SUMMARY
Gut mucosal barrier breakdown and inflammation have been associated with high levels of flagellin, the principal bacterial flagellar protein. Although several gut commensals can produce flagella, flagellin levels are low in the healthy gut, suggesting the existence of control mechanisms. We find that mice lacking the flagellin receptor Toll-like receptor (TLR) 5 exhibit a profound loss of flagellin-specific immunoglobulins (Ig) despite higher total Ig levels in the gut. Ribotyping of IgA-coated cecal microbiota showed Proteobacteria evading antibody coating in the TLR5−/− gut. A diversity of microbiome members over-expressed flagellar genes in the TLR5−/− host. Proteobacteria and Firmicutes penetrated small intestinal villi, and flagellated bacteria breached the colonic mucosal barrier. In vitro, flagellin-specific Ig inhibited bacterial motility and down-regulated flagellar gene expression. Thus, innate-immunity directed development of flagellin-specific adaptive immune responses can modulate the microbiome’s production of flagella in a three-way interaction that helps to maintain mucosal barrier integrity and homeostasis.
INTRODUCTION
The human gut contains 10–100 trillion bacterial cells, which in the healthy state reside in the lumen and on the outside of the mucosal barrier separating host cells from gut contents. Breaching of this barrier by microbial cells can lead to inflammation and tissue damage. The adult human is estimated to secrete 3–6 grams of immunoglobulin A (IgA) into the gut daily (Delacroix et al., 1982), and this IgA coats a large fraction of the resident microbes (van der Waaij et al., 1996), thereby staving off damaging inflammatory responses (Salim and Soderholm, 2011; Turner, 2009). IgA’s role in barrier defense is generally assumed to be immune exclusion, in which the IgA binds microbial surface antigens, and promotes the agglutination of microbial cells and their entrapment in mucus and physical clearance (Hooper and Macpherson, 2010; Mantis et al., 2011). In this view, bacteria are largely passive objects that are trapped; however, their ability to alter surface antigen presentation raises the possibility that they may actively participate in antibody binding and barrier defense.
A few bacteria have been shown to modulate the degree of IgA binding by halting production of the inducing antigen (Lonnermark et al., 2012; Mantis et al., 2011). Although most studies have been conducted with pathogens, a behavioral response to IgA coating has also been observed in a commensal gut bacterium. Bacteroides thetaiotaomicron was monoassociated to germfree RAG1−/− mice producing a single antibody raised against one of the bacterium’s capsular polysaccharides: its response to this antibody milieu was to downregulate the epitope’s expression (Peterson et al., 2007). If a wide diversity of microbiota responds to IgA binding by altering the gene expression of surface epitopes, this collective behavior could have a significant role in how IgA interacts with bacteria in host barrier defense.
Mucosal barrier breakdown and inflammation in the gut have been associated with high levels of flagellin, the principal protein comprising bacterial flagella (McCole and Barrett, 2003; Sanders, 2005). A wide diversity of gut commensals including members of the phyla Firmicutes and Proteobacteria, though not Bacteroidetes, have the capacity to produce flagella (Lozupone et al., 2012). As these are the dominant phyla in the human gut, motility-related genes are readily recovered in healthy gut metagenomes (Kurokawa et al., 2007; Turnbaugh et al., 2006). But despite the gut microbiome’s genomically encoded capacity to produce flagella, levels of flagellin protein are low in the healthy gut (Verberkmoes et al., 2009), suggesting that some control occurs to render commensal gut bacteria generally non-motile.
Here, we investigate the relationships between innate and adaptive immunity and the production of flagella by complex microbiota, and the importance of this three-way interaction in host barrier defense. We used mice deficient in Toll-like receptor (TLR) 5 to determine the impact of anti-flagellin antibodies upon the composition, gene expression, and localization of the microbiota. Although traditionally thought a component of the innate system, TLR5 acts as both a specific sensor in the innate immune system and as its own adjuvant (Letran et al., 2011). Loss of innate immune recognition of flagellin is associated with reduced levels of anti-flagelllin antibodies specifically (Gewirtz et al., 2006; Sanders et al., 2006). Thus, the TLR5−/− mouse model is useful for asking how deficiency in a specific suite of antibodies (i.e., against flagellin) impacts the microbiota and barrier defense. Our results indicate that anti-flagellin antibodies induce the downregulation of flagellar motility genes by a wide variety of gut bacteria. In the absence of these specific antibodies, the host fails to contain the microbiota, which breach the mucosal barrier.
RESULTS
Reduced levels of anti-flagellin antibodies in TLR5−/− gut
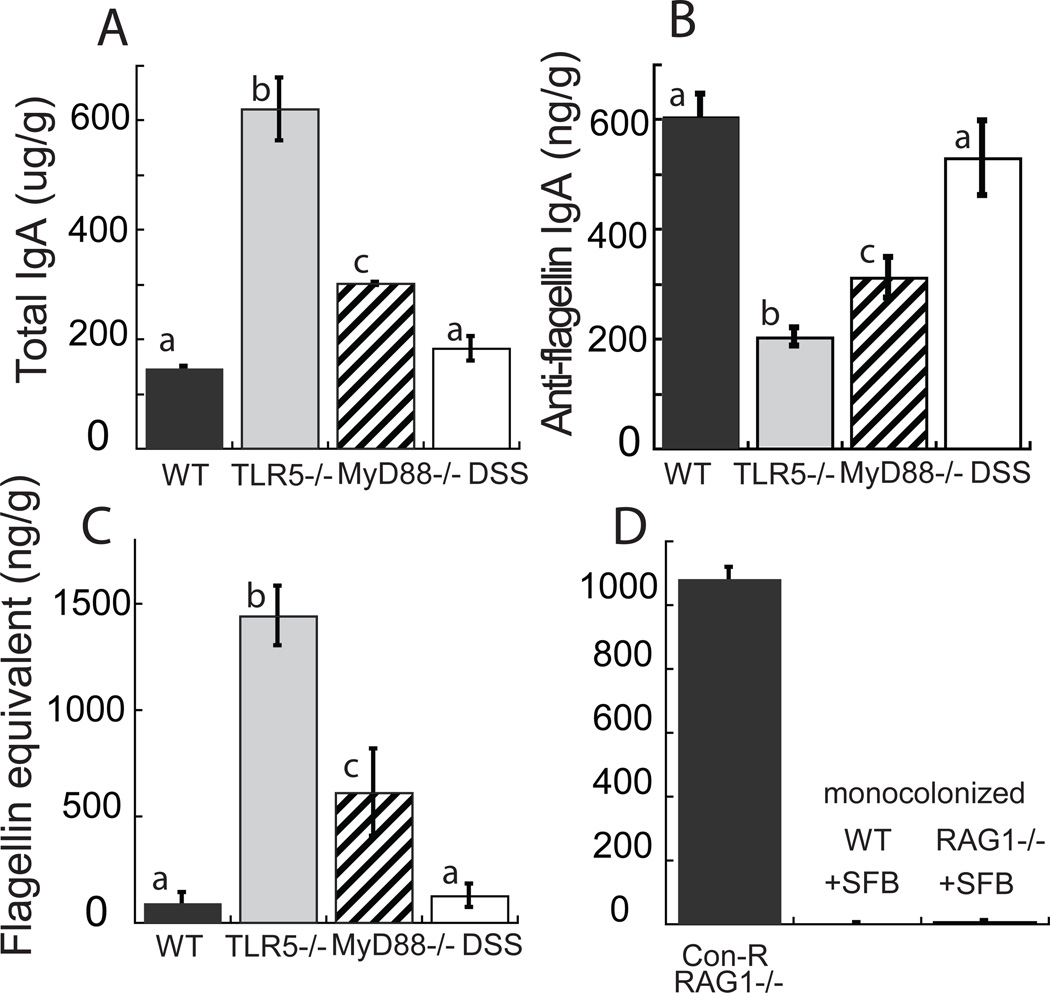
We measured antibody levels in cecum and stool by ELISA. As expected, TLR5−/− mice exhibited lower levels of anti-flagellin IgA in their ceca and fecal pellets compared to WT mice, despite higher level of total IgA constant over time (Figs. 1A, B, see Fig. S1 for IgG patterns). To assess if the impaired production of anti-flagellin antibodies was related to loss of TLR5 signaling or with inflammation that has been reported for TLR5−/− intestines (Vijay-Kumar et al., 2010), we also measured total and anti-flagellin IgA and IgG in MyD88−/− mice which have impaired, but not completely eliminated, TLR5 signaling (i.e., partial via TRIF (Choi et al., 2010)), and in WT mice with intact TLR5 signaling but treated with dextran sulfate sodium (DSS) to induce an inflamed state. Levels of anti-flagellin IgA and IgG levels were intermediate in MyD88−/− mice (Fig. 1B, S1). In contrast, DSS-treated WT mice displayed WT levels of anti-flagellin antibodies (Fig. 1B). These results are consistent with loss of TLR5 signaling leading to reduced anti-flagellin IgA production irrespective of inflammation.
Figure 1. Gut flagellin load is inversely proportional to anti-flagellin IgA levels.
(A) Total IgA levels, (B) anti-flagellin IgA levels (anti-flagellin IgG equivalents), and (C) flagellin (Salmonella Typhimurium flagellin equivalents) load for WT, TLR5−/−, MyD88−/−, and DSS-treated WT B57Bl/6 mice. (D) Flagellin amounts for conventionally raised RAG1−/− mice (Con-R), and WT and RAG1−/− mice monocolonized with SFB (+SFB). The y-axis label for C applies to D also. Columns represent means ± s.e.m. N= 8 mouse per group; lower-case letters next to the bars indicate significance: bars with different letters indicate significantly different means at P < 0.05 using a two-tailed t-test corrected for multiple comparisons. Relates to Figure S1.
High levels of flagellin in the TLR5−/− gut
We assessed levels of bioactive flagellin in the ceca and stool of mice using a TLR5 reporter cell assay standardized to flagellin from the proteobacterium Salmonella Typhimurium. Flagellin bioactivity was significantly higher in the ceca of TLR5−/− mice compare to WT mice (Figs. 1C, S1A). For both WT and TLR5−/− mice, cecal and fecal levels were equivalent, and the levels fairly constant over time, as were the corresponding anti-flagellin antibody levels (Fig. S1D, E). We observed that flagellin bioactivity levels were high in ceca of TLR5−/− mice housed at four different institutions (Fig. S1F). The ratio of bioactive flagellin in TLR5−/− to WT mice was equivalent if samples were boiled or not, indicating that interference by immunoglobulins or other proteins were not the cause of low flagellin levels in the WT mice (see Supplementary Methods).
Elevated levels of flagellin bioactivity were also present in MyD88−/− mice, but not in DSS-treated WT mice (Fig. 1C), suggesting an association with impaired TLR5 signaling but not inflammation alone. To ensure that the elevated levels of flagellin bioactivity in the TLR5−/− gut were related to a loss of antibodies rather than another consequence of impaired TLR5-signaling, we also measured flagellin bioactivity in conventionally-raised RAG1−/− mice, which are broadly deficient in all aspects of adaptive immunity due to inability to rearrange Ig genes. As predicted from the lack of antibodies in the gut, conventionally-raised RAG1−/− mice also exhibited high levels of flagellin in the gut (Fig. 1D). RAG1−/− and MyD88−/− mice have been reported to harbor an expanded population of segmented filamentous bacteria (SFB) when these are present in the microbiome (Larsson et al., 2012; Suzuki et al., 2004), and SFB are reported to genomically encode flagella (Prakash et al., 2011). However, the cecal contents derived from RAG1−/− mice mono-colonized with SFBs did not stimulate TLR5 signaling (Fig. 1D).
A wide diversity of commensal microbes express flagella-related genes in the TLR5−/− gut
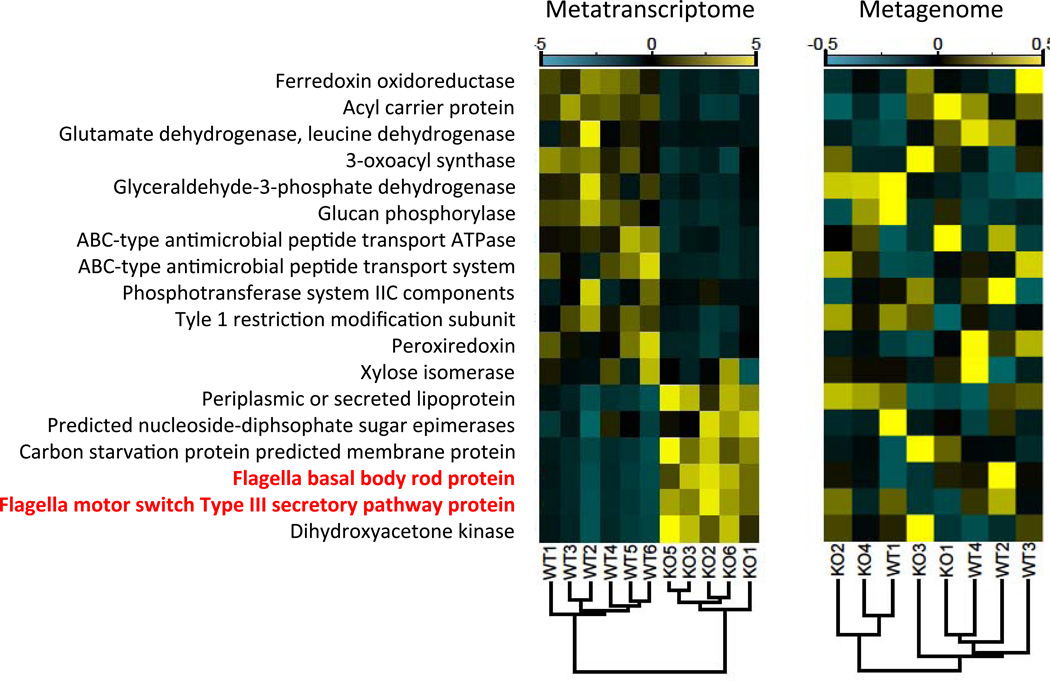
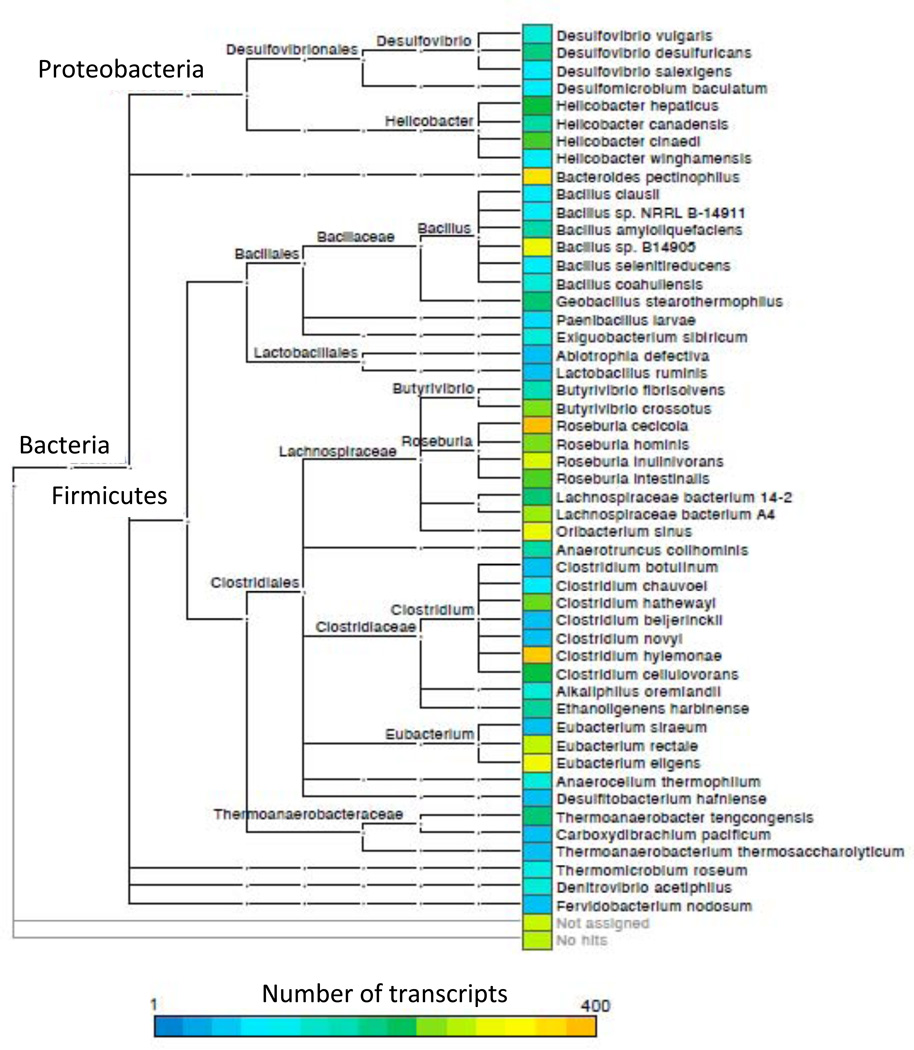
Using a combined shotgun metatranscriptomics and metagenomics approach, we identified a diversity of bacterial species that upregulated flagella-related gene expression in the TLR5−/− cecum. We designed custom probes to selectively remove rRNA from total cecal RNA (average percentage of non-ribosomal RNA was 53.6% ± 6.3%) prior to cDNA synthesis and Illumina sequencing (Stewart et al., 2010), and we shotgun sequenced the cDNA as well as bulk community DNA obtained from the cecum (Table S1). Unsupervised hierarchical clustering of samples based on the enrichment or depletion of conserved orthologous groups (COGs) of genes segregated the metatranscriptomes, but not the metagenomes, by host genotype (Fig. 2). A similar clustering of genotypes based on the taxonomic assignments of the sequence reads also failed to differentiate the genotypes (Fig. S2). These results indicate that the species composition and the functional capacities of the microbiomes were similar between TLR5−/− and WT, but the gene expression patterns differed substantially (Fig. 2). Three of the six COGs upregulated in TLR5−/− microbiomes were motility-related. Taxonomic assignments of the gene transcripts indicated that the majority of the sequence reads mapping to flagella-related genes in the TLR5−/− microbiome belonged to commensal members of the Firmicutes phylum (Fig. 3). These included commensal genera such as Roseburia, Eubacterium, and Clostridium. In addition, a subset of the flagella-related transcripts mapped to Proteobacteria (e.g., Desulfovibrio spp., Fig. 3). In addition to an enrichment in flagellar motility in TLR5−/−mice, COGs analysis in the TLR5−/− metatranscriptome indicated a depletion in carbohydrate metabolism (Fig. 2).
Figure 2. Flagella-motility genes are upregulated in the TLR5−/− microbiome despite similar encoding capacity.
Metatranscriptome and metgenome analyses for TLR5−/− and WT cecal microbiomes. Left panel, metatranscriptomes: cDNAs assigned to the 18 most significant COG categories were normalized and hierarchically clustered. Functional categories are indicated to the left of the panels; flagella-associated gene functions are highlighted in red. Dendrogram (bottom) depicts the uncentered correlation similarity metric. Correlation coefficients are represented by color ranging from blue (−5× depletion) to yellow (5× enrichment). Right panel, metagenomes: metagenomic reads from the same 18 COG categories were identically processed, with the exception that correlation coefficients are an order of magnitude less (−0.5× to 0.5×). Relates to Figure S2 and Table S1.
Figure 3. Metatranscriptomic reads annotated as flagellin are phylogenetically diverse.
Taxonomic assignments of flagellin gene transcripts are shown to the species level, with abundance of reads corresponding to the key at bottom. Hierarchical classification and the Subsystems database in MG-RAST was used to annotate function. We isolated reads with an annotation of flagellin and assigned taxonomy using BLASTX with default arguments. Note that Bacteroides pectinophilus is a Firmicute based on 16S rRNA gene sequence analysis but misclassified in the phylum Bacteroidetes in the MG-RAST database. Relates to Figure S3, which shows stimulation of TLR5 by flagellins from different species, and to Table S2.
Flagella of gut commensal bacteria can stimulate TLR5
Flagellin produced by different bacterial species can differ substantially in structure and ability to stimulate TLR5 (Andersen-Nissen et al., 2005; Smith et al., 2003), but there is little information available on the ability of flagellin produced by gut commensal microbes to stimulate TLR5. We purified flagellins from cultured gut bacteria and tested their ability to stimulate TLR5. We found that flagella harvested from several species of gut commensal Firmicutes (Clostridium scindens, C. ramosum, C. bolteae, C. bartletti, Roseburia inulivorans, R. intestinalis) could stimulate TLR5, as could several members of the Proteobacteria (Providencia stuartii, Citrobacter amalonaticus, S. Typhimurium) (Fig. S3A, Table S2). In accord with these observations, a multiple sequence alignment constructed with flagellin gene peptide sequences from diverse bacteria, including gut commensal Firmicutes, indicated that they contain the amino acid residues previously shown to be necessary for TLR5 stimulation (Fig. S3B) (Andersen-Nissen et al., 2005).
Levels of E. coli flagellin in a tri-associated gnotobiotic mouse model are decoupled from its population size
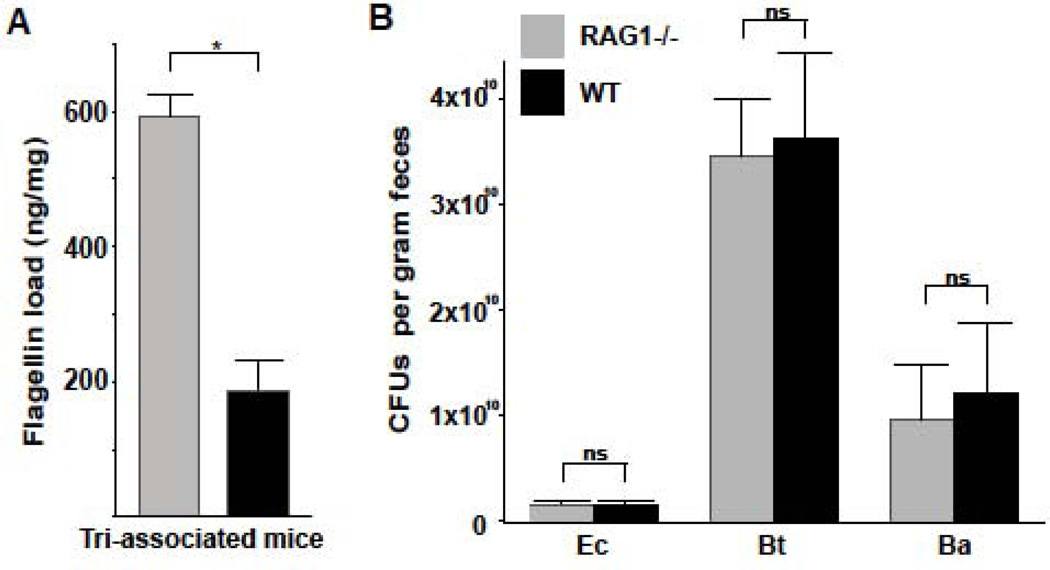
To assess how bacterial community structure was related to levels of flagellin bioactivity in the gut, we used a simplified gnotobiotic system in which we could control the composition of the microbiota and quantify its population levels. Germ-free RAG1−/− and WT mice were colonized with one motile (E. coli MG1655) and two non-motile (Bifidobacterium adolescentis FST-1, Bacteroides thetaiotaomicron VPI-5482) species. We found that levels of E. coli flagellin in the ceca of tri-associated RAG1−/− mice were significantly greater than in WT mice (Fig. 4A). CFU counts and denaturing gel electrophoresis analysis indicated that the three species reached similar population levels in both mouse genotypes (Figs. 4B, S4). Thus, the high flagellin load in the TLR5−/− mouse gut occurred despite highly similar microbial community composition and structure.
Figure 4. The population sizes for three species of bacteria colonizing gnotobiotic WT and RAG1−/− mice are equivalent despite elevated flagellin production in RAG1−/− mice.
(A) Flagellin load (Salmonella Typhimurium flagellin equivalents measured by cell reporter assay) in ceca of gnotobiotic WT and RAG1−/− mice colonized with E. coli (Ec), Bacteroides thetaiotaomicron (Bt), and Bifidobacterium adolescentis (Ba). (B) CFU counts of bacteria cultured from the ceca. Bars are means ± s.e.m., *P < 0.001; two-tailed t-test; ns, non-significant. Relates to Figure S4.
IgA coats a different suite of gut bacteria in TLR5−/− compared to WT mice
To evaluate how an IgA repertoire depleted in anti-flagellin Ig’s interacts with complex bacterial populations, we assessed the proportion of bacterial biomass coated by IgA in TLR5−/− and WT by sorting cecal bacteria into IgA-coated (IgA+) and uncoated (IgA−) pools using fluorescence-activated cell sorting (FACS) (van der Waaij et al., 1996). We found that the proportion of IgA+ bacteria was similar for TLR5−/− and WT mice (Fig 5A, 21.7%±5.9 vs. 18.8%±4.2, n=4/group, n.s.). When incubated with a bacterial biomass previously unexposed to IgA (derived from RAG1−/− mice), IgA from both genotypes also bound bacteria equivalently (55.7%±7.8% and 50.1%±7.8% of sorted cells, respectively, n=4/group, n.s.).
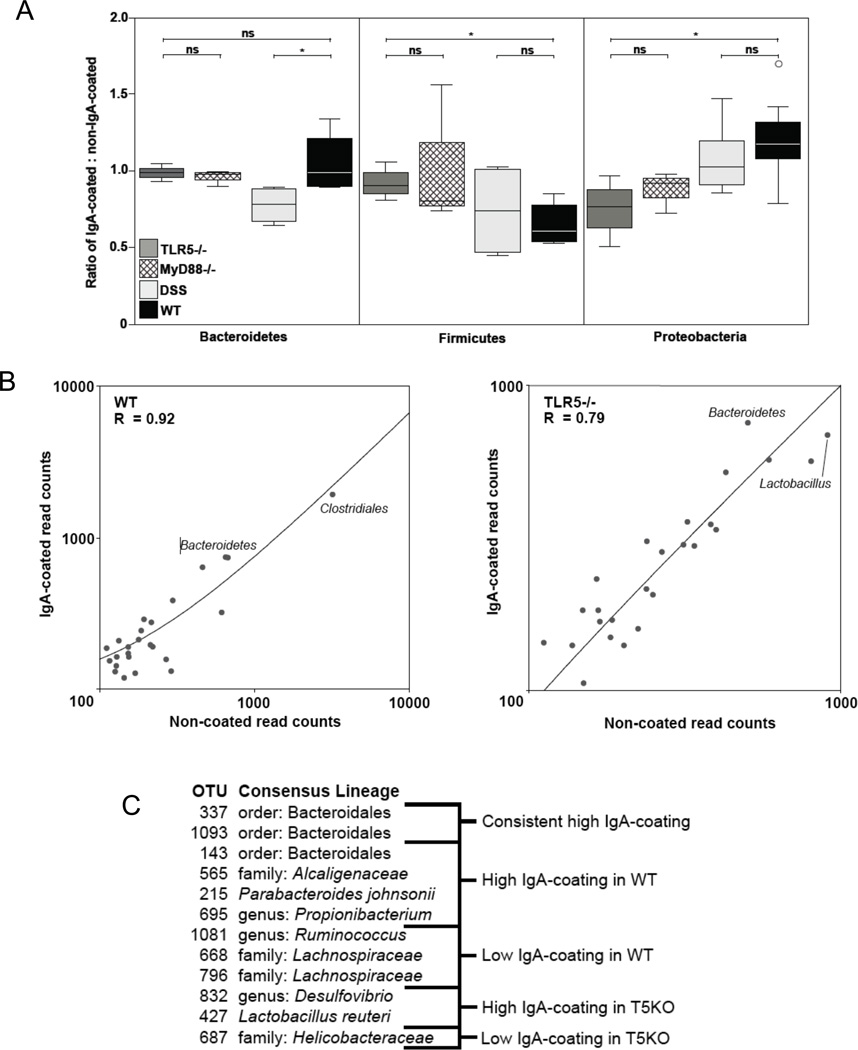
Figure 5. Effect of TLR5 signaling on IgA coating of bacterial populations.
(A) Box plots represent the ratio of IgA-coated to non-coated reads for each OTU within the specified phyla, such that a value=1 means the OTUs within a phyla are coated or non-coated with IgA at equal frequency. An open circle represents an outlier. N= 5–8 mice/group. *P < 0.05, two-tailed t-test; n.s., nonsignificant. (B) The frequency of IgA-coated (closed circles) and non-coated (open squares) cells are displayed for the 25 most abundant OTUs in either TLR5−/− or wildtype mice. The two most abundant OTUs for each genotype are indicated by their consensus taxonomy. (C) OTUs with a significant Cook’s distance (and therefore outliers from the trend of equal frequencies of IgA-coating and non-coating) are categorized as having high IgA-coating or low IgA-coating for both TLR5−/− and WT mice. The most specific consensus lineage is shown. Relates to Figure S5.
We next asked if the differing TLR5−/− and WT IgA repertoires result in a different suite of bacteria coated with IgA. After sorting IgA+ and IgA− microbial cells derived from ceca of TLR5−/− and WT mice, we extracted DNA from the two pools and pyrosequenced 16S rRNA gene sequence amplicons using Bacteria-specific primers. We repeated this for MyD88−/− and DSS-treated WT mice as controls to analyze effect of TLR5 signaling and inflammation, respectively. Compared to WT levels, TLR5−/− mice were associated with an overcoating of Firmicutes by IgA and an undercoating of Proteobacteria (Fig. 5A), even though the total sorted microbiota were similar in phylum composition for WT and TLR5−/− mice (Fig. S5). Patterns of bacterial coating were similar for TLR5−/− and MyD88−/− mice, and those for DSS-treated mice were similar to WT (with the exception of Bacteroidetes), which points again to a role for TLR5 signaling rather than inflammation in driving these patterns.
At the genus-level, the frequencies of specific taxa in the IgA+ and IgA− populations were generally equivalent within WT or TLR5−/− gut microbiomes (Fig. 5B). This is consistent with a lack of selection by IgA for specific taxa. However, some taxa deviated from this neutral pattern, with differences between gentoypes (Fig. 5C). Members of the Bacteroidetes phylum were consistently enriched in the IgA+ populations in both WT and TLR5−/− mice. In the TLR5−/− mouse alone, over-coated genera included Desulfovibrio and Lactobacillus. Members of the Helicobacter genus were also under-coated in the TLR5−/− host (Fig. 5C).
Anti-flagellin antibodies can be cross-reactive for the flagella of different bacteria
Since flagella from phylogenetically diverse bacterial species stimulate TLR5 due to conserved regions of the flagellin, we reasoned that antibodies raised against flagella from one species could have the potential to be cross-reactive with flagella from other unrelated species. Indeed, we observed cross reactivity of anti-flagellin IgG raised against single flagellin types (R. hominis Fla1 and R. hominis Fla2) for flagella harvested from different bacterial species (Proteobacteria: P. stuartii, S. Typhimurium, Citrobacter amalonaticus; Firmcutes: R. inulinivorans, R. intestinalis, C. scindens, C. ramosum, C. bartletii; Fig. S6).
Anti-flagellin antibodies immobilize bacteria in vitro
We next conducted in vitro assays to assess how anti-flagellin antibodies could impact bacterial motility. Anti-flagellin antibodies totally inhibited E. coli motility, whereas anti-LPS antibodies only partially inhibited motility (Fig. 6A). Furthermore, we applied two anti-flagellin antibody treatments (“FliC”, raised against flagellin from Salmonella, and “Fla2”, an antiserum raised against R. hominis flagellin FLa2) and a control antibody (anti-mouse IgG) to liquid cultures of R. intestinalis and Clostridium ramosum. Movies of R. intestinalis and C. ramosum cultures with and without antibody show qualitatively fewer actively moving rods and overall less movement with both anti-flagellin antibody treatments (http://ecommons.library.cornell.edu/handle/1813/31547).
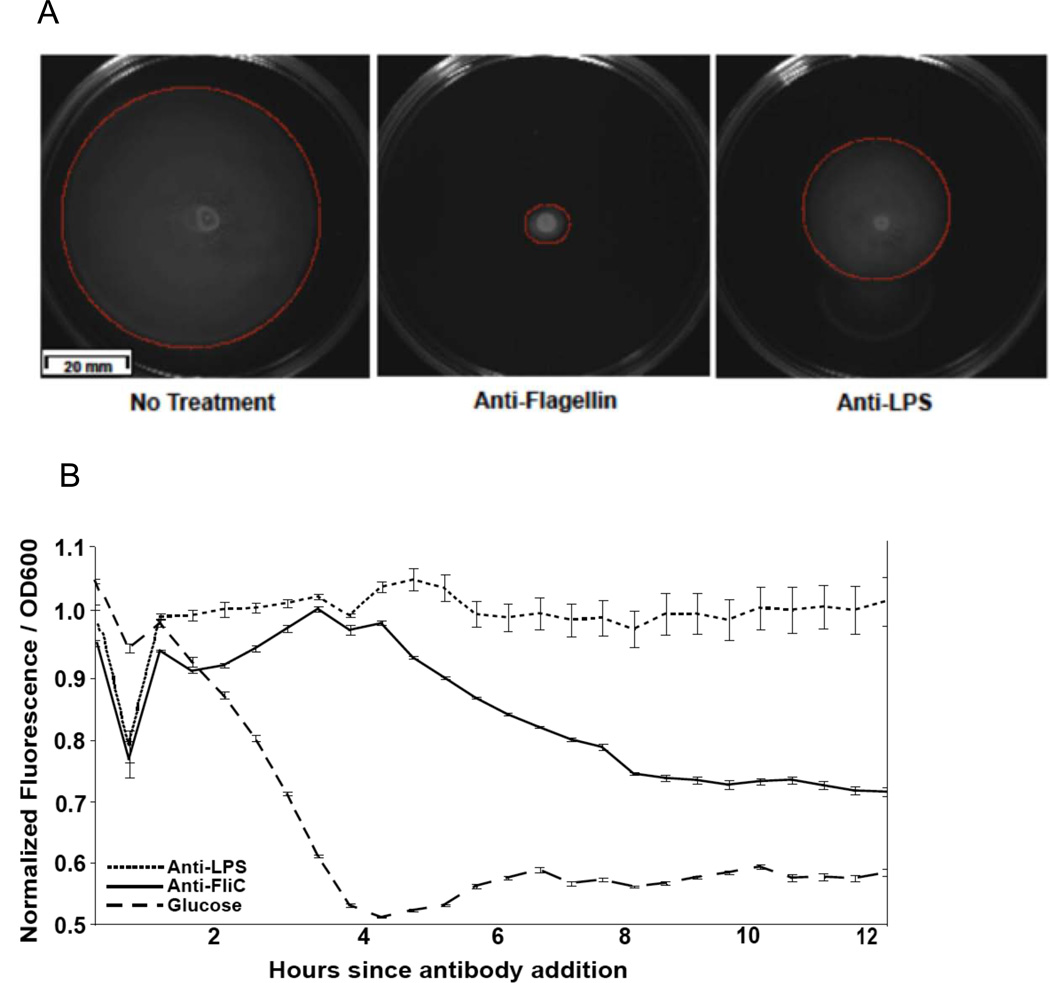
Figure 6. The addition of anti-flagellin antibody represses bacterial motility.
(A) Motility plates containing 0.3% agar stab inoculated with E. coli with or without the addition of antibody (anti-flagellin IgG). Plates incubated at 37°C for 14 hours. Red circles highlight the extent of bacterial movement through the agar. (B) GFP FliC-reporter E. coli PHL628 treated with antibodies or glucose. Means ± s.e.m.’s. for ratios of normalized GFP fluorescence:OD are plotted, n=3/group. Relates to figure S6.
Anti-flagellin antibodies induce down-regulation of flagella genes in E. coli
To verify that antibodies could impact the production of flagella directly, we constructed a flagellin gene (FliC) reporter E. coli K12 strain with GFP fused to the FliC gene. Monitoring of GFP levels over time indicated that the addition of anti-LPS antibodies had no effect on E. coli flagella gene expression, whereas anti-flagellin antibodies reduced the expression of flagellin significantly within 2 hours of addition (Fig. 6B).
Flagellated bacteria breach the mucosal barrier in the TLR5−/− mouse small and large intestines
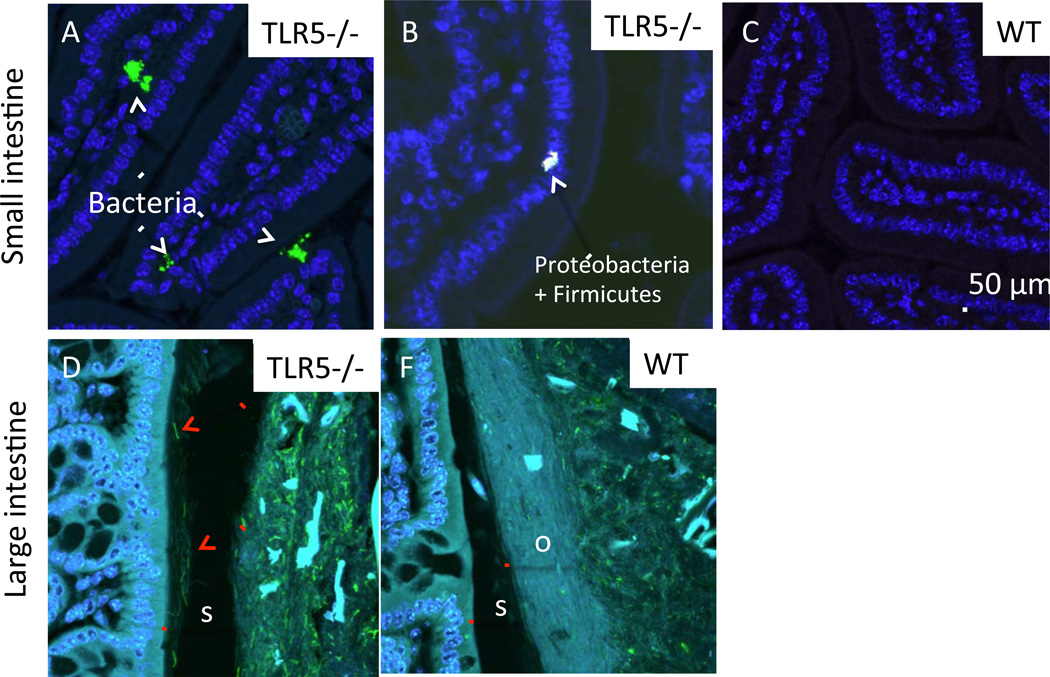
Using fluorescence in situ hybridization (FISH) with a fluorescently labeled oligonucleotide probe (EUB338) specific to bacteria in general, we observed bacterial cells penetrating the crypts and the villi in the small intestine of TLR5−/− (Fig. 7A). Using FISH probes specific for members of the γ-Proteobacteria and Firmicutes (GAM42a and a mix of LGC354 a,b and c, respectively), we identified members of both groups making up the small clusters of bacteria penetrating the villi (Fig. 7B). No bacteria were observed penetrating villi of WT small intestines (Fig. 7C). In the large intestine, labeling of flagellin using a fluorescent antibody showed that flagellated bacterial cells were present in the inner mucus layer in the TLR5−/− but not WT intestine, resulting in direct contact between bacteria and epithelial cells (Fig. 7D, E, S7).
Figure 7. Flagellated bacteria breach the large intestinal mucous barrier and penetrate small intestinal villi of TLR5−/− mice.
In all panels, host tissue is on the left, and mucus is on the right. For each pair of images in A, B, and C, the brightness of the images was increased in both panels simultaneously. (A) Flagellin imaged using immunohistochemistry with fluorescently labeled anti-flagellin antibody (GREEN) highlights flagellated rods in the s-layer of the mucus of TLR5−/− (arrows point to examples). Sections are counterstained with Hoechst 33342 (BLUE) to visualize host tissue. The outer (o) mucus layer in the WT also appears to contain less flagellin that in TLR5−/−. “s” indicates the inner, thicker s-layer of the mucus, “o” indicates the outer o-layer where bacteria are normally found. (B) Fluorescent in situ hybridization using the universal probe EUB338 (GREEN) shows clusters of bacteria penetrating the intestinal villi of the TLR5−/− small intestine. For all panels, the scale bar is 50 µm. Relates to Figure S7.
DISCUSSION
Our results suggest that TLR5-mediated innate immune recognition of flagellin is necessary for generation of mucosal antibodies directed against flagella and that such antibodies play a key role in managing the microbiota. In the absence of this innate-adaptive signaling pathway, bacterial motility goes unchecked and results in breach of the mucosal barrier, which is associated with great risk of developing chronic inflammation. Thus, while commensal bacteria can dynamically respond to mucosal antibodies and avoid clearance, the normal maintenance of flagellin-directed immune pressure helps contain the microbiota and promote health of the host.
The high flagellin bioactivity and low anti-flagellin antibody status of the TLR5−/− microbiome suggests that the high flagellin bioactivity fails to drive a commensurate anti-flagellin antibody response in the TLR5−/− host. In the WT mouse, mucosal IgA induction has been shown to function as a stepwise response to bacterial load (Hapfelmeier et al., 2010). A proportional antibody:antigen relationship likely applies to antigens that are constitutively expressed, such as LPS, but not to optional antigens such as flagella. It is likely that the TLR-mediated feedback between antibody and antigen described here is unique to TLR5, since ligands to other TLRs are constitutively expressed.
The metatranscritome data indicate that many different types of gut commensal bacteria upregulated their flagellar motility genes in the TLR5−/− hosts, even though the underlying species composition and functional gene profiles of the microbiome were similar for TLR5−/− and WT mice. Flagella have been shown to promote adherence of taxa such as Clostridium difficile in the gut and to aid colonization (Tasteyre et al., 2001). Whether flagella perform similar roles for commensal bacteria remains to be explored. Regardless of the role of flagella in the gut, our results indicate that the immune environment is a key regulator of their production. We showed that in vitro, two species of commensal Firmicutes were readily immobilized by anti-flagella antibodies, and that the addition of anti-flagellin antibodies to E coli grown in vitro induced down-regulation of its flagellin gene within a few hours of addition. Studies in pure culture (using E. coli and Salmonella) have previously shown that anti-flagella antibodies can inhibit motility (Forbes et al., 2012; Suo et al., 2009); our results extend these findings to a phylogenetically diverse variety of commensal microbes comprising the microbiome.
Anti-flagella antibodies function directly by immobilizing bacteria, but our results show that they also function also indirectly by inducing down-regulation of flagellin genes expression. When LPS antibodies were added to E. coli cultures, we observed less inhibition of motility compared to anti-flagellin antibodies, and no change in flagellin gene expression. This raises the question of how anti-flagellin antibodies induce down-regulation of flagellin gene expression. One possibility is that the total immobilization is sensed and bacteria respond with down-regulation of the flagellar genes. Alternatively, anti-flagellin immunoglobulins may act as a signal to down-regulate the flagellin genes. We note that although our in-vitro experiments utilized flagellin-specific IgG, the same responses can be expected with IgA.
Our results corroborate earlier findings that TLR5 can recognize the flagellins from a wide range of species, and support the notion that antibodies against flagellins can be cross-reactive across bacterial species. Although highly specific flagellar antigens have been used to differentiate Enterobacterial serotypes for a century, serological cross-reactivity against flagellar antigens have also been reported across species (Datta et al., 2008; Ibrahim et al., 1985; Kovach et al., 2011) and across serovars/genotypes (Biswas et al., 2010; Saha et al., 2007). Conserved epitopes are unmasked when the flagellum filament is de-polymerized (Ibrahim et al., 1985), which is also required for TLR5 activation (Smith et al., 2003). The TLR5-binding site of flagellin is conserved across phyla (Andersen-Nissen et al., 2005; Erridge et al., 2010), such that antibodies that bind this region of flagellin are likely to be cross-reactive across species. Phase variation of flagellar antigens has been described for species of Proteobacteria (van der Woude and Baumler, 2004). Other antigens, such as polysaccharides, can evade antibody recognition by phase variation (Peterson et al., 2007), so bacteria may use phase variation in flagellins to similarly evade antibody recognition. However, the region of flagellin that is phase variable is not the same as the conserved regions that activate TLR5, and if the antibodies recognize this region, then phase variation is not likely to affect antibody binding.
The loss of TLR5 signaling was also associated with an altered profile of IgA-coated microbiota. If IgA coated the various species of bacteria in the gut indiscriminately, we would expect a 1:1 ratio of IgA-coated and uncoated cells. This is largely what we observed in both WT and TLR5−/− mice for the Bacteroidetes phylum, whose representatives in the gut do not possess genes for the production of flagella. For the Fimicutes and Proteobacteria phyla, which include flagellated gut species, the patterns were opposite, with TLR5−/− generally under-coating Proteobacteria and over-coating Firmicutes compared to WT. Comparisons with DSS-treated WT and MyD88−/− mice indicated that these patterns were due to loss of TLR5−/− signaling rather than inflammation per se. These patterns suggest that deficiency in anti-flagellin antibodies is compensated by alternate antibodies for the Firmicutes, but not for the Proteobacteria. Despite the high levels of antibody coating of the Firmicutes in the TLR5−/− host, we nevertheless observed barrier breach by Firmicutes in the TLR5−/− small intestine. Thus, consistent with our in vitro observation that anti-LPS antibodies did not inhibit E. coli’s motility, these alternate antibodies appear to be insufficient to contain the microbiota.
The hypermotile phenotype of the TLR5−/− host microbiome is a form of dysbiosis, one that is unrelated to microbial community composition. We had previously reported an altered microbiota for TLR5−/−mice compared to WT based on 16S rRNA gene sequence profiles (Carvalho et al., 2012; Vijay-Kumar et al., 2010). Based on these analyses, the main differences between the TLR5−/− and WT included a higher between-mouse variation in diversity, and greater volatility (reduced stability) in microbiota over time. Levels of Proteobacteria were particularly volatile, and slightly elevated compared to WT (Carvalho et al., 2012). In contrast to the TLR5−/− microbiome’s temporal volatility, we found that the levels of flagellin and anti-flagellin antibodies were stable over time. The notion that the high-flagellin microbiome phenotype is robust to underlying community diversity is further supported by our observations that TLR5−/− mice from several different facilities, and stemming from two independent derivations (Flavell vs. Akira), all had high flagellin loads. It is increasingly clear that the species composition of mouse gut microbiotas can differ between facilities, and even between cages within a facility. Together these results indicate that microbiomes can vary widely in species content but still show high levels of flagella production in the right immune milieu.
Our experiment with the tri-associated gnotobiotic RAG1−/− and WT mice, in which community composition and structure were identical for both genotypes and only the E. coli could produce flagella, clearly indicated that the levels of flagella were independent of E. coli’s population size. What is interesting to note is that the production of flagella did not appear to impose a fitness cost on E. coli in this highly simplified model. Flagella are considered to be energetically expensive and glucose induces downregulation of these genes. Thus, other factors besides energy expenditure likely limited E. coli population levels in this system.
Expression of flagella genes is one mechanism that can lead to the observed higher levels of flagellin in the TLR5−/− gut, another is selection for motile over nonmotile strains within populations. Previous work has shown that motility in E. coli MG1655 is selected against in the mouse gut, although a portion of the cells retains motility (Gauger et al., 2007). Mono-colonization of E. coli into WT and MyD88−/− germfree mice has been shown to result in selection for non-motile mutants within 10 days of inoculation in both mouse genotypes (De Paepe et al., 2011; Giraud et al., 2008). If antibodies against flagella are important in this selection process, inoculation of E. coli into germfree RAG1−/− or TLR5−/− mice should fail to select non-motile mutants. Roseburia cecicola monoassociated in gnotobiotic WT mice has also been shown to lose motility, but only in mono-association, not when associated with four other species (Stanton and Savage, 1984). Thus, motility is likely important for bacterial fitness in the context of microbe-microbe interactions in the complex microbiome. This might explain why so many different kinds of bacteria maintain flagella genes in their genomes despite the host selection against flagellar motility. How, when and where flagella are essential for bacterial fitness in the mammalian gut remains to be explored.
We propose that greater flagellar motility leads directly to barrier breach. Imaging of bacteria in the TLR5−/−gut showed penetration of intestinal villi in the small intestine, which we did not observe in the WT mice. Consistent with the metatranscriptome data indicating that members of both Firmicutes and Proteobacteria upregulated their flagella genes, we observed both phyla in the small intestinal villi of TLR5−/− mice. We also observed flagellated rods in the normally bacteria-free thicker (s) mucus layer of the large intestine, clearly adjacent to host epithelial cells. Although the probes are unlikely to detect all flagellated species, we nevertheless observed a qualitatively stark difference between the genotypes. Proteobacterial breach of the slayer has been previously reported for the TLR5−/− mouse (Carvalho et al., 2012), and in the MyD88−/− mouse (Vaishnava et al., 2011), and appears to be associated with inflammation (Johansson et al., 2013). Thus, in the absence of TLR5 signaling, a large bacterial load, much of which can be flagellated, comes into contact with host tissues both in the small and large intestines, and likely participates in perpetrating inflammation.
In summary, we suggest that anti-flagellin antibodies are key to mucosal barrier protection and homeostasis. Our findings further suggest that mucosal antibodies quench motility by immobilizing flagellated bacteria, but additionally by inducing the down-regulation of motility-related genes. Thus, we propose that mucosal immunoglobulins function physically on flagella to bind them as well as a signaling molecule to alter gut microbial gene expression and ultimately control bacterial behavior and localization.
EXPERIMENTAL PROCEDURES
Experimental procedures are outlined in brief here: for more details, see Supplement. All animal experiments were approved by the local IACUCs. All mice used in this study were C57BL/6 strain, and were housed at different facilities. The tri-associated gnotobiotic mice were maintained at the University of Nebraska Lincoln, and their populations monitored by plate counts and by denaturing gradient gel electrophoresis (DGGE, see Supplemental Methods). Bacterial strains used in the study are listed in Table S2.
Measurement of antibodies using ELISA
We measured total IgA, total IgG, flagellin-specific IgA, and flagellin-specific IgG in fecal and cecal samples by ELISA as previously described (Sitaraman et al., 2005). Details of the ELISA, including the sandwich ELISA used for the flagellin-specifc IgA measures, are in the Supplemental Methods.
Flagellin quantification
We quantified flagellin using HEK-Blue-hTLR5 cells (Invivogen, San Diego, CA). Between 10–100 mg of either cecal or fecal material were diluted by homogenizing for 10 seconds using a Mini-Beadbeater-24 without the addition of beads (BioSpec, Bartlesville, OK), centrifuging the samples at 8,000g for 2 min, followed by serial dilutions. After 20–24 h of incubation, we applied cell culture supernatant to HEK-Blue Detection medium (Invivogen, San Diego, CA) and measured alkaline phosphatase activity at 620 nm every 30 min for 3 h on a Synergy H1 multiple detection microplate reader (Biotek Instruments Inc., Winooski, VT). Results are representative of 1 repeat of the experiment and both experiments included 1 technical replicate per sample. We used purified Salmonella Typhimurium strain 14028 flagellin (Enzo Life Sciences, Inc., Farmingdale, NY) in standard curves. As a control for TLR5 specificity, we included 1 additional replicate per sample, per plate, to which we added anti-hTLR5 neutralizing antibody (5 µg/ml) (Invivogen, San Diego, CA).
Flagellin purification and production
Flagella were sheared using 80 mL of each culture (strains listed in Table S2) by vortex agitation. Staphylococcus epidermidis was used as an aflagellate control. Cultures were centrifuged at 2,754×g for 30 minutes at 4°C and supernatants centrifuged again at 2,754×g for 1.5 hours at 4°C. The resulting pellets were resuspended in PBS and centrifuged once more at 3,442×g for 30 minutes at 4°. To reduce endotoxin levels in the flagellar purification of Gram-negative bacteria (Citrobacter amalonaticus and Providencia stuartii), we detoxified using polymyxin B using Dtoxi-Gel AffinityPak Pre-packed Columns (Thermo Scientific, Rockford, Illinois). Recombinant Fla1 from R. hominis was produced by cloning the fla1 gene with a His tag into an E. coli expression vector (see Supplemental Methods for details).
Western blot
Purified flagellin or cell pellets (R. hominis, and E. coli cell pellets with recombinant Fla1, or supernatant from those cells) were prepared by adding Laemmli sample buffer with β-mercaptoethanol and boiled for 10 minutes. Proteins were separated on SDS/10% PAGE gels, transferred to nitrocellulose membrane, and blocked for 1 hour in Tris-buffered saline (TBS) solution containing 5% milk. The membrane was probed overnight at 4° C with primary antibodies anti-Fla2 and anti-Fla1 (Covalab, Cambridge, United Kingdom).
Metatranscriptomic analysis
We enriched for nonribosomal RNA using a modification of a technique previously described (Stewart et al., 2010). We divided cecal contents in half and extracted bulk RNA and bulk DNA in parallel using the Powersoil RNA Isolation kit and Powersoil DNA isolation kit, respectively, as described by the manufacturer (MoBio Laboratories Ltd, Carlsbad, CA). We generated sample-specific ribonucleotide probes targeting bacterial 16S and 23S rRNA genes by PCR amplifying these gene sequences from bulk DNA using the universal primers 27F and 1492R for 16S and 189F and 2490R for 23S. We separately converted 16S and 23S rRNA gene sequence amplicons to biotinylated antisense rRNA probes, which hybridized to complementary rRNA molecules in the total RNA sample. Next, we converted rRNA-subtracted samples to double-stranded cDNA, amplified via in vitro transcription, and converted back to double-stranded cDNA. We used the LSU and SSU reference databases from SILVA (http://www.arb-silva.de/) to separate Illumina reads with >70% similarity to a database rRNA sequence. Using this approach, we identified 40.2% of the reads as ribosomal and removed them from downstream analysis. We used MG-RAST (Meyer et al., 2008) with the default quality filtering. COG relative abundance data for protein-coding reads were summarized using MG-RAST (e-value < 10−5; ID > 50%; length > 20 aa). We uploaded functional assignments to Cluster 3.0 and centered and normalized counts before hierarchical clustering using the uncentered correlation similarity metric. To assign taxonomy to sequences annotated as flagellin, we used hierarchichal classification and the Subsystems database in MG-RAST to annotate function using default cutoff parameters. We isolated reads with an annotation of flagellin and assigned taxonomy using BLASTX (Altschul et al., 1990).
Metagenomic analysis
DNAs were sequenced at the Core Laboratories Center at Cornell University on the Roche 454 FLX platform and at the DNA Sequencing Lab (Columbia University Medical center) using the Illumina HiSeq 2000 platform. We used MG-RAST (Meyer et al., 2008) with the default quality filtering and without identical read dereplication. Taxonomy assignments (LCA) and COG relative abundance data for protein-coding reads were summarized using MG-RAST.
Sorting of IgA-coated bacteria
Cecal preparations were incubated with and without FITC-labeled goat F(ab’)2 anti-mouse IgA and propidium iodide. We performed flow cytometry with BD-Biosciences FACS Aria high-speed flow cytometer/cell sorting using an Argon laser operating at 15 mW and 488 nm. We used standard ELITE software comprising the Immuno-4 program to determine the percentage of stained events. The discriminator was set on PI fluorescence as a specific probe for bacteria. We analyzed a portion of each sample incubated with PBS (background fluorescence) and a portion incubated with FITC-labeled goat F(ab’)2 anti-mouse IgA. Both measurements were performed with 10,000 events. We collected a minimum of 500,000 cells of each category (IgA-bound and non-bound).
16S gene sequence analysis
For quality filtering 16S rRNA gene sequence data we discarded sequences <200 bp or >1,000 bp, and ones containing >0 primer mismatches, uncorrectable barcodes, >0 ambiguous bases, or homopolymer runs in excess of 6 bases using the open source software package Quantitative Insights into Microbial Ecology (QIIME) (Caporaso et al., 2010). We checked sequences for chimeras (UCHIME) and assigned to operational taxonomic units (OTUs) using Otupipe (Edgar et al., 2011) with a 97% threshold of pairwise identity, and then classified them taxonomically using the Greengenes reference database (McDonald et al., 2012). We rarified samples to 7,000 reads per sample, calculated the ratio of IgA+ : IgA- for each OTU, and calculated Cook’s distance in R to find OTUs with IgA coating ratios that diverged significantly from the mean (Ferrari and Cribari-Neto, 2004).
Observations of bacterial motility
Clostridium ramosum and Roseburia intestinalis were cultured for 48 hours in M2GSC medium in strictly anaerobic conditions at 37°C. 10 µl of culture was mixed with either 1 µl of anti-flagellin antibody, anti-mouse antibody as a specificity control, or water and applied to a glass slide and sealed with a glass cover slip and acrylic sealant. Slide preparations were completely dried and sealed before exiting the anaerobic chamber. Timelapse microscopy was performed on slides 30 minutes after the addition of antibody with a Metamorph imaging system. We made time series of the cultures with a spacing of 0.3 seconds over the course of 35 frames. The videos are named as follows: bacterial species, antibody used (or water in the case of “control”), and the amount of time lapsed between slide preparation and imaging (30 minutes). The antibodies used were either anti-FliC (monoclonal antibody made against E. coli flagellin), anti-Fla2 (rabbit antiserum made against Roseburia hominis flagellin), or a polyclonal anti-mouse antibody (used as a control non-flagellin specific antibody). Details of the plate motility assays are in the Supplemental Methods.
Monitoring flagellin expression of E. coli bioreporter
Details on the construction of the reporter strain are in Supplemental Methods. We grew E. coli in minimal salts medium (MSM) containing 0.2% glucose (as an inhibitor of flagellin expression) and ampicillin (15 mg/L) at 37°C to an OD600 of 0.8±0.2. We then diluted the culture 1:100 in MSM containing either 0.2% glucose or 0.2% casamino acids. We divided cultures into 100 µl aliquots in 96 well clear flat-bottom plates (Costar, Corning, NY) and incubated at 37°C with periodic shaking on a Synergy H1, multiple detection microplate reader (Biotek Instruments Inc., Winooski, VT). OD600 and fluorescence of the fliC promoter fusion (Abs/Em: 488/509 nm) were measured every 30 min over the course of 15 hours. We added antibodies after 2 hours of growth at a concentration of (5 µg/ml) (Forbes et al., 2008). The fluorescence values were normalized to growth. Results are representative of 3 repeats of the experiment, each of which included 1 technical replicate per sample.
Microscopy
Samples of intestine were preserved in Carnoy solution and paraffin-embedded for sectioning. FISH for bacterial groups was performed using flueorescently-labeled oligonucleotide probes specific for all Bacteria, or specific subgroups (See Supplemental Methods). Flagellin and IgA were visualized using fluorescently labeled antibodies (FITC-conjugated anti-flagellin IgG, and anti-IgA IgA).
Statistics
We performed statistical analysis and linear regression analysis in Microsoft Excel 2011, StatPlus 2009, R version 2.11.1, and IBM SPSS Statistics. Statistical tests were 1-way ANOVA, unpaired student ‘t’-test, or generalized mixed linear model, as indicated. Significance level were p<0.05 unless otherwise indicated. All multiple comparison testing was accounted for with Bonferroni correction.
Data Deposition
16S rRNA gene sequence data are deposited under Study ID#1891 in the QIIME-DB database (qiime.org). Transcriptome and metagenome data are deposited under Project names TLR5KO_Transcritptome, TLR5KO_Metagenome (Illumina data) and TLR5KO_454_Metagenome (454 data) in the MG-RAST database (metagenomics.anl.gov).
Supplementary Material
Hightlights.
TLR5−/− mice have reduced levels of anti-flagellin antibodies in the gut
Flagella-related genes of commensal microbes are upregulated in the gut of TLR5−/− mice
Antibodies against flagellin induce downregulation of flagella genes
Flagellated bacteria penetrate TLR5−/− small intestinal villi and breach mucosal barrier
ACKNOWLEDGMENTS
We thank S. McSorley, D. Hackam, F. Carvalho, and L. Junker. This work was supported by NIH grants R01DK093595 and DP2OD007444, and Fellowships to R. E. L. from The Hartwell Foundation, the Arnold and Mabel Beckman Foundation, and the Lucile and David Packard Foundation. The Cornell University cytometry core is supported in part by the ESSCF, NYS-DOH, Contract #123456.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andersen-Nissen E, Smith KD, Strobe KL, Barrett SL, Cookson BT, Logan SM, Aderem A. Evasion of Toll-like receptor 5 by flagellated bacteria. Proc Natl Acad Sci U S A. 2005;102:9247–9252. doi: 10.1073/pnas.0502040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas D, Herrera P, Fang L, Marquardt RR, Ricke SC. Cross-reactivity of anti-Salmonella egg-yolk antibodies to Salmonella serovars. J Environ Sci Health Part B. 2010;45:790–795. doi: 10.1080/10934521003709016. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho FA, Koren O, Goodrich JK, Johansson ME, Nalbantoglu I, Aitken JD, Su Y, Chassaing B, Walters WA, Gonzalez A, et al. Transient inability to manage Proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe. 2012;12:139–152. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YJ, Im E, Chung HK, Pothoulakis C, Rhee SH. TRIF mediates Toll-like receptor 5-induced signaling in intestinal epithelial cells. J Biol Chem. 2010;285:37570–37578. doi: 10.1074/jbc.M110.158394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Janes ME, Simonson JG. Immunomagnetic separation and coagglutination of Vibrio parahaemolyticus with anti-flagellar protein monoclonal antibody. Clin Vacc Immunol. 2008;15:1541–1546. doi: 10.1128/CVI.00141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paepe M, Gaboriau-Routhiau V, Rainteau D, Rakotobe S, Taddei F, Cerf-Bensussan N. Trade-off between bile resistance and nutritional competence drives Escherichia coli diversification in the mouse gut. PLoS Genet. 2011;7:e1002107. doi: 10.1371/journal.pgen.1002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacroix DL, Hodgson HJ, McPherson A, Dive C, Vaerman JP. Selective transport of polymeric immunoglobulin A in bile. Quantitative relationships of monomeric and polymeric immunoglobulin A, immunoglobulin M, and other proteins in serum, bile, and saliva. J Clin Invest. 1982;70:230–241. doi: 10.1172/JCI110610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erridge C, Duncan SH, Bereswill S, Heimesaat MM. The induction of colitis and ileitis in mice is associated with marked increases in intestinal concentrations of stimulants of TLRs 2, 4, and 5. PloS One. 2010;5:e9125. doi: 10.1371/journal.pone.0009125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari SLP, Cribari-Neto F. Beta regression for modelling rates and proportions. J Appl Stat. 2004;31:799–815. [Google Scholar]
- Forbes SJ, Eschmann M, Mantis NJ. Inhibition of Salmonella enterica serovar typhimurium motility and entry into epithelial cells by a protective antilipopolysaccharide monoclonal immunoglobulin A antibody. Infect Immun. 2008;76:4137–4144. doi: 10.1128/IAI.00416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SJ, Martinelli D, Hsieh C, Ault JG, Marko M, Mannella CA, Mantis NJ. Association of a protective monoclonal IgA with the O antigen of Salmonella enterica serovar Typhimurium impacts type 3 secretion and outer membrane integrity. Infect Immun. 2012;80:2454–2463. doi: 10.1128/IAI.00018-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauger EJ, Leatham MP, Mercado-Lubo R, Laux DC, Conway T, Cohen PS. Role of motility and the flhDC Operon in Escherichia coli MG1655 colonization of the mouse intestine. Infect Immun. 2007;75:3315–3324. doi: 10.1128/IAI.00052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gewirtz AT, Vijay-Kumar M, Brant SR, Duerr RH, Nicolae DL, Cho JH. Dominant-negative TLR5 polymorphism reduces adaptive immune response to flagellin and negatively associates with Crohn's disease. Am J Physiol Gastrointest Liver Physiol. 2006;290:G1157–G1163. doi: 10.1152/ajpgi.00544.2005. [DOI] [PubMed] [Google Scholar]
- Giraud A, Arous S, De Paepe M, Gaboriau-Routhiau V, Bambou JC, Rakotobe S, Lindner AB, Taddei F, Cerf-Bensussan N. Dissecting the genetic components of adaptation of Escherichia coli to the mouse gut. PLoS Genet. 2008;4:e2. doi: 10.1371/journal.pgen.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M, Cahenzli J, Velykoredko Y, Balmer ML, Endt K, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328:1705–1709. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper L, Macpherson A. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- Ibrahim GF, Fleet GH, Lyons MJ, Walker RA. Immunological relationships between Salmonella flagellins and between these and flagellins from other species of Enterobacteriaceae. Med Microbiol Immunol. 1985;174:101–113. doi: 10.1007/BF02123231. [DOI] [PubMed] [Google Scholar]
- Johansson ME, Gustafsson JK, Holmen-Larsson J, Jabbar KS, Xia L, Xu H, Ghishan FK, Carvalho FA, Gewirtz AT, Sjovall H, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2013;61:1124–1131. doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach Z, Kaakoush NO, Lamb S, Zhang L, Raftery MJ, Mitchell H. Immunoreactive proteins of Campylobacter concisus, an emergent intestinal pathogen. FEMS Immunol Med Microbiol. 2011;63:387–396. doi: 10.1111/j.1574-695X.2011.00864.x. [DOI] [PubMed] [Google Scholar]
- Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, Takami H, Morita H, Sharma VK, Srivastava TP, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson E, Tremaroli V, Lee YS, Koren O, Nookaew I, Fricker A, Nielsen J, Ley RE, Backhed F. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut. 2012;61:1124–1131. doi: 10.1136/gutjnl-2011-301104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letran SE, Lee SJ, Atif SM, Uematsu S, Akira S, McSorley SJ. TLR5 functions as an endocytic receptor to enhance flagellin-specific adaptive immunity. Eur J Immunol. 2011;41:29–38. doi: 10.1002/eji.201040717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonnermark E, Nowrouzinan F, Adlerberth I, Ahrne S, Wold A, Friman V. Oral and faecal lactobacilli and their expression of mannose-specific adhesins in individuals with and without IgA deficiency. Int J Med Microbiol. 2012;302:53–60. doi: 10.1016/j.ijmm.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Faust K, Raes J, Faith JJ, Frank DN, Zaneveld J, Gordon JI, Knight R. Identifying genomic and metabolic features that can underlie early successional and opportunistic lifestyles of human gut symbionts. Genome Res. 2012;22:1974–1984. doi: 10.1101/gr.138198.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantis NJ, Rol N, Corthesy B. Secretory IgA's complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011;4:603–611. doi: 10.1038/mi.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCole DF, Barrett KE. Epithelial transport and gut barrier function in colitis. Curr Opin Gastroenterol. 2003;19:578–582. doi: 10.1097/00001574-200311000-00011. [DOI] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, et al. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson D, McNulty N, Guruge J, Gordon J. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- Prakash T, Oshima K, Morita H, Fukuda S, Imaoka A, Kumar N, Sharma VK, Kim SW, Takahashi M, Saitou N, et al. Complete genome sequences of rat and mouse segmented filamentous bacteria, a potent inducer of th17 cell differentiation. Cell Host Microbe. 2011;10:273–284. doi: 10.1016/j.chom.2011.08.007. [DOI] [PubMed] [Google Scholar]
- Saha S, Takeshita F, Matsuda T, Jounai N, Kobiyama K, Matsumoto T, Sasaki S, Yoshida A, Xin KQ, Klinman DM, et al. Blocking of the TLR5 activation domain hampers protective potential of flagellin DNA vaccine. J Immunol. 2007;179:1147–1154. doi: 10.4049/jimmunol.179.2.1147. [DOI] [PubMed] [Google Scholar]
- Salim SY, Soderholm JD. Importance of disrupted intestinal barrier in inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17:362–381. doi: 10.1002/ibd.21403. [DOI] [PubMed] [Google Scholar]
- Sanders CJ, Yu Y, Moore DA, Williams IR, Gewirtz AT. Humoral immune response to flagellin requires T cells and activation of innate immunity. J Immunol. 2006;177:2810–2818. doi: 10.4049/jimmunol.177.5.2810. [DOI] [PubMed] [Google Scholar]
- Sanders DS. Mucosal integrity and barrier function in the pathogenesis of early lesions in Crohn's disease. J Clin Pathol. 2005;58:568–572. doi: 10.1136/jcp.2004.021840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Andersen-Nissen E, Hayashi F, Strobe K, Bergman MA, Barrett SL, Cookson BT, Aderem A. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat Immunol. 2003;4:1247–1253. doi: 10.1038/ni1011. [DOI] [PubMed] [Google Scholar]
- Stanton TB, Savage DC. Motility as a factor in bowel colonization by Roseburia cecicola, an obligately anaerobic bacterium from the mouse caecum. J Gen Microbiol. 1984;130:173–183. doi: 10.1099/00221287-130-1-173. 1984. [DOI] [PubMed] [Google Scholar]
- Stewart FJ, Ottesen EA, DeLong EF. Development and quantitative analyses of a universal rRNA-subtraction protocol for microbial metatranscriptomics. ISME J. 2010;4:896–907. doi: 10.1038/ismej.2010.18. [DOI] [PubMed] [Google Scholar]
- Suo Z, Yang X, Avci R, Deliorman M, Rugheimer P, Pascual DW, Idzerda Y. Antibody selection for immobilizing living bacteria. Analyt Chem. 2009;81:7571–7578. doi: 10.1021/ac9014484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, Fagarasan S. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci U S A. 2004;101:1981–1986. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasteyre A, Barc MC, Collignon A, Boureau H, Karjalainen T. Role of FliC and FliD flagellar proteins of Clostridium difficile in adherence and gut colonization. Infect Immun. 2001;69:7937–7940. doi: 10.1128/IAI.69.12.7937-7940.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Waaij LA, Limburg PC, Mesander G, van der Waaij D. In vivo IgA coating of anaerobic bacteria in human faeces. Gut. 1996;38:348–354. doi: 10.1136/gut.38.3.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Woude MW, Baumler AJ. Phase and antigenic variation in bacteria. Clin Microbiol Rev. 2004;17:581–611. doi: 10.1128/CMR.17.3.581-611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verberkmoes NC, Russell AL, Shah M, Godzik A, Rosenquist M, Halfvarson J, Lefsrud MG, Apajalahti J, Tysk C, Hettich RL, et al. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 2009;3:179–189. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.