Abstract
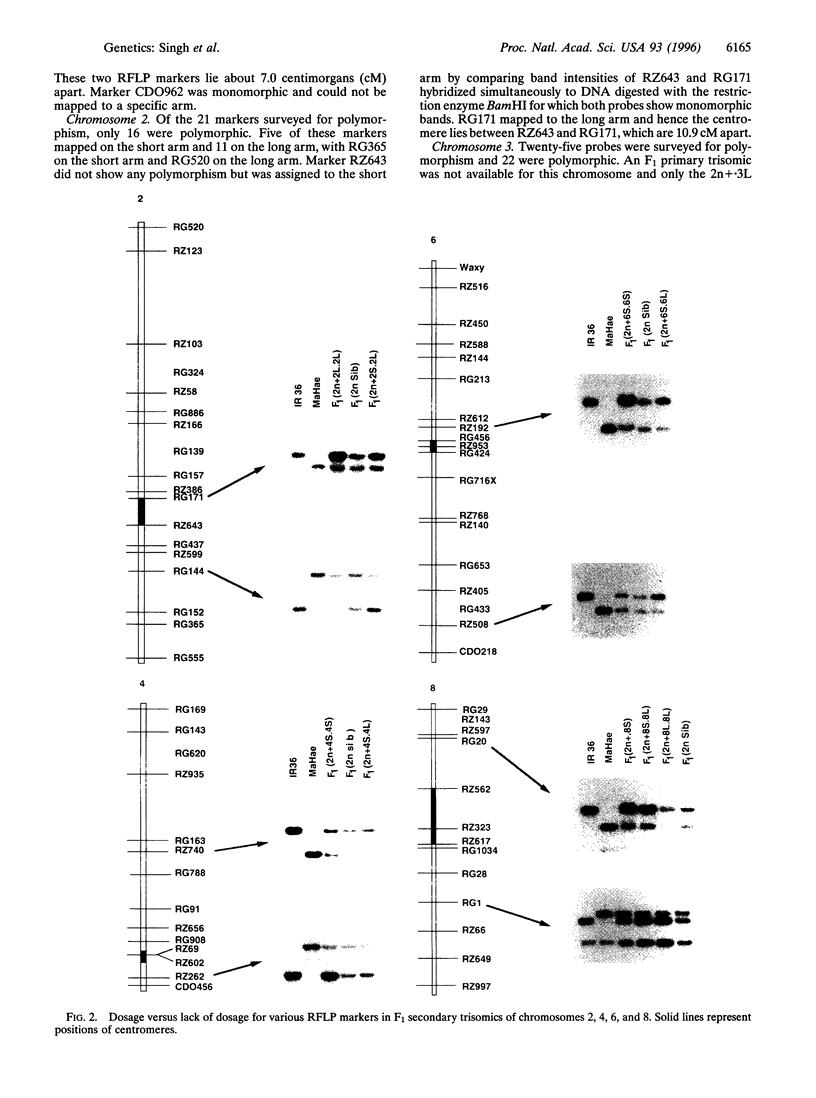
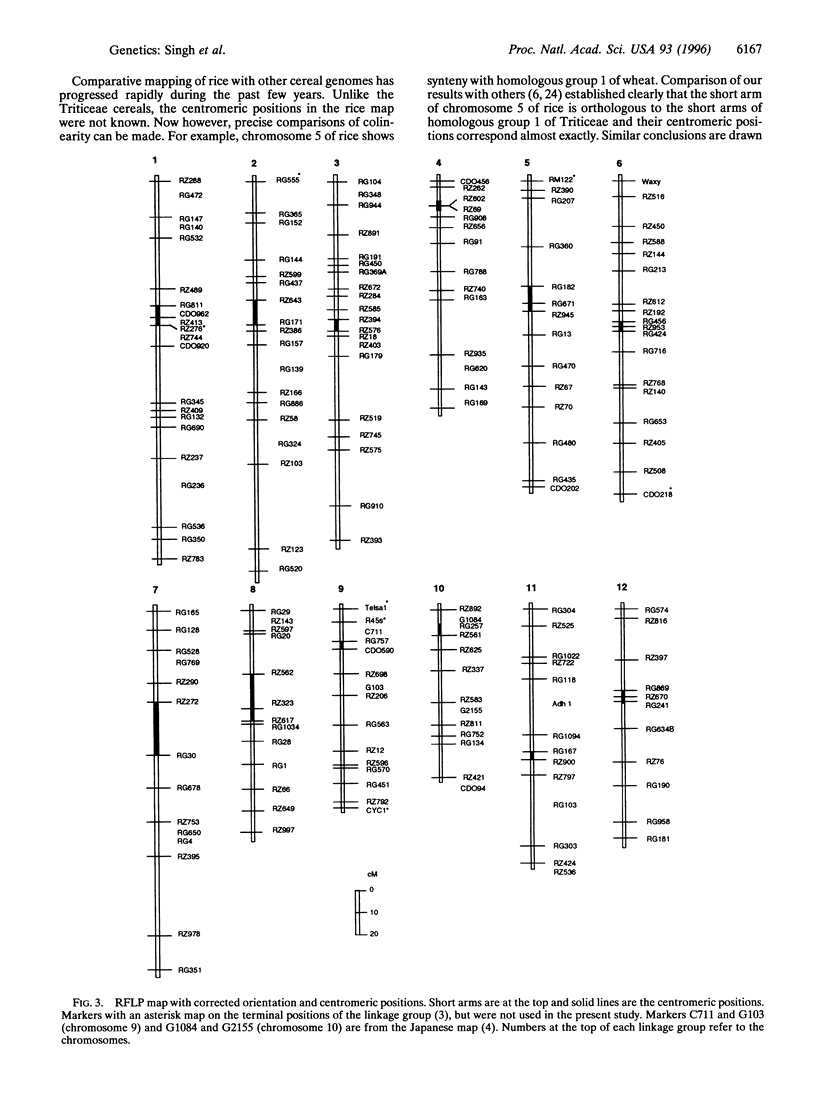
Rice has become a model cereal plant for molecular genetic research. Rice has the most comprehensive molecular linkage maps with more than 2000 DNA markers and shows synteny and colinearity with the maps of other cereal crops. Until now, however, no information was available about the positions of centromeres and arm locations of markers on the molecular linkage map. Secondary and telotrisomics were used to assign restriction fragment length polymorphism markers to specific chromosome arms and thereby to map the positions of centromeres. More than 170 restriction fragment length polymorphism markers were assigned to specific chromosome arms through gene dosage analysis using the secondary and telotrisomics and the centromere positions were mapped on all 12 linkage groups. The orientations of seven linkage groups were reversed to fit the "short arm on top" convention and the corrected map is presented.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahn S., Anderson J. A., Sorrells M. E., Tanksley S. D. Homoeologous relationships of rice, wheat and maize chromosomes. Mol Gen Genet. 1993 Dec;241(5-6):483–490. doi: 10.1007/BF00279889. [DOI] [PubMed] [Google Scholar]
- Causse M. A., Fulton T. M., Cho Y. G., Ahn S. N., Chunwongse J., Wu K., Xiao J., Yu Z., Ronald P. C., Harrington S. E. Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics. 1994 Dec;138(4):1251–1274. doi: 10.1093/genetics/138.4.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gustafson J. P., Dillé J. E. Chromosome location of Oryza sativa recombination linkage groups. Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8646–8650. doi: 10.1073/pnas.89.18.8646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khush G. S., Singh R. J., Sur S. C., Librojo A. L. Primary trisomics of rice: origin, morphology, cytology and use in linkage mapping. Genetics. 1984 May;107(1):141–163. doi: 10.1093/genetics/107.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata N., Nagamura Y., Yamamoto K., Harushima Y., Sue N., Wu J., Antonio B. A., Shomura A., Shimizu T., Lin S. Y. A 300 kilobase interval genetic map of rice including 883 expressed sequences. Nat Genet. 1994 Dec;8(4):365–372. doi: 10.1038/ng1294-365. [DOI] [PubMed] [Google Scholar]
- Moore G., Devos K. M., Wang Z., Gale M. D. Cereal genome evolution. Grasses, line up and form a circle. Curr Biol. 1995 Jul 1;5(7):737–739. doi: 10.1016/s0960-9822(95)00148-5. [DOI] [PubMed] [Google Scholar]
- Singh K., Multani D. S., Khush G. S. Secondary trisomics and telotrisomics of rice: origin, characterization, and use in determining the orientation of chromosome map. Genetics. 1996 May;143(1):517–529. doi: 10.1093/genetics/143.1.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. L., Holsten T. E., Song W. Y., Wang H. P., Ronald P. C. Construction of a rice bacterial artificial chromosome library and identification of clones linked to the Xa-21 disease resistance locus. Plant J. 1995 Mar;7(3):525–533. doi: 10.1046/j.1365-313x.1995.7030525.x. [DOI] [PubMed] [Google Scholar]