OGDH2 (336KA) reverses the hypoxic drop in αKGDH activity, stimulates glutamine oxidation, and reduces glutamine-dependent lipid synthesis. 336KA OGDH2 expressing cells require exogenous lipids or citrate for growth in hypoxia in vitro and fail to grow as model tumors in immune-deficient mice. Reversal of hypoxic mitochondrial function may provide a novel target for the development of next-generation anti-cancer agents targeting tumor metabolism.
Keywords: OGDH, alpha-ketoglutarate, proteolysis, ubiquitination, lipid synthesis
Introduction
Glutamine is the most abundant amino acid in blood, and is second only to glucose as a carbon source for energy production and anabolic processes. Recently glutamine was identified as being essential for fuelling mitochondrial metabolism in rapidly dividing cancer cells transformed with either c-MYC (Gao et al., 2009; Wise et al., 2008) or KRAS (Son et al., 2013). Inhibiting glutamine metabolism has been proposed as a novel anti-cancer therapy (Wise and Thompson, 2010).
Mitochondrial glutamine metabolism can follow either oxidative or reductive pathways (Holleran et al., 1995). Mitochondrial utilization of glutamine begins with a two-step conversion of glutamine to α-ketoglutarate (αKG), typically by glutaminase and glutamate dehydrogenase. αKG can be either oxidized by α-ketoglutarate dehydrogenase (αKGDH) to succinate (standard TCA cycle reaction), or reductively carboxylated by isocitrate dehydrogenase (reverse TCA cycle) to isocitrate and then to citrate. The reductive cycle has been shown to be favored in cells where HIF-1α is stabilized (Metallo et al., 2012b; Wise et al., 2011), or in cells with compromised electron transport capacity (Mullen et al., 2012). Glutamine-derived citrate can be transported to the cytoplasm to generate acetyl CoA for anabolic processes such as fatty acid synthesis (Gameiro et al., 2013b; Metallo et al., 2012b).
αKGDH consists of an E1 (oxogluterate dehydrogenase, OGDH), an E2 (dihydrolipoamide Ssuccinyltransferase DLST), and an E3 (dihydrolipoamide dehydrogenase, DLD) component that collectively convert αKG to succinyl-CoA and NADH (Patel and Harris, 1995). The three enzyme (E1-E2-E3) dehydrogenase complex structure is conserved in the pyruvate dehydrogenase (PDH) and branched chain α-ketoacid dehydrogenase complexes (BCKDH). PDH serves as the major entry point for glucose-derived pyruvate into the mitochondrial TCA cycle (Patel and Korotchkina, 2001). Recent work has found that HIF1 activation inhibits PDH which blocks glucose oxidation (Kim et al., 2006; Papandreou et al., 2006). In humans, there are three major splice variants of the OGDH gene, V1 (114 kDa), V2 (48 kDa) and V3 (114 kDa), although the functional differences of these enzymes remain unreported.
Environmental hypoxia has a major effect on gene expression, largely by induction of the HIFα family of transcription factors through protein stabilization (Epstein et al., 2001). However, protein destabilization by hypoxia-regulated ubiquitination (Vinas-Castells et al., 2010) or SUMOlation (Comerford et al., 2003) has also been reported. The E3 ubiquitin ligase SIAH2 destabilizes several critical proteins in hypoxia (Qi et al., 2008). In this work, we connect the crucial HIF-1 and SIAH2 regulatory systems that together control the metabolism of glutamine. We further show that this circuit is necessary for the growth of cells in hypoxia and as model tumors.
Results
HIF1 stabilization is necessary and sufficient to reduce mitochondrial glutamine oxidation and αKGDH activity
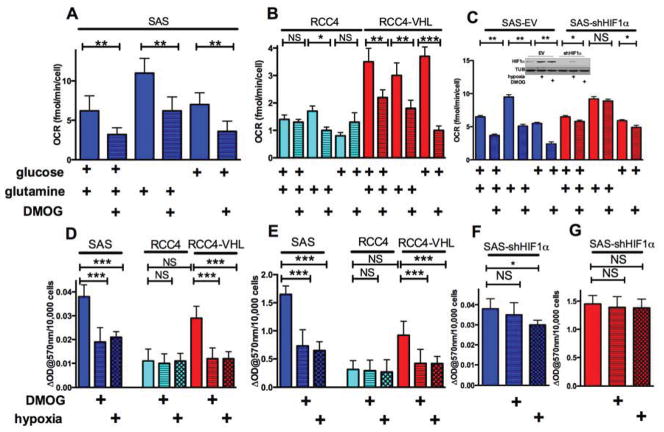
Hypoxia inhibits mitochondrial glucose oxidation (Denko, 2008; Kim et al., 2006), so we asked if it could also inhibit glutamine oxidation. We found reduced mitochondrial oxygen consumption (OCR) in the head and neck tumor cell line SAS when they were treated with the HIF-stabilizing PHD inhibitor DMOG. This decrease was detected in complete basal media, glucose-only media, or glutamine-only media (fig 1A). We also found that VHL-negative renal clear cell cancer (RCC4) cells that have constitutive HIF1 activation also show reduced glucose and glutamine oxidation. Re-introduction of VHL into these cells restores the hypoxic regulation of HIF-1, increases normoxic mitochondrial OCR and restores its reduction in response to DMOG (fig 1B). These experiments determined that HIF1α stabilization was sufficient to inhibit glutamine oxidation. We next asked if HIF1α was necessary using ShRNA knockdown. Figure 1C shows that HIF1α is essential, because its knockdown in SAS cells blocked DMOG-dependent reduction in glutamine oxidation.
Figure 1. Hypoxia down-regulates glutamine oxidation, see also figure S1.
Panel A. Mitochondrial oxygen consumption (OCR) in control and DMOG treated SAS cells (500 μM 16h). OCR was measured in basal DMEM without serum, containing only 5 mM glucose, or only 2 mM glutamine or both as indicated.
Panel B. OCR in VHL-deficient RCC4 (constitutively active HIF1) and RCC4 cells with VHL reintroduced, treated as in A.
Panel C. OCR in empty vector SAS or ShHIF1α SAS treated as in A.
Panel D. Pyruvate dehydrogenase J(PDH) enzyme activity in SAS, RCC4 and RCC4-VHL cells in control conditions, or after 16h of 0.5% oxygen, or 500 μM DMOG.
Panel E. αKGDH enzyme activity in SAS, RCC4 and RCC4-VHL cells, treated as in D.
Panel F PDH activity in ShHIF1α SAS cells, treated as in D.
Panel F αKGDH activity in ShHIF1a SAS cells, treated as in D.
Data are represented 3 independent replicates ± S.D.
Glutamine oxidation can be inhibited at any of the steps involved in its uptake or conversion to αKG. However, addition of αKG failed to restore mitochondrial OCR in DMOG (fig S1A), suggesting that the block is at αKGDH itself. Therefore we measured αKGDH enzyme activity in SAS cells, and found approximately 60% reduction upon HIF stabilization, similar to the reduction in PDH activity (fig 1D and 1E). RCC4 cells have low level αKGDH activity that is insensitive to hypoxia, and VHL reintroduction increases normoxic αKGDH activity and restores its hypoxic response (fig 1E). Similar HIF-responsive reductions in αKGDH and PDH activity were seen in RKO colorectal cancer and miapaca2 pancreatic cancer cells (fig S1B and S1C). However, in the SAS cells with ShRNA to HIF1α there was greatly reduced decrease in PDH (fig 1F) and no decrease in αKGDH activity (fig 1F). There was very modest hypoxic regulation of glutamate dehydrogenase (fig S1D), NADP-dependent isocitrate dehydrogenase (fig S1E), or carboxylase (fig S1F), suggesting that these activities were not rate limiting.
OGDH2 is marked for proteolysis in hypoxia by SIAH2-dependent ubiquitination
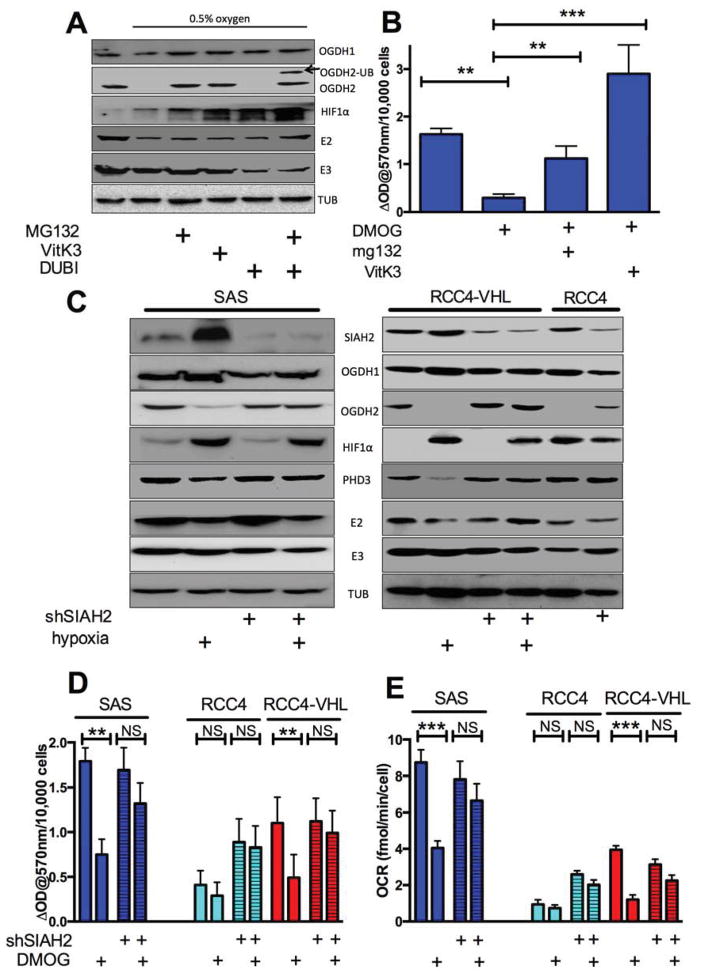
HIF-1 control of αKGDH suggested that there is a HIF-1 responsive gene whose product interacts with αKGDH to inhibit it. However, gene expression studies have not identified such a gene. We therefore examined the αKGDH protein subunits. There was modest hypoxia-dependent decrease in the level of E2 (DSLT) but no change in E3 (DLD), or in the larger splice variants of OGDH (114 kDa proteins 1/3). However, we found that hypoxia caused the complete loss of the small protein derived from splice variant OGDH2 (fig 2A). Treatment of hypoxic SAS cells with the proteasome inhibitor MG132 restored OGDH2 protein levels (fig 2A). Similarly, hypoxia specifically reduced OGDH2 in RKO and MiaPaca2 cells, and RCC4 cells have very low levels of OGDH2 that could be increased with MG132 (Fig S2A).
Figure 2. SIAH2 E3 ligase is responsible for OGDH2 protein degradation after HIF1 stabilization, see also figure S2.
Panel A. Western blot of SAS protein extracts from control, 0.5% O2 conditions, and 0.5% O2 with either 10 μM MG132, 50 μM VitK3 or 10 μM DUBI and MG132 as indicated.
Panel B. αKGDH activity in SAS cells treated with either 500μM DMOG or DMOG in combination with 10μM MG132 or 50μM VitK3.
Panel C. Western blot analysis of the indicted proteins in SAS, RCC4 and RCC4-VHL cells with and without ShRNA-SIAH2 as indicated cultured in normoxia or 16h 0.5% oxygen.
Panel D. αKGDH activity in control and ShRNA-SIAH2 SAS, RCC4 and RCC4-VHL cells ± 16h 500μM DMOG.
Panel E. Mitochondrial OCR in control and ShRNASIAH2 SAS, RCC4 and RCC4-VHL cells 16h ± 500μM DMOG measured in basal, serum-free media containing only glutamine. Data from b, d and e are two independent experiments in triplicate ± S.D.
αKGDH E2 has been reported to be marked for proteolysis by the E3 ubiquitin ligase SIAH2 upon mitochondrial disruption (Habelhah et al., 2004), so we examined if SIAH2 could be responsible for the hypoxic destruction of OGDH2. We found that the SIAH2 inhibitor Vitamin K3 (Shah et al., 2009) could also restore OGDH2 protein (fig 2A). Furthermore, treatment of hypoxic cells with both MG132 and the de-ubiquitinating enzyme inhibitor nsc632839 resulted in the appearance of variant of OGDH2 7kDa larger, suggesting the addition of ubiquitin (fig 2B). Larger, polyubiquitinated species could not be detected due to non-specific antibody reactivity at 60 kDa. We also found that MG132 or VitK3 could restore hypoxic αKGDH activity (fig 2B). These treatments had no effect on OGDH2 protein levels in normoxic conditions (fig S2B).
In order to genetically connect SIAH2 ubiquitination to HIF-dependent decrease of OGDH2 protein, we reduced SIAH2 protein with stable ShRNA in SAS, RCC4 and RCC4VHL cells (fig 2C, top row). This treatment decreased both the baseline and hypoxia-inducible SIAH2 protein, and blocked HIF-dependent reduction of OGDH2 and the SIAH2 target protein PHD3 (Nakayama et al., 2004). This genetic manipulation also blocked the HIF-dependent decrease in αKGDH activity and mitochondrial OCR in the SAS cells (fig 2D and 2E). We also found that stable knockdown of SIAH2 in VHL-deficient RCC4 cells resulted in increased normoxic OGDH2 protein levels, αKGDH activity and mitochondrial OCR that were all insensitive to hypoxia or DMOG (fig 2C, 2D and 2E). Finally, SIAH2 knockdown in the RCC4 VHL cells resulted in no change in normoxic OGDH2 protein or activity, but blocked the reduction of these measures in DMOG (figure 2C, 2D and 2E).
HIF-dependent change in glutamine metabolism enhances the growth of tumors in vivo
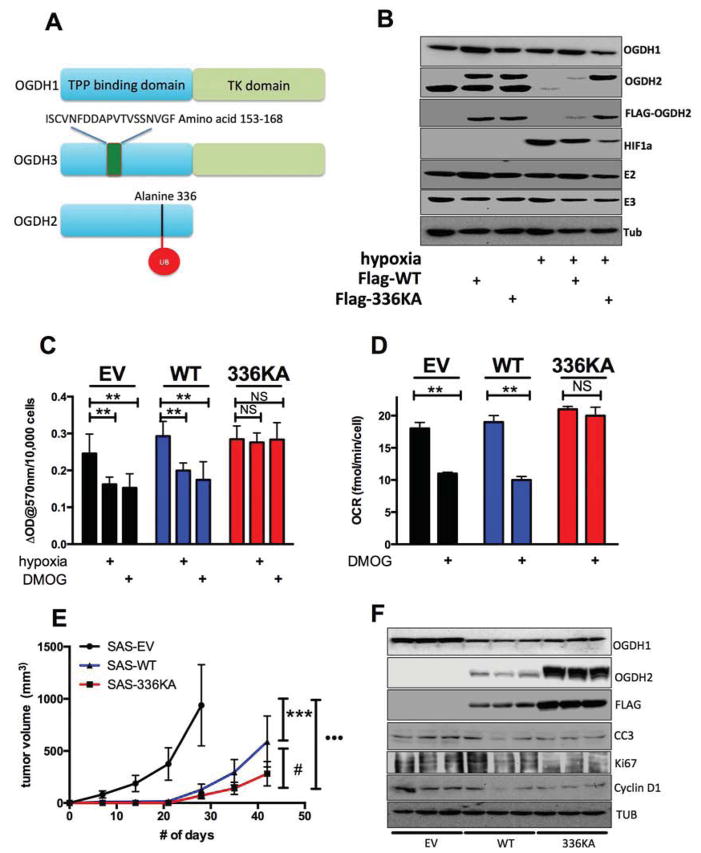
To determine the functional consequence of hypoxic SIAH2 dependent proteolysis of OGDH2, we needed to specifically block it while allowing SIAH2 to mark other proteins (Nakayama et al., 2009). We therefore identified the specific ubiquitinated lysine residue on OGDH2 using mass spectrometry so we could mutate it and block the SIAH2-dependent destruction. We immunoprecipitated Flag-OGDH2 from hypoxic cells treated with MG132 and DUBI and analysed the precipitate at the OSUCCC proteomic core facility. 24 mitochondrial proteins were identified as co-purifying with OGDH2, including the E2 subunit of αKGDH (DSLT), and the E1β subunit of PDH (Table S1). One peptide of OGDH2 was ubiquitinated at residue lysine 336 (fig S2C). We converted lysine 336 to alanine by site-directed mutagenesis and stably introduced the wild-type or mutant proteins into SAS cells. The modified protein showed significant resistance to hypoxic degradation (fig 3B second and third rows), and did show high molecular weight poly-ubiquitinated species when treated with hypoxia, MG132 and DUBI (fig S3A). The cells expressing the 336KA OGDH2 protein were also resistant to HIF-dependent reduction of αKGDH activity (fig 3C) and OCR (fig 3D). 336KA OGDH2 showed similar function when introduced into RKO and MiaPaca2 cells (fig S3B–I).
Figure 3. Hypoxia-resistant αKGDH activity is not compatible with tumor growth, see also figure S3 and table S1.
Panel A. Different splice variants of OGDH. OGDH 1/3 are 99% identical except at amino acid positions 153–169 (represented by
 ). OGDH2 is identical to OGDH1 until aa 403.
). OGDH2 is identical to OGDH1 until aa 403.
Panel B. Western blot of lysates from SAS cells stably expressing empty vector, Flag-WT OGDH2 and Flag-336KA OGDH2 exposed to normoxia or 0.5% oxygen for 16h. Lysates were probed for proteins as indicated. Note both anti-Flag and anti-OGDH2 antibodies show OGDH2 336KA is resistant to hypoxic degradation.
Panel C. αKGDH activity in SAS cell lines described in B, treated with either control, 16h 500μM DMOG, or 0.5% oxygen.
Panel D. Mitochondrial OCR in SAS cell lines described in B treated with either control, 500μM DMOG, or 0.5% oxygen and measured in glutamine-only media.
Panel E. Tumor volume of SAS cells expressing empty vector, OGDH-WT and OGDH-336KA grown in nude mice (n=8–10/group). Statistically significant growth differences exist between all three groups.
Panel F. Western blot analysis of lysates of tumors harvested after growth as described in E. Note decrease in markers of proliferation as tumors grow more slowly. Data from c and d are mean ± S.D and tumor volumes are mean ± S.E.
We could then determine the importance of reductive glutamine metabolism for the growth of model tumors. Cells expressing either wild-type or hypoxia-resistant OGDH2 were injected into immune-deficient mice for growth as tumors. Overexpression of the wild-type protein had a modest inhibitory effect on tumor growth (fig 3E), possibly because high level expression resulted in incomplete degradation (fig 3B and 3F). Expression of the hypoxia-resistant 336KA protein had profound inhibitory effect (fig 3E). While both OGDH2-modified cell line tumors grew slower than the parental tumors, the cells expressing 336KA OGDH2 grew significantly slower than the cells expressing WT OGDH2. Similar tumor growth inhibition of the two proteins was seen in RKO and MiaPaca2 cells (figure S3B–I). Analysis of lysates from these tumors showed that WT Flag-OGDH2 was expressed at intermediate levels in vivo, while 336KA Flag-OGDH2 was expressed at much higher levels (figures 3F, and S3E and S3I). Furthermore, analysis of the tumor lysates for markers of proliferation (Ki67 and cyclin D) indicated that there was reduced proliferation in the336KA OGDH2 tumors. There was no increase in the apoptotic marker, cleaved caspase 3. These results suggest that reductive carboxylation of glutamine is necessary for the proliferation of tumor cells in vivo.
Cellular growth in hypoxia requires glutamine-derived lipid production
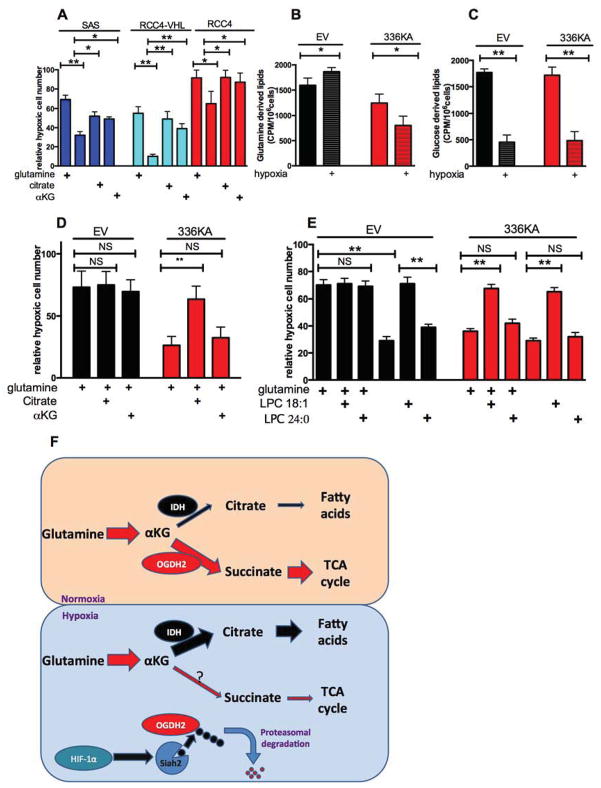
The tumors expressing 336KA OGDH2 appeared to have reduced proliferation, so we examined the effect of forced glutamine oxidation on the proliferation of tumor cells in vitro. Reductive carboxylation of glutamine in hypoxia has been proposed to be a mechanism to generate citrate for lipid synthesis when glucose entry into the TCA cycle is reduced (Le et al., 2012; Metallo et al., 2012a; Wise et al., 2011). Removal of glutamine from the media causes significant growth inhibition in hypoxia that can be rescued with addition of αKG (Wise et al., 2011) or citrate (fig 4A). We therefore directly measured the incorporation of either glucose or glutamine into newly synthesized fatty acids with 14C-labelled precursors. Cells expressing either empty vector, or 336KA OGDH2 were treated with hypoxia and then pulsed with radio-labelled glucose or glutamine. Glutamine uptake was slightly reduced by hypoxia in all lines, while glucose uptake was increased (fig S4A–D). Fatty acid production was measured by determining hexane-extractable counts from the pulsed cells. Both lines showed hypoxic decrease in glucose incorporation into lipids (fig 4C). However, there was a specific reduction in glutamine incorporation into lipids in the 336KA-OGDH2 expressing cells. Glutamine-derived lipids were increased by hypoxia in empty vector cells, but further reduced in 336KA cells (fig 4B). Consistent results were observed in RCC4 and RCC4VHL cells (fig S4E–F).
Figure 4. OGDH 336KA inhibits cell growth under hypoxia by reducing glutamine derived lipid production, see also figure S4.
Panel A: Relative hypoxic proliferation of SAS, RCC4 and RCC4VHL cells grown for 72 h in the indicated medias, presented as a percentage of normoxic growth. Hypoxic growth requires either glutamine (2mM), or glutamine derivatives αKG (2mM) or citrate (2mM).
Panel B Hexane soluble lipids derived from a 1h pulse of 0.5 μCi 14C-glutamine in SAS cells expressing either empty vector or OGDH2-336KA, grown for 16h in normoxia or hypoxia.
Panel C. Hexane soluble lipids derived from a 1h pulse of 0.5 μCi 14C-glucose in SAS cells as described in B.
Panel D. Relative hypoxic proliferation of SAS cells expressing empty vector or OGDH2-336KA after 72 hours in basal glutamine-containing media and 10% charcoal-stripped serum supplemented either 2mM citrate or 2mM αKG. Note WT cells have maximal hypoxic proliferation with glutamine, but 336KA OGDH2 expressing cells require citrate.
Panel E Relative hypoxic proliferation of SAS cells described in D, in basal media with 10% charcoal-stripped serum. Supplementation with absorbable lysophospholipid rescues the glutamine dependence of the WT OGDH2 cells, and the citrate dependence of the 336KA OGDH2 expressing cells. Data from panels A–E represents mean +/− S.D.
Panel F. A model illustrating how hypoxic degradation of OGDH2 shifts the fate of αKG from energy production to the production of lipids.
The 336KA OGDH2 cells allowed us to determine if the inability to generate lipids from glutamine would inhibit hypoxic proliferation in vitro. We found that the 336KA OGDH2 expressing cells grew as fast as the empty vector cells in normoxia but had a reduced growth rate in hypoxia, even in complete media with glutamine and 10% charcoal stripped serum (to remove the serum lipids) (fig 4D). This 336KA-dependent growth defect was due to an inability to produce mitochondrial metabolites, because hypoxic growth of these cells could be rescued by addition of exogenous citrate, but not exogenous αKG (which is simply oxidized instead of being used to make citrate) (fig 4D). In order to establish that the growth defect was due to a lack of citrate to produce lipids, we added the desaturated lysophospholipid (C18:1) that is rapidly taken up by cells (Kamphorst et al., 2013). This lipid could rescue the glutamine-dependent growth of WT OGDH2 cells, and the citrate-dependent growth of the 336KA OGDH2 expressing cells. The saturated lysophospholipid (C24:0) that is poorly taken up by cells could not rescue either cell (fig 4E and S4G).
Discussion
Microenvironmental hypoxia is a significant stress that produces compensatory metabolic changes in tumor cells. The cell conserves oxygen by reducing its use for mitochondrial energy production. However, reduction of TCA cycle reactions leads to a reduced supply of mitochondrial intermediates. Under hypoxic pressure, cells use alternative fuels such as glutamine to generate citrate and support proliferation (fig 4F). Cells can use this compensation until the oxygen level becomes too low, at which point they die (Papandreou et al., 2005).
Recent publications have described how low levels of intracellular citrate and high levels of α ketoglutarate are associated with HIF-dependent reductive carboxylation of glutamine (Fendt et al., 2013; Gameiro et al., 2013a). These elegant tracer studies are entirely consistent with our results and complement our current findings. Their conclusions focussed on the changes in the metabolites during hypoxia, but did not identify how these changes occurred. We now propose that hypoxia causes a decrease in glucose-derived citrate due to decreased PDH activity, and also increases α ketoglutarate levels due to decreased αKGDH activity. These changes in substrate concentrations drive the reverse reaction at isocitrate dehydrogenase.
The biochemical regulation of the mammalian αKGDH complex is understudied. The E1 component of the related PDH contains two proteins, the α protein with a thiamine pyrophosphate binding domain (TBD) and the β protein with a transketolase-like domain (TK). In PDH the E1 subunit functions as a heterotetramer of α2β2. The full length αKGDH E1 (variants 1 or 3) OGDH protein contains both a TBD and TK domain, and functions as a homodimer. The smaller OGDH variant 2 contains just the TBD. However, PDHE1β co-purified with OGDH2 (table S1). This suggests that OGDH2 can function in the αKGDH complex by adding transketolase activity using the PDH version found in the E1β enzyme. Careful biochemical purification and analysis will be needed to determine if this is indeed the case.
The mechanism by which OGDH2 is specifically degraded in hypoxia is interesting because lysine 336 is conserved on the full length OGDH1/3. There may be variable accessibility to the lysine based on the 3-dimensional structure of the whole versus half protein, or by function of its interactions with PDH E1β. It is also not yet clear how an intramitochondrial protein could be degraded by the cytoplasmic proteasome, although recent work has shown that such a mechanism exists (Azzu and Brand, 2010).
Fatty acid desaturation and cholesterol synthesis require molecular oxygen as an enzymatic substrate, indicating why these processes should be regulated under hypoxia. Recent work has shown that hypoxic cells preferentially take up unsaturated lysophospholipids from the growth media, and the level of activity of the sterol desaturase is also down-regulated by hypoxia (Kamphorst et al., 2013). Here we show that de novo lipogenesis is also required for the growth of cells in hypoxia in vitro, and in the hypoxia that exists within the tumor microenvironment. Glutamine appears to play a crucial role in hypoxic lipogenesis because glucose flux into the mitochondria is limited by reduced PDH activity. However, it is not apparent why blocking the hypoxic down-regulation of pyruvate dehydrogenase (and stimulate glucose flux into the mitochondria) can also block the growth of model tumors (Hitosugi et al., 2011; McFate et al., 2008).
Methods
Cells
MiaPaca2 pancreatic cancer and RKO colorectal cancer cells were from ATCC. SAS head and neck cancer, RCC4 renal cancer and RCC4-VHL cells were gifts from Dr. Le and Dr. Giaccia of Stanford University. All cells were cultured in DMEM with 10% FBS, 25 mM glucose and 4mM L-glutamine, unless stated otherwise, metabolic experiments were conducted in 5 mM glucose. Hypoxia (0.5% oxygen) was generated in a Hypoxygen H35 workstation. Cells were regularly tested for mycoplasma. For lipid dependent experiments, DMEM was supplemented with 10% charcoal stripped FBS (Invitrogen). For tumor growth, 5 × 106 cells were injected subcutaneously into the flank of nude mice and growth of tumors monitored with calipers, volume calculated using w2 × l × 0.52
Dehydrogenase assay
For αKGDH activity assay, 104 treated cells were permeabilized with 0.5% triton-X100 and incubated 1mM MgCl2, 0.1mM CaCl2, 0.05mM EDTA, 0.2% tritonX-100, 0.3mM ThDP, rotenone, 3mM αKG, 3mM NAD+, 1mM Co-A, 0.75mM nitroblue tetrazolium and 0.05mM phenazine methosulfate in 50mM Tris pH7.6 at 37°C for 40min. Cells were then solubilised in 10% SDS, 0.01N HCl over-night. OD at 570 was determined(Molecular devices). Slight modifications were made for PDH; IDH and GDH activities (see supplementary material)
Oxygen consumption rate (OCR)
1–2×104 cells were plated in 96-well Seahorse Assay plates and the next morning culture media was replaced with bicarbonate free DMEM with/without glucose/glutamine and oxygen consumption rate was measured.
Mass spectrometry
Carboxy-tagged Flag OGDH2 expression plasmid was purchased from Origene (Rockville MD). Flag-OGDHV2 cells were cultured for 16h in 0.5% oxygen and Mg132 and DUBI for 3h. Precipitated Flag OGDH2 was analysed on a Thermo Finnigan LTQ orbitrap mass spectrometer equipped with a microspray source (Michrom Bioresources Inc, Auburn, CA), data collected was searched by Mascot Daemon (Matrix Science). Modifications identified were methionine oxidation (variable), deamidation (variable), ubiquitination (variable) and carbamidomethyl cysteine (fixed).
Lipid synthesis
5×104 cells were plated in 12-well plates in 0.5% oxygen or 0.5 mM DMOG in DMEM (5mM glucose 1 mM glutamine) 16h later 0.5 μCi 14C-L-glutamine (0.5 μCi/mM) or glucose (0.1μCi/mM) was added for 1 hour. Cells were rinsed with PBS and lipids were extracted with 500ul of hexane:isopropanol (3: 1) for 30 min. Total extractable counts are reported.
Statistics
Statistical comparisons were made using a 2 tailed Students T test and values indicated with:
* 0.05>p>0.01
** 0.01>p>0.001
*** 0.001>p
Supplementary Material
Highlights.
HIF-1 activation reduces mitochondrial OCR while generating anabolic precursors
Hypoxia reduces glutamine oxidation through SIAH2-dependent proteolysis of OGDH2
Active OGDH2 makes cells dependent on exogenous lipids for hypoxic growth
Expression of non-degradable OGDH2 blocks the growth of model tumors
Acknowledgments
This work was supported by the NCI (NCD). The authors would like to thank the proteomics core at OSUCCC. They would also like to thank Drs Ioanna Papandreou, Amato Giaccia, Ze’ev Ronai, Naduparambil Jacob, Deliang Guo and members of the Denko lab for their helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Cited Literature
- Azzu V, Brand MD. Degradation of an intramitochondrial protein by the cytosolic proteasome. J Cell Sci. 2010;123:578–585. doi: 10.1242/jcs.060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comerford KM, Leonard MO, Karhausen J, Carey R, Colgan SP, Taylor CT. Small ubiquitin-related modifier-1 modification mediates resolution of CREB-dependent responses to hypoxia. Proc Natl Acad Sci U S A. 2003;100:986–991. doi: 10.1073/pnas.0337412100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nature Reviews Cancer. 2008;8:705–713. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- Fendt SM, Bell EL, Keibler MA, Olenchock BA, Mayers JR, Wasylenko TM, Vokes NI, Guarente L, Vander Heiden MG, Stephanopoulos G. Reductive glutamine metabolism is a function of the alpha-ketoglutarate to citrate ratio in cells. Nat Commun. 2013;4:2236. doi: 10.1038/ncomms3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gameiro PA, Laviolette LA, Kelleher JK, Iliopoulos O, Stephanopoulos G. Cofactor balance by nicotinamide nucleotide transhydrogenase (NNT) coordinates reductive carboxylation and glucose catabolism in the tricarboxylic acid (TCA) cycle. J Biol Chem. 2013a;288:12967–12977. doi: 10.1074/jbc.M112.396796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gameiro PA, Yang J, Metelo AM, Perez-Carro R, Baker R, Wang Z, Arreola A, Rathmell WK, Olumi A, Lopez-Larrubia P, et al. In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell Metabolism. 2013b;17:372–385. doi: 10.1016/j.cmet.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–U100. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habelhah H, Laine A, Erdjument-Bromage H, Tempst P, Gershwin ME, Bowtell DD, Ronai Z. Regulation of 2-oxoglutarate (alpha-ketoglutarate) dehydrogenase stability by the RING finger ubiquitin ligase Siah. J Biol Chem. 2004;279:53782–53788. doi: 10.1074/jbc.M410315200. [DOI] [PubMed] [Google Scholar]
- Hitosugi T, Fan J, Chung TW, Lythgoe K, Wang X, Xie J, Ge Q, Gu TL, Polakiewicz RD, Roesel JL, et al. Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Mol Cell. 2011;44:864–877. doi: 10.1016/j.molcel.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleran AL, Briscoe DA, Fiskum G, Kelleher JK. Glutamine metabolism in AS-30D hepatoma cells. Evidence for its conversion into lipids via reductive carboxylation. Mol Cell Biochem. 1995;152:95–101. doi: 10.1007/BF01076071. [DOI] [PubMed] [Google Scholar]
- Kamphorst JJ, Cross JR, Fan J, de Stanchina E, Mathew R, White EP, Thompson CB, Rabinowitz JD. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc Natl Acad Sci U S A. 2013;110:8882–8887. doi: 10.1073/pnas.1307237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metabolism. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang HX, et al. Glucose-Independent Glutamine Metabolism via TCA Cycling for Proliferation and Survival in B Cells. Cell Metab. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFate T, Mohyeldin A, Lu H, Thakar J, Henriques J, Halim ND, Wu H, Schell MJ, Tsang TM, Teahan O, et al. Pyruvate dehydrogenase complex activity controls metabolic and malignant phenotype in cancer cells. J Biol Chem. 2008;283:22700–22708. doi: 10.1074/jbc.M801765200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012a;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang JJ, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012b;481:380–U166. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Frew IJ, Hagensen M, Skals M, Habelhah H, Bhoumik A, Kadoya T, Erdjument-Bromage H, Tempst P, Frappell PB, et al. Siah2 regulates stability of prolyl-hydroxylases, controls HIF1alpha abundance, and modulates physiological responses to hypoxia. Cell. 2004;117:941–952. doi: 10.1016/j.cell.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Qi J, Ronai Z. The ubiquitin ligase Siah2 and the hypoxia response. Mol Cancer Res. 2009;7:443–451. doi: 10.1158/1541-7786.MCR-08-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metabolism. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Papandreou I, Krishna C, Kaper F, Cai D, Giaccia AJ, Denko NC. Anoxia is necessary for tumor cell toxicity caused by a low-oxygen environment. Cancer Res. 2005;65:3171–3178. doi: 10.1158/0008-5472.CAN-04-3395. [DOI] [PubMed] [Google Scholar]
- Patel MS, Harris RA. Mammalian alpha-keto acid dehydrogenase complexes: gene regulation and genetic defects. FASEB J. 1995;9:1164–1172. doi: 10.1096/fasebj.9.12.7672509. [DOI] [PubMed] [Google Scholar]
- Patel MS, Korotchkina LG. Regulation of mammalian pyruvate dehydrogenase complex by phosphorylation: complexity of multiple phosphorylation sites and kinases. Exp Mol Med. 2001;33:191–197. doi: 10.1038/emm.2001.32. [DOI] [PubMed] [Google Scholar]
- Qi J, Nakayama K, Gaitonde S, Goydos JS, Krajewski S, Eroshkin A, Bar-Sagi D, Bowtell D, Ronai Z. The ubiquitin ligase Siah2 regulates tumorigenesis and metastasis by HIF-dependent and -independent pathways. Proc Natl Acad Sci U S A. 2008;105:16713–16718. doi: 10.1073/pnas.0804063105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M, Stebbins JL, Dewing A, Qi J, Pellecchia M, Ronai ZA. Inhibition of Siah2 ubiquitin ligase by vitamin K3 (menadione) attenuates hypoxia and MAPK signaling and blocks melanoma tumorigenesis. Pigment Cell Melanoma Res. 2009;22:799–808. doi: 10.1111/j.1755-148X.2009.00628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, Perera RM, Ferrone CR, Mullarky E, Shyh-Chang N, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinas-Castells R, Beltran M, Valls G, Gomez I, Garcia JM, Montserrat-Sentis B, Baulida J, Bonilla F, de Herreros AG, Diaz VM. The hypoxia-controlled FBXL14 ubiquitin ligase targets SNAIL1 for proteasome degradation. J Biol Chem. 2010;285:3794–3805. doi: 10.1074/jbc.M109.065995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. P Natl Acad Sci USA. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, Platt JM, DeMatteo RG, Simon MC, Thompson CB. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A. 2011;108:19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.