Abstract
A breed known for its versatility, the American Quarter Horse (QH), is increasingly bred for performance in specific disciplines. The impact of selective breeding on the diversity and structure of the QH breed was evaluated using pedigree analysis and genome-wide SNP data from horses representing 6 performance groups (halter, western pleasure, reining, working cow, cutting, and racing). Genotype data (36 037 single nucleotide polymorphisms [SNPs]) from 36 Thoroughbreds were also evaluated with those from the 132 performing QHs to evaluate the Thoroughbred’s influence on QH diversity. Results showed significant population structure among all QH performance groups excepting the comparison between the cutting and working cow horses; divergence was greatest between the cutting and racing QHs, the latter of which had a large contribution of Thoroughbred ancestry. Significant coancestry and the potential for inbreeding exist within performance groups, especially when considering the elite performers. Relatedness within performance groups is increasing with popular sires contributing disproportionate levels of variation to each discipline. Expected heterozygosity, inbreeding, F ST, cluster, and haplotype analyses suggest these QHs can be broadly classified into 3 categories: stock, racing, and pleasure/halter. Although the QH breed as a whole contains substantial genetic diversity, current breeding practices have resulted in this variation being sequestered into subpopulations.
Key words: breeds, coancestry, equine, performance, relatedness, selection
The American Quarter Horse (QH) had its beginnings in the colonial United States, where horses primarily of English descent were used for transportation and agriculture (Denhardt 1967; Laune 1973; Hendricks 2007). Those early founders of what would become the QH breed continued to evolve as Spanish-derived bloodlines from Cherokee and Chickasaw ponies were incorporated to increase the horses’ quickness (Denhardt 1967; Laune 1973). The unmatched ability of the colonial QH to run one-quarter of a mile resulted in the horses earning the title of “American Quarter Running Horse.” In the mid 1700s, the QH found influence from the English Thoroughbred (TB) (Denhardt 1967; Hendricks 2007), an influence that continues today. The result of breeding for speed and ruggedness was the American QH, a short distance sprinter and ranch horse that was valuable to the development of the American West.
The American QH was formally recognized as a breed with the establishment of the American Quarter Horse Association (AQHA) in 1940 and the publication of the first stud book in 1941, which included fewer than 600 horses (Denhardt 1941). The breed grew rapidly and as of 2011, 2.64 million of the estimated 10.15 million horses in the United States were registered QHs (FAO Statistics Division 2013 [last accessed 22 Apr 2013]); American Quarter Horse Association 2010). The popularity of the QH in the United States is also illustrated by the fact that in 2011, over 3000 AQHA approved shows and 8450 QH races were held, with the racing industry alone paying over 129 million dollars in purses (American Quarter Horse Association 2010).
Recent studies of the QH show that the genetic diversity of the breed is moderate to high compared with other modern breeds, owing to a large and expanding population, diverse founding stock, and continued admixture with the TB (Luís et al. 2007; Leroy et al. 2009; McCue et al. 2012; Petersen et al. 2013). Genetic diversity of the QH is also demonstrated by the range of performance types found within the breed. These performance categories represent racing and cattle horses in addition to horses bred solely for physical conformation or for smooth, controlled movement (Table 1). Although horses of each type are all members of the QH breed, horses specialized for disciplines such as cutting and racing have significant differences in morphometric characters (Meira et al. 2013; Supplementary Material online) and contributing bloodlines (Fletcher 1945; Tunnell et al. 1983). These characteristics, coupled with disease allele frequency differences among the performance types (Tryon et al. 2009), suggest that while diversity within the breed as a whole may be high, breeding for specific performance abilities has created substructure within the population.
Table 1.
Primary characteristics of the 6 performance groups of the QH evaluated in the study as described by the American Quarter Horse Association
| Performance type | Characteristics |
|---|---|
| Halter | Horses are led before judges so that lameness and quality of movement can be evaluated. Horses are judged on conformation including balance, structural correctness, breed and sex characteristics, and degree of muscling. |
| Western pleasure | Horses are evaluated on quality of movement under saddle at the walk, jog, and lope, while staying quiet and calm, travelling on a loose rein. |
| Reining | Judges the horse on movements under saddle, mastery of a prescribed maneuver and attitute as he is guided through a specific pattern. The horse is required to perform stops, spins, rollbacks, lead changes, and circles at a lope. |
| Working cow (reined cow horse) | Combines reining ability and cow sense. The competition consists of 2 parts: Prescribed reined work and actual cow work. Judging is based on good manners, smoothness, cow sense, and ease of reining. |
| Cutting | Horse and rider must move quietly into a herd of cattle, cut 1 cow from the herd, drive it to the center of the arena, and “hold” it away from the herd. The horse is scored on its ability to keep the cow from returning to the herd, cow sense, attentiveness, and courage. |
| Racing | Horses race against one another at distances between 220 and 870 yards. The classic distance is 440 yards (1/4 mile). |
This study was designed to quantitatively evaluate population structure within 6 popular and diverse performance groups of the QH breed. Both pedigree and genotype analyses were completed on horses following varied sampling schemes within and across performance groups. Horses included in genotype analysis were selected to eliminate sibships, thus capturing a more comprehensive illustration of total diversity within each discipline. Pedigree analysis on these same horses allowed for a comparison of similar statistics calculated on the 2 types of data. In addition, pedigree analysis was completed on 3 additional sample sets that represented random horses within each performance group regardless of relationship, the top money earners from 2009–10 and 3 sets of individuals selected across performance groups. Analysis of these additional cohorts provided a comparison to the data collected in the genotyped horses and allowed for a better estimation of the potential effects of inbreeding on future generations. Finally, within each performance group, the influence of the TB was investigated through haplotype sharing and cluster analyses. These results illustrate the consequences of selective breeding for performance type in a large, domestic horse population, will allow for informed investigations of phenotypes specific to QHs within particular disciplines, and will be useful for QH breeders and managers by providing a baseline upon which to monitor changes in genetic diversity over time.
Materials and Methods
QH Sample Collection
All individuals included in this study were selected from among the 200 top performers of the 6 performance groups as determined by money (racing, working cow (reined cow horse), cutting, and reining) or points (western pleasure and halter) earned in 2009 and 2010. In the categories of cutting, reining, and working cow horse, these records were obtained from Equi-stat (Cowboy Publishing Group). AQHA performance records were queried to identify the top western pleasure (hereafter “pleasure”) performers, and the top racing QHs were found using AQHARacing.com. Halter horses, which compete in gender-specific classes, were selected to equally represent stallions, geldings (castrated males), and mares.
Within each performance group, 3 subsets of individuals were assembled from the top 200 performers as follows: 1) A reference cohort of 24 individuals randomly selected from within each performance group after eliminating full- and half-sibships (hereafter referred to as reference (REF)), 2) a random cohort selected from within each performance group regardless of pedigree-based relationships (hereafter referred to as random (RA)), and 3) a cohort of top winners within each performance group (hereafter referred to as top performers (TP)). Finally, 3 separate random samples of 20 individuals were drawn from the combined set of REF, RA, and TP horses to represent diversity expected across performance groups; these sets of individuals are hereafter referred to as across group (AG) cohorts: AG1, AG2, and AG3.
Pedigree Analysis
Pedigree analyses were performed for each performance group within each sample (REF, RA, and TP) as well as on the 3 AG cohorts. For each individual, a 4-generation pedigree was input into Endog (Gutiérrez and Goyache 2005) for analysis of individual inbreeding (Wright 1922), coancestry, and average relatedness, as well as calculation of pairwise F ST (Caballero and Toro 2000, 2002), and generation intervals (James 1977). Putative significance of F ST values was determined empirically by declaring the minimum F ST observed between AG samples as the threshold for significance. Analysis of variance (ANOVA), followed by Tukey’s HSD test, were implemented in R (http://cran.r-project.org) to evaluate if inbreeding and coancestry values of individuals within the REF cohorts were significantly different than those found within the RA, TP, or AG cohorts and were also used to compare values across performance groups within samples. Average relatedness values were examined to identify the 3 sires with the greatest influence on each performance group.
Genotype Analysis
Hair root samples from 25 horses from the REF cohort of each performance group were obtained from the Veterinary Genetic Laboratory (University of California Davis). Hairs were cut 0.5–1.0cm above the root, and DNA was isolated from the roots using Qiagen’s Gentra Puregene Kit with the following modifications: 750 μl of isopropanol was used instead of 300 μl, centrifuge time was extended to 15min at 4 °C, and the pellet was washed twice. The 24 samples from each performance group with the highest quality DNA were submitted to Geneseek (Lincoln, NE) for SNP genotyping on the Illumina Equine SNP70 BeadChip; in the pleasure group, 23 horses passed DNA quality standards and were genotyped.
QH Genotype Quality Filtering
As one means to assure samples of the correct individuals from the REF cohorts were obtained, the gender of each sample was determined based upon rates of heterozygosity on the X chromosome (ECAX) in Plink (Purcell et al. 2007) using the command --sex-check. In the case of potentially mismatched samples as determined by gender matching, additional information (prior genetic test results and registry information) was obtained; any sample with questionable identity was removed from all analyses. Horses with missing data at a level greater than 2 standard deviations from the mean were also removed from analyses.
Of those genotyped, 6 QHs were removed from the analysis due to potential misidentification: 4 were eliminated after subsequent records check found that hair from the incorrect horse was obtained, one was misidentified due to a name change, and one had mismatching sex (genotype vs. pedigree records). Five additional individuals were removed due to missing data at a level greater than 2 times the standard deviation from the mean (≥8.5%). After all quality control measures, genotype data from 132 QHs remained. The number of horses from each performance group included in subsequent analyses (RA and TP samples) was set to equal the number of horses in the corresponding performance group of the REF sample. This allowed for inclusion of the maximal amount of genotype data possible while also allowing for across cohort comparisons within performance groups.
TB Genotypes
Equine SNP50 Beadchip (Illumina) genotypes were available from 36 TBs born from 1973 to 2005 (median 1994.5) and represented horses randomly sampled in the United States, the United Kingdom, and Ireland; data from these horses were also reported in McCue et al. (2012) and Petersen et al. (2013).
SNP Pruning
To allow for a uniform analysis across breeds, only loci found on both versions of the Illumina Equine Beadchip were considered, and data from the X and Y chromosome were removed. SNPs failing to genotype at a rate of at least 95% (2808) and those with a minor allele frequency (maf) of less than 0.05 (4663) across all individuals as determined by Plink (Purcell et al. 2007) were eliminated from all analyses. After quality control, 36 037 autosomal SNPs remained with an overall genotyping rate of 0.987.
The genotype data were then pruned for between-SNP linkage disequilibrium (LD) greater than R 2 = 0.4 using the Plink command --indep (multiple regression) considering 100bp windows, moving 25 loci per window. This data set of 14 580 SNPs was used for calculations of expected heterozygosity (H e), F ST, and AMOVA in Arelquin (Excoffier et al. 2005), with significance of F ST and analysis of molecular variance (AMOVA) determined by 20 000 permutations. Estimates of individual inbreeding were calculated from identity by state in PLINK (--het). Correlation between the genotype-based and pedigree-based inbreeding values was described using Pearson’s correlation coefficient.
Cluster Analysis
The LD-pruned data set was also used to perform Bayesian cluster analysis across all REF QH and TB samples in Structure (Pritchard et al. 2000; Falush et al. 2003). Structure analyses consisted of 30 000 burn-in and 70 000 MCMC repetitions, run in 5 replicates each of K = 1–10 using the admixture model and assuming correlated allele frequencies. The number of clusters that best described the data was inferred by attempting to maximize the likelihood and minimize the variance of lnP(X|D) across replicates. CLUMPP (Jakobsson and Rosenberg 2007) was used to determine average membership to each of the clusters across the 5 replicates using the LargeK Greedy algorithm; output was visualized using Distruct (Rosenberg 2004). Structure was run in a similar manner considering only the 6 performance groups of the QH using a data set of 16 785 loci pruned for R 2 = 0.4. Principal components analysis (PCA) was performed using snpStats in R (http://cran.r-project.org) on the LD-pruned autosomal data set as well as the original data pruned for genotyping rate of 95% or greater, and maf of 0.05, but not for LD.
Haplotype Richness and Sharing
Haplotype richness was evaluated on 20 individuals from each REF cohort in 5 different, randomly selected 10Mb autosomal regions. Genotypes of all individuals within the selected regions were phased with FastPhase (Scheet and Stephens 2006), using a data set consisting of autosomal markers shared across genotyping platforms, pruned for maf of 0.01, and genotyping rate of 0.05. FastPhase was configured to perform 20 runs (-T20) with K = 40 clusters, as determined to be most likely after testing values of 15 to 40 in increments of 5; performance group and breed identification were used to define subpopulations to inform the analysis. Resulting haplotypes were evaluated in 500kb, nonoverlapping, sliding windows across each chromosomal region, considering only windows with 5 or more SNPs. Haplotype richness was defined as the sum of unique haplotypes observed across all 500kb windows per performance group or breed. A private haplotype was defined as a 500kb haplotype that occurred at a frequency greater than zero in only one performance group or breed. Haplotype sharing of the REF QH samples with the TB was evaluated by quantifying the number and proportion of total haplotypes found in the TB that were also found in each QH cohort.
Data Archiving
In fulfillment of data archiving guidelines, we will deposit the primary data underlying these analyses at animalgenome.org one year after publication.
Results
QH Pedigree Analysis
Pedigree Composition
The full, 4-generation pedigree for all 132 QHs in the REF sample contained 2185 individuals, 112 of which were TBs (Table 2). The number of horses in the pedigree of each performance group of the REF sample ranged from 295 (cutting) to 480 (racing) or from 14.8 to 21.8 horses per proband. Across samples, significantly fewer horses were observed in the RA and TP samples than in the REF sample (P = 0.003 and P < 0.001, respectively); additionally, significantly fewer individuals were in the TP compared with the RA samples (P = 0.009). The number of horses per proband found in all REF cohorts was lower than that observed in the 3 AG replicates, which contained 21.6–23.2 horses per proband; the only exception was AG3, which had fewer individuals than the racing cohort. Similar to total pedigree size, the number of sires responsible for proband individuals was significantly fewer in the TP sample compared with the RA sample (P = 0.011) and in both compared with the REF sample (P = 0.001 for each) (Table 2).
Table 2.
Summary of pedigree analysis from 4-generation pedigrees of the REF, RA, TP, and AG samples
| Sample | Cohort (N) | ||||||
|---|---|---|---|---|---|---|---|
| Reference | Halter (23) | Pleasure (22) | Reining (23) | Working cow (22) | Cutting (20) | Racing (22) | All (132) |
| Total pedigree size | 477 | 455 | 425 | 393 | 295 | 480 | 2185 |
| Sires of proband | 23 | 22 | 23 | 22 | 20 | 22 | 132 |
| Thoroughbred | 20 | 6 | 1 | 3 | 1 | 84 | 112 |
| Random | |||||||
| Total pedigree size | 411 | 373 | 408 | 311 | 255 | 362 | 1919 |
| Sires of proband | 22 | 18 | 18 | 17 | 14 | 19 | 106 |
| Thoroughbred | 19 | 6 | 1 | 1 | 1 | 81 | 106 |
| Top performers | |||||||
| Total pedigree size | 365 | 342 | 382 | 301 | 236 | 360 | 1797 |
| Sires of proband | 17 | 16 | 18 | 13 | 9 | 18 | 86 |
| Thoroughbred | 0 | 8 | 1 | 2 | 1 | 85 | 94 |
| Across group | AG1 (20) | AG2 (20) | AG3 (20) | ||||
| Total pedigree size | 464 | 457 | 432 | ||||
| Sires of proband | 20 | 18 | 20 | ||||
| Thoroughbred | 10 | 35 | 18 | ||||
Given is the total number of horses in each pedigree, the number of sires represented by the proband individuals, and the representation of Thoroughbreds. N represents the number of proband individuals in each performance group.
Across performance groups, the proband of the cutting and working cow TP samples had 4 sires in common, whereas the reining and working cow cohorts had 1 sire shared between proband individuals. In the RA sample, 1 sire was shared between proband each in the halter and pleasure cohorts and in the cutting and working cow cohorts. No dams were shared by proband horses across cohorts within any sample.
Within the REF cohorts, TBs were most common in the racing QH (3.82 TB per horse genotyped), followed by the halter QH (0.87; Table 2). In the RA and TP samples, TBs were also most prevalent in the racing QH.
Average Relatedness
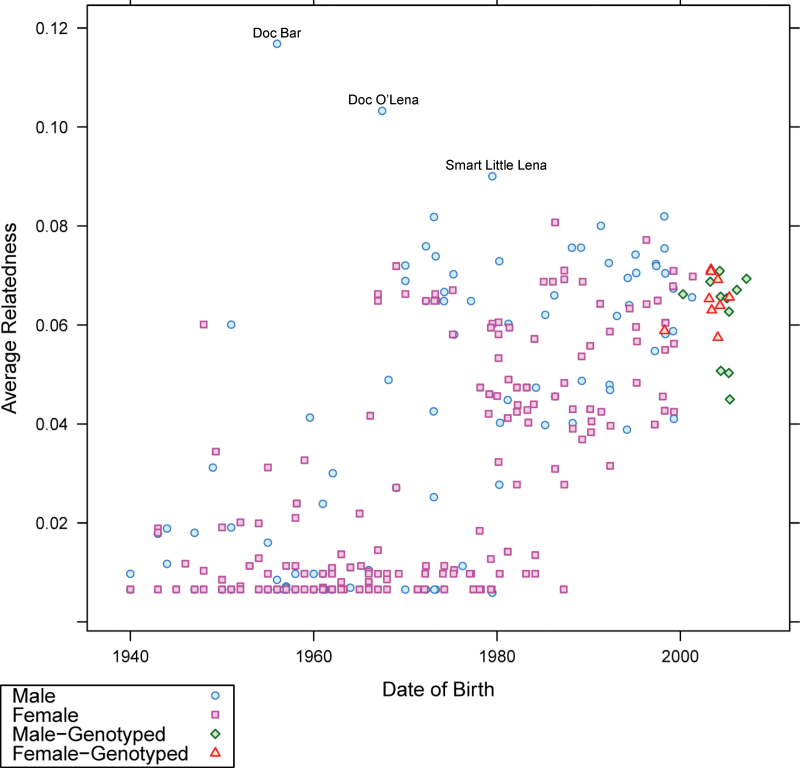
Average relatedness of all horses in the pedigree of each REF cohort was plotted by year of birth (Figure 1; Supplementary Material online). In each performance group, average relatedness showed a positive increase with time with skewed contributions from popular sires/dams apparent as outliers within each figure. The sires with the highest average relatedness to the others in the pedigree of each performance group were Doc Bar (cutting, working cow, and reining), Dash For Cash (racing), Zippo Pine Bar (pleasure), and Kid Clu (halter) (Table 3). The paternal line of each of these sires can be traced back to the Thoroughbred stallion, Three Bars (Figure 2). Within the working cow, cutting, and reining horses, similar levels of average relatedness were observed to the same 5 sires (Table 3), whereas the halter and pleasure groups had 3 sires in common. However, the top 3 sires in the racing horses were unique to that performance group.
Figure 1.
Average relatedness by birth date, accounting for completeness of pedigree, for all horses in the REF cutting horse cohort. An increase in average relatedness is observed with time. Popular sires and dams appear as outliers, and sires showing the highest average relatedness are noted.
Table 3.
Average relatedness of the top 3 sires in each performance group to others in the pedigrees of the REF horses, and their contribution to the other performance groups
| Halter | Pleasure | Reining | Working cow | Cutting | Racing | |
|---|---|---|---|---|---|---|
| Conclusive | 0.054 | 0 | 0 | 0 | 0 | 0 |
| Dash For Cash | 0 | 0 | 0 | 0 | 0 | 0.061 |
| Doc Bar | 0.004 | 0.006 | 0.061 | 0.078 | 0.117 | 0 |
| Doc O Lena | 0 | 0 | 0.05 | 0.07 | 0.103 | 0 |
| Docs Remedy | 0 | 0 | 0.04 | 0.056 | 0.074 | 0 |
| First Down Dash | 0 | 0 | 0 | 0 | 0 | 0.06 |
| Genuine Doc | 0 | 0 | 0.043 | 0.049 | 0.065 | 0 |
| Im Kiddin | 0.051 | 0 | 0 | 0 | 0 | 0 |
| Kid Clu | 0.058 | 0 | 0 | 0 | 0 | 0 |
| Mr Conclusion | 0.049 | 0 | 0 | 0 | 0 | 0 |
| Mr Eye Opener | 0 | 0 | 0 | 0 | 0 | 0.045 |
| Pine Interest | 0 | 0.038 | 0 | 0 | 0 | 0 |
| Smart Little Lena | 0 | 0 | 0.039 | 0.059 | 0.006 | 0 |
| Zippo Pat Bars | 0.012 | 0.039 | 0 | 0 | 0 | 0 |
| Zippo Pine Bar | 0.014 | 0.057 | 0 | 0 | 0 | 0 |
Bold type indicates the sire with the highest value per performance group.
Figure 2.
Pedigree showing the relationship among 4 of the most influential sires of REF horses in each performance group to Three Bars, a Thoroughbred stallion, and AQHA Hall of Fame inductee. Year of birth is noted in parenthesis; circles denote unique dams.
Generation Intervals
The overall generation interval of the REF sample was 10.5 years, ranging from 9.5 in the halter horses to 11.8 in the cutting horses (Table 4). The generation interval of the halter horses was significantly less than that of all samples other than the racing QH, whereas those of the cutting, reining, and working cow groups were significantly higher than all others. Median age of the proband individuals within the REF cohorts shows that the pleasure horses were significantly older than the halter and racing horses, which were significantly younger than all other samples (Table 4).
Table 4.
Generation interval calculated across 4-generation pedigrees for the REF cohorts as well as the range and median year of birth of proband individuals
| Generation interval | Proband year of birth | ||
|---|---|---|---|
| Range | Median | ||
| Halter | 9.5c | 1998–2008 | 2006ac |
| Pleasure | 10.3ad | 1996–2007 | 2003b |
| Reining | 11.4b | 2000–2007 | 2005ab |
| Working cow | 10.9ab | 1997–2007 | 2005ab |
| Cutting | 11.8b | 1998–2007 | 2004ab |
| Racing | 9.9cd | 2004–2008 | 2007c |
| All | 10.5 | 1996–2008 | 2005 |
Values sharing the same superscript are not significantly different from one another (generation interval tested independent of year of birth).
FST
Among performance groups, pairwise F ST calculated from the REF cohorts (0.012–0.033) showed all values were greater than those calculated between the AG cohorts (0.013–0.014) with the exception of the cutting and working cow horses (0.012). Across the 3 types of samples, the divergence observed among performance groups in the REF horses was less than that observed in the RA cohorts (data not shown) and from the TP cohorts (Table 5).
Table 5.
Pedigree-based F ST values among cohorts within the TP (top) and REF samples (bottom)
| Halter | Pleasure | Reining | Working cow | Cutting | Racing | |
|---|---|---|---|---|---|---|
| Halter | 0.031 | 0.027 | 0.035 | 0.045 | 0.033 | |
| Pleasure | 0.018 | 0.026 | 0.033 | 0.044 | 0.033 | |
| Reining | 0.021 | 0.021 | 0.017 | 0.029 | 0.027 | |
| Working cow | 0.024 | 0.024 | 0.013 | 0.022 | 0.036 | |
| Cutting | 0.031 | 0.030 | 0.018 | 0.012 | 0.046 | |
| Racing | 0.023 | 0.023 | 0.022 | 0.026 | 0.033 |
Bold type indicates a nonsignificant difference between cohorts.
Inbreeding
Mean individual inbreeding estimates from the 4-generation pedigrees ranged from 0.009 (reining) to 0.037 (cutting) in the REF sample (Table 6). Regardless of pedigree sample (REF, RA, and TP), mean individual inbreeding values were highest in the cutting horses and lowest in the reining horses. Significance between cohorts within samples is found in Table 6 and Supplementary Material online; no significant difference was observed among the 3 AG cohorts. The highest observed inbreeding value was 0.188. Within each of the QH performance groups, the REF horses were, as individuals, not significantly more or less inbred than the RA, TP, or AG samples (Supplementary Material online).
Table 6.
Pedigree-based estimates of individual inbreeding for the REF sample
| Estimated inbreeding | |||
|---|---|---|---|
| Min | Max | Average | |
| Halter | 0.000 | 0.188 | 0.026ab |
| Pleasure | 0.000 | 0.078 | 0.015b |
| Reining | 0.000 | 0.047 | 0.009b |
| Working Cow | 0.000 | 0.066 | 0.020ab |
| Cutting | 0.008 | 0.079 | 0.037a |
| Racing | 0.000 | 0.047 | 0.016b |
Means sharing the same superscript with samples are not significantly different from one other.
Coancestry
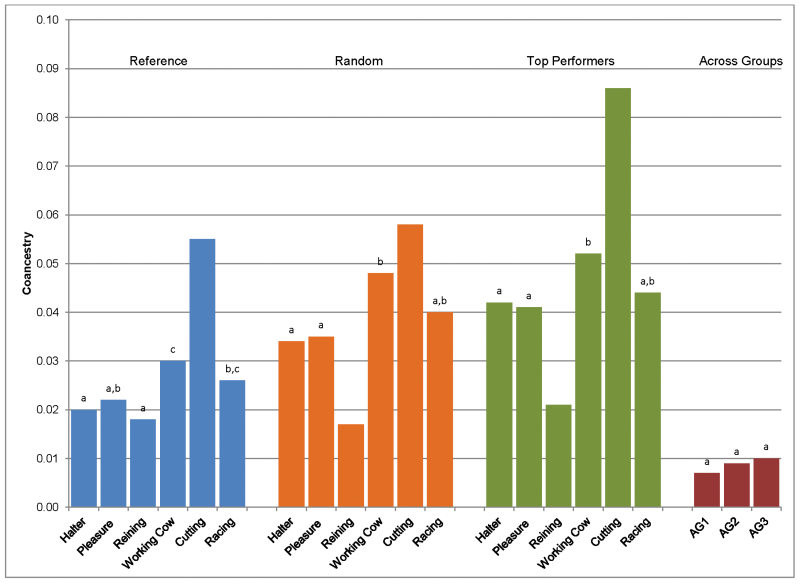
The average pedigree-based coefficients of coancestry for the REF horses ranged from 0.018 in the reining horses to 0.055 in the cutting group (Figure 3). Among performance groups in all samples (REF, RA, and TP), coancestry was significantly greater in the cutting horses than all others. The reining horses showed lower coancestry than other performance groups regardless of sample and significantly lower coancestry than the racing QH. As anticipated due to sampling methods, coancestry was significantly greater in the RA and TP samples relative to the REF sample and highest in TP (Figure 3). An exception is found in the reining horses, where coancestry was not significantly different across the 3 samples or when comparing the REF to RA cutting horses. Coancestry was not significantly different across the AG cohorts; however, coancestry of AG samples was significantly less than that of all performance groups in all samples with the exception of the RA reining cohort.
Figure 3.
Mean pairwise coancestry within each performance group pedigree among proband individuals of each sample. Superscripts shared between performance groups within each sample are not significantly different.
Genotype Analysis
QH Among Group Diversity
Pairwise F ST values based upon SNP data show that performance groups are significantly different from one another with the exception of the comparison between the cutting and working cow QH (F ST = 0.000; Supplementary Material online). Divergence among the performance groups was greatest between the cutting and racing QHs (F ST = 0.074).
AMOVA showed that a significant component of variance (3.44%, P < 0.001) was accounted for among the performance groups with 96.56% of variance found within individuals. When variation at the level of the individual was considered, AMOVA found a negative component of variance (−2.91%) among individuals within populations. The global F ST for the QH REF sample was 0.035.
QH Cluster Analysis
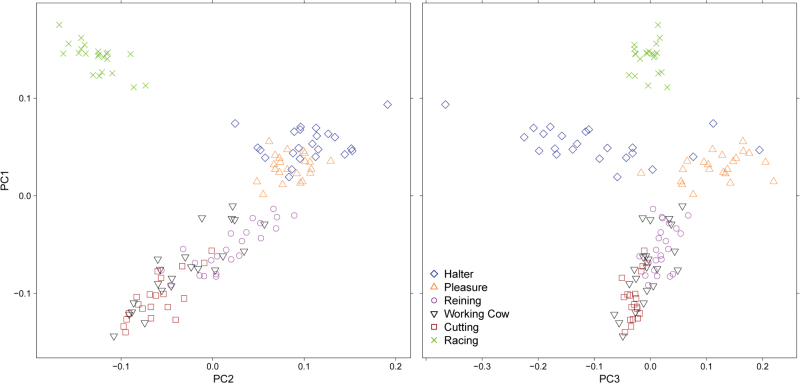
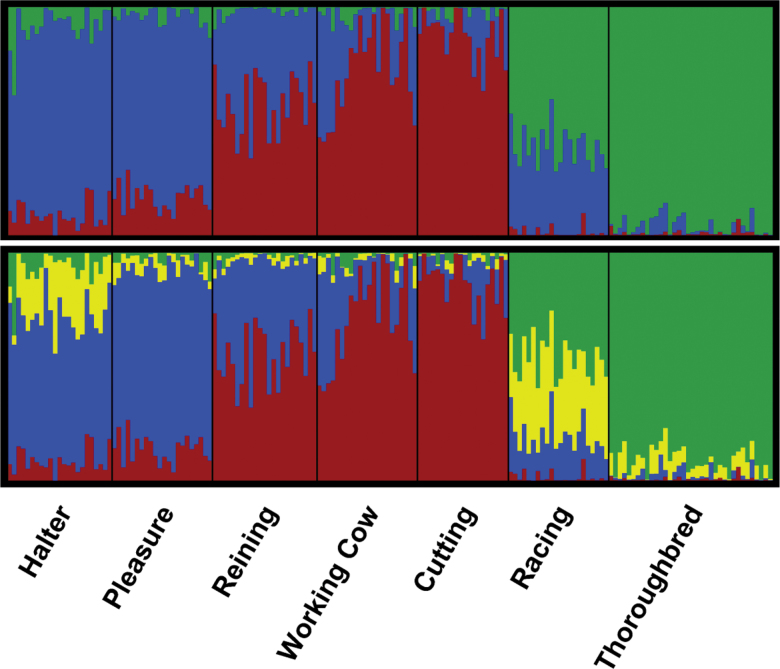
Likelihood values from Bayesian cluster analysis of the QH REF sample were similar from K = 1 to 6 (Supplementary Material online). The standard deviation among runs at each value of K increased substantially at K = 7, whereas at K = 3, the standard deviation among runs was lower, with similar likelihood as observed at higher values of K. Assuming K = 3, clustering was observed between the pleasure and halter cohorts, as well as between the working cow and cutting cohorts; both the working cow and cutting QHs shared assignment with the reining cohort. The racing QH assigned strongly to a unique cluster (Supplementary Material online). PCA using 36 037 SNPs also showed the racing QHs separate from the remainder of the population in PC1 versus PC2 (Figure 4). The halter and pleasure QHs clustered together until PC3. Working cow horses were distributed among the reining and cutting performance groups. Results of PCA were similar regardless of if the data were pruned for LD (data not shown).
Figure 4.
Principal component analysis (PCs 1 versus 2 and 1 versus 3) for the 6 performance groups of the Quarter Horse.
QH within Group Diversity
Measures of H e within the QH performance groups showed the least diversity within the racing and cutting cohorts, whereas the reining, pleasure, and halter cohorts were most diverse, sharing H e values of 0.346 (Table 7).
Table 7.
Expected heterozygosity (H e) and individual estimates of inbreeding (f) based upon SNP genotype data
| N | H e | Individual inbreeding (f) | |||
|---|---|---|---|---|---|
| Min | Max | Average | |||
| Halter | 23 | 0.346 | −0.040 | 0.269 | 0.030 |
| Pleasure | 22 | 0.346 | −0.049 | 0.076 | 0.014 |
| Reining | 23 | 0.346 | −0.018 | 0.079 | 0.024 |
| Working cow | 22 | 0.344 | −0.028 | 0.120 | 0.027 |
| Cutting | 20 | 0.334 | −0.011 | 0.091 | 0.039 |
| Racing | 22 | 0.334 | −0.029 | 0.127 | 0.033 |
| All QH | 132 | 0.352 | −0.049 | 0.269 | 0.028 |
| Thoroughbred | 36 | 0.330 | −0.041 | 0.098 | 0.034 |
| Total | 168 | 0.354 | |||
Number of horses used in the analyses is also provided (N).
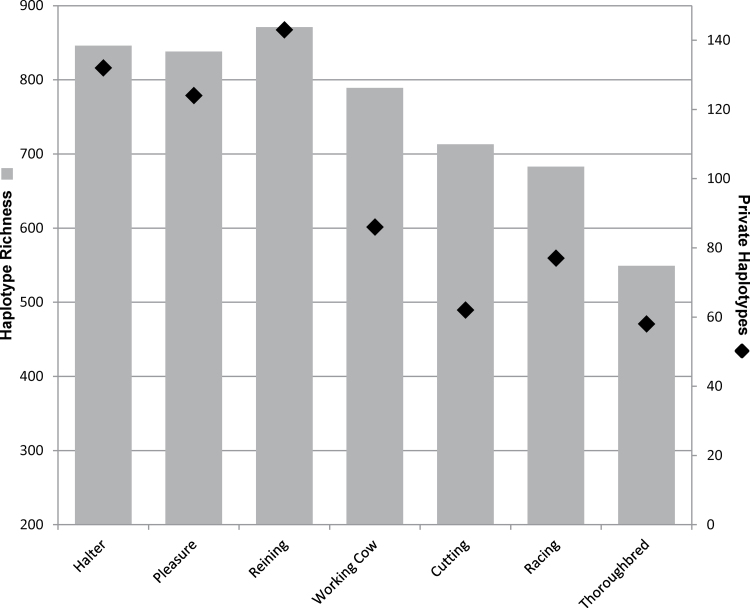
Haplotype analysis across 5 chromosomal regions included a total of 69 individual 500kb windows and 710 SNPs. An average of 25.8 haplotypes was observed per window (range 5–68). Haplotype richness results largely follow that observed for H e; the greatest number of haplotypes were found in the reining cohort and least in the racing and cutting horses (Figure 5). Private haplotypes, signifying uniqueness within each performance group, were most abundant in the reining group, whereas relatively less abundant in the cutting QHs (Figure 5).
Figure 5.
The total number of unique haplotypes observed across 69, 500kb windows of the genome for the REF QH performance groups and the Thoroughbred (gray bars, left axis). Number of private haplotypes observed per sample (diamond, right axis).
Estimates of individual inbreeding (f) calculated from excess homozygosity showed mean inbreeding in the REF QH of 0.028, ranging among the performance groups from 0.014 (pleasure) to 0.039 (cutting) (Table 7). The highest individual estimated f value was found in a halter horse (0.269) who was the result of a mating between parents who shared a sire; that sire, himself, was the result of a mating between half-siblings. This was the same individual with the maximum observed inbreeding value as determined by pedigree. Individual inbreeding estimates of the REF horses was significantly correlated to inbreeding calculated from pedigree data (R = 0.70, P < 2.2×10−16).
Relationship to TBs
The TB was most divergent from the cutting QH (F ST = 0.092) followed by the working cow (F ST = 0.079) and reining cohorts (F ST = 0.077) (Supplementary Material online); the TB was most similar to the racing QH (F ST = 0.034). All pairwise comparisons between QH performance groups and the TB were significantly greater than zero.
The inclusion of the TB in Structure analyses resulted in likelihood scores plateauing from K = 1 to 7 with no ideal value of K clearly apparent (Supplementary Material online). The standard deviation among runs increased almost 4-fold between K = 3 and 4, with a diminishing increase in likelihood; therefore, both values of K were examined. At K = 3, the racing QH shared a majority (61.3%) assignment to the same cluster as the TB, whereas the remainder of the racing QH variation was assigned to the cluster represented by the pleasure and halter horses (Figure 6; Supplementary Material online). Forcing K = 4 found the new cluster comprising 34.7% of assignment in the racing QH, with moderate representation (20.7%) in the halter horse and a small portion of assignment (6.4%) in the TB (Figure 6; Supplementary Material online). PCA similarly showed the distinction of the TB from the QH while also supporting a closer association of the TB to the racing cohort than to the other performance groups (Supplementary Material online).
Figure 6.
Structure output assuming 3 (K = 3) (top) and 4 (K =4) clusters for all Quarter Horse REF individuals and the Thoroughbred. Each vertical line represents 1 individual with the proportion of assignment to each of the 3 clusters denoted on the y-axis.
Diversity in the TB (H e = 0.330) was less than that found in any QH REF cohort or the entire QH sample considered as a whole (H e = 0.352) (Table 7). Similarly, haplotype richness in the TB was less than that observed in any QH cohort (Figure 5). Genotype-based estimates of f in the TB averaged 0.034, with a maximum of 0.098 (Table 7).
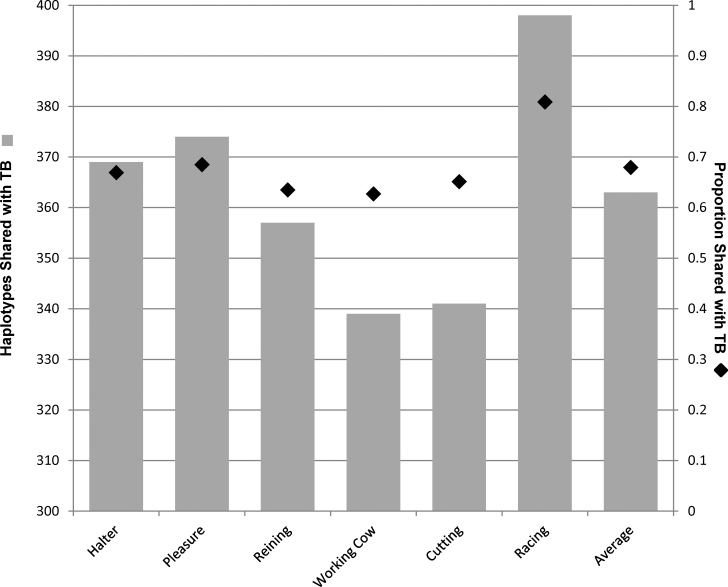
Considering only the presence or absence of unique haplotypes, 398 of the 549 haplotypes observed in the TB were also found in the racing QH (Figure 7). Taking into account the frequency of each of these haplotypes across all individuals, 80.8% (2232) of the haplotypes examined in the racing cohort (total N = 2760 [20 individuals × 2 chromosomes × 69 windows]) were also present in the TB. The total proportion of haplotypes shared with the TB was similar among the other 5 performance groups (62.6% to 68.4%) and less than that of the racing QH.
Figure 7.
Sum of the count of haplotypes of the Quarter Horse REF cohorts with shared identity to those found in the Thoroughbred (gray bar, left axis). Proportion of the total haplotypes observed within each cohort that corresponds to a haplotype also present in the Thoroughbred (diamonds, right axis).
Discussion
The expansion of the QH population since inception of the breed association, as well as the diversity present in the founding stock, has allowed for the maintenance of genetic diversity within the breed. It is likely that this genetic diversity is responsible for the versatility in performance abilities that are touted as a hallmark of the QH breed. However, regardless of whole-breed diversity, the presence of population subdivision was suggested in early pedigree analyses of the breed (Fletcher 1945; Tunnell et al. 1983). Pedigree studies have since been supplemented with genetic data and morphological analyses, adding further support to the hypothesis that there is population structure within the QH based upon performance type (Tryon et al. 2009; Meira et al. 2013). Significant population structure within the breed could be of concern if the resulting divergence is the result of or accompanied by an increase in inbreeding, which has the potential to lead to an increased incidence of undesirable traits. This study represents the first evaluation of 6 diverse performance groups of the QH utilizing both genome-wide SNP and pedigree data. These results not only describe diversity within and relationships among QHs but consider diversity of the top performing horses that represent the best of the best, as well as the influence of the TB.
To investigate whether detectable subpopulation structure is present in the modern QH, it was necessary to identify individuals representative of each performance group. Therefore, horses that were actively competing in each discipline were selected. The 6 performance groups studied represent the most popular and phenotypically diverse categories within the breed. In addition, these 6 groups have differing histories. Although QHs were bred for racing and used to work cattle before the formal establishment of the breed, disciplines such as western pleasure and halter classes have much more recent roots, allowing less time to evolve as a distinct population.
F ST analyses of SNP and pedigree data both show that all performance groups, excluding the comparison between the cutting and working cow cohorts, are significantly divergent from one another; and, within these 6 performance groups, the distinction of the racing QH was clear. The racing QH consistently showed significant divergence from the other performance groups with the greatest divergence from the stock-type horses, especially the cutting horses, where quickness and “cow sense” are more important than outright speed. In addition to selective breeding for bloodlines or popular sires (see below), this distinctiveness can be attributed to a significant contribution of TB bloodlines into the racing QH lineages. Pedigree analysis as well as haplotype sharing confirmed a similarity of the TB to the racing QH at a level not observed with the other QH performance groups. As the TB was instrumental in development of the QH, and both groups are bred for racing, this result was not unexpected.
Apart from the racing QH, notable clustering of samples and corresponding low, but significant divergence was observed among what are often classified as the “stock-type” horses: the cutting, reining, and working cow samples. Of the performance groups studied, the most recently derived, the working cow horse, represents a competition that combines the critical skills required of reining and cutting horses. The intersection of these required traits is plainly visible in the SNP analyses where the working cow horses were minimally divergent from and distributed among individuals of reining and cutting performance groups in F ST and PCA, respectively. Common sires were also found between these 3 categories with each showing a notable contribution of variation from the stallion Doc Bar and his son, Doc O’Lena.
Although still showing signs of significant divergence, the halter and pleasure horses also showed similarities in cluster analyses, supported by the fact that popular sires in the pleasure sample were also observed in the halter pedigree. As discussed in regard to the cutting, working cow, and reining horses above, the reasoning behind these relationships likely stems from shared similarities of desired phenotypes. In the halter and western pleasure samples, speed and quickness are not required, and horses are instead selected to have smooth, slow movements, be of large size, and be visually appealing.
Overall, the analyses of divergence and population structure across the 6 performance groups, as well as similarities in desirable performance characteristics, point toward the presence of 3 major types of QH in the samples studied: stock horses (cutting, reining, and working cow), halter/pleasure horses, and racing horses. Pedigree analyses complementary to the genetic data showed that popular sires shared within these 3 groups were rarely shared between. However, the pedigrees still reflect the common roots of the QH among performance classes.
Popular sires within the 4-generations studied are indicated by disproportionate values of average relatedness. These sires not only reflect trends in breeding that are specific to performance groups but also illustrate how the diversity present in the founding population has been selected upon to create these divergent classes of QHs. An example of 1 sire who contributed to all 6 performance groups, although through varying paths, is clear through the study of Three Bars (1940–1968). Three Bars, a TB stallion later inducted into the AQHA Hall of Fame, also serves as an example of the significance of the TB in the development of the QH breed. As illustrated in Figure 2, the sires contributing the largest proportion of variance to each performance group all trace back to Three Bars.
The pedigree-based distance between the top sires and Three Bars is related to differences observed among generation intervals of the performance groups. In a similar study, generation intervals of the QH were reported to range from 8.1 to 10.3, varying based upon the time point sampled (Tunnell et al. 1983). This is similar to the range observed in the Franches-Montagnes (7.8 years) (Poncet et al. 2006), Lusitano (10.5 years) (Vicente et al. 2012), and Hungarian TB (11.4 years) (Bokor et al. 2013). The overall generation interval found across all REF samples in this study was 10.5, with large variance that can be attributed to the ability of horses to reproduce from as early as the age of 2, well into their 20s. Differences in generation intervals between samples in this study are likely a result of the difference in the age of optimal performance for the horses in each category. For example, racing QHs compete as 2-year olds and are often retired by the age of 6, whereas halter horses compete as weanlings through adulthood. Although successful individuals in both racing and halter may be slated for breeding at a very young age, the introduction of new horses from the remaining performance groups into the breeding herd likely does not take place until after they have had sufficient training under saddle to find success and are retired from showing. The differences in generation interval statistics among cohorts are also reflected in the median age of proband individuals.
Although the removal of close relationships in the genotyped animals was helpful to more completely assay the diversity representative of each performance group, sampling in such a manner also biased indices of coancestry downward, which could lead to inaccurate conclusions about the potential for inbreeding. To more accurately evaluate inbreeding in each performance group, the additional within-performance group samples were considered. The AG cohorts in particular allowed for a better comparison of overall diversity of the breed relative to that within each performance group.
Pedigree analysis of the RA, TP, and AG samples showed the expected result of sampling bias in the REF horses: the RA and TP samples had significantly higher coancestry than the REF sample where sibships were avoided. In contrast, horses within the AG cohorts had lower coancestry than the REF, RA, or TP cohorts. Increased coancestry and relatedness in the TP sample compared with the REF horses are also apparent in a decrease in the number of total horses in the full pedigree. Exceptions include the reining horses where coancestry of the RA and TP cohorts did not differ from that of the REF individuals, as well as the cutting REF cohort, which was not significantly different from the RA cohort. This result in the reining horses may stem from the greater diversity within this sample as a whole (see below). However, in both cases, the number of horses in the pedigree decreased stepwise when comparing the REF cohort to the RA cohort to the TP cohort. This demonstrates that the best of the best in each performance group represents an even smaller gene pool than a random sample of individuals within the same performance group.
Coancestry between a pair of individuals, the probability that 2 alleles drawn at random are identical by descent, reflects the coefficient of inbreeding that would be observed in an offspring if they were to mate. Using these data, estimates of coancestry suggest that breeding among a pair of proband individuals within a REF cohort could result in up to a 1.9-fold increase in the current mean level of inbreeding (based upon the reining cohort). Considering the coancestry within the TP sample, the projected increase in inbreeding resulting from mating between proband individuals increases as high as 4-fold, projected in the racing performance group. However, those estimates include matings of half-siblings, which breeders should avoid. The projected increase in inbreeding as a result of mating among proband individuals also assumes that the horses studied will be actively contributing to the future gene pool. In actuality, the geldings and likely some other proband individuals will not contribute genetic material to the next generation. However, it was shown that a performance prospect is more desirable if the horse’s sire or dam had previously produced a successful offspring or if the second or third sire or dam were champions or producers of champions (Lansford et al. 1998). Thus, the genetic variation of winning horses is expected to be perpetuated by the continued breeding and increased desirability of the horses’ parents, grandparents, and siblings; this preferential breeding could also result in an increase in inbreeding in future generations. In contrast to the performance group cohorts, pedigree data showed that breeding between individuals in any AG cohort would result in a 46% to 64% decrease in individual inbreeding compared with that observed in the proband.
As expected, individual inbreeding estimates did not differ significantly between the REF, RA, and TP samples. Although the sampling scheme of the REF horses was biased, removing related individuals did not affect the content of individual pedigrees. Pedigree-based inbreeding within the REF horses averaged 2.1%, driven downward by low, average inbreeding (0.9%) in the reining cohort. Inbreeding based upon genotype data is correlated to but generally higher than the pedigree-based estimates even given that the genotype-based estimates can be negative. Discrepancies in genotype versus pedigree inbreeding values may be attributed to several characteristics of the data, one being the depth of the pedigrees. If relationships among founders are not accounted for, inbreeding can be underestimated (MacCluer et al. 1983); in this case, it is evident that there are relationships among individuals that are found in the fourth and final generation studied in this analysis. However, also based upon 4-generation pedigrees, inbreeding in 1945 was estimated to be 1.7% (Fletcher 1945), similar to that found in the AG sample (mean = 1.5%), but lower than that observed if considering the mean across performance groups of the REF, RA, and TP samples (2.1%, 2.5%, and 2.5%, respectively). These estimates of inbreeding in the QH are still substantially lower than that reported in Hungarian TBs (9.6%) (Bokor et al. 2013) although that analysis was performed with exhaustive pedigrees. AMOVA, showing a negative component of variance among individuals within the performance groups, also supports the lack of variation within performance groups.
Inbreeding leads to a loss of diversity, and conversely, in populations of limited size, a loss of diversity due to a population bottleneck or founder effect results in increased inbreeding. Therefore, it is not a surprise that the mean estimate of individual inbreeding from the genotypes and pedigrees was greatest in the cutting group, where the lowest within-performance group diversity was observed. Additionally, as relatedness and inbreeding rises within performance groups, divergence among these groups is also expected to increase. This is illustrated by pedigree-based F ST values that are greater in comparisons of the TP and RA samples relative to the REF samples. Within the 6 performance groups studied, average relatedness is increasing over time, suggesting that divergence between these groups will increase if current breeding practices are maintained. However, in light of these data, expected heterozygosity suggests that genetic diversity remains within the breed and is available for supplementation of subpopulations that may be at risk for inbreeding depression.
With H e similar to the cutting cohort, the racing QHs show lower diversity measures despite a relatively high contribution of TB outcrossing in the pedigree. Based upon the results of cluster and haplotype sharing analyses, the racing QH has high similarity to the TB as a result of both historical and recent admixture. However, the addition of TB ancestry may do little to increase the diversity of the racing QH. Results herein as well as in prior studies show that the TB breed comprises relatively low genetic diversity (Cunningham et al. 2001; Glowatzki-Mullis et al. 2005; McCue et al. 2012; Bokor et al. 2013; Petersen et al. 2013). Along with the fact that the TB contributed significantly to the founding of the QH, the current outcrossing to the racing QH may not be adding detectable, novel genetic material. On the other hand, introgression from the TB cannot explain the low diversity observed in the cutting horse, which shows little modern influence from the breed. The relatively low diversity of the cutting performance group appears to be attributable to a founder-type effect resulting from selection for a few popular bloodlines.
Diversity was almost identical across the halter, pleasure, reining, and working cow groups as assessed by H e, and this result was largely mirrored by calculation of haplotype richness. However, the reining, pleasure, and halter cohorts showed increased uniqueness relative to the other performance groups as measured by the incidence of private haplotypes. The relatively high number of private haplotypes in the reining sample despite similar diversity between samples supports its distinctiveness from the working cow and other performance groups. Diversity within performance groups was lower than that observed when all QH samples were combined. However, in all cases, the QH performance groups showed higher diversity than that found in the sample of the TB. This is especially notable as the sample of TBs represented diversity assayed over 32 years, across 2 continents, and is likely further biased upward due to the ascertainment of SNPs largely based upon the TB reference genome.
Conclusions
Both genotype and pedigree data in this study reveal that although the QH breed as a whole contains substantial genetic diversity, variation is partitioned into subpopulations defined by performance type. The versatility of the QH is still present, but while 1 horse may have had multiple talents at the founding of the breed, individuals are now specialized to perform more specific tasks. Current analyses show that with the exception of the working cow and cutting groups, each performance group is genetically distinct from one another and in a broad sense can be classified into one of 3 categories: stock horses, halter/pleasure horses, and racing horses. These data also show evidence of increased inbreeding over time, suggesting that continuation of the current breeding trends may result in further divergence among the subpopulations. Although these data do not consider diversity of nonperforming, recreational QHs, the impact of selection for top performers undoubtedly influences the overall structure and direction of the breed.
Supplementary Material
Supplementary material can be found at http://www.jhered.oxfordjournals.org/.
Funding
American Quarter Horse Foundation grant, “Selective Breeding Practices in the American Quarter Horse: Impact on Health and Disease” 2011–2012; USDA-NIFA (2008-35205-18766, 2009-55205-05254, 2012-67015-19423). National Institute of Health–National Institute of Arthritis and Musculoskeletal and Skin Disease (NIAMS) (1K08AR055713-01A2 M.E.M.) and (2T32AR007612 to J.L.P.); the University of Minnesota College of Veterinary Medicine/Merial Veterinary Summer Scholars’ Program (K.D.C.).
Supplementary Material
Acknowledgments
Quarter Horse hair root samples were graciously supplied by the Veterinary Genetics Laboratory at the University of California Davis with assistance from MCT Penedo and M Le. E. W. Hill, M. M. Binns, and E. Bailey provided information on the Thoroughbred samples. J. P. Gutierrez is acknowledged for discussions of pedigree analyses. AMD Schreier and two reviewers provided helpful comments on a previous version of the manuscript. Images of performing Quarter Horses were courtesy of The American Quarter Horse Journal.
References
- American Quarter Horse Association 2010. 2012 Annual Report. Amarillo (TX): American Quarter Horse Association [Google Scholar]
- Bokor A, Jonas D, Ducro B, Nagy I, Bokor J, Szabari M. 2013. Pedigree analysis of the Hungarian Thoroughbred population. Livest Sci. 151:1–10 [Google Scholar]
- Caballero A, Toro MA. 2000. Interrelations between effective population size and other pedigree tools for the management of conserved populations. Genet Res. 75:331–343 [DOI] [PubMed] [Google Scholar]
- Caballero A, Toro MA. 2002. Analysis of genetic diversity for the management of conserved subdivided populations. Conserv Gen. 3:289–299 [Google Scholar]
- Cunningham EP, Dooley JJ, Splan RK, Bradley DG. 2001. Microsatellite diversity, pedigree relatedness and the contributions of founder lineages to thoroughbred horses. Anim Genet. 32:360–364 [DOI] [PubMed] [Google Scholar]
- Denhardt R. 1941. American Quarter Horse Stud Book and Registry: Vol 1 No 1. Amarillo (TX): American Quarter Horse Association [Google Scholar]
- Denhardt RM. 1967. Quarter horses: a story of two centuries. Norman (OK): University of Oklahoma Press [Google Scholar]
- Excoffier L, Laval G, Schneider S. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 1:47–50 [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 164:1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JL. 1945. A genetic analysis of the American quarter horse. J Hered. 36:346–352 [DOI] [PubMed] [Google Scholar]
- Glowatzki-Mullis ML, Muntwyler J, Pfister W, Marti E, Rieder S, Poncet PA, Gaillard C. 2005. Genetic diversity among horse populations with a special focus on the Franches-Montagnes breed. Anim Genet. 37:33–39 [DOI] [PubMed] [Google Scholar]
- Gutiérrez JP, Goyache F. 2005. A note on ENDOG: a computer program for analysing pedigree information. J Anim Breed Genet. 122:172–176 [DOI] [PubMed] [Google Scholar]
- Hendricks BL. 2007. International Encyclopedia of horse breeds. Norman (OK): University of Oklahoma Press [Google Scholar]
- Jakobsson M, Rosenberg NA. 2007. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 23:1801–1806 [DOI] [PubMed] [Google Scholar]
- James JW. 1977. A note on selection differential and generation length when generations overlap. Anim Prod. 24:109–112 [Google Scholar]
- Lansford NH, Jr, Freeman DW, Topliff DR, Walker OL. 1998. Hedonic pricing of race-bred yearling Quarter Horses produced by Quarter Horse sires and dams. J Agribusiness. 16:169–185 [Google Scholar]
- Laune P. 1973. America’s quarter horses. Garden City (NY): Doubleday [Google Scholar]
- Leroy G, Callède L, Verrier E, Mériaux JC, Ricard A, Danchin-Burge C, Rognon X. 2009. Genetic diversity of a large set of horse breeds raised in France assessed by microsatellite polymorphism. Genet Sel Evol. 41:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luís C, Juras R, Oom MM, Cothran EG. 2007. Genetic diversity and relationships of Portuguese and other horse breeds based on protein and microsatellite loci variation. Anim Genet. 38:20–27 [DOI] [PubMed] [Google Scholar]
- MacCluer JW, Boyce AJ, Dyke B, Weitkamp LR, Pfennig DW, Parsons CJ. 1983. Inbreeding and pedigree structure in Standardbred horses. J Hered. 74:394–399 [DOI] [PubMed] [Google Scholar]
- McCue ME, Bannasch DL, Petersen JL, Gurr J, Bailey E, Binns MM, Distl O, Guérin G, Hasegawa T, Hill EW, et al. 2012. A high density SNP array for the domestic horse and extant Perissodactyla: utility for association mapping, genetic diversity, and phylogeny studies. PLoS Genet. 8:e1002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meira CT, Curi RA, Vasconcelos Silva JA, II, Monteiro Correa MJ, Nunes de Oliveira H, Silveira da Mota MD. 2013. Morphological and genomic differences between cutting and racing lines of quarter horses. J Equine Vet Sci. 33:244–249 [Google Scholar]
- Petersen JL, Mickelson JR, Cothran EG, Andersson LS, Axelsson J, Bailey E, Bannasch D, Binns MM, Borges AS, Brama P, et al. 2013. Genetic diversity in the modern horse illustrated from genome-wide SNP data. PLoS One. 8:e54997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncet PA, Pfister W, Muntwyler J, Glowatzki-Mullis ML, Gaillard C. 2006. Analysis of pedigree and conformation data to explain genetic variability of the horse breed Franches-Montagnes. J Anim Breed Genet. 123:114–121 [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics. 155:945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Tood-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, De Bakker PIW, Daly MJ, et al. 2007. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA. 2004. DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes. 4:137–138 [Google Scholar]
- Scheet P, Stephens M. 2006. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 78:629–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryon RC, Penedo MCT, McCue ME, Valberg SJ, Mickelson JR, Famula TR, Wagner ML, Jackson M, Hamilton MJ, Nooteboom S, et al. 2009. Evaluation of allele frequencies of inherited disease genes in subgroups of American Quarter Horses. J Am Vet Med Assoc. 234:120–125 [DOI] [PubMed] [Google Scholar]
- Tunnell JA, Sanders JO, Williams JD, Potter GD. 1983. Pedigree analysis of four decades of Quarter Horse breeding. J Anim Sci. 57:585–593 [DOI] [PubMed] [Google Scholar]
- Vicente AA, Carolino N, Gama LT. 2012. Genetic diversity in the Lusitano horse breed assessed by pedigree analysis. Livest Sci. 148:16–25 [Google Scholar]
- Wright S. 1922. Coefficients of inbreeding and relationship. Am Nat. 56:330–338 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.