Abstract
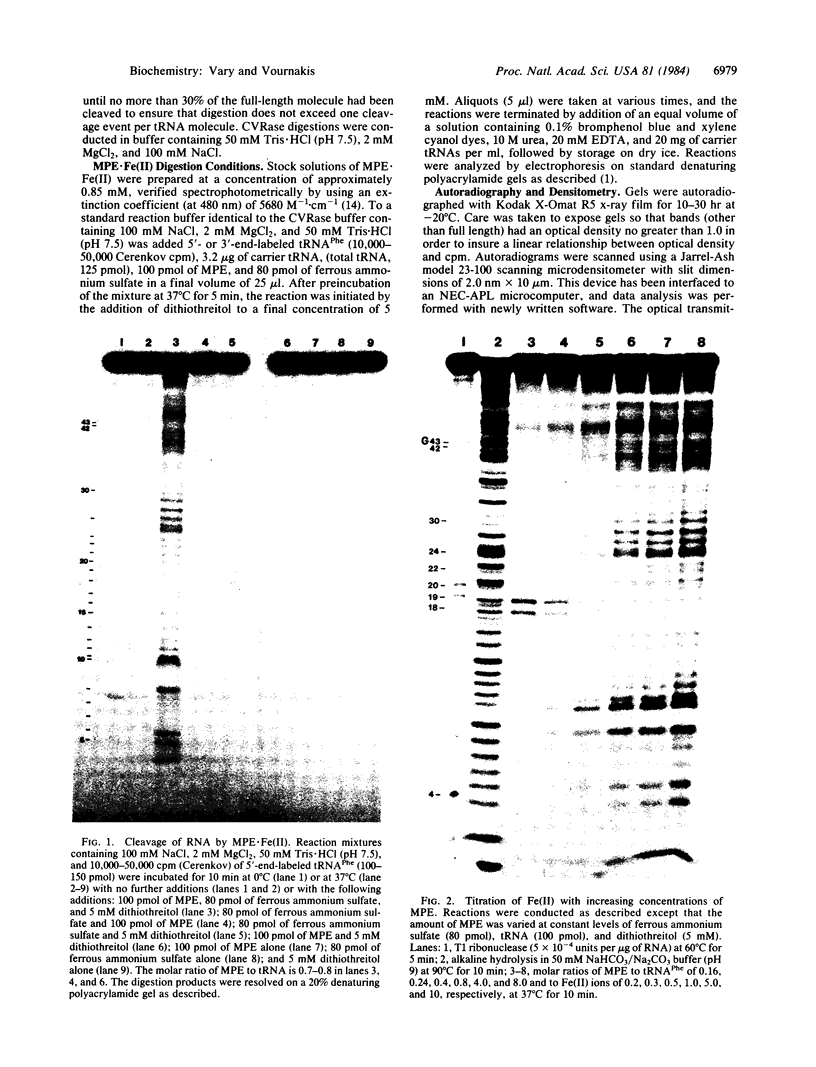
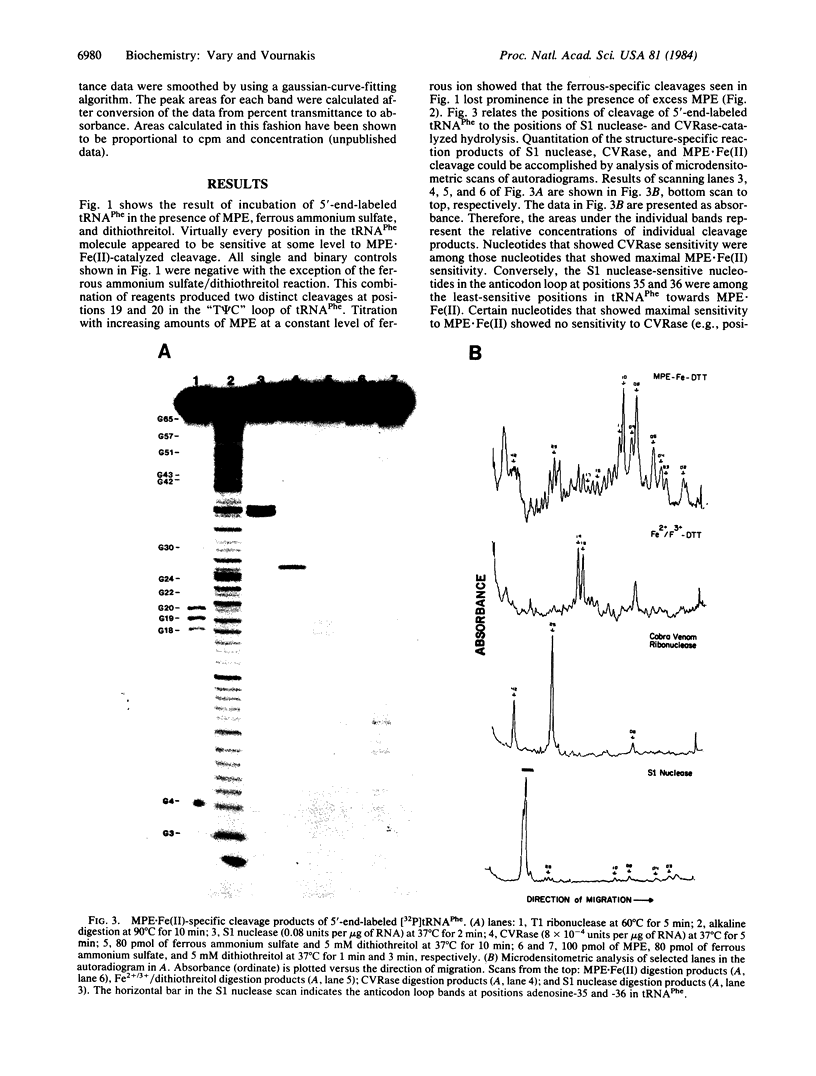
Methidiumpropyl-EDTA.Fe(II) [MPE.Fe(II)] in the presence of dithiothreitol, is shown to cleave phenylalanine-accepting tRNA (tRNAPhe) in a structure-specific fashion. Molar ratios of MPE.Fe(II) to tRNAPhe of less than 1 preferentially cleave phosphodiester bonds known to occur in double-stranded regions of the tRNAPhe molecule. Microdensitometric analysis of autoradiograms of MPE.Fe(II) cleavage products following gel electrophoresis reveals a correspondence between preferred sites of MPE.Fe(II) cleavage and sites in tRNAPhe most sensitive to cobra venom ribonuclease, a double-strand-specific endoribonuclease. Conversely, sites of cleavage by the single-strand-specific S1 nuclease correspond to those nucleotides that are least susceptible to MPE.Fe(II) hydrolysis. Sensitive helical regions in tRNAPhe include the dihydrouracil and the "T psi C" stems, which cannot be detected by cobra venom ribonuclease because of steric constraints. Phosphodiester bonds within the T psi C and dihydrouracil loop regions, which are not detected by S1 nuclease under rigorously controlled digestion conditions, are revealed by inference from their relative insensitivity to MPE.Fe(II). These results demonstrate the utility of MPE.Fe(II) as a general small molecular weight probe of RNA structure, having a greater accessibility to base-paired regions than do the more bulky enzymic probes.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England T. E., Bruce A. G., Uhlenbeck O. C. Specific labeling of 3' termini of RNA with T4 RNA ligase. Methods Enzymol. 1980;65(1):65–74. doi: 10.1016/s0076-6879(80)65011-3. [DOI] [PubMed] [Google Scholar]
- Jones C. R., Kearns D. R. Identification of a unique ethidium bromide binding site on yeast tRNAPhe by high resolution (300 MHz) nuclear magnetic resonance. Biochemistry. 1975 Jun 17;14(12):2660–2665. doi: 10.1021/bi00683a016. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Quigley G. J., Suddath F. L., McPherson A., Sneden D., Kim J. J., Weinzierl J., Rich A. Three-dimensional structure of yeast phenylalanine transfer RNA: folding of the polynucleotide chain. Science. 1973 Jan 19;179(4070):285–288. doi: 10.1126/science.179.4070.285. [DOI] [PubMed] [Google Scholar]
- Liebman M., Rubin J., Sundaralingam M. Nonintercalative binding of ethidium bromide to nucleic acids: crystal structure of an ethidium--tRNA molecular complex. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4821–4825. doi: 10.1073/pnas.74.11.4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J. W., Tinoco I., Jr Intercalation of ethidium ion into DNA and RNA oligonucleotides. Biopolymers. 1984 Feb;23(2):213–233. doi: 10.1002/bip.360230205. [DOI] [PubMed] [Google Scholar]
- Pavlakis G. N., Lockard R. E., Vamvakopoulos N., Rieser L., RajBhandary U. L., Vournakis J. N. Secondary structure of mouse and rabbit alpha- and beta-globin mRNAs: differential accessibility of alpha and beta initiator AUG codons towards nucleases. Cell. 1980 Jan;19(1):91–102. doi: 10.1016/0092-8674(80)90391-8. [DOI] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A., Douthwaite S., Garrett R. A., Noller H. F. A "bulged" double helix in a RNA-protein contact site. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7331–7335. doi: 10.1073/pnas.78.12.7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A., Herr W. Chemical probing of the tRNA--ribosome complex. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2273–2277. doi: 10.1073/pnas.78.4.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley G. J., Teeter M. M., Rich A. Structural analysis of spermine and magnesium ion binding to yeast phenylalanine transfer RNA. Proc Natl Acad Sci U S A. 1978 Jan;75(1):64–68. doi: 10.1073/pnas.75.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troutt A., Savin T. J., Curtiss W. C., Celentano J., Vournakis J. N. Secondary structure of Bombyx mori and Dictyostelium discoideum 5S rRNA from S1 nuclease and cobra venom ribonuclease susceptibility, and computer assisted analysis. Nucleic Acids Res. 1982 Jan 22;10(2):653–664. doi: 10.1093/nar/10.2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C. C., Jain S. C., Sobell H. M. Visualization of drug-nucleic acid interactions at atomic resolution. I. Structure of an ethidium/dinucleoside monophosphate crystalline complex, ethidium:5-iodouridylyl (3'-5') adenosine. J Mol Biol. 1977 Aug 15;114(3):301–315. doi: 10.1016/0022-2836(77)90252-2. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Hertzberg R. P., Dervan P. B. Map of distamycin, netropsin, and actinomycin binding sites on heterogeneous DNA: DNA cleavage-inhibition patterns with methidiumpropyl-EDTA.Fe(II). Proc Natl Acad Sci U S A. 1982 Sep;79(18):5470–5474. doi: 10.1073/pnas.79.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vary C. P., Vournakis J. N. RNase H-catalyzed site-specific deadenylylation of rabbit alpha- and beta- globin mRNAs. Secondary structure of 3'-noncoding regions. J Biol Chem. 1984 Mar 10;259(5):3299–3307. [PubMed] [Google Scholar]
- Vary C. P., Vournakis J. N. Secondary structure of eukaryotic messenger RNA. Biochem Soc Symp. 1982;47:61–78. [PubMed] [Google Scholar]
- Vournakis J. N., Celantano J., Finn M., Lockard R. E., Mitra T., Pavlakis G., Troutt A., van den Berg M., Wurst R. M. Sequence and structure analysis of end-labeled RNA with nucleases. Gene Amplif Anal. 1981;2:267–298. [PubMed] [Google Scholar]
- Winkle S. A., Rosenberg L. S., Krugh T. R. On the cooperative and noncooperative binding of ethidium to DNA. Nucleic Acids Res. 1982 Dec 20;10(24):8211–8223. doi: 10.1093/nar/10.24.8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurst R. M., Vournakis J. N., Maxam A. M. Structure mapping of 5'-32P-labeled RNA with S1 nuclease. Biochemistry. 1978 Oct 17;17(21):4493–4499. doi: 10.1021/bi00614a021. [DOI] [PubMed] [Google Scholar]






