Abstract
Matrix metalloproteinases (MMPs), as the enzymes to degrade extracellular matrix proteins, play a major role on cell behaviors. Among them, MMP-9 usually catalyzes the degradation of proteins with the dominant cleavage at G/L site. Recent high-throughput screening suggests that S/L is a new major site for the cleavage when the substrates of MMP-9 are oligopeptides. Here we examine the cleavage sites of the N-terminal substituted short oligopeptides as the substrates of MMP-9. As the first example of such study of N-substituted small peptides, our results suggest that the substitute group at the N-terminal and the length of peptides significantly affect the position of the cleavage site on the oligopeptides, which provides a useful insight for the design of small peptide derivatives as the substrates of MMP-9.
Keywords: N-terminal substitution, oligopeptide, MMP-9, proteolytic sites
INTRODUCTION
In the past decade, molecular self-assembly has become a powerful strategy in nanotechnology1. Peptides, as one key, universal, biomolecular building blocks of life, have also provided a versatile platform for the design of self-assembling nanomaterials due to their specifically structures and properties. Several research groups2–4 already have pioneered the use of the self-assembly of peptides to develop nanobiomaterials in the applications ranging from cell culture,5–10 drug delivery,11–13 to biosensors.14,15 Several factors such as pH,3,16 temperature,17 light,18 and enzymes6,19 have been explored to trigger the self-assembly of small molecular peptides. Particularly, enzyme-catalyzed ones exhibit superior advantages such as high selectivity and substrate specificity, and the ability to proceed under mild aqueous conditions.20 Like themolysin and subtilisin,21–23 MMP also can trigger the self-assembly of small molecular peptides to form nanobiomaterial.24–26 To further expand the applications of the substrates of MMP for enzyme-instructed molecular self-assembly that allow the formation of biomaterials in vivo, we choose to investigate the parameters (e.g., N-terminal substitution and lengths of peptides) that affect the hydrolysis of small peptide derivatives catalyzed by MMP.
Matrix metalloproteinases (MMPs), as a class of zinc dependent secreted endopeptidases, are capable of degrading all kinds of extracellular matrix (ECM) proteins and play essential roles in normal physiological processes.27–29 For example, MMP-2 and MMP-9 have been previously described as the important enzymes related to the invasiveness and metastatic potency of human malignant tumors including breast, prostate, and ovarian carcinoma.30–32 Recently, MMP catalyzed hydrolysis also has been explored for molecular imaging and drug delivery.33–36 To develop a new approach for inhibiting cancer cells based on the overexpression of MMPs, we aim to use the overexpressed MMPs as the catalysts for the generation of molecular aggregates to interfere with the extracellular microenvironment of cancer cells as a possible means for blocking metastasis. One way to generate such molecular aggregates is to use MMPs to instruct the molecular self-assembly of small molecules via the enzymatic cleavage of peptide derivatives.25 Thus, the important first step should be to determine the cleavage sites on the substrates during the proteolysis catalyzed by MMPs so that one can use proper substrates for achieving intended effects. In this work, we focus on the substrates of MMP-9 because its relevance with cancer metastasis.
MMP-9, also historically referred as gelatinases because of its ability to cleave gelatins in vivo, catalyzes the proteolytic cleavage of the substrates dominantly at G/L site of proteins, such as α1(I) and α1(XI) collagens.37,38 Recent advances in high throughput screening, however, reveal S/L as a new major cleavage site when the substrates of MMP-9 are oligopeptides.39,40 This result suggests that the cleavage sites of the proteolysis catalyzed by MMP-9 depend significantly on the structures of the substrates, and raises an important question how the length and the modification of the terminal of the oligopeptides would affect the position of the cleavage site(s).
To address the above question, we designed two group oligopeptides containing a supposed G/L or S/L cleavage site and having increased lengths and different N-terminal substitution such as acetyl (Ac), fluorenylmethoxycarbonyl (Fm), pyrene (Py), and naphthalene (Np) to study the influence of N-terminal substitution and length of peptide on the site of cleavage of the oligopeptides. We choose PLG(S)/LRSK as the selected core sequence of the substrates (P3-P2-P1-P1′-P2′-P3′) in this study because (i) proline at the P3 position maximizes the specificity of MMP-9 to the substrates.38–41 (ii) RSK, as the hydrophilic residues, increases the solubility of the substrates in water to confer a drastic difference of solubility between the substrates and the cleavage products.
Our results show that the cleavage sites of heptapeptides catalyzed by MMP-9 largely differ from the cleavage sites of nona- and decapeptides catalyzed by MMP-9. Except peptide 1c (a pyrene N-terminated peptide), none of the major cleavage occurs at the site G/L or S/L (the conventional cleavage sites of the peptides catalyzed by MMP-9), but at GL/R or L/SL sites of the heptapeptides. For the nonapeptides, the most common cleavages, catalyzed by MMP-9, occur at L/GL or L/SL site. However, when the length of peptide increases to decapeptides, the cleavage proceeds at L/GL or L/SL site. The sites of the cleavage of the peptides in this work are largely different from the sites of the cleavage on dodecapeptides (S/L)39,40 and on proteins (G/L) when the protease is MMP-9. Apparently, the group of N-terminal substitution has limit effect on the sites of the cleavage for the cases of nona- and decapeptides. These results suggest that the cleavage sites on the short oligopeptides depend on the length of peptide, which provides a useful insight for the design of small peptidic substrates or inhibitors of MMP-9.
MATERIALS AND METHODS
Peptide synthesis
All the compounds were prepared by solid-phase peptide synthesis (SPPS) Using 2-chlorotrityl chloride resin, N-Fmoc-protected amino acids, and DIPEA/HBTU (N,N-diisopropylethylamine/O-benzotriazole-N,N,N',N'-tetramethyl-uronium-hexafluoro-phosphate) as the coupling reagent, we synthesized the peptides and the peptide derivatives by Fmoc SPPS protocol42 with side chains properly protected if necessary. After the removal of Fmoc-protecting group by 20% piperidine and the release of the products by trifluoroacetic acid (TFA), we used reverse phase high-performance liquid chromatography (HPLC) to purify the products and liquid chromatography-mass spectrometry (LC-MS) and NMR to characterize the products.
Digestion of peptides by MMP-9
The digestion of the peptides or the peptide derivatives follows a simple protocol carried out in a glass vial (1 mL): a peptide is dissolved in water at a 10 mM concentration, and 50 μL of 10 mM stock solution is diluted into 0.95 mL Tris-HCl buffer (50 mM of Tris-HCl, 300 mM of NaCl, 5 mM of CaCl2, pH7.5) to give the final concentration of 500 μM, then 10 U of MMP-9 is added to start hydrolysis reaction. The mixture was incubated at 37 °C. At a desired time interval, an aliquot of 0.05 mL was taken from the reaction mixture for HPLC analysis.
Determination of hydrolytic products
The hydrolysis samples were analyzed on a Waters 2489 HPLC. Runs were performed on a C18 column (150 × 4.6-mm I.D., 5-μm particle size) using a linear AB gradient (2 % acetonitrile/min) and a flow rate of 0.6 mL/min, where eluent A was 0.1 % aqueous TFA, and eluent B was 0.1 % TFA in acetonitrile. The cleavage products were confirmed by LC-MS on Waters Acquity Ultra Performance LC with waters MICROMASS detector.
RESULTS AND DISCUSSION
Peptide design
Figure 1 shows the peptides and the peptide derivatives investigated in this work. Based on the core sequence, the different substitution groups such as fluorene (Fm), pyrene (Py), and naphthalene (Np) serve as the hydrophobic groups to block the N-terminal of peptides because these aromatic groups promote peptides to form nanofibers and hydrogels in water due to the aromatic-aromatic interactions.43–45 In addition, we attached one or two phenylalanine residues to the N-terminal to increase the ability of the N-terminal hydrolysis products to self-assemble in water and to form hydrogels. In order to minimize the steric hinder, originated from the relatively large aromatic groups at the N-terminal, that may inhibit MMP-9, we used a glycine residue as the linker between the proline and phenylalanine.
Figure 1.
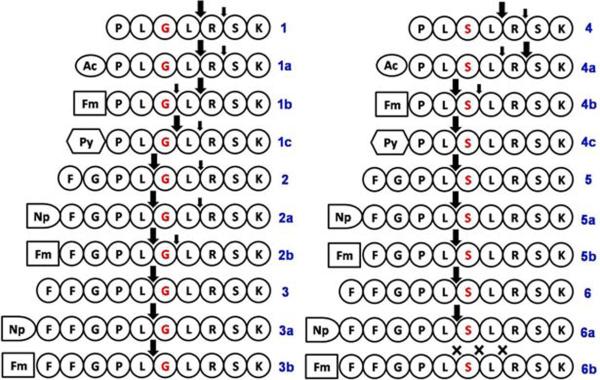
The oligopeptide sequences and the experimentally determined cleavage sites when they are treated by MMP-9 (the large arrows indicate the major cleavage sites).
Digestion of the oligopeptides containing PLGLRSK
Figure 2 shows the result of the digestion of the oligopeptides containing the core sequence of PLGLRSK (peptide 1) according to the detailed hydrolysis profiles (Figures S1–S10). As shown in Figure 2, during the first 6 hours of the digestion, except the peptides 1c, 3a and 3b, the other substrates hydrolyze up to more than 50%, according to that the amounts of residual substrates are 46% (1), 27% (1a), 24% (1b), 68% (1c), 37% (2), 42% (2a), 46 %(2b), 26% (3), 83% (3a) and 95% (3b), respectively. Without the N-terminal substitution, the increase of the length of the peptide leads to increased rate of hydrolysis, as indicated by that there is less residual substrate of 3 (26 %) than those of 1 (46 %) and 2 (37%). While the N-terminal substitution by acetyl and fluorene leads to more hydrolysis of the heptapeptide derivatives (1a and 1b) than that of 1, significant amounts of the substrates still remain. Generally, the larger N-terminal substitutions (e.g., in the cases of 1c, 2b, and 3b) result in lower hydrolysis rate than that of unsubstituted or acetylated peptides. These data indicate that the N-terminal substitution is an effective way for modulating the hydrolytic rates of the oligopeptides that are the substrates of MMP-9.
Figure 2.
The histogram of the percentage of the hydrolysis products from the oligopeptides (containing the G/L site) incubated with MMP-9 for 6 h and 72 h. The red bar shows the percentage of the remaining substrate, the other bars show the percentage of the N-substituted products due to the cleavage at the indicated site.
The products after 72 h of hydrolysis (Figure 2) offer useful insights not only on the sites of cleavage, but also the distribution of the final products. For example, after 72 h hydrolysis, while the larger size heptapeptides (1b and 1c) are undetectable, small amount of larger size nonapeptides (2b) remains (10 %), and the larger size decapeptides 3a and 3b still dominate at 41 % and 68 %, respectively. Interestingly, peptides 1b, 1c, and 2b hydrolyze to yield detectable amounts of products resulted from cleavage at the site G/L, which are the cpnventional cleavage site of the peptides that are hydrolyzed by the catalysis of MMP-9.37,38,46 This result indicates that cleavage at G/L site is a slow process for heptapeptides and nonapeptides that have fluorene or pyrene N-terminal substitution. While the major cleavage occurs at L/R site of 1, 1a, or 1b, G/L site of 1c, the major cleavages for nona- and decapeptides (2, 2a, 2b, 3, 3a, and 3b) are at L/G sites. Specifically, the major hydrolysis products and their amounts are PLGL from 1 (84 %), Ac-PLGL from 1a (71 %), Fm-PLGL from 1b (57 %), Py-PLG from 1c (59 %), FGPL from 2 (96 %), Nap-FGPL from 2a (77 %), Fm-FGPL from 2b (64 %), FFGPL from 3 (100 %), Nap-FFGPL from 3a (60 %), and Fm-FFGPL from 3b (28 %), respectively. Without the N-terminal substitution, the increase of the length of the peptide leads to more exclusive cleavage products and higher efficiency of cleavage, as indicated by that 1 is hydrolyzed at the cleavage sites of L/R and R/S with 6% of 1 remaining, and 3 breaks at L/G sites with no residual substrate of 3. Based on more detailed analysis of the rate of hydrolysis (Figure S1–S10), the peptide 2a exhibits the fastest rate of hydrolysis and efficiency of conversion because it undergoes complete digestion in 12h. These results indicate that the N-terminal substitution also can modulate the specificity and the efficiency of the conversion of the oligopeptides that are the substrates of MMP-9. Compared to the products formed after 6h hydrolysis, the amounts of secondary hydrolysis products (L/R) of peptide 2 and 3 decrease significantly, and the major hydrolysis products (L/G) obviously increase, suggesting that the secondary hydrolysis products of FGPLGL or FFGPLGL likely further hydrolyze to give FGPL or FFGPL after 72 h hydrolysis.
Digestion of oligopeptides containing PLSLRSK
Recent advances in high throughput screening reveal S/L as a new major cleavage site when the substrates of MMP-9 are oligopeptides.39,40 Therefore, we also synthesized the peptides containing the core sequence of PLSLRSK (peptide 4) and tested their hydrolysis catalyzed by MMP-9. Figure 3 shows the results according to the detailed hydrolysis profiles (Figures S11–S19), which indicate that the peptides containing the S/L site display the similar pattern of proteolytic cleavage as the peptides containing G/L site. For example, during 6 hours of the initial reaction, the amounts of residual substrates are 50 % (4), 33 % (4a), 58 % (4b), 86 % (4c), 7 % (5), 10 % (5a), 72 % (5b), 11 % (6), 56 % (6a) and 99 % (6b), respectively. Similarly, without the N-terminal substitution, the increase of the length of the peptide leads to more hydrolysis, as evidenced by that the amount of the residual substrate of 5 (7 %) or 6 (11%) is much less than that of 4 (50 %). With the N-terminal substitution, the major cleavage site of the heptapeptide is at L/R (4), R/S (4a), or L/S (4c) instead of S/L. Like the peptides containing G/L site, the amounts of residual substrates of 4c, 5b, and 6b also indicate that the larger N-terminal substitutions result in slower rates of hydrolysis than those of unsubstituted or acetylated peptides.
Figure 3.
The histogram of the percentage of the hydrolysis products from the oligopeptides (containing the S/L site) incubated with MMP-9 for 6 h and 72 h. The red bar shows the percentage of the remaining substrate, the other bars show the percentage of the N-substituted products due to the cleavage at the indicated site.
After being hydrolyzed for 72 h, except that the peptides containing the large size N-terminal substitution still exist as the substrates at the concentrations of 18 %, 19 %, 30 % and 91 % for 4c, 5b, 6a, and 6b, respectively, most of the substrates remain only less than 10% (4, 4a, and 4b), or undergo complete digestion (5, 5a, and 6). Based on more detailed analysis of the rate of hydrolysis (Figure S11–S19), the peptide 5a, which has the same length and structure with 2a, undergoes complete cleavage at L/S site in 8h, exhibiting the fastest rate of hydrolysis. However, as shown in Fig S10, the hydrolysis of peptides 3b and 6b turns out to be very slow, less than 30% of 3b and less than 10% of 6b are hydrolyzed after 72h, suggesting that the big hydrophobic N-terminal substitution inhibits the interaction of substrates with MMP-9.
Figure 3 also reveals that the major hydrolytic products, after 72 h of digestion, for the tested S/L containing peptides are PLSL, Ac-PLSLR, Fm-PL, Py-PL, FGPL, Nap-FGPL, Fm-FGPL, FFGPL, Nap-FFGPL, and Fm-FFGPL resulted from the digestion of 74% of 4, 53% of 4a, 51% of 4b, 75% of 4c, 100% of 5, 100% of 5a, 81% of 5b, 100% of 6, 70% of 6a, 7% of 6b, respectively. Except the major cleavage occurs at L/R site of 4 or R/S site of 4a, the others peptides cleave at L/S site. Like the peptides containing G/L site, without the N-terminal substitution, the increase of the lengths of the peptides leads to more exclusive cleavage products and higher efficiency of cleavage, as evidenced by that 4 hydrolyzes at the cleavage sites of L/R and R/S with 5 % remaining substrate, and 6 only cleaves at L/S site with no residual substrate after 72h. Interestingly, being different with the peptides containing G/L site, most peptides containing S/L site (e.g. 4c, 5, 5a, 5b, 6, 6a) give more exclusive products of hydrolysis, that is, they likely are hydrolyzed to give a single product with the cleavage at L/S site. But the amounts of residual substrates of 4c, 5b, 6b also indicate that the larger N-terminal substitutions result in slower conversion than those unsubstituted peptides. The above results further indicate that the N-terminal substitution likely affect more on the efficiency of hydrolysis than on the sites of the cleavage of the hydrolysis catalyzed by MMP-9.
In conclusion, in this study of the influence of length and N-terminal substitution on the cleavage site of the short oligopeptides, two group relatively short oligopeptide substrates of MMP-9 appear to hydrolyze in a quite different manner with the cleavage of proteins that are the substrates of MMP-9. These results also indicate that, while the N-terminal substitution modulates more on the hydrolytic rate the oligopeptides, the length of the oligopeptide is a more important parameter that affect both in the hydrolytic rate and the cleavage sites. One possible reason for slow digestion of the oligopeptides with large groups at their N-terminal could be that these molecules form aggregates, suggesting that more hydrophilic groups may help address this limitation. In addition, the major cleavage sites of most peptides in this study are different with the previously reported cleavage sites, G/L or S/L,39,40 which may be resulted from the different structures of the substrates both on the sequence, length, and N-terminal substitution. This information will be useful for designing the substrates of MMP-9 for enzyme-instructed molecular self-assembly in water. Moreover, the influence of the N-terminal substitution suggests that it is also worthwhile to explore the influence of C-substitution and side-chain substitution on the cleavage sites of short oligopeptides.
Supplementary Material
Acknowledgment
This work is partially supported by NIH (R01CA142746) and a scholarship from the Chinese Scholarship Council (2011617538 for Y.B.H.)
References
- 1.Whitesides GM, Mathias JP, Seto CT. Science. 1991;254:1312–1319. doi: 10.1126/science.1962191. [DOI] [PubMed] [Google Scholar]
- 2.Zhang S, Holmes T, Lockshin C, Rich A. Proc Natl Acad Sci U S A. 1993;90:3334–3338. doi: 10.1073/pnas.90.8.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schneider JP, Pochan DJ, Ozbas B, Rajagopal K, Pakstis L, Kretsinger J. J Am Chem Soc. 2002;124:15030–15037. doi: 10.1021/ja027993g. [DOI] [PubMed] [Google Scholar]
- 4.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Science. 2004;303:1352–1355. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 5.Zhou M, Smith AM, Das AK, Hodson NW, Collins RF, Ulijn RV, Gough JE. Biomaterials. 2009;30:2523–2530. doi: 10.1016/j.biomaterials.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 6.Yang Z, Liang G, Xu B. Acc Chem Res. 2008;41:315–326. doi: 10.1021/ar7001914. [DOI] [PubMed] [Google Scholar]
- 7.Sinthuvanich C, Haines-Butterick LA, Nagy KJ, Schneider JP. Biomaterials. 2012;33:7478–7488. doi: 10.1016/j.biomaterials.2012.06.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sur S, Matson JB, Webber MJ, Newcomb CJ, Stupp SI. ACS Nano. 2012;6:10776–10785. doi: 10.1021/nn304101x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumada Y, Zhang S. PLoS One. 2010;5:e10305. doi: 10.1371/journal.pone.0010305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui H, Webber MJ, Stupp SI. Biopolymers. 2010;94:1–18. doi: 10.1002/bip.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chow LW, Wang LJ, Kaufman DB, Stupp SI. Biomaterials. 2010;31:6154–6161. doi: 10.1016/j.biomaterials.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao F, Ma ML, Xu B. Chem Soc Rev. 2009;38:883–891. doi: 10.1039/b806410p. [DOI] [PubMed] [Google Scholar]
- 13.Yuan H, Fales AM, Vo-Dinh T. J Am Chem Soc. 2012;134:11358–11361. doi: 10.1021/ja304180y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Nakata E, Hamachi I. Chembiochem. 2009;10:2560–2577. doi: 10.1002/cbic.200900249. [DOI] [PubMed] [Google Scholar]
- 15.Wink T, van Zuilen SJ, Bult A, van Bennkom WP. Analyst. 1997;122:43R–50R. doi: 10.1039/a606964i. [DOI] [PubMed] [Google Scholar]
- 16.Rajagopal K, Lamm MS, Haines-Butterick LA, Pochan DJ, Schneider JP. Biomacromolecules. 2009;10:2619–2625. doi: 10.1021/bm900544e. [DOI] [PubMed] [Google Scholar]
- 17.Hughes M, Frederix PWJM, Raeburn J, Birchall LS, Sadownik J, Coomer FC, Lin IH, Cussen EJ, Hunt NT, Tuttle T, Webb SJ, Adams DJ, Ulijn RV. Soft Matter. 2012;8:5595–5602. [Google Scholar]
- 18.Haines LA, Rajagopal K, Ozbas B, Salick DA, Pochan DJ, Schneider JP. J Am Chem Soc. 2005;127:17025–17029. doi: 10.1021/ja054719o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giano MC, Pochan DJ, Schneider JP. Biomaterials. 2011;32:6471–6477. doi: 10.1016/j.biomaterials.2011.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao Y, Yang Z, Kuang Y, Ma ML, Li J, Zhao F, Xu B. Biopolymers. 2010;94:19–31. doi: 10.1002/bip.21321. [DOI] [PubMed] [Google Scholar]
- 21.Guilbaud JB, Vey E, Boothroyd S, Smith AM, Ulijn RV, Saiani A, Miller AF. Langmuir. 2010;26:11297–11303. doi: 10.1021/la100623y. [DOI] [PubMed] [Google Scholar]
- 22.Hirst AR, Roy S, Arora M, Das AK, Hodson N, Murray P, Marshall S, Javid N, Sefcik J, Boekhoven J, van Esch JH, Santabarbara S, Hunt NT, Ulijn RV. Nat Chem. 2010;2:1089–1094. doi: 10.1038/nchem.861. [DOI] [PubMed] [Google Scholar]
- 23.Toledano S, Williams RJ, Jayawarna V, Ulijn RV. J Am Chem Soc. 2006;128:1070–1071. doi: 10.1021/ja056549l. [DOI] [PubMed] [Google Scholar]
- 24.Akers WJ, Xu B, Lee H, Sudlow GP, Fields GB, Achilefu S, Edwards WB. Bioconjug Chem. 2012;23:656–663. doi: 10.1021/bc300027y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang ZM, Ma ML, Xu B. Soft Matter. 2009;5:2546–2548. [Google Scholar]
- 26.Tauro JR, Lee BS, Lateef SS, Gemeinhart RA. Peptides. 2008;29:1965–1973. doi: 10.1016/j.peptides.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Egeblad M, Werb Z. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 28.Malemud CJ. Front Biosci. 2006;11:1696–1701. doi: 10.2741/1915. [DOI] [PubMed] [Google Scholar]
- 29.Coussens LM, Fingleton B, Matrisian LM. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 30.Nelson AR, Fingleton B, Rothenberg ML, Matrisian LM. J Clin Oncol. 2000;18:1135–1149. doi: 10.1200/JCO.2000.18.5.1135. [DOI] [PubMed] [Google Scholar]
- 31.Sier CF, Kubben FJ, Ganesh S, Heerding MM, Griffioen G, Hanemaaijer R, van Krieken JH, Lamers CB, Verspaget HW. Br J Cancer. 1996;74:413–417. doi: 10.1038/bjc.1996.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramos-DeSimone N, Hahn-Dantona E, Sipley J, Nagase H, French DL, Quigley JP. J Biol Chem. 1999;274:13066–13076. doi: 10.1074/jbc.274.19.13066. [DOI] [PubMed] [Google Scholar]
- 33.Xia ZY, Xing Y, So MK, Koh AL, Sinclair R, Rao JH. Analytical Chemistry. 2008;80:8649–8655. doi: 10.1021/ac801562f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olson ES, Aguilera TA, Jiang T, Ellies LG, Nguyen QT, Wong EH, Gross LA, Tsien RY. Integr Biol. 2009;1:382–393. doi: 10.1039/b904890a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ouyang MX, Huang H, Shaner NC, Remacle AG, Shiryaev SA, Strongin AY, Tsien RY, Wang YX. Cancer Research. 2010;70:2204–2212. doi: 10.1158/0008-5472.CAN-09-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albright CF, Graciani N, Han W, Yue E, Stein R, Lai ZH, Diamond M, Dowling R, Grimminger L, Zhang SY, Behrens D, Musselman A, Bruckner R, Zhang MZ, Jiang X, Hu D, Higley A, DiMeo S, Rafalski M, Mandlekar S, Car B, Yeleswaram S, Stern A, Copeland RA, Combs A, Seitz SP, Trainor GL, Taub R, Huang P, Oliff A. Molecular Cancer Therapeutics. 2005;4:751–760. doi: 10.1158/1535-7163.MCT-05-0006. [DOI] [PubMed] [Google Scholar]
- 37.Xia T, Akers K, Eisen AZ, Seltzer JL. Biochim Biophys Acta. 1996;1293:259–266. doi: 10.1016/0167-4838(95)00259-6. [DOI] [PubMed] [Google Scholar]
- 38.Seltzer JL, Akers KT, Weingarten H, Grant GA, McCourt DW, Eisen AZ. J Biol Chem. 1990;265:20409–20413. [PubMed] [Google Scholar]
- 39.Kridel SJ, Chen E, Kotra LP, Howard EW, Mobashery S, Smith JW. J Biol Chem. 2001;276:20572–20578. doi: 10.1074/jbc.M100900200. [DOI] [PubMed] [Google Scholar]
- 40.Turk BE, Huang LL, Piro ET, Cantley LC. Nat Biotechnol. 2001;19:661–667. doi: 10.1038/90273. [DOI] [PubMed] [Google Scholar]
- 41.Schechter I, Berger A. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 42.Carreno C, Mendez ME, Kim YD, Kim HJ, Kates SA, Andreu D, Albericio F. J Pept Res. 2000;56:63–69. doi: 10.1034/j.1399-3011.2000.00711.x. [DOI] [PubMed] [Google Scholar]
- 43.Shi J, Gao Y, Yang Z, Xu B. Beilstein J Org Chem. 2011;7:167–172. doi: 10.3762/bjoc.7.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma M, Kuang Y, Gao Y, Zhang Y, Gao P, Xu B. J Am Chem Soc. 2010;132:2719–2728. doi: 10.1021/ja9088764. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, Yang Z, Yuan F, Gu H, Gao P, Xu B. J Am Chem Soc. 2004;126:15028–15029. doi: 10.1021/ja044401g. [DOI] [PubMed] [Google Scholar]
- 46.Seltzer JL, Weingarten H, Akers KT, Eschbach ML, Grant GA, Eisen AZ. J Biol Chem. 1989;264:19583–19586. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




