Abstract
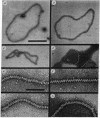
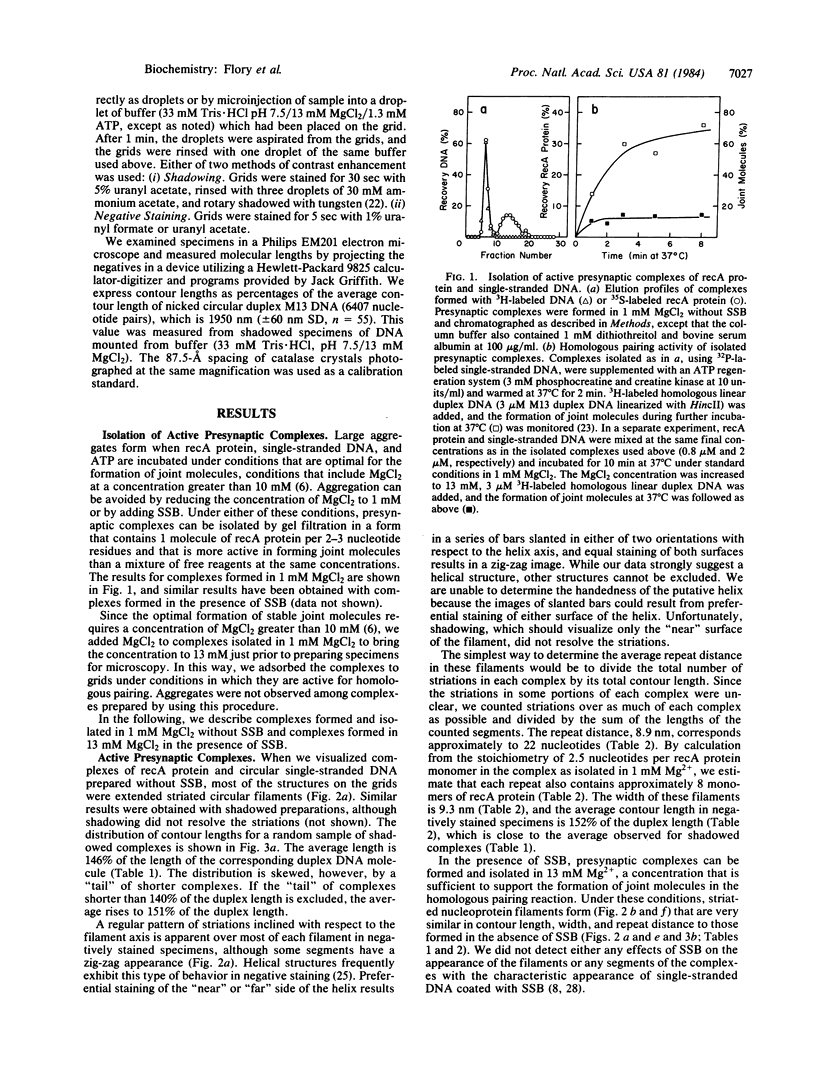
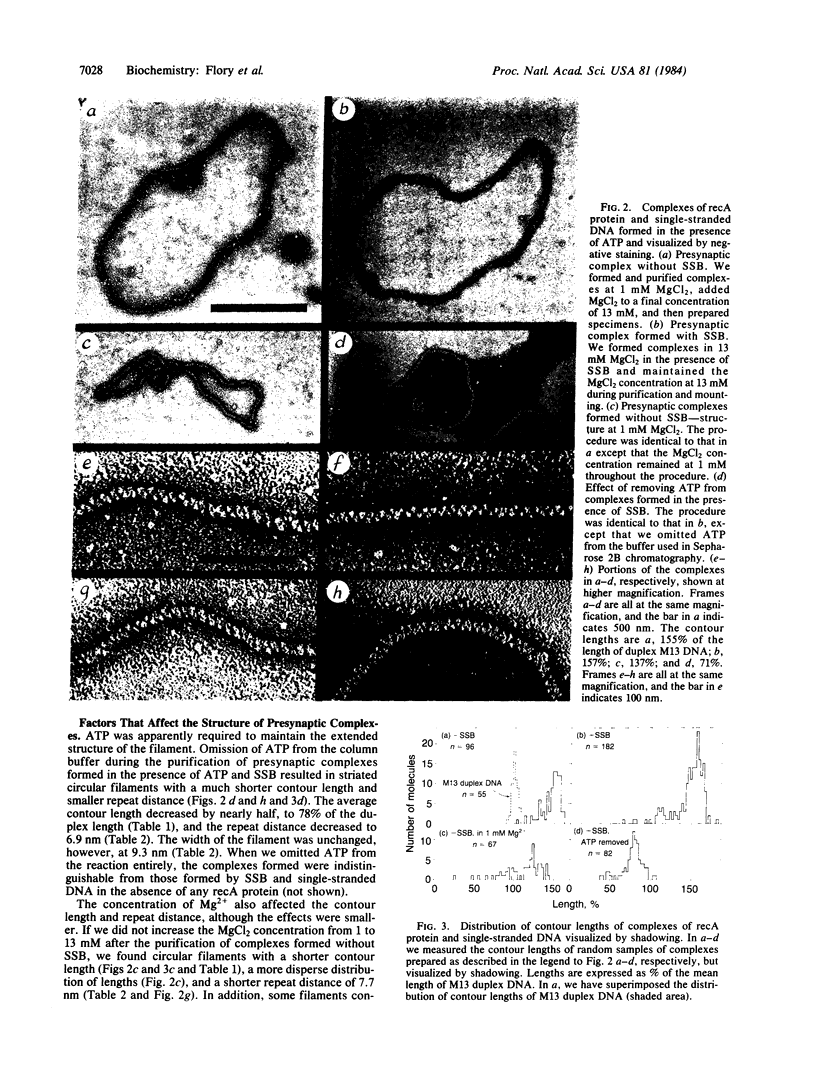
In the presence of ATP, recA protein forms a presynaptic complex with single-stranded DNA that is an obligatory intermediate in homologous pairing. Presynaptic complexes of recA protein and circular single strands that are active in forming joint molecules can be isolated by gel filtration. These isolated active complexes are nucleoprotein filaments with the following characteristics: (i) a contour length that is at least 1.5 times that of the corresponding duplex DNA molecule, (ii) an ordered structure visualized by negative staining as a striated filament with a repeat distance of 9.0 nm and a width of 9.3 nm, (iii) approximately 8 molecules of recA protein and 20 nucleotide residues per striation. The widened spacing between bases in the nucleoprotein filament means that the initial matching of complementary sequences must involve intertwining of the filament and duplex DNA, unwinding of the latter, or some combination of both to equalize the spacing between nascent base pairs. These experiments support the concept that recA protein first forms a filament with single-stranded DNA, which in turn binds to duplex DNA to mediate both homologous pairing and subsequent strand exchange.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bianchi M., DasGupta C., Radding C. M. Synapsis and the formation of paranemic joints by E. coli RecA protein. Cell. 1983 Oct;34(3):931–939. doi: 10.1016/0092-8674(83)90550-0. [DOI] [PubMed] [Google Scholar]
- Chrysogelos S., Griffith J. Escherichia coli single-strand binding protein organizes single-stranded DNA in nucleosome-like units. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5803–5807. doi: 10.1073/pnas.79.19.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrysogelos S., Register J. C., 3rd, Griffith J. The structure of recA protein-DNA filaments. 2 recA protein monomers unwind 17 base pairs of DNA by 11.5 degrees/base pair in the presence of adenosine 5'-O-(3-thiotriphosphate). J Biol Chem. 1983 Oct 25;258(20):12624–12631. [PubMed] [Google Scholar]
- Clark A. J. Recombination deficient mutants of E. coli and other bacteria. Annu Rev Genet. 1973;7:67–86. doi: 10.1146/annurev.ge.07.120173.000435. [DOI] [PubMed] [Google Scholar]
- Cox M. M., Lehman I. R. recA protein-promoted DNA strand exchange. Stable complexes of recA protein and single-stranded DNA formed in the presence of ATP and single-stranded DNA binding protein. J Biol Chem. 1982 Jul 25;257(14):8523–8532. [PubMed] [Google Scholar]
- Cox M. M., Soltis D. A., Livneh Z., Lehman I. R. On the role of single-stranded DNA binding protein in recA protein-promoted DNA strand exchange. J Biol Chem. 1983 Feb 25;258(4):2577–2585. [PubMed] [Google Scholar]
- Cunningham R. P., Shibata T., DasGupta C., Radding C. M. Single strands induce recA protein to unwind duplex DNA for homologous pairing. Nature. 1979 Sep 20;281(5728):191–195. doi: 10.1038/281191a0. [DOI] [PubMed] [Google Scholar]
- Di Capua E., Engel A., Stasiak A., Koller T. Characterization of complexes between recA protein and duplex DNA by electron microscopy. J Mol Biol. 1982 May 5;157(1):87–103. doi: 10.1016/0022-2836(82)90514-9. [DOI] [PubMed] [Google Scholar]
- Dunn K., Chrysogelos S., Griffith J. Electron microscopic visualization of recA-DNA filaments: evidence for a cyclic extension of duplex DNA. Cell. 1982 Apr;28(4):757–765. doi: 10.1016/0092-8674(82)90055-1. [DOI] [PubMed] [Google Scholar]
- Flory J., Radding C. M. Visualization of recA protein and its association with DNA: a priming effect of single-strand-binding protein. Cell. 1982 Apr;28(4):747–756. doi: 10.1016/0092-8674(82)90054-x. [DOI] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonda D. K., Radding C. M. By searching processively RecA protein pairs DNA molecules that share a limited stretch of homology. Cell. 1983 Sep;34(2):647–654. doi: 10.1016/0092-8674(83)90397-5. [DOI] [PubMed] [Google Scholar]
- Griffith J. D., Christiansen G. Electron microscope visualization of chromatin and other DNA-protein complexes. Annu Rev Biophys Bioeng. 1978;7:19–35. doi: 10.1146/annurev.bb.07.060178.000315. [DOI] [PubMed] [Google Scholar]
- Griffith J. D. Chromatin structure: deduced from a minichromosome. Science. 1975 Mar 28;187(4182):1202–1203. doi: 10.1126/science.187.4182.1202. [DOI] [PubMed] [Google Scholar]
- Iwabuchi M., Shibata T., Ohtani T., Natori M., Ando T. ATP-dependent unwinding of the double helix and extensive supercoiling by Escherichia coli recA protein in the presence of topoisomerase. J Biol Chem. 1983 Oct 25;258(20):12394–12404. [PubMed] [Google Scholar]
- KLUG A., BERGER J. E. AN OPTICAL METHOD FOR THE ANALYSIS OF PERIODICITIES IN ELECTRON MICROGRAPHS, AND SOME OBSERVATIONS ON THE MECHANISM OF NEGATIVE STAINING. J Mol Biol. 1964 Dec;10:565–569. doi: 10.1016/s0022-2836(64)80081-4. [DOI] [PubMed] [Google Scholar]
- Kahn R., Radding C. M. Separation of the presynaptic and synaptic phases of homologous pairing promoted by recA protein. J Biol Chem. 1984 Jun 25;259(12):7495–7503. [PubMed] [Google Scholar]
- Kobayashi I., Ikeda H. On the role of recA gene product in genetic recombination: an analysis by in vitro packaging of recombinant DNA molecules formed in the absence of protein synthesis. Mol Gen Genet. 1978 Oct 25;166(1):25–29. doi: 10.1007/BF00379725. [DOI] [PubMed] [Google Scholar]
- McEntee K., Weinstock G. M., Lehman I. R. Binding of the recA protein of Escherichia coli to single- and double-stranded DNA. J Biol Chem. 1981 Aug 25;256(16):8835–8844. [PubMed] [Google Scholar]
- McEntee K., Weinstock G. M., Lehman I. R. Initiation of general recombination catalyzed in vitro by the recA protein of Escherichia coli. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2615–2619. doi: 10.1073/pnas.76.6.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniyappa K., Shaner S. L., Tsang S. S., Radding C. M. Mechanism of the concerted action of recA protein and helix-destabilizing proteins in homologous recombination. Proc Natl Acad Sci U S A. 1984 May;81(9):2757–2761. doi: 10.1073/pnas.81.9.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani T., Shibata T., Iwabuchi M., Watabe H., Iino T., Ando T. ATP-dependent unwinding of double helix in closed circular DNA by recA protein of E. coli. Nature. 1982 Sep 2;299(5878):86–89. doi: 10.1038/299086a0. [DOI] [PubMed] [Google Scholar]
- Radding C. M., Flory J., Wu A., Kahn R., DasGupta C., Gonda D., Bianchi M., Tsang S. S. Three phases in homologous pairing: polymerization of recA protein on single-stranded DNA, synapsis, and polar strand exchange. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):821–828. doi: 10.1101/sqb.1983.047.01.094. [DOI] [PubMed] [Google Scholar]
- Radding C. M. Homologous pairing and strand exchange in genetic recombination. Annu Rev Genet. 1982;16:405–437. doi: 10.1146/annurev.ge.16.120182.002201. [DOI] [PubMed] [Google Scholar]
- Shibata T., Cunningham R. P., DasGupta C., Radding C. M. Homologous pairing in genetic recombination: complexes of recA protein and DNA. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5100–5104. doi: 10.1073/pnas.76.10.5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T., Cunningham R. P., Radding C. M. Homologous pairing in genetic recombination. Purification and characterization of Escherichia coli recA protein. J Biol Chem. 1981 Jul 25;256(14):7557–7564. [PubMed] [Google Scholar]
- Shibata T., DasGupta C., Cunningham R. P., Williams J. G., Osber L., Radding C. M. Homologous pairing in genetic recombination. The pairing reaction catalyzed by Escherichia coli recA protein. J Biol Chem. 1981 Jul 25;256(14):7565–7572. [PubMed] [Google Scholar]
- Shibata T., Ohtani T., Iwabuchi M., Ando T. D-loop cycle. A circular reaction sequence which comprises formation and dissociation of D-loops and inactivation and reactivation of superhelical closed circular DNA promoted by recA protein of Escherichia coli. J Biol Chem. 1982 Dec 10;257(23):13981–13986. [PubMed] [Google Scholar]
- Stasiak A., Di Capua E. The helicity of DNA in complexes with recA protein. Nature. 1982 Sep 9;299(5879):185–186. doi: 10.1038/299185a0. [DOI] [PubMed] [Google Scholar]
- Williams J. G., Shibata T., Radding C. M. Escherichia coli recA protein protects single-stranded DNA or gapped duplex DNA from degradation by RecBC DNase. J Biol Chem. 1981 Jul 25;256(14):7573–7582. [PubMed] [Google Scholar]
- Williams R. C. Use of polylysine for adsorption of nuclei acids and enzymes to electron microscope specimen films. Proc Natl Acad Sci U S A. 1977 Jun;74(6):2311–2315. doi: 10.1073/pnas.74.6.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A. M., Bianchi M., DasGupta C., Radding C. M. Unwinding associated with synapsis of DNA molecules by recA protein. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1256–1260. doi: 10.1073/pnas.80.5.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]