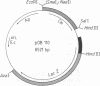
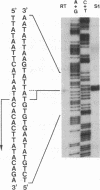
Abstract
The spo0B gene is one of the genes involved in initiation of sporulation of Bacillus subtilis. This gene, previously cloned into the pHV33 shuttle vector, is expressed in Escherichia coli and B. subtilis. We have determined the sequence of 1118 base pairs (bp) of the DNA insert carrying the spo0B gene. The promoter sequence of this gene shows the canonical T-A-T-A-A-T region at 10 bp from the transcriptional start (-10 region) but an unusual sequence, T-T-T-T-C-T-, in the -35 region. The nucleotide sequence shows an open reading frame encoding a 192-amino-acid polypeptide of Mr 22,542, which is close to the molecular weight of the spo0B product synthesized in E. coli minicells. To investigate the regulation of the spo0B gene under a variety of physiological conditions, we constructed an in-frame fusion between the spo0B promoter proximal region and the lacZ gene of E. coli. This hybrid gene was subsequently integrated into the B. subtilis chromosome, and the beta-galactosidase activity was measured. It was found that the spo0B gene is preferentially expressed during exponential growth; it is not induced by exhaustion of the growth medium nor repressed by glucose.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bonamy C., Szulmajster J. Cloning and expression of Bacillus subtilis spore genes. Mol Gen Genet. 1982;188(2):202–210. doi: 10.1007/BF00332676. [DOI] [PubMed] [Google Scholar]
- Brehm S. P., Le Hegarat F., Hoch J. A. Deoxyribonucleic acid-binding proteins in vegetative Bacillus subtilis: alterations caused by stage O sporulation mutations. J Bacteriol. 1975 Nov;124(2):977–984. doi: 10.1128/jb.124.2.977-984.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976 Jul 5;104(3):541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Dubnau E., Ramakrishna N., Cabane K., Smith I. Cloning of an early sporulation gene in Bacillus subtilis. J Bacteriol. 1981 Aug;147(2):622–632. doi: 10.1128/jb.147.2.622-632.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Débarbouillé M., Raibaud O. Expression of the Escherichia coli malPQ operon remains unaffected after drastic alteration of its promoter. J Bacteriol. 1983 Mar;153(3):1221–1227. doi: 10.1128/jb.153.3.1221-1227.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari F. A., Lang D., Ferrari E., Hoch J. A. Molecular cloning of the spo0B sporulation locus in bacteriophage lambda. J Bacteriol. 1982 Nov;152(2):809–814. doi: 10.1128/jb.152.2.809-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman M. Z., Chamberlin M. J. Developmental and genetic regulation of Bacillus subtilis genes transcribed by sigma 28-RNA polymerase. Cell. 1983 Nov;35(1):285–293. doi: 10.1016/0092-8674(83)90231-3. [DOI] [PubMed] [Google Scholar]
- Gilman M. Z., Wiggs J. L., Chamberlin M. J. Nucleotide sequences of two Bacillus subtilis promoters used by Bacillus subtilis sigma-28 RNA polymerase. Nucleic Acids Res. 1981 Nov 25;9(22):5991–6000. doi: 10.1093/nar/9.22.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henner D. J., Hoch J. A. The Bacillus subtilis chromosome. Microbiol Rev. 1980 Mar;44(1):57–82. doi: 10.1128/mr.44.1.57-82.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika H., Kobayashi Y., Kawamura F., Saito H. Cloning of sporulation gene spoOB of Bacillus subtilis and its genetic and biochemical analysis. J Bacteriol. 1981 May;146(2):494–505. doi: 10.1128/jb.146.2.494-505.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick R., Pero J. Cascades of Sigma factors. Cell. 1981 Sep;25(3):582–584. doi: 10.1016/0092-8674(81)90164-1. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McLaughlin J. R., Murray C. L., Rabinowitz J. C. Unique features in the ribosome binding site sequence of the gram-positive Staphylococcus aureus beta-lactamase gene. J Biol Chem. 1981 Nov 10;256(21):11283–11291. [PubMed] [Google Scholar]
- Moran C. P., Jr, Lang N., LeGrice S. F., Lee G., Stephens M., Sonenshein A. L., Pero J., Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186(3):339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- Mulligan M. E., Hawley D. K., Entriken R., McClure W. R. Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):789–800. doi: 10.1093/nar/12.1part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot P. J., Coote J. G. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976 Dec;40(4):908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna N., Dubnau E., Smith I. The complete DNA sequence and regulatory regions of the Bacillus licheniformis spoOH gene. Nucleic Acids Res. 1984 Feb 24;12(4):1779–1790. doi: 10.1093/nar/12.4.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Schaeffer P., Millet J., Aubert J. P. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci U S A. 1965 Sep;54(3):704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira S. K., Chou J., Richaud F. V., Casadaban M. J. New versatile plasmid vectors for expression of hybrid proteins coded by a cloned gene fused to lacZ gene sequences encoding an enzymatically active carboxy-terminal portion of beta-galactosidase. Gene. 1983 Nov;25(1):71–82. doi: 10.1016/0378-1119(83)90169-5. [DOI] [PubMed] [Google Scholar]
- Shimotsu H., Kawamura F., Kobayashi Y., Saito H. Early sporulation gene spo0F: nucleotide sequence and analysis of gene product. Proc Natl Acad Sci U S A. 1983 Feb;80(3):658–662. doi: 10.1073/pnas.80.3.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Stragier P., Richaud F., Borne F., Patte J. C. Regulation of diaminopimelate decarboxylase synthesis in Escherichia coli. I. Identification of a lysR gene encoding an activator of the lysA gene. J Mol Biol. 1983 Aug 5;168(2):307–320. doi: 10.1016/s0022-2836(83)80020-5. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Trowsdale J., Sheflett M., Hoch J. A. New cluster of ribosomal genes in Bacillus subtilis with regulatory role in sporulation. Nature. 1978 Mar 9;272(5649):179–181. doi: 10.1038/272179a0. [DOI] [PubMed] [Google Scholar]
- Wong S. L., Price C. W., Goldfarb D. S., Doi R. H. The subtilisin E gene of Bacillus subtilis is transcribed from a sigma 37 promoter in vivo. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1184–1188. doi: 10.1073/pnas.81.4.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber P., Losick R. Use of a lacZ fusion to study the role of the spoO genes of Bacillus subtilis in developmental regulation. Cell. 1983 Nov;35(1):275–283. doi: 10.1016/0092-8674(83)90230-1. [DOI] [PubMed] [Google Scholar]