Abstract
We are concerned with multiple imputation of the ratio of two variables, which is to be used as a covariate in a regression analysis. If the numerator and denominator are not missing simultaneously, it seems sensible to make use of the observed variable in the imputation model. One such strategy is to impute missing values for the numerator and denominator, or the log-transformed numerator and denominator, and then calculate the ratio of interest; we call this ‘passive’ imputation. Alternatively, missing ratio values might be imputed directly, with or without the numerator and/or the denominator in the imputation model; we call this ‘active’ imputation. In two motivating datasets, one involving body mass index as a covariate and the other involving the ratio of total to high-density lipoprotein cholesterol, we assess the sensitivity of results to the choice of imputation model and, as an alternative, explore fully Bayesian joint models for the outcome and incomplete ratio. Fully Bayesian approaches using Winbugs were unusable in both datasets because of computational problems. In our first dataset, multiple imputation results are similar regardless of the imputation model; in the second, results are sensitive to the choice of imputation model. Sensitivity depends strongly on the coefficient of variation of the ratio's denominator. A simulation study demonstrates that passive imputation without transformation is risky because it can lead to downward bias when the coefficient of variation of the ratio's denominator is larger than about 0.1. Active imputation or passive imputation after log-transformation is preferable. © 2013 The Authors. Statistics in Medicine published by John Wiley & Sons, Ltd.
Keywords: missing data, multiple imputation, ratios, compatibility
1 Introduction
Missing values of covariates are a common problem in regression analyses. Missing data are classified as being missing completely at random (MCAR) if missingness does not depend on observed or unobserved data, missing at random (MAR) if missingness does not depend on unobserved data given observed data, or missing not at random if missingness depends on missing data even given the observed data 1. Amongst methods that attempt to deal with missing data, rather than discarding them, multiple imputation (MI) can provide valid inference under MAR and has become popular in practice since its inception over 30 years ago 2.
Briefly, MI works as follows. Missing values are replaced with imputed values, drawn from their posterior predictive distribution under a model given the observed data. We term this model the imputation model. The process is repeated M > 1 times, giving M imputed datasets with no missing values. Each imputed dataset is analysed using the model that would have been used had the missing values been observed. We call this model the analysis model. The M estimates of each parameter of interest are then combined using ‘Rubin's rules’ 3. When the imputation model is correctly specified, Rubin's rules can provide standard errors and confidence intervals that fully incorporate uncertainty due to missing data.
MI is an attractive tool for analyses with missing data: The nuisance issue of modelling missing data is neatly separated from the analyses of substantive interest; the imputation model can make use of auxiliary variables that it would be undesirable to include as covariates in the analysis model (such as post-baseline measurements in a randomised controlled trial); the same M imputed datasets can be used for a variety of substantive analyses; and the imputation model can be tailored to reflect possible departures from MAR, which is helpful for sensitivity analysis.
Ratios are commonly used as covariates in regression analyses; examples are body mass index (BMI = Weight in kg ÷ (Height in m) 2) 4, waist–hip ratio 5, urinary albumin-to-creatinine ratio (Albumin concentration in mg/g ÷ Creatinine concentration in mg/g) 6, and what we refer to as ‘cholesterol ratio’ (Total cholesterol in mg/dL ÷ HDL in mg/dL) 7.
An individual's ratio measurement may be missing for one of the three reasons:
The denominator is missing.
The numerator is missing.
Both components are missing.
For both 1 and 2, the ratio is semi-missing rather than fully missing; that is, one of the two components is observed. Ratio missingness due to more than one of these reasons for different observations in the same dataset means it is not obvious how best to impute the ratio. A mixture of reasons 1 and 2 is particularly awkward.
One reasonable question at this stage is, ‘Why use a ratio covariate?’ There are mathematical arguments against their use 8. Senn and Julious claim that ratios are always poor candidates for parametric analysis unless the components, and therefore the ratio, follow a lognormal distribution or the ratio's coefficient of variation (CV) is small 9. We make three points. First, applied researchers do use ratios, and we are unlikely to persuade them to stop, especially because the use of certain ratios is well established; we should be pragmatic and try to guide practitioners on how to analyse datasets involving incomplete ratio covariates. Second, arguments against ratios assume that a ratio is not the correct functional form for a covariate, but it may be. Third, ratios are not used by accident: A ratio may be of genuine substantive interest when its separate components are not. For example, BMI is widely used because it measures weight-for-height and as such is regarded as a proxy measure of body fat. Substantive interest is in the influence of body fat on outcome, not weight or height. Weight alone may be considered a measure of body fat, but BMI is measured with less error because it aims to remove the effect of height (although it may not do so completely or accurately). It is our opinion that when researchers propose a relationship they believe, such as the influence of a ratio on outcome, this should not be cast aside lightly. The substantive question should not be altered for statistical convenience unless we have little choice.
We assume that the aims of analysis are unbiased estimation of a parameter describing the association between a ratio and some outcome, confidence intervals with the ascribed coverage and fully efficient parameter estimation. There may be other covariates in the analysis model, and primary interest may be in one of these, but the properties of the ratio parameter estimator are important nonetheless. There has been no previous methodological work on MI for a ratio covariate, although White et al. 10 and Bartlett et al. 11 allude to the issue, but practitioners are imputing ratio covariates nonetheless 12. We aim to highlight issues with imputing an incomplete ratio covariate and to identify imputation strategies that are practicable for applied statisticians.
Despite the positive features listed previously, MI is neither the only approach to dealing with missing covariates, nor necessarily the best approach for any given analysis. Joint models for the outcome and covariates may be superior because they make use of the full likelihood in a coherent way. In this paper, we also investigate results for fully Bayesian joint models.
The remainder of this paper is as follows. In Section 2, we introduce and describe our two motivating datasets; in Section 3, we consider candidate models for imputing incomplete ratios. Section 4 presents two case studies, contrasting the different imputation models (for the datasets introduced in Section 2). Section 5 presents a simulation study in a simpler setting than our case studies; and Section 6 is a discussion.
2 Datasets: Aurum and EPIC-Norfolk
For both of our datasets, regression analyses involving a ratio as a covariate have previously been published 4,7. The analysis models used in our example analyses are not the same as the original articles because of the following: (i) we want to keep the analysis models and imputation models relatively simple, and (ii) we do not wish to make any substantive claims about these data. Therefore, we have chosen to use analysis models resembling but not matching those used in the earlier publications 4,7.
For both datasets, the analysis model is the Cox model,
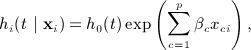 |
(1) |
where H0(t) is the nonparametric baseline hazard function at time t, hi(t | xi) is the hazard for the ith individual and xci is the value of the cth covariate in the ith individual. Survival (or censoring) times are assumed to be fully observed.
2.1 The Aurum cohort
The Aurum dataset comes from a South African cohort study of 1350 HIV-infected participants starting antiretroviral therapy. Participants were recruited from 27 centres in five provinces between February 2005 and June 2006 and followed to March 2007. Information was recorded on a range of baseline characteristics, and participants were followed up for death. The aim of the work by Russell et al. 4 was to estimate the influence of hæmoglobin on mortality using a Cox model. Of the participants, 1348 had a recorded time of death/censoring, with 185 deaths occurring within the follow-up time. We restrict our analysis to these 1348 individuals.
The analysis model is (1) with p = 6, where x1, … ,x6 are age in years, sex, hæmoglobin in g/mL, viral load in copies per mL, CD4 count in cells per μL and BMI. Table 1 provides a summary of these covariates and of weight and height. We give transformations of the covariates used in the analysis model, and summarise the transformed measure in the final column. Note that 381 (28%) patients are missing a weight and/or height measurement, but only five of these have height missing when weight is observed. Five of the covariates are continuous, and one (sex, which is complete) is categorical. Hæmoglobin, weight, height 2 and BMI appear to be approximately normal on the transformed scale, while (log) viral load and (square root of) CD4 count do not. We focus on the estimation of β3 and β6, the log hazard ratios for hæmoglobin and BMI, respectively, (hæmoglobin was the focus of the original publication 4).
Table 1.
Aurum summary of covariates and of the analysis model and components of body mass index (BMI); n = 1348.
| Covariate | Frequency missing (%) | Mean (SD) or frequency (%) | |
|---|---|---|---|
| x1 | Age (years) | 0 (0%) | 37 (9) |
| x2 | Sex: male | 0 (0%) | 542 (40%) |
| x3 | Hæmoglobin (g/mL) | 143 (11%) | 11.4 (2.3) |
| x4 | *Viral load (copies per mL) | 162 (12%) | 4.8 (0.8)† |
| x5 | *CD4 count (cells per μL) | 94 (7%) | 8.9 (4.5)† |
| x6 = a1 / a2 | BMI (kg/m 2) | 381 (28%) | 21.9 (4.9) |
| a1 | ‡Weight (kg) | 376 (28%) | 58 (12) |
| a2 | ‡Height (m 2) | 275 (20%) | 2.7 (0.3)† |
Transformation used for viral load is log 10(x4); transformation used for CD4 count is 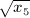 . These are standard transformations in HIV research, and we use them in the imputation models and the analysis models.
. These are standard transformations in HIV research, and we use them in the imputation models and the analysis models.
†Summarised on transformed scale.
‡Only enters into the analysis model via BMI.
2.2 The EPIC-Norfolk cohort
The European Prospective Investigation Into Cancer and Nutrition (EPIC)-Norfolk study is a large cohort study designed to investigate the link between dietary factors and cancer. Dietary and non-dietary factors were collected at baseline, and participants were followed up for cancer and non-cancer outcomes. We use some of the non-dietary characteristics as covariates and time to death as the outcome.
The analysis model is (1) with p = 6, where x1, … ,x6 are age, sex, smoking status, systolic blood pressure, diastolic blood pressure and cholesterol ratio. We summarise these six covariates and total cholesterol and HDL in Table 2; none are transformed. In total, 2155 (9%) participants are missing a total cholesterol and/or HDL measurement. Total cholesterol is always missing when HDL is missing. Incomplete covariates are all continuous and appear approximately normal, except for HDL, which is positively skewed. We focus on the estimation of β6, the log hazard ratio for cholesterol ratio.
Table 2.
EPIC-Norfolk summary of covariates of the analysis model and of components of cholesterol ratio; n = 22 754.
| Covariate | Frequency missing (%) | Mean (SD) or frequency (%) | |
|---|---|---|---|
| x1 | Age (years) | 0 (0%) | 59 (9) |
| x2 | Sex: male | 0 (0%) | 10145 (45%) |
| x3 | Smoking status: ever smoked | 0 (0%) | 11971 (53%) |
| x4 | Systolic blood pressure (mm Hg) | 52 (<1%) | 135 (18) |
| x5 | Diastolic blood pressure (mm Hg) | 52 (<1%) | 82 (11) |
| x6 = a1 / a2 | Cholesterol ratio | 2155 (9%) | 4.7 (1.6) |
| a1 | †Total cholesterol (mg/dl) | 1514 (7%) | 6.2 (1.2) |
| a2 | †HDL (mg/dl) | 2155 (9%) | 1.4 (0.4) |
†Only enters into the analysis model via cholesterol ratio.
3 Methods and models
3.1 Model for analysis
The analysis model is the Cox model (1) with p covariates (x1, … ,xp) made up of the ratio xp = a1 / a2 and p − 1 other covariates (x1, … ,xp − 1), which we denote (z,w) where z are incomplete and w are complete (in both example datasets, we have z and w).
3.2 Models for missing data
We list candidate models for the covariates in Table 3 (note the Label column, which we henceforth use to refer to models). For MI, the outcome must be explicitly included as a covariate in the imputation model 13. In Table 3, we denote outcome by f(yi). For the Cox model, f(yi) involves a censoring indicator and the Nelson–Aalen estimate of the cumulative hazard function to the survival time (an approximation to the cumulative baseline hazard function H0(t) 14), included as separate covariates in the imputation model. When the analysis model is linear or logistic regression, f(yi) = yi.
Table 3.
Candidate imputation models for xi.
| Imputation model | Label | Relationship to analysis model |
|---|---|---|
| (zi,xpi | f(yi),wi) ∼ MVN | M1 | Compatible |
| (zi,xpi,a1i | f(yi),wi) ∼ MVN | M2 | Semi-compatible |
| (zi,xpi,a2i | f(yi),wi) ∼ MVN | M3 | Semi-compatible |
| (zi,xpi,a1i,a2i | f(yi),wi) ∼ MVN | M4 | Semi-compatible |
| †(zi,a1i,a2i | f(yi),wi) ∼ MVN | M5 | Incompatible |
| ‡(zi,ln(a1i),ln(a2i) | f(yi),wi) ∼ MVN | M6 | Incompatible |
†Passive imputation of 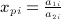 is required.
is required.
‡Passive imputation of xpi = exp[ln(a1i) − ln(a2i)] is required.
3.3 Compatibility in relation to active and passive imputation
Multiple imputation can provide an approximation to fitting a joint model if the models for imputation and analysis are compatible 15, where a joint model may be either maximum likelihood or Bayesian (if the joint model is Bayesian, compatibility also requires that priors are non-zero over the entire parameter space). Considering whether or not the models M1–M6 are compatible with the analysis model helps us to formulate hypotheses and understand future results.
By ‘compatible’, we mean that a joint model exists that implies both the imputation model and the analysis model as conditional models. This does not mean that the joint model is correct, but that the analysis model and imputation model are both implied by it, and so the MI procedure is coherent. Appendix A describes how to tell if models are compatible and works through two examples of imputation models where one is compatible and the other is not (A.1 and A.2, respectively).
Compatibility is related the concept of ‘congeniality’, and the term congeniality is often used to mean compatibility 16,17,10. Congeniality requires that the joint model from which the imputation and analysis models can be derived is Bayesian. Further, the researcher's incomplete and complete data procedures must be specified, and the inferences must be asymptotically equivalent to a Bayesian model. We refer interested readers to Meng 18.
Non-compatibility of models is not always problematic; Meng 18 and Rubin 19 have both shown that there can be some benefit to using imputation models that correctly draw on information not used by the analysis model. Collins, Schafer and Kam demonstrate via simulation that auxiliary variables (i.e. variables that are in the imputation model but not the analysis model) are unlikely to be harmful, and may be of benefit by making the MAR assumption more plausible, while ‘restrictive’ imputation strategies can lead to problems 20.
We therefore distinguish between two types of non-compatibility: If there is a special case of the imputation model that is compatible with the analysis model, as when it includes auxiliary variables, then the imputation model is termed ‘semi-compatible’ (following Liu et al. 21); otherwise, the imputation model is simply termed ‘incompatible’. In previous work, imputation models that are compatible or semi-compatible appear to perform well even when misspecified 22,23, but this is not necessarily true for imputation models that are incompatible 23,20. We hypothesise that imputation models that are compatible or semi-compatible will be more robust to modest degrees of misspecification than models that are incompatible.
Imputation of a ratio is performed either actively or passively. Of the imputation models listed in Table 3, only M1 is compatible with the analysis model. Of the remaining models M2–M4, which use active imputation, are semi-compatible because they include a1 and/or a2, which do not appear in the analysis model, as auxiliary variables in the imputation model; models M5 and M6, which use passive imputation, are incompatible with the analysis model because xp is present in the analysis model but not in the imputation model, while a1 and a2 are present in the imputation model but not in the analysis model. We expect models M5 and M6 to be prone to bias and poor coverage, despite making use of all the observed data when imputing the ratio.
3.4 Motivation for missing data models
The choice of a model listed in Table 3 might be motivated by the way it makes use of observed information in a1,a2, which will depend on the pattern of missingness.
Model M1 may be a good approach when a1,a2 are missing simultaneously. If a1 is only missing when a2 is missing, M2 may be used because model M2 makes use of observed a1 values when imputing the ratio, and there is no information in a2 about missing values of a1 that might be used to improve imputation of xp. (Conversely, if a2 is only missing when a1 is missing, M3 may be attractive.) Note that M2 and M3 do not respect the deterministic relationship xp = a1 / a2.
Model M4 makes use of information on a1,a2 by imputing both alongside xp; this may be motivated by having a1, a2 or both missing. This is similar to the approach advocated by von Hippel 22, which has been termed just another variable 10,23. As with M2 and M3, the model ignores the deterministic relationship xp = a1 / a2 and assumes multivariate normality. This will appear a bizarre assumption; it is clearly wrong because the distributions of two of these variables must define the distribution of the third, yet software does not know this and will sample without complaint. If the assumption made by M4 is uncomfortable, we may be attracted to M5 or M6.
Model M5 is incompatible with the analysis model (Appendix A.1) and requires xp to be imputed passively from imputed values of a1 / a2. The components a1,a2 are not auxiliary but completely determine the values of xp. The ratio of a1 and a2, which are both normal, is expected to be heavy tailed.
M6 alters the problem by transforming xp into a linear function of its logged components and passively imputing it. Model M6 guarantees that imputed values of a1,a2 are positive, as with all observed ratios. While this may be desirable, it is important to remember that our primary goal is valid inference, and we are not trying to recreate the missing values 19,24. The cosmetics of this model should therefore be a secondary consideration.
We have omitted from Table 3 the imputation model (zi,ln(xpi) | f(yi)) ∼ MVN. We do not consider this because ln(xp) = ln(a1) − ln(a2) where ln(a1) and ln(a2) are normal, and the sum of two normal distributions is normal. Model M6 is therefore equivalent to imputing ln(xp) but makes more use of the observed data when components are not simultaneously missing. The only setting where modelling ln(xp) alone is appropriate is if (a1,a2) are always either both observed or both missing. In this case, the model would then be equivalent to M6.
To summarise our discussion of the models in Table 3, there are conceptual problems with each one: Model M1 is compatible with the analysis model but does not use information on observed a1 or a2 when the other component is missing; M2–M4 are likely to be misspecified; and M5 and M6, the two models that make use of all the observed information on a1 and a2 and respect the relationship xp = a1 / a2, are incompatible with the analysis model.
3.5 Software and details of imputation
We used Stata 12's misuite for MI in our case studies and simulations in Section 5 25,26. We performed multiple imputation using mi impute mvn, and implemented Rubin's rules using mi estimate.
Advice on the number of imputations typically suggests that a small number (fewer than 10) is sufficient 16. This idea comes from comparing the length of confidence intervals based on M imputations to intervals based on ∞ imputations. Our view on choosing the number of imputations, described in White et al. 10, is slightly different, being based on the reproducibility of analyses. To achieve negligible Monte Carlo error from our MI analyses, we use M = 300 imputations for the Aurum case study and M = 100 for EPIC-Norfolk. Note that we are not advocating such large values of M in general.
Our imputation models, all of which are based on a multivariate normal model, used a burn-in of 1000 iterations of the MCMC chain. Thereafter, we stored imputed datasets at every 10th iteration of the chain.
4 Case studies
This section presents the results for MI. However, in analyses with missing data, Bayesian models are widely regarded as a sensible alternative if there is reason to be suspicious of MI results. We outline and present Bayesian analyses of the Aurum and Epic datasets, corresponding to the MI approaches presented in this section, in Appendix B.
4.1 Imputing body mass index in the Aurum cohort
The MI procedures took between 2 min, 7 s (M1) and 2 min, 44 s (M6) to impute 300 times, fit the analysis model in each imputed dataset and use Rubin's rules to combine estimates.
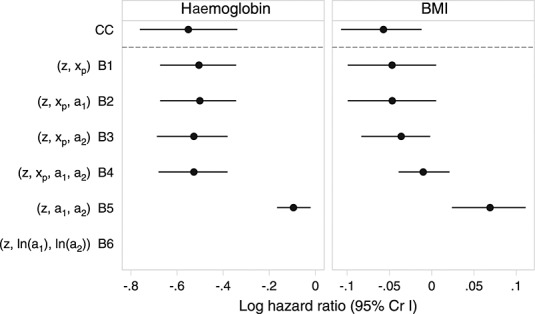
Figure 1 shows estimates resulting from different imputation models. There is very little difference in the point estimates or width of confidence intervals; all returned essentially the same result. The number of imputations meant Monte Carlo error was negligible, at a maximum reaching 1/50th of the estimated standard error. The relative efficiency versus infinite M was > 0.999 for all models. For both hæmoglobin and BMI, the MI estimates gave a slight change in the point estimate and a small reduction in the width of confidence intervals as compared to complete cases.
Figure 1.
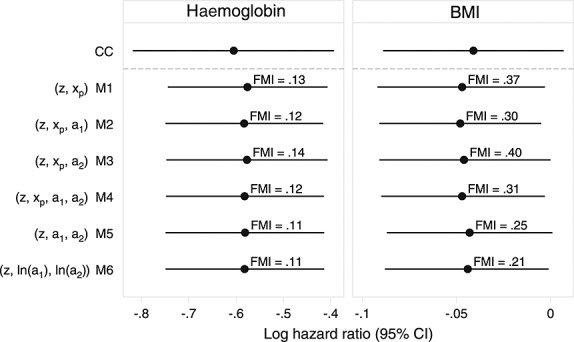
Results from analyses of Aurum data under different models for imputing body mass index (BMI). The estimated fraction of missing information (FMI) is given next to multiple imputation analyses.
4.2 Imputing cholesterol ratio in the EPIC-Norfolk cohort
For MI of the EPIC-Norfolk data, we used M = 100. We used a smaller number of imputations than in Aurum because only 9% of individuals were missing cholesterol ratio. MI took between 19 min, 2 s (M1) and 21 min, 0 s (M5) to impute 100 times, analyse each imputed dataset and combine estimates using Rubin's rules. The relative efficiency versus infinite M was > 0.999 for all models except M5, where relative efficiency was 0.991.
There was consistency between estimates from models that impute cholesterol ratio directly (Figure 2). Monte Carlo error for point estimates was negligible (around 0.0005, less than 1/50th of the standard error) for all models except M5 where it was 0.003. MI models are less consistent than in the Aurum MI analyses but would in five of six cases give similar substantive conclusions. These estimates are also very similar to complete-cases analysis and, interestingly, the imputation model that passively imputes cholesterol ratio through log-total cholesterol and log-HDL. However, the estimate after the standard passive imputation approach (M5) is much closer to the null, with wider confidence intervals.
Figure 2.
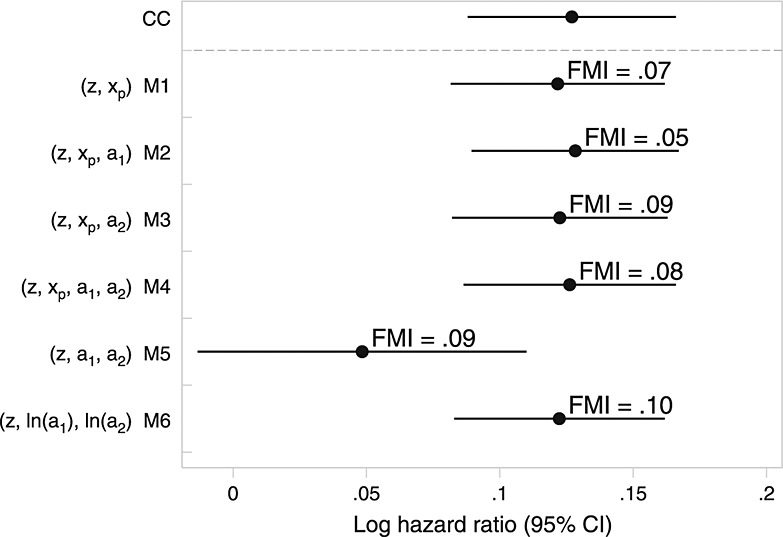
Results from analyses of EPIC-Norfolk data under different models for cholesterol ratio. The estimated fraction of missing information (FMI) is given next to multiple imputation analyses.
Figure 3 demonstrates the problem with model M5 in the EPIC-Norfolk data, plotting imputed values of cholesterol ratio from a single, typical, imputed dataset under models M1–M6 alongside 2155 randomly selected observed values. The largest observed value of cholesterol ratio was 15.7. Note that for model M5, some imputed values were very large or very small; plotting these extreme values distorted the y-axis, and so we have censored the y-axis below − 3 and above +20, ranking and listing the values of imputed HDL and cholesterol ratio values outside of this range.
Figure 3.
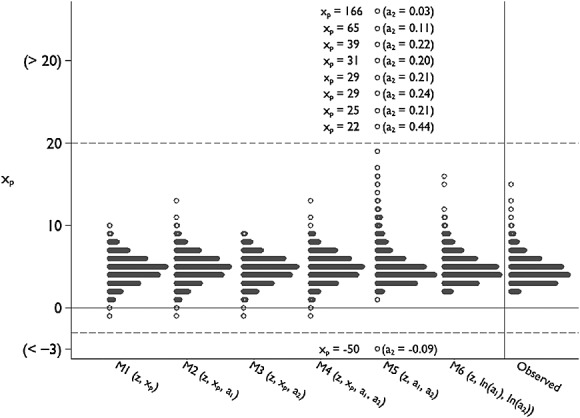
Dotplot of imputed cholesterol ratio for single (typical) imputed datasets in EPIC-Norfolk under models M1–M6. Imputed values of xp < 3 or xp > 20 are not plotted but represented according to rank; imputed values of (xp,a1) are listed.
The problem with M5 arises because the mean and SD of HDL are 1.42 and 0.42, respectively, meaning its coefficient of variation (CV) is 0.30, resulting in a danger of a2 being imputed close to zero or even negatively. This CV is far larger than in the Aurum data, where CV(height 2) = 0.11 and imputed values are never close to zero (data not shown).
Figure 3 also highlights the difference between the other imputation models. Imputation on the log scale (M6) is the only model to guarantee that a1,a2 and xp are positive. Further, the imputed values closely resemble the observed. M1–M4 can and did impute some xp < 0; these models all assume xp ∼ N, and so the distribution of imputed values is symmetrical about its mean. By looking at Figure 3, model M6 appears to be appealing, while from a statistical inference perspective (Figure 2), there appears to be little to choose between M6 and M1–M4. From all perspectives, M5 is a poor choice.
4.2.1 Predictive mean matching
A natural question about model M5 that arises from Figure 3 is whether removing the high-leverage points could reduce the bias. For example, a truncated normal imputation model could be used to invoke the constraint xp > 0, which would remove the negative outliers of model M5.
A better alternative, which can also remove the positive outliers, is predictive mean matching (PMM) 27,28,10. Briefly, the imputation model is fitted, and for each individual with a missing value, the k individuals (‘donors’) with observed values with the closest predicted mean are identified. One of these is selected at random and their value ‘donated’ as the imputed value. This ensures that imputed values are within the range of observed values.
To improve model M5, PMM is most easily implemented in a chained equations procedure 10. Imputation of a1 and a2 uses PMM, and xp is passively imputed. The largest possible imputed value of xp is then the ratio of the largest observed value of a1 to the smallest observed value of a2 (and vice versa for the smallest imputed value of xp).
We used this imputation model on the EPIC-Norfolk data, using k = 10 and storing imputed values after 10 cycles of chained equations. This reduced the bias of model M5, giving an estimated log-hazard ratio of 0.119 (95% CI 0.079–0.159). See Appendix C for the full results.
5 Simulation study
5.1 Design
We performed a simulation study designed to investigate models M1–M6 in a simpler setting than the two case studies. With xp as the only covariate and a continuous outcome y, we investigated the performance of the imputation models and how this varied with the strength of xp–y association and the CV of the ratio's denominator, CV(a2). This affects the distribution of xp, and we hypothesise that when CV(a2) is large, model M5 will be biased. An imputed value of a2i may be very small, meaning the corresponding value of xpi will be large, and possibly outside the range of observed xp. The xpi will thus have high leverage. For such values, there are unlikely to be appropriately large or small y to preserve the true xp–y relationship, which leads us to expect bias towards no association.
Scenarios investigated include two values of CV(a2): 0.1, taken from height 2 in the Aurum data, and 0.3, taken from HDL in the EPIC-Norfolk data; we vary these factorially with R2 values of 0.1 and 0.3. We performed all simulations using Stata 12 25. Our simulation procedures were as follows:
Simulate n = 500 complete values of ln(a1),ln(a2) to follow a bivariate normal distribution. In our first scenario, the mean, standard deviation and correlation are taken from ln(weight) and ln(height 2) in the Aurum data: ln(a1) has mean 4 and SD 0.21, ln(a2) has mean 0.97 and SD 0.11, and Corr(ln(a1),ln(a2)) = 0.22. This gives CV(a2) = 0.1.
Generate complete xp = exp(ln(a1) − ln(a2)), meaning that xp follows a lognormal distribution. For the ratios and components in our two example datasets, the lognormal distribution seems to be a suitable choice.
Simulate y ∼ N(β0 + β1xp, σ2). We used the same value of β1 (arbitrarily 2) throughout to make bias comparable across all simulation settings. To vary the strength of association, we altered σ2 to achieve the desired R2.
Simulate binary indicators of response, R1 and R2, for a1 and a2, respectively. Each R is generated independently from the model logit{P(R = 1)} = γ0 + γ1y. Under MCAR, γ1 = 0. Under MAR, γ1 is chosen so that ROC analysis of y versus an indicator of response R produces a mean area under the curve of 0.65. This is to achieve the same degree of MAR across scenarios. We then alter γ0 so that P(R1 = 1) = P(R2 = 1) = 0.75. Because γ1 has the same sign for both R1 and R2 and both depend on y, the probability of a1,a2 being missing simultaneously is slightly larger under MAR than MCAR. This means that the overall proportion of observations missing xp is slightly smaller under MAR (42% missing xp) than MCAR (44% missing xp).
Set a1i to missing if R1i = 0, a2i to missing if R2i = 0 and xpi to missing if R1i = 0 or R2i = 0.
Impute xp five times using each of the models M1–M6 (Table 3).
Fit the correct analysis model to each imputed dataset, and combine the results using Rubin's rules.
We used 5000 replicates of this process under each combination of simulation settings. Interest is in β1. We calculated bias, coverage of 95% confidence intervals and efficiency of 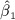 (expressed by the empirical standard error, SD
(expressed by the empirical standard error, SD 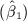 over all replications 29) under models M1–M6, with analysis of complete data (i.e. before any data are set to missing) and complete cases (dropping observations with missing xp) also provided for reference.
over all replications 29) under models M1–M6, with analysis of complete data (i.e. before any data are set to missing) and complete cases (dropping observations with missing xp) also provided for reference.
5.2 Results
Table 4 summarises the results of our simulation study. Results of the complete data and complete cases analyses are both as expected. Complete data are always unbiased with 95% coverage and the smallest empirical standard error of all methods. Complete cases are unbiased under MCAR but biased under MAR. Coverage is correspondingly low, and efficiency is lower than complete data.
Table 4.
Simulation results: bias, coverage and efficiency of different imputation models.
| Bias (β1 = 2) |
Empirical SE |
Coverage |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| R2 | CV (a2) | Imputation model | MCAR | MAR | MCAR | MAR | MCAR | MAR | |
| 0.1 | 0.1 | Complete data | 0.000 | 0.273 | 95.2 | ||||
| Complete cases | 0.003 | − 0.172 | 0.366 | 0.352 | 95.1 | 92.6 | |||
| x | M1 | − 0.005 | − 0.004 | 0.368 | 0.386 | 93.8 | 94.9 | ||
| x,a1 | M2 | − 0.001 | 0.002 | 0.333 | 0.345 | 94.6 | 94.7 | ||
| x,a2 | M3 | − 0.009 | − 0.003 | 0.363 | 0.383 | 94.6 | 94.9 | ||
| x,a1,a2 | M4 | − 0.005 | 0.005 | 0.330 | 0.342 | 94.7 | 95.0 | ||
| a1,a2 | M5 | − 0.017 | − 0.016 | 0.328 | 0.337 | 94.8 | 95.0 | ||
| ln(a1),ln(a2) | M6 | − 0.016 | − 0.034 | 0.329 | 0.332 | 94.9 | 95.1 | ||
| 0.1 | 0.3 | Complete data | 0.006 | 0.267 | 95.3 | ||||
| Complete cases | 0.001 | − 0.168 | 0.359 | 0.351 | 95.3 | 92.9 | |||
| x | M1 | − 0.009 | 0.005 | 0.358 | 0.385 | 94.7 | 94.9 | ||
| x,a1 | M2 | − 0.007 | 0.014 | 0.348 | 0.372 | 94.9 | 94.9 | ||
| x,a2 | M3 | − 0.001 | 0.031 | 0.334 | 0.362 | 95.4 | 95.0 | ||
| x,a1,a2 | M4 | − 0.001 | 0.038 | 0.325 | 0.346 | 95.0 | 94.7 | ||
| a1,a2 | M5 | − 0.562 | − 0.665 | 0.350 | 0.334 | 94.3 | 92.6 | ||
| ln(a1),ln(a2) | M6 | − 0.038 | − 0.064 | 0.313 | 0.318 | 95.8 | 95.4 | ||
| 0.3 | 0.1 | Complete data | 0.003 | 0.137 | 95.2 | ||||
| Complete cases | 0.001 | − 0.139 | 0.183 | 0.188 | 95.5 | 88.5 | |||
| x | M1 | − 0.005 | 0.031 | 0.171 | 0.187 | 95.3 | 94.0 | ||
| x,a1 | M2 | − 0.003 | 0.026 | 0.159 | 0.171 | 95.8 | 95.0 | ||
| x,a2 | M3 | − 0.007 | 0.029 | 0.170 | 0.188 | 95.2 | 93.8 | ||
| x,a1,a2 | M4 | − 0.003 | 0.026 | 0.159 | 0.171 | 95.9 | 94.6 | ||
| a1,a2 | M5 | − 0.016 | 0.000 | 0.158 | 0.168 | 96.1 | 95.3 | ||
| ln(a1),ln(a2) | M6 | − 0.016 | − 0.031 | 0.158 | 0.163 | 96.2 | 95.6 | ||
| 0.3 | 0.3 | Complete data | − 0.002 | 0.137 | 95.0 | ||||
| Complete cases | − 0.006 | − 0.143 | 0.184 | 0.192 | 94.9 | 88.5 | |||
| x | M1 | − 0.009 | 0.054 | 0.174 | 0.196 | 94.2 | 93.0 | ||
| x,a1 | M2 | − 0.012 | 0.057 | 0.172 | 0.193 | 94.8 | 93.3 | ||
| x,a2 | M3 | − 0.010 | 0.076 | 0.170 | 0.191 | 94.3 | 91.5 | ||
| x,a1,a2 | M4 | − 0.009 | 0.080 | 0.167 | 0.187 | 94.2 | 91.8 | ||
| a1,a2 | M5 | − 0.580 | − 0.814 | 0.287 | 0.300 | 94.3 | 93.3 | ||
| ln(a1),ln(a2) | M6 | − 0.051 | − 0.070 | 0.162 | 0.164 | 95.1 | 94.6 | ||
SE, standard error; CV, coefficient of variation; MCAR, missing completely at random; MAR, missing at random.
M1 is mainly unbiased, but there is a small upward bias under MAR and R2 = 0.3, and coverage is slightly low when data are MAR. This is perhaps because it assumes normality for xp when it is actually lognormal. M1 also tends to be inefficient compared to other imputation models, as would be expected, regardless of the missingness mechanism.
With this general pattern of missingness, M3 is usually more biased than M2, although coverage tends to be similar (except where CV(a2) = 0.3 and R2 = 0.3). Efficiency of M2 and M3 seems to depend on CV(a2) and R2. Model M4 has similar bias to M2 and M3; at worst, this reaches about 4% with both large CV(a2) and R2. Empirical standard errors for M4 are at least as small as M2 and M3, while coverage tends to be good except when both CV(a2) and R2 are 0.3.
Model M5 performs well in the two scenarios when CV (a2) = 0.1. There is a small downward bias, but efficiency and coverage are both good compared with other methods. However, when CV(a2) = 0.3, we observe unacceptable bias towards the null and lower efficiency than other methods, although coverage is still over 90%. When considered alongside bias, this coverage implies that while the empirical standard error is large, the estimated standard errors are even larger, reducing the effect of the large bias on coverage and implying low power.
M6 is more biased than M5 when CV(a2) = 0.1 but much less so when CV(a2) = 0.3. Across all of our settings, it is more efficient than M1–M5 and with coverage close to 95%. If the small bias seems acceptable, then this is the best imputation model.
6 Discussion
We have presented the results of two case studies involving commonly used ratios and a simulation study based in part on these datasets. A key message is the caution against passive imputation of a1 and a2 without prior transformation. Superficially, the approach appears to make more use of the available data; however, it is often inefficient and can suffer from large bias. Our analysis of the EPIC-Norfolk data demonstrated this problem in practice. However, in our Aurum case study, the use of passive imputation appeared to make little difference to the substantive results compared to active imputation. Our simulation study confirmed that problems arise when CV(a2) is large. Note that a ratio with very small CV(a2) is unlikely to be used in applied work (unless CV(a1) is also very small) because as CV (a2) → 0, xp becomes a function of a1 divided by a constant. We therefore recommend that incomplete ratios be imputed actively or passively after log transformation as in model M6.
In considering models for missing data, joint models for the covariates and outcome are attractive because they use the full data likelihood in a coherent way. In our two case studies, we attempted to fit fully Bayesian joint models and summarise posterior distributions for parameters of interest. Computational problems prevented this approach from being useful. In one dataset, some of the models did not appear to converge to any true posterior distribution (or if they did, results were extraordinarily sensitive to the choice of model for the ratio). In the other dataset, it was not possible to load the observed data into Winbugs, and so the attempt was abandoned.
Compatibility is a useful concept for considering whether various imputation models are sensible. We hypothesised that models M1 and M2–M4 would perform well because of being compatible and semi-compatible respectively, while models M5 and M6 would perform poorly because of being incompatible. In our simulations, M1–M4 did tend to perform well despite being misspecified, and model M5 did often perform poorly. In our EPIC-Norfolk example, where model M5 gave nonsense results, problems could be identified by inspecting the imputed values of xp.
Model M6 was surprisingly as good as any other model considered throughout. Despite being more robust than M5, we know it is not completely ‘safe’. In our simulation study, the imputation model assumed (log(a1),log(a2) | y) ∼ N, and because log(xp) = log(a1) − log(a2), this implies (log(xp) | y) ∼ N. The imputation model therefore has mean function log(xp) = α0 + α1y, while the analysis model has mean function y = β0 + β1xp. In further simulations, we noted that M6 was still robust when R2 = 0.5 and CV(a2) = 0.3 (results not shown). We can provide no guarantee for greater values other than that this model will eventually fall apart. However, it is our experience that associations stronger than R2 = 0.5 are rare in medical applications.
Some of the issues with model M5 could have been alleviated by using partly parametric imputation techniques such as PMM 30 or local residual draws 28. In practice, this requires a switch to the chained equations approach rather than a multivariate imputation model. Because a parametric model is used only to identify suitable donors, this makes it impossible to think about compatibility. We investigated PMM in the problematic EPIC-Norfolk dataset and found model M5 much improved. PMM may therefore be a useful adjunct to a suitably chosen imputation model.
In evaluating methods, we have focused on bias, coverage and efficiency. For those interested in accurate prediction, efficiency may be more important and coverage less so or even unimportant 31. It is worth noting that precision is also lower for model M5. Therefore, if passive imputation is to be used for a ratio in prediction settings, it should be performed on the log scale.
We have considered the imputation of ratio covariates. Some similar issues arise when the analysis model contains any nonlinear function, for example, interactions and squares. The difference is that in both cases, the main effects and their interaction, or the variable and its square, are included in the analysis model. In the case of squares, a measurement and its square will also be observed or missing simultaneously. Imputation is then complicated by the fact that the analysis model contains both the untransformed variable and a nonlinear function as covariates, rather than just the nonlinear function, as in the case of ratios. This makes issues around compatibility somewhat more complicated. See von Hippel 22, Seaman et al. 23 and Bartlett et al. 11 for recent work on imputation of squares and interactions.
Bartlett et al. proposed the use of rejection sampling when producing imputations and showed it to be useful for imputing squares and interactions; this may therefore be a good approach for imputing ratios. By explicitly involving the analysis model in the specification of the imputation model, each imputation model used in the chained equations is compatible with the imputation model 11. However, the method is more time intensive than any imputation models investigated here, and it is yet to become available in standard software packages. It also sacrifices one of the advantages of MI: separation of missing data issues from substantive analyses. However, this may be necessary and has already been partly conceded when we tailor imputation models to be compatible with the analysis model.
Appendix A. Compatibility
Section 3.3 models are compatible if a joint model exists that implies both as conditionals. How can we tell whether there is a joint model underpinning both the imputation model and the analysis model? Arnold et al. give a theorem that is restated here for clarity 32.
Theorem 1
Given two conditional densities f(x | y) and g(y | x), a joint density exists if and only if {(x,y) : f(x | y) > 0} = {(x,y) : g(y | x) > 0}, and there exist functions u(x) and v(y) such that, first,
| (2) |
and, second, u(x) is integrable.
Here, u(x) is a marginal density for x, and v(y) is a marginal density for y. Later, we posit an analysis model and check compatibility against two different imputation models using (2).
We distinguish between two kinds of non-compatibility:
Semi-compatibility: There is a special case of the imputation model that is compatible with the analysis model.
Incompatibility: There is no case of the imputation model that is compatible with the analysis model.
That is, if setting certain parameters of the imputation model to 0 yields a compatible model, the imputation model is drawing on more information than the analysis model and is richer rather than the same, hence semi-compatible. If parameters of the imputation model cannot be set to 0 to identify a compatible model, the imputation model is using different information to or less information than the analysis model. Previous work has shown that incompatibility can be harmless or beneficial 18–20. When the analysis model is correctly specified, these are examples of using semi-compatible imputation models, while incompatible imputation models are always harmful when the analysis model is correctly specified.
Appendices A.1 and A.2 work through two simple examples. For both, the analysis model involves only the ratio as a covariate. Appendix A.1 uses model M5 and is shown to be incompatible; A.2 uses model M1 and is shown to be compatible.
Instead of dividing the densities, we subtract the log-densities. For clarity, we omit the intercept terms α0 and β0 from the imputation model and the analysis model, respectively, assuming both equal zero. Note that because neither parameter involves a1,a2 nor y, this does not impact on compatibility.
A.1 Imputation model incompatible with the analysis model
Suppose the proposed analysis model is a linear regression of y on the ratio a1 / a2. The log-density for this is
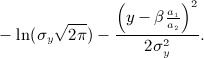 |
(3) |
The proposed imputation model is a bivariate normal model,
which has log-density
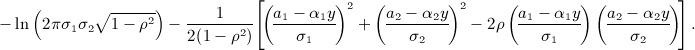 |
(4) |
The imputation model (4) is of the form b(a1,a2) + c(y) + d1(a1y) + d2(a2y) and the analysis model (A.1) is of the form 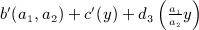 . Subtracting one from the other, we cannot express the result as u(a1,a2) − v(y), indicating that they are incompatible.
. Subtracting one from the other, we cannot express the result as u(a1,a2) − v(y), indicating that they are incompatible.
A.2 Imputation model compatible with the analysis model
The proposed analysis model is as in (A.1), and so the log-density is given by (3). However, the imputation model involves a linear regression of 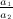 on y. The log-density is
on y. The log-density is
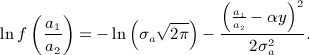 |
(5) |
Subtracting (5) from (3), we obtain
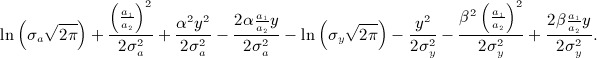 |
(6) |
By setting 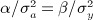 , we can express (6) without any terms involving both (a1,a2) and y, indicating that for any choice of (
, we can express (6) without any terms involving both (a1,a2) and y, indicating that for any choice of (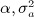 ), there are values of (
), there are values of (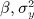 ) for which the proposed imputation model is compatible with the analysis model.
) for which the proposed imputation model is compatible with the analysis model.
Appendix B. Bayesian models for an incomplete ratio
It is conceptually natural to model missing covariates using Bayesian methods. The problem discussed in Section 3.3, that the imputation model and the analysis model may not correspond to any joint model, does not exist for Bayesian models, where the model for missing data and the analysis model are joint. The compatibility between the missing data model and the analysis model is thus assured.
The practical disadvantage of fully Bayesian models for an incomplete ratio and/or its components is computation. Bayesian models are also in general more computationally demanding than MI. Further, the imputation models described previously could be implemented fairly automatically using a choice of software, while the Bayesian models require knowledge of Winbugs 33 and/or the ability to code the models manually in another package.
Here, we explore whether Bayesian models, by working with the full joint likelihood, will provide more coherent results than MI. In our example datasets, we aim to obtain posterior means and credible intervals under various models.
B.1 Models, software and priors
A Bayesian model combines model (1) with a model for the incomplete covariates given the complete covariates. We list candidate Bayesian models for the covariates in Table 5 (again, note the Label column, where the number corresponds to the imputation model with equivalent motivation). Section 3.5 and Appendix B.2 give details of how the Cox model is fit. In contrast to MI, no explicit conditioning on the outcome is required for Bayesian models.
Table B.1.
Candidate fully Bayesian models for xi.
| Model for covariates | Label |
|---|---|
| (zi,xpi | wi) ∼ MVN | B1 |
| (zi,xpi,a1i | wi) ∼ MVN | B2 |
| (zi,xpi,a2i | wi) ∼ MVN | B3 |
| (zi,xpi,a1i,a2i | wi) ∼ MVN | B4 |
| (zi,a1i,a2i | wi) ∼ MVN | B5 |
| (zi,ln(a1i),ln(a2i) | wi) ∼ MVN | B6 |
Note that, except for the lack of issues around compatibility, the critique of the imputation models given in section 3.4 with equivalent labels applies equally to the Bayesian models given in Table 5. That is, models B1–B3 may ignore some of the observed data, while B2–B4 are likely to be misspecified to some degree.
To fit Bayesian joint models in our case studies, we used Winbugs 1.4.3 33. Because we are dealing with the Cox model, we used the method outlined in the Winbugs manuals (Leuk: survival analysis using Cox regression in Examples Volume I) to specify the models 34.
We used vague prior distributions for all parameters (see B.2 for details).
B.2 Details on Bayesian analyses
Below, we give Winbugs code used to demonstrate the setup of the fully Bayesian Cox model where xp is modelled and a1,a2 are ignored (this is the model denoted B1 in Table 3). Models B2–B6 differ only in that they simply specify the models for BMI, weight and height 2 differently.
The data file is made up of the covariates age sex hb logvl sqcd4 bmi, a vector of length N indicating death fail, a vector of length N of survival times for all individuals obst, and a vector of length T of distinct failure times t. Note that the data must be sorted in ascending order of obstbefore being passed to Winbugs. All covariates are centred at their mean.
 |
The priors for regression coefficients are ∼ N(0,100). The prior for dL0, the baseline intensity, requires slightly more explanation. This is modelled as dL0 ∼ Γ(cr{t(j + 1) − t(j)},c), that is, a gamma distribution with mean r{t(j + 1) − t(j)}and variance r{t(j + 1) − t(j)} / c. The expression {t(j + 1) − t(j)}is the time increment between the jth and j + 1th failure times; in the Aurum data, the mean time increment was 8 days. Note that r is not invariant to the scale of t, although c is. We used c = 0.1 and r = 0.1. A change of time scale would require r to be altered to obtain an equivalent prior distribution.
B.3 Results
Fitting the Bayesian models in Winbugs was troublesome.
For the Aurum data, all MCMC chains ran slowly, and some stalled persistently. The simplest models (for example B1) took 5–10 h to produce 5000 iterations of the MCMC sampler. Model B5 took 10 days to produce 1000 iterations and would only update under a very specific set of initial values. Winbugs stalled repeatedly, and the need to set the model updating again inflated the run time. We present results for model B5 but do not claim the MCMC sampling converged to the true posterior distribution. Results for model B6 are absent because Winbugs was unable to sample at all; the reason for this was unclear. Winbugs ran a lot faster when fitting models that imputed missing values of xp actively, that is, B1–B4.
Figure 4 presents results for the Aurum data (contrasting with the results obtained via MI in Figure 1). Posterior distributions obtained from different fully Bayesian analyses give diverse results. For hæmoglobin, posterior means for all models except B5 are slightly closer to 0 than any of the MI models, and the 95% credible intervals tend to be slightly shorter than the MI confidence intervals. This may in part be the effect of the prior for the hazard, as seen in the comparison of Bayesian and frequentist analysis of complete cases. Under model B5, the posterior distribution for the log hazard ratio had mean much closer to zero with smaller posterior variance than under other models.
Figure B.1.
Results from analyses of Aurum data under different Bayesian models for body mass index (BMI).
For BMI, posterior means from B1–B5 are very variable. B1 and B2 largely agree with the MI and (Bayesian) complete cases estimates, although the intervals are longer than those obtained after MI. Posterior means from B3 and B4 are closer to 0 and have shorter credible intervals than MI models or the other Bayesian models. For B4, this perhaps reflects the incorrect assumption made about the joint distribution of xp,a1,a2 (this is surprising because the issue does not appear to affect model M4). Model B5 shows an effect in the opposite direction to all other estimates. This was the model that was very difficult to run in Winbugs. As noted previously, we do not claim B5 ever converged to the true posterior density.
For the EPIC-Norfolk data, it was not possible to compile any of the fully Bayesian models in Winbugs, even for complete cases. We tried compiling the complete cases model for subsets of the data of gradually increasing size (starting with n = 1000); model compilation failed beyond n > 4000. The EPIC-Norfolk dataset is too large for Winbugs, and so attempts to fit the fully Bayesian models were abandoned. This is a setting where a fully Bayesian analysis is impractical to any but the most dedicated.
Appendix C. Results for EPIC-Norfolk after imputation using predictive mean matching
As described in Section 4.2.1, we re-ran the imputation models for EPIC-Norfolk using PMM. Figure 5 gives the full results analogous to those given in Figure 2. Note that, with the exception of model M5, there is less consistency between models than between the models that did not use PMM. Note also that the fraction of missing information is uniformly greater for the models that use PMM.
Figure C.1.
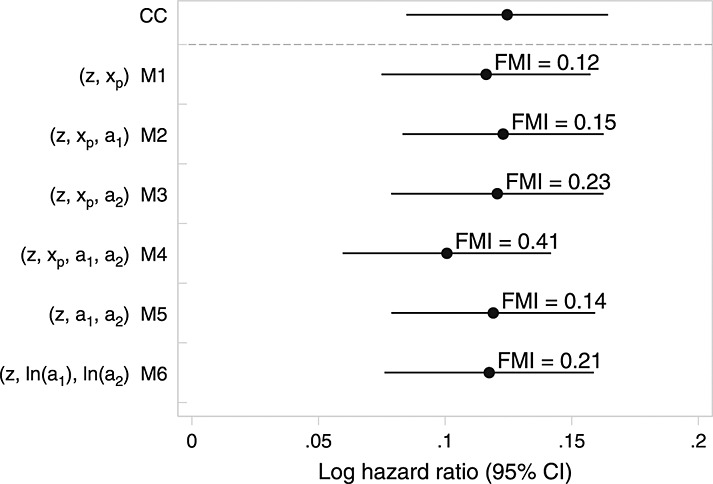
Results from analyses of EPIC-Norfolk data under different models for cholesterol ratio using predictive mean matching. The estimated fraction of missing information (FMI) is given next to multiple imputation analyses.
Acknowledgments
We are grateful to the Aurum Institute and to the EPIC-Norfolk team for allowing us to use their data and to three anonymous reviewers for their comments and suggestions. Tim Morris is supported by a UK MRC studentship A731-5QL14, Ian White and Shaun Seaman are supported by UK MRC Unit Programme U105260558 and grant MC-US-A030-0015, Patrick Royston is supported by a UK MRC grant A735-5QP21, and Angela Wood is supported by UK MRC grant G0701619.
References
- Rubin DB. Inference and missing data. Biometrika. 1976;63:581–592. [Google Scholar]
- Rubin DB. 1978. pp. 20–28. Multiple imputations in sample surveys: a phenomenological Bayesian approach to nonresponse Proceedings of the Survey Research Methods Section, American Statistical Association.
- Rubin DB. Multiple Imputation for Nonresponse in Surveys. New York: John Wiley and Sons; 1987. [Google Scholar]
- Russell E, Charalambous S, Pemba L, Churchyard G, Grant A, Fielding K. Low haemoglobin predicts early mortality among adults starting antiretroviral therapy in an HIV care programme in South Africa: a cohort study. BMC Public Health. 2010;10(1):433+. doi: 10.1186/1471-2458-10-433. DOI: 10.1186/1471-2458-10-433. http://dx.doi.org/10.1186/1471-2458-10-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf S, Hawken S, Ôunpuu S, Bautista L, Franzosi MG, Commerford P, Lang CC, Rumboldt Z, Onen CL, Lisheng L. Obesity and the risk of myocardial infarction in 27,000 participants from 52 countries: a case-control study. The Lancet. 2005;366(9497):1640–1649. doi: 10.1016/S0140-6736(05)67663-5. DOI: 10.1016/s0140-6736(05)67663-5. http://dx.doi.org/10.1016/s0140-6736(05)67663-5. [DOI] [PubMed] [Google Scholar]
- Wang TJ, Gona P, Larson MG, Tofler GH, Levy D, Newton-Cheh C, Jacques PF, Rifai N, Selhub J, Robins SJ, Benjamin EJ, D'Agostino RB, Vasan RS. Multiple biomarkers for the prediction of first major cardiovascular events and death. New England Journal of Medicine. 2006;355(25):2631–2639. doi: 10.1056/NEJMoa055373. DOI: 10.1056/nejmoa055373. http://dx.doi.org/10.1056/nejmoa055373. [DOI] [PubMed] [Google Scholar]
- Arsenault BJ, Rana JS, Stroes ESG, Despres JP, Shah PK, Kastelein JJP, Wareham NJ, Boekholdt SM, Khaw KT. Beyond low-density lipoprotein cholesterol: respective contributions of non-high-density lipoprotein cholesterol levels, triglycerides, and the total cholesterol/high-density lipoprotein cholesterol ratio to coronary heart disease risk in apparently healthy men and women. Journal of the American College of Cardiology. 2010;55(1):35–41. doi: 10.1016/j.jacc.2009.07.057. DOI: 10.1016/j.jacc.2009.07.057. http://dx.doi.org/10.1016/j.jacc.2009.07.057. [DOI] [PubMed] [Google Scholar]
- Allison DB, Paultre F, Goran MI, Poehlman ET, Heymsfield SB. Statistical considerations regarding the use of ratios to adjust data. International journal of obesity and related metabolic disorders: journal of the International Association for the Study of Obesity. 1995;19(9):644–652. http://view.ncbi.nlm.nih.gov/pubmed/8574275. [PubMed] [Google Scholar]
- Senn S, Julious S. Measurement in clinical trials: a neglected issue for statisticians? Statistics in Medicine. 2009;28(26):3189–3209. doi: 10.1002/sim.3603. DOI: 10.1002/sim.3603. http://dx.doi.org/10.1002/sim.3603. [DOI] [PubMed] [Google Scholar]
- White IR, Royston P, Wood AM. Multiple imputation using chained equations: issues and guidance for practice. Statistics in Medicine. 2011;30(4):377–399. doi: 10.1002/sim.4067. DOI: 10.1002/sim.4067. http://dx.doi.org/10.1002/sim.4067. [DOI] [PubMed] [Google Scholar]
- Bartlett JW, Seaman SR, White IR, Carpenter JR. 2013. Multiple imputation of covariates by fully conditional specification: accommodating the substantive model http://arxiv.org/abs/1210.6799.
- Hippisley-Cox J, Coupland C, Vinogradova Y, Robson J, May M, Brindle P. Derivation and validation of QRISK, a new cardiovascular disease risk score for the United Kingdom: prospective open cohort study. BMJ. 2007;335(7611):136+. doi: 10.1136/bmj.39261.471806.55. DOI: 10.1136/bmj.39261.471806.55. http://dx.doi.org/10.1136/bmj.39261.471806.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moons K, Donders R, Stijnen T, Harrel F. Using the outcome for imputation of missing predictor values was preferred. Journal of clinical epidemiology. 2006;59(10):1092–1101. doi: 10.1016/j.jclinepi.2006.01.009. DOI: 10.1016/j.jclinepi.2006.01.009. http://dx.doi.org/10.1016/j.jclinepi.2006.01.009. [DOI] [PubMed] [Google Scholar]
- White IR, Royston P. Imputing missing covariate values for the Cox model. Statistics in Medicine. 2009;28(15):1982–1998. doi: 10.1002/sim.3618. DOI: 10.1002/sim.3618. http://dx.doi.org/10.1002/sim.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer JL. Multiple imputation in multivariate problems when the imputation and analysis models differ. Statistica Neerlandica. 2003;57(1):19–35. DOI: 10.1111/1467-9574.00218. http://dx.doi.org/10.1111/1467-9574.00218. [Google Scholar]
- Schafer JL. Multiple imputation: a primer. Statistical Methods in Medical Research. 1999;8(1):3–15. doi: 10.1177/096228029900800102. DOI: 10.1177/096228029900800102. http://dx.DOI.org/10.1177/096228029900800102. [DOI] [PubMed] [Google Scholar]
- Andridge RR. Quantifying the impact of fixed effects modeling of clusters in multiple imputation for cluster randomized trials. Biometrical journal. Biometrische Zeitschrift. 2011;53(1):57–74. doi: 10.1002/bimj.201000140. DOI: 10.1002/bimj.201000140. http://dx.DOI.org/10.1002/bimj.201000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng XL. Multiple-imputation inferences with uncongenial sources of input. Statistical Science. 1994;9:538–558. [Google Scholar]
- Rubin DB. Multiple imputation after 18+ years. Journal of the American Statistical Association. 1996;91:473–489. [Google Scholar]
- Collins LM, Schafer JL, Kam CM. A comparison of inclusive and restrictive strategies in modern missing data procedures. Psychological methods. 2001;6(4):330–351. DOI: 10.1037//1082-989X.6.4.330-351. http://dx.DOI.org/10.1037//1082-989X.6.4.330-351. [PubMed] [Google Scholar]
- Liu J, Gelman A, Hill J, Su YS. 2012. On the Stationary Distribution of Iterative Imputations http://arxiv.org/abs/1012.2902.
- Von Hippel PT. How to impute squares, interactions, and other transformed variables. Sociological Methodology. 2009;39:265–291. DOI: 10.1111/j.1467-9531.2009.01215.x. http://dx.DOI.org/10.1111/j.1467-9531.2009.01215.x. [Google Scholar]
- Seaman SR, Bartlett JW, White IR. Multiple imputation of missing covariates with non-linear effects and interactions: an evaluation of statistical methods. BMC medical research methodology. 2012;12(1):46+. doi: 10.1186/1471-2288-12-46. DOI: 10.1186/1471-2288-12-46. http://dx.DOI.org/10.1186/1471-2288-12-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer JL, Graham JW. Missing data: our view of the state of the art. Psychological Methods. 2002;7(2):147–177. DOI: 10.1037//1082-989x.7.2.147. http://dx.DOI.org/10.1037//1082-989x.7.2.147. [PubMed] [Google Scholar]
- StataCorp. Stata Statistical Software: Release 12. College Station, TX: Stata Press; 2011. [Google Scholar]
- 4905 Lakeway Drive, College Station, Texas 77845: Stata Press; 2011. Stata multiple-imputation reference manual release 12 StataCorp LP, [Google Scholar]
- Little RJA. Models for nonresponse in sample surveys. Journal of the American Statistical Association. 1982;77:237–250. [Google Scholar]
- Schenker N, Taylor JMG. Partially parametric techniques for multiple imputation. Computational Statistics & Data Analysis. 1996;22(4):425–446. DOI: 10.1016/0167-9473(95)00057-7. http://dx.DOI.org/10.1016/0167-9473(95)00057-7. [Google Scholar]
- White IR. simsum: analyses of simulation studies including Monte Carlo error. Stata Journal. 2010;10(3):369–385. [Google Scholar]
- Little RJA. Missing-data adjustments in large surveys. Journal of Business & Economic Statistics. 1988;6:287–296. [Google Scholar]
- Copas JB. Regression, prediction and shrinkage. Journal of the Royal Statistical Society. Series B (Methodological) 1983;45(3):311–354. DOI: 10.2307/2345402. http://dx.DOI.org/10.2307/2345402. [Google Scholar]
- Arnold BC, Castillo E, Sarabia JM. Conditionally specified distributions: an introduction. Statistical Science. 2001;16(3):249–265. http://www.jstor.org/stable/2676688. [Google Scholar]
- Lunn DJ, Thomas A, Best N, Spiegelhalter D. WinBUGS - a Bayesian modelling framework: concepts, structure, and extensibility. Statistics and Computing. 2000;10(4):325–337. DOI: 10.1023/a:1008929526011. http://dx.DOI.org/10.1023/a:1008929526011. [Google Scholar]
- Clayton D. 1994. Bayesian analysis of frailty models Technical Report MRC Biostatistics Unit, Cambridge,


