Abstract
Multiple substance dependence (MSD) trait comorbidity is common, and MSD patients are often severely affected clinically. While shared genetic risks have been documented, so far there has been no published report using the linkage scan approach to survey risk loci for MSD as a phenotype. A total of 1,758 individuals in 739 families [384 African American (AA) and 355 European American (EA) families] ascertained via affected sib-pairs with cocaine or opioid or alcohol dependence were genotyped using an array-based linkage panel of single-nucleotide polymorphism markers. Fuzzy clustering analysis was conducted on individuals with alcohol, cannabis, cocaine, opioid, and nicotine dependence for AAs and EAs separately, and linkage scans were conducted for the output membership coefficients using Merlin-regression. In EAs, we observed an autosome-wide significant linkage signal on chromosome 4 (peak lod = 3.31 at 68.3 cM; empirical autosome-wide P = 0.038), and a suggestive linkage signal on chromosome 21 (peak lod = 2.37 at 19.4 cM). In AAs, four suggestive linkage peaks were observed: two peaks on chromosome 10 (lod = 2.66 at 96.7 cM and lod = 3.02 at 147.6 cM] and the other two on chromosomes 3 (lod = 2.81 at 145.5 cM) and 9 (lod = 1.93 at 146.8 cM). Three particularly promising candidate genes, GABRA4, GABRB1, and CLOCK, are located within or very close to the autosome-wide significant linkage region for EAs on chromosome 4. This is the first linkage evidence supporting existence of genetic loci influencing risk for several comorbid disorders simultaneously in two major US populations.
Keywords: comorbidity, multiple substance dependence, fuzzy clustering, chromosome 4
INTRODUCTION
Substance dependence (SD) is a chronic psychiatric illness [Leshner, 1997; McLellan et al., 2000]. This illness has led to substantial injury, loss of life, and disability in societies worldwide. Developing effective treatments for SD is particularly crucial because of the exceptionally high rate of relapse [McGovern et al., 2005] and the tremendous disease burden to society. Dependence on multiple substances, which is frequently observed clinically, complicates the development of effective treatments. Multiple substance use is common among alcohol and drug users and the average percentage of individuals using multiple substances is around 56% [SAMSHA, 2009]. Individuals with comorbid DSM-IV alcohol and drug use disorders outnumber patients with a drug use or alcohol use disorder alone [Stinson et al., 2005] among treatment seekers. This treatment seeking pattern may be a reflection of disease severity and the damage that multiple substance dependence(MSD) inflicts on an individual’s health and well being. Indeed, MSD patients are often severely affected.
Research from adoption, twin and family studies has shown that there is a genetic diathesis in the development of SD [Cloninger, 1987; Mirin et al., 1991; Pickens et al., 1991; Rounsaville et al., 1991; Luthar and Rounsaville, 1993; Merikangas et al., 1998; Tsuang et al., 1998; Kendler et al., 2003; Agrawal and Lynskey, 2008]. Investigations on the genetic liability for SD, in particular MSD, suggest that there are common as well as substance-specific genetic factors [Metten and Crabbe, 1994, 1999; Pickens et al., 1995; Bierut et al., 1998; Tsuang et al., 1998; Grucza and Bierut, 2006; Belknap et al., 2008; Huizink et al., 2010]. And indeed, there are numerous known examples of specific genes known to affect multiple substance use disorders [Gelernter et al., 2006b; Luo et al., 2006, 2007; Yang et al., 2008; Sherva et al., 2010].
Current evidence suggests that copy number variants (CNVs) and rare single-nucleotide polymorphisms (SNPs) might be implicated in the risk for many complex diseases [Walsh et al., 2008; Johansen et al., 2010]. Genomewide association studies are unable to capture most of these rare genetic variants. An adequately powered linkage study design has the advantage of detecting diverse inherited genetic effects that segregate in families, including common variants, rare variants and CNVs. Since the current cost of conducting a large-scale whole genome sequencing study is still high, genetic linkage approaches remain valuable and informative to identify candidate regions for targeted resequencing studies.
Prior linkage studies have identified genomic susceptibility regions for single SD diagnoses [Gelernter et al., 2004, 2005, 2006a, 2007; Han et al., 2010; Panhuysen et al., 2010]. In this study, we proceed from the well-supported hypothesis that substance dependence traits share common genetic liability to some varying extent. Given the potential for shared genetic liability, investigation of dependence on multiple substances makes it possible to gain substantial phenotypic information and statistical power to map-shared susceptibility regions. In this autosomal linkage scan, we used dependence on three illicit drugs (cannabis, cocaine, and opioid) and two legal substances (alcohol and nicotine) as a phenotype. We used fuzzy clustering analysis [Kaufman and Rousseeuw, 1990] to reduce the dimensions and derive a more homogeneous quantitative trait to identify a genetic component common to multiple SDs. Fuzzy clustering yields a membership coefficient that contains more information than the standard hard clustering approaches.
MATERIALS AND METHODS
Recruitment
Study subjects were recruited at four recruitment sites including University of Connecticut Health Center (UConn, Farmington, CT), Yale University School of Medicine (APT Foundation, New Haven, CT), Medical University of South Carolina (MUSC, Charleston, SC), and McLean Hospital (Harvard Medical School; Belmont, MA). The number of families recruited at each site is: for AA, Yale 153, UConn 140, McLean 43, MUSC 48, and for EA, Yale 101, UConn 152, McLean 45, and MUSC 52. We screened the families based on the belief that at least two siblings in each family would meet diagnostic criteria for cocaine dependence (CD) or opioid dependence(OD) or alcohol dependence (AD) for inclusion in small nuclear family linkage studies. OD families were recruited primarily at the UConn and Yale sites, while CD families were recruited at all sites. Probands with an axis I clinical diagnosis of a major psychotic disorder (e.g., schizophrenia or schizoaffective disorder) were excluded from the study. We also recruited other siblings and parents whenever available regardless of their affection status to increase the power to detect linkage. Each subject provided written informed consent prior to participation. We complied with the Code of Ethics of the World Medical Association (Declaration of Helsinki). The institutional review board at each site approved the study, and certificates of confidentiality for the work were issued by the National Institute on Drug Abuse and the National Institute on Alcohol Abuse and Alcoholism.
Diagnosis and Study Subjects
We interviewed and assessed subjects using the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA) for psychiatric diagnosis as described previously [Gelernter et al., 2005; Pierucci-Lagha et al., 2005; Pierucci-Lagha et al., 2007]. The five substance dependence (SD) traits included in this study are dependence on alcohol (AD), cocaine (CD), cannabis (CanD), opioids (OD), and nicotine (ND). The diagnosis of each SD trait was based on DSM-IV diagnostic criteria [APA, 1994]. Our study sample was approximately equally divided by sex and subjects ranged in age from 17 to 66 (mean ± SD = 38.9 ± 7.8). Within each SD trait, there were more males among the EA affecteds, who were also younger than AA affecteds. Complete demographic data on study subjects are presented in Table I.
TABLE I.
Characteristics of the Study Sample and the Distribution of the Five Phenotypes in African Americans and European Americans
| AAs | EAs | Total | |
|---|---|---|---|
| No. of families | 384 | 355 | 739 |
| No. of probandsa | 384 | 354 | 738 |
| No. of fathers | 16 | 24 | 40 |
| No. of mothers | 59 | 72 | 131 |
| No. of sibs | 504 | 402 | 906 |
| No. of half-sibs | 51 | 12 | 63 |
| No. of genotyped individuals | 952 | 806 | 1758 |
| Mean age in years (SD) | |||
| Proband generation only | 40.72 (6.37) | 36.64 (8.67) | 38.88 (7.78) |
| AD | 40.70 (6.75) | 37.86 (8.33) | 39.36 (7.66) |
| CD | 40.88 (6.25) | 36.90 (8.57) | 39.13 (7.62) |
| CanD | 39.91 (6.68) | 34.38 (9.30) | 36.97 (8.62) |
| ND | 40.68 (6.77) | 37.04 (9.23) | 38.82 (8.32) |
| OD | 42.73 (6.30) | 37.15 (8.46) | 39.03 (8.23) |
| Males (%) | |||
| All genotyped individuals | 44.85 | 52.73 | 48.46 |
| Proband generation only | 45.20 | 54.76 | 49.52 |
| AD | 46.95 | 59.47 | 52.85 |
| CD | 46.12 | 53.59 | 49.40 |
| CanD | 59.82 | 65.86 | 63.03 |
| ND | 42.83 | 52.52 | 47.78 |
| OD | 53.49 | 57.06 | 55.86 |
| Percentage of affecteds (%) | |||
| AD | 55.9 | 62.6 | 58.9 |
| CD | 85.7 | 81.4 | 83.8 |
| CanD | 26.4 | 37.3 | 31.3 |
| ND | 60.1 | 73.9 | 66.4 |
| OD | 27.4 | 65.3 | 44.6 |
| Number of comorbid SD (%)b | |||
| 0 | 7.2 | 4.9 | 6.2 |
| 1 | 12.7 | 6.6 | 10.1 |
| 2 | 26.8 | 14.2 | 21.4 |
| 3 | 28.0 | 28.8 | 28.4 |
| 4 | 19.6 | 31.5 | 24.7 |
| 5 | 5.7 | 14.0 | 9.2 |
AAs, African Americans; EAs, European Americans; ND, nicotine dependence; AD, alcohol dependence; CD, cocaine dependence; OD, opioid dependence; CanD, cannabis dependence.
One family in the EAs was included through alcohol dependence such that there were no proband in terms of cocaine dependence or opioid dependence for this family.
Missing SD was excluded from this calculation.
Genotyping and Quality Control
For most subjects, we obtained DNA from immortalized cell lines, but for a small proportion of the subjects, DNA was obtained directly from blood or saliva. A total of 6,008 SNPs were genotyped by the Center for Inherited Disease Research (CIDR) using the Illumina Linkage IVb Marker Panel (http://www.cidr.jhmi.edu). An additional 266 individuals were genotyped at Yale (Keck Center) using the 6,090 SNP Illumina Infinium-12 Human Linkage Marker Panel. The analyses were limited to the autosomal SNPs, and 5,636 and 5,735 SNPs were used from the first and second panels, respectively. Among the SNPs in the two panels, there were 4,518 SNPs in common across the two platforms, and the common SNPs were used for the following quality control and analyses. Allele frequencies were calculated and HWE was examined in each population by means of PLINK software [Purcell et al., 2007] using a set of unrelated subjects (355 EA and 384 AA subjects were randomly selected, one per family). Any SNP that had a genotyping rate ≤0.95, a minor allele frequency (MAF) ≤0.1, or was not in Hardy–Weinberg equilibrium (HWE) (P≤0.01) was excluded from the analysis. We used PedCheck [O’Connell and Weeks, 1998] and Merlin [Abecasis et al., 2002] to identify Mendelian inconsistencies. We used the Merlin “—error” option to detect error-prone genotypes based on the estimated probability of double-crossover events. The Pedigree Relationship Statistical Test (PREST) [McPeek and Sun, 2000] was applied to verify family relationships.
In total, 1,366 (1360), 322 (70), and 46 (40) SNPs failed to pass the quality control including the criteria of genotyping rate, MAF and HWE in AAs (EAs). We limited our analyses to the 4,133 (4,395) remaining autosomal markers in AAs (EAs). In the linkage analysis, we set these problematic genotypes as missing. Pedigree errors were detected in five EA families and two AA families by PREST. Among these families, the relationships in one AA family and five EA families were corrected based on the shared IBD patterns. The re-assigned family relationships were verified by running PREST again. An AA family with an unresolved relationship problem was excluded from further analysis.
Population Assignment by Individual Genetic Profile
Populations with cryptic genetic clusters could contribute differential linkage signals [Lewis et al., 2008; Li et al., 2008; Myles et al., 2008; Lei et al., 2009; Rebbeck et al., 2009] (although not false positive linkage results). We used ancestry informative markers (AIMs) and analyzed them in STRUCTURE, which implements a Bayesian clustering method, to infer genetic ancestry for each subject [Falush et al., 2003, 2007; Pritchard et al., 2000]. A total of 1,574 AIMs were selected from the SNP linkage panel. The selection of AIMs was based on the marker characteristic inferred from the Hapmap CEU (European) and YRI (African) samples. The following criteria were used to select AIMs: (i) the absolute allele-frequency difference (d) between the two HapMap populations > 0.2, (ii) pair-wise SNP r2 < 0.1 within each population, and (iii) HWE testing with P-value > 0.01 within each population. A combination of 10,000 burn-ins and 10,000 collected iterations was implemented in the STRUCTRUE analysis. Individual inferred population was based on >50% estimated ancestry in that population. We identified families as either EA or AA based on the predominant classification for each family.
Fuzzy Clustering Analysis
We used fuzzy clustering analysis [Kaufman and Rousseeuw, 1990] to reduce the phenotype dimensions and to obtain more detailed information of data structures, rather than use each categorical phenotype alone, or typical “hard” clustering methods. The five SD traits, including AD, CD, CanD, OD and ND, for the 1,758 study subjects were the input data for the fuzzy clustering analysis. In a typical (non-fuzzy) partition, each subject is assigned to only one cluster. As a result, this approach is referred to as “hard clustering” because a clear-cut decision is made on each subject’s cluster membership. In contrast, fuzzy clustering allows for some ambiguity of belonging (i.e., cluster membership). In this approach, coefficients (probabilities ranging from 0 to 1) of cluster membership are derived for each subject. Given the substantial comorbidity of SDs and the possible shared genetic liability, fuzzy cluster analysis has the advantage of providing correlated data structures that are drawn from inherent comorbidity and/or underlying shared genetic liability, and provides a way to reduce and mine data. The membership coefficients can be considered more homogeneous quantitative traits for linkage analysis. The fuzzy cluster analysis was implemented using an R clustering algorithm known as the “fanny” algorithm [Kaufman and Rousseeuw, 1990]. The result of fuzzy clustering by the fanny algorithm can be summarized by a silhouette value for each subject [Rousseeuw, 1987]. The silhouette value (“Ŝ”, which ranges from −1 to 1) for each subject is a measure of how similar that subject is to subjects in the same cluster compared to subjects in other clusters. Subjects with a large value of “Ŝ” (almost 1) are very well clustered, a small “Ŝ” (around 0) means that the subject lies between two clusters, and subjects with a negative “Ŝ” are probably placed in the wrong cluster. Our study of anxiety disorders was the first to successfully implement fuzzy clustering in a genome scan [Kaabi et al., 2006].
Linkage Analyses
Linkage analyses were carried out using the model-free regression method implemented in Merlin-regression [Sham et al., 2002]. This method is based on a modified Haseman–Elston method that regresses the estimated identity-by-descent sharing between relative pairs on the squared sums and squared differences of trait values of the relative pairs. Given that marker–marker linkage disequilibrium (LD) inflates multi-point linkage signals [Abecasis and Wigginton, 2005], SNPs were grouped by LD into clusters using the Merlin “—rsq” option of pairwise r2 > 0.10 to reduce linkage bias. We repeated the analyses with r2 thresholds of 0.05, 0.2, and 0.3 to evaluate the robustness of the linkage results.
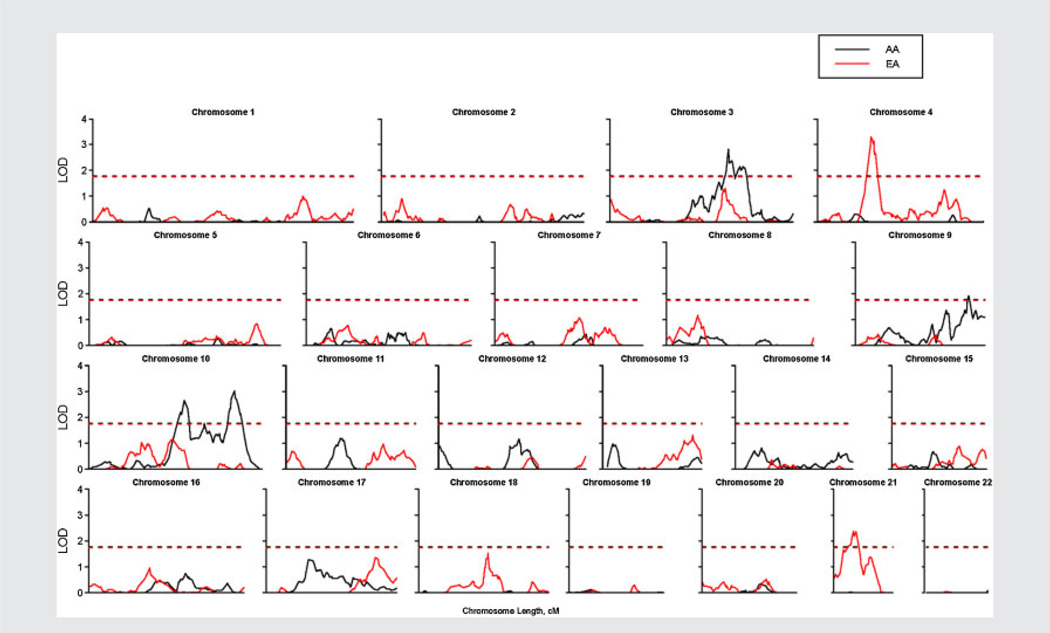
Empirical thresholds for autosome-wide suggestive and significant linkages were determined by Monte Carlo simulations under the null hypothesis of random linkage via a gene-dropping algorithm in Merlin. We simulated 1,000 data sets conditional on the observed family structure, marker spacing, allele frequencies, and missing data pattern [Sawcer et al., 1997; Kruglyak and Daly, 1998]. A marker set with low LD of r2 < 0.1 was selected for each pair of markers (3,675 SNPs in AAs and 3,760 SNPs in EAs) to reduce computational burden. Regression-based linkage analysis, the same approach used for the observed data, was then carried out for each simulated dataset and the highest lod score for each chromosome was retained. The autosome-wide suggestive linkage threshold is defined as the maximum lod score expected once by chance per genome scan [Lander and Kruglyak, 1995], and is set as the 1,000th highest lod score out of 22,000 lod scores for each of the 22 autosomal chromosomes from the 1,000 simulations. The autosome-wide significant threshold is set as the 95th percentile of the 22,000 lod scores. The empirical “suggestive” and “significant” thresholds for AA (EA) are 1.77 (1.74) and 3.22 (3.19), respectively (Table II). The autosomal empirical significance of an observed lod score was determined by counting how often the entire genomehad a maximum lod score greater than or equal to the observed lod score from the above 1,000 simulated estimates.
TABLE II.
Summary of Linkage Analysis for lod Scores Exceeding the Empirically Derived Suggestive Linkage Thresholds for African Americans and European Americans
| AA | EA | |||||
|---|---|---|---|---|---|---|
| Population | ||||||
| Chromosome | 3 | 9 | 10 | 10 | 4 | 21 |
| Cytogenetic location | 3q21.3 | 9q34.2 | 10q26.13 | 10q22.2 | 4q12 | 21q21.2 |
| Peak location (cM) | 145.53 | 146.83 | 147.64 | 96.74 | 68.33 | 19.44 |
| Peak lod score | 2.81 | 1.93 | 3.02 | 2.66 | 3.31 | 2.37 |
| Pointwise P-value | 0.0002 | 0.0015 | 0.0001 | 0.0002 | 0.00005 | 0.0005 |
| Autosome-wide P-value | 0.114 | 0.526 | 0.077 | 0.146 | 0.038 | 0.228 |
| Number of families | 384 | 384 | 384 | 384 | 355 | 355 |
| Suggestive Threshold | 1.77 | 1.77 | 1.77 | 1.77 | 1.74 | 1.74 |
| Significant Threshold | 3.22 | 3.22 | 3.22 | 3.22 | 3.19 | 3.19 |
| lod scores in EA or AA at the same location | 0.835 | 0 | 0.009 | 0.731 | 0 | 0.006 |
The bolded values indicate that the linkage peak reaches autosome-wide significance.
RESULTS
Comorbidity of MSD and the Fuzzy Clustering Analysis
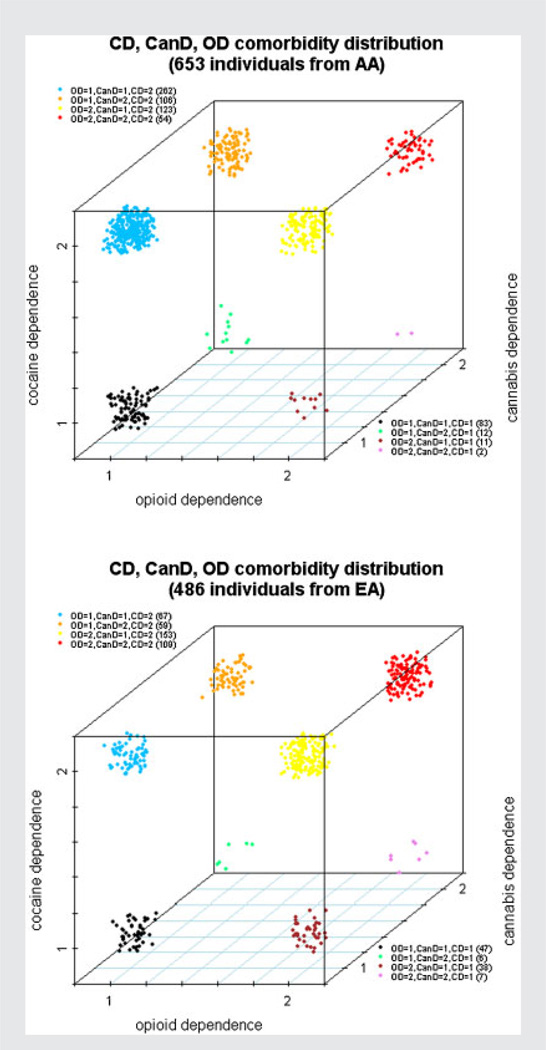
The rate of affection with multiple forms of substance dependence was high among the family members recruited for our genetic studies of CD and OD. The rate of comorbidity (i.e., at least two SD disorders) was 80.1% and 88.5% in AAs and EAs, respectively. The break-down by the number of SD disorders is shown at the bottom of Table I. In our sample, the rate of OD was much higher in EAs than AAs (65.3% vs. 27.4%); the difference was not as great for AD (62.6% vs. 55.9%). CD was less common in EAs (81.4% vs. 85.7%). There were no other significant differences between the AA and EA samples on the characteristics listed in Table I. The complex comorbidity of SD disorders is depicted in a 3-D plot for dependence on the three illegal drugs, cannabis, cocaine, and opioids, regardless of alcohol and nicotine (Fig. 1). The complete dissection of all five studied SD disorders is shown in 3-D plots as well (Supplementary Fig. S1).
Figure 1.
The comorbidity distribution for the three illegal drugs, cocaine dependence, cannabis dependence, and opioid dependence regardless of the dependence on the two legal substances, alcohol or nicotine, in African Americans and European Americans
The fanny algorithm, with the number of clusters pre-selected to be two, resulted in two average silhouette values of 0.6259 and 0.6257 for the AAs and EAs, respectively, implying that the clustering patterns were very similar in the two population groups; subjects were clustered with modest similarity, on average, within the cluster each subject belonged to, compared to the other cluster. Mean (SD) of the cluster membership coefficients are 0.5014 (0.3370) in AAs and 0.5001 (0.3364) in EAs. The means are close to the theoretical 0.5 due to the constraint of summation to one for two clusters. For AAs, there are 475 subjects in one cluster, and 477 in the other. For EAs, subjects were equally clustered with 403 subjects in each cluster. The two clusters that incorporated the first two principal components of the five SDs explained 60.8% and 60.5% of the total phenotypic variation for AAs and EAs, respectively. The membership coefficient was used as a quantitative trait in the subsequent linkage analysis (Fig. 2).
Figure 2.
Lod scores on 22 autosomal chromosomes resulting from quantitative linkage analyses using Merlin regression including African American and European American families. The dashed lines denote the thresholds of lod scores for the autosome-wide “suggestive” linkage for the two populations. (The two thresholds appear as one dashed line since they are very close.)
Linkage Analysis of the Fuzzy Clustering Membership Coefficients
Results are summarized in Table II. In EAs, we detected a genome-wide significant linkage signal in chromosome 4 with a peak lod = 3.31 at 68.3 cM (point-wise P = 0.00005, empirical genome-wide P = 0.038). The 1-lod score support interval centered on this linkage peak extends from 66.6 to 74.02 cM. A suggestive linkage signal in chromosome 21 was also observed with a peak lod = 2.37 at 19.4 cM. In AAs, two suggestive linkage peaks were observed in chromosome 10 with a peak lod = 2.66 at 96.7 cM and a peak lod = 3.02 at 147.6 cM; these flank our previously reported linkage peak near 117.2 cM for alcohol dependence in AAs. In addition, another suggestive linkage region was identified in chromosome 3 with a peak lod = 2.81 at 145.5 cM and in chromosome 9 with a peak lod = 1.93 at 146.8 cM.
DISCUSSION
In this linkage scan we identified several loci predisposing to comorbid dependence on multiple substances using a fuzzy clustering approach to derive a measure of common factors among the SD disorder phenotypes. This general measure of substance dependence was derived from five substance dependence traits including alcohol, cocaine, cannabis, opioid and nicotine, and explained about 60% of the total variability among the MSDs in the two US populations under study. We identified an autosome-wide significant linkage peak in EAs on chromosome 4q12 and obtained suggestive evidence for linkage with loci on chromosomes 3, 9 and 10 in AAs and on chromosome 21 in EAs. The two suggestive linkage peaks (peak locations at 147.6 and 96.7 cM) identified on chromosome 10 for the common component of MSD in AAs in the current study approximate our previously reported linkage signals for alcohol dependence on chromosome 10 at 117.2 cM in AAs [Gelernter et al., 2009] and at 137.7 cM in EAs [Panhuysen et al., 2010].
Linkage analysis using the derived measure of MSD as phenotype could increase power to detect shared risk loci due to pleiotropically severe affection, compared to analysis of an individual SD disorder, because each single SD does not fully reflect the clinical manifestation of these patients. The derived measure of MSD, which extracts the common component of the multiple phenotypes within each individual, reflects a more homogeneous trait corresponding to the underlying shared genetic risk loci. This measure was derived by fuzzy clustering. In comparison to hard clustering, fuzzy clustering preserves much more of the data structure and allows for diagnostic complexities often observed in real data. We pre-selected a solution with two clusters for the study based on the following considerations. First, if the two clusters could explain 100% of the five substance dependence traits, then the new clustering traits would be better phenotypes. For that reason, the key to selecting an appropriate number of clusters relied on the percentage of variation that the clusters could explain. In the exploratory stage, we observed over 60% variation in the five substance dependence traits could be explained by these two clusters. Second, the goal of implementing fuzzy clustering is to reduce the phenotypic dimensions such that the subsequent linkage analysis could be carried out in a conventional software package. We used the coefficients of fuzzy cluster membership as the trait for the subsequent linkage analysis. The membership coefficients of all clusters sum to one. A pre-selected number of clusters at two results in two membership coefficients such that one coefficient determines the other coefficient due to the constraint of summation to one, and this coefficient was used for the subsequent linkage analysis. Simulation studies showed that fuzzy clustering is more powerful than the principal component approach for multivariate continuous traits and more powerful than the joint linkage method [Mangin et al., 1998] in the presence of pleiotropy [Kaabi and Elston, 2003]. On the other hand, fuzzy clustering could be disadvantageous in studies of very large numbers of clusters because the amount of information may be overwhelming.
Three particularly promising candidate genes, GABRA4, GABRB1 and CLOCK, are located within or very close to the autosome-wide significant linkage region on chromosome 4. The two GABA receptor-encoding genes, GABRA4 and GABRB1, are located ~5 Mb upstream of the 1-lod support interval, and are part of a gene cluster encoding alpha 4, alpha 2 and gamma 1 subunits of the gamma-aminobutyric acid (GABA) alpha receptors, which are ligand-gated chloride channels that mediate inhibition in synaptic signal transduction in the mammalian central nervous system. Functional evidence shows that α4-containing GABAA receptors in the nucleus accumbens mediate alcohol intake [Rewal et al., 2009].
The CLOCK gene, located within the 1-lod support interval of the linkage peak on chromosome 4, encodes one of many proteins that regulate circadian rhythm. Addiction to various drugs has been found to disrupt circadian rhythms [Johanson et al., 1999; Schierenbeck et al., 2008; Acuna-Goycolea et al., 2010]. The CLOCK gene also plays an important role in regulating dopaminergic transmission and cocaine reward [McClung et al., 2005], and circadian-related genes are implicated in cocaine-induced locomotor sensitization in the day/night cycle [Abarca et al., 2002].
The autosome-wide significant linkage peak on chromosome 4 was observed in EAs only. Notably, association of alcoholism with GABRA2, about 10.7 Mb from the linkage peak we identified on chromosome 4 for the current study, has been documented and replicated in both EAs and AAs [Edenberg et al., 2004; Lappalainen et al., 2005; Covault et al., 2008; Enoch et al., 2009; Enochet al., 2010; Ittiwut et al., 2011]. A significant association of alcohol dependence with a haplotype block that extends from the intergenic region between GABRA2 and GABRAG1 to GABRG1 intron 3 was observed in EAs but not in AAs [Covault et al., 2008]. Covault et al. [2008] also observed different comorbid MSD patterns in this study sample between AAs and EAs; for example, the percentage of the OD affecteds is much higher in EAs than in AAs (65.3% vs. 27.4%, Table I). This difference is likely due to the ascertainment scheme and not due to inherent differences between populations. Thus, the distinct comorbid MSD pattern or ascertainment differences in EAs and AAs might contribute to the linkage peak being identified only in EAs.
In summary, we identified an autosome-wide linkage peak for MSD in EAs and several suggestive linkage peaks for the two major US populations based on a dense SNP map. Replication of the linkage findings in other samples, including those from other populations, is warranted, as is a focused analysis of the genes located in the linkage regions implicated here. These novel linkage regions can provide useful information in the search for genes involved in susceptibility to MSD, perhaps for targeted sequencing studies to find both disease associated common and rare variants.
ACKNOWLEDGMENTS
The authors thank the volunteer families and individuals who participated in this study. The Center for Inherited Disease Research (CIDR) genotyped most of the array-based linkage SNPs. CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University (contract number N01-HG-65403). Additional genotyping was supported in part by a Yale CTSA and NIH Neuroscience Microarray Consortium award, U24 NS051869-02S1. We are grateful to Ann Marie Lacobelle and Greg Kay for their excellent technical assistance. This study was supported by National Institute on Drug Abuse (NIDA) grants K01 DA24758, R01 DA12690, R01 DA12849, R01 DA18432, R01 AA11330, and the VA Connecticut REAP center, a VA MERIT grant, and the VA Connecticut MIRECC Center.
Footnotes
Additional supporting information may be found in the online version of this article.
Conflict of interest: Drs. Yang, Han, and Gelernter report no competing interests. Dr. Kranzler reports consulting arrangements with Alkermes Inc, Ortho-McNeil Pharmaceuticals, Elbion Pharmaceuticals, Solvay Pharmaceuticals, Sanofi-Aventis Pharmaceuticals, and Gilead Sciences, Inc. and has received research support from Merck and Company, Bristol-Myers Squibb Company, and Ortho-McNeil Pharmaceuticals. Dr. Farrer received a research grant from Eisai Pharmaceuticals and consultant fees from Novartis Pharmaceuticals.
REFERENCES
- Abarca C, Albrecht U, Spanagel R. Cocaine sensitization and reward are under the influence of circadian genes and rhythm. Proc Natl Acad Sci USA. 2002;99(13):9026–9030. doi: 10.1073/pnas.142039099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Wigginton JE. Handling marker-marker linkage disequilibrium: Pedigree analysis with clustered markers. Am J Hum Genet. 2005;77(5):754–767. doi: 10.1086/497345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acuna-Goycolea C, Obrietan K, van den Pol AN. Cannabinoids excite circadian clock neurons. J Neurosci. 2010;30(30):10061–10066. doi: 10.1523/JNEUROSCI.5838-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Lynskey MT. Are there genetic influences on addiction: Evidence from family, adoption and twin studies. Addiction. 2008;103(7):1069–1081. doi: 10.1111/j.1360-0443.2008.02213.x. [DOI] [PubMed] [Google Scholar]
- APA. Diagnostic and Statistical Manual of Mental Disorders IV. Washington DC: American Psychiatric Association; 1994. [Google Scholar]
- Belknap JK, Metten P, Beckley EH, Crabbe JC. Multivariate analyses reveal common and drug-specific genetic influences on responses to four drugs of abuse. Trends Pharmacol Sci. 2008;29(11):537–543. doi: 10.1016/j.tips.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Dinwiddie SH, Begleiter H, Crowe RR, Hesselbrock V, Nurnberger JI, Jr, Porjesz B, Schuckit MA, Reich T. Familial transmission of substance dependence: Alcohol, marijuana, cocaine, and habitual smoking: A report from the Collaborative Study on the Genetics of Alcoholism. Arch Gen Psychiatry. 1998;55(11):982–988. doi: 10.1001/archpsyc.55.11.982. [DOI] [PubMed] [Google Scholar]
- Cloninger CR. Neurogenetic adaptive mechanisms in alcoholism. Science. 1987;236(4800):410–416. doi: 10.1126/science.2882604. [DOI] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Jensen K, Anton R, Kranzler HR. Markers in the 5’-region of GABRG1 associate to alcohol dependence and are in linkage disequilibrium with markers in the adjacent GABRA2 gene. Neuropsychopharmacology. 2008;33(4):837–848. doi: 10.1038/sj.npp.1301456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, Crowe RR, Goate A, Hesselbrock V, Jones K, et al. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74(4):705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, Hodgkinson CA, Yuan Q, Albaugh B, Virkkunen M, Goldman D. GABRG1 and GABRA2 as independent predictors for alcoholism in two populations. Neuropsychopharmacology. 2009;34(5):1245–1254. doi: 10.1038/npp.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, Hodgkinson CA, Yuan Q, Shen PH, Goldman D, Roy A. The influence of GABRA2, childhood trauma, and their interaction on alcohol, heroin, and cocaine dependence. Biol Psychiatry. 2010;67(1):20–27. doi: 10.1016/j.biopsych.2009.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164(4):1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol Ecol Notes. 2007;7(4):574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Panhuysen C, Weiss RD, Brady K, Poling J, Farrer L. Dense genomewide linkage scan for alcohol dependence in African Americans: Significant linkage on chromosome 10. Biol Psychiatry. 2009;65(2):111–115. doi: 10.1016/j.biopsych.2008.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Liu X, Hesselbrock V, Page GP, Goddard A, Zhang H. Results of a genomewide linkage scan: Support for chromosomes 9 and 11 loci increasing risk for cigarette smoking. Am J Med Genet Part B. 2004;128B(1):94–101. doi: 10.1002/ajmg.b.30019. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Panhuysen C, Weiss R, Brady K, Hesselbrock V, Rounsaville B, Poling J, Wilcox M, Farrer L, Kranzler HR. Genomewide linkage scan for cocaine dependence and related traits: Significant linkages for a cocaine-related trait and cocaine-induced paranoia. Am J Med Genet Part B. 2005;136B(1):45–52. doi: 10.1002/ajmg.b.30189. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Panhuysen C, Weiss R, Brady K, Poling J, Krauthammer M, Farrer L, Kranzler HR. Genomewide linkage scan for nicotine dependence: Identification of a chromosome 5 risk locus. Biol Psychiatry. 2007;61(1):119–126. doi: 10.1016/j.biopsych.2006.08.023. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Panhuysen C, Wilcox M, Hesselbrock V, Rounsaville B, Poling J, Weiss R, Sonne S, Zhao H, Farrer L, et al. Genomewide linkage scan for opioid dependence and related traits. Am J Hum Genet. 2006a;78(5):759–769. doi: 10.1086/503631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Yu Y, Weiss R, Brady K, Panhuysen C, Yang BZ, Kranzler HR, Farrer L. Haplotype spanning TTC12 and ANKK1, flanked by the DRD2 and NCAM1 loci, is strongly associated to nicotine dependence in two distinct American populations. Hum Mol Genet. 2006b;15(24):3498–3507. doi: 10.1093/hmg/ddl426. [DOI] [PubMed] [Google Scholar]
- Grucza RA, Bierut LJ. Co-occurring risk factors for alcohol dependence and habitual smoking: Update on findings from the Collaborative Study on the Genetics of Alcoholism. Alcohol Res Health. 2006;29(3):172–178. [PMC free article] [PubMed] [Google Scholar]
- Han S, Gelernter J, Luo X, Yang BZ. Meta-analysis of 15 genome-wide linkage scans of smoking behavior. Biol Psychiatry. 2010;67(1):12–19. doi: 10.1016/j.biopsych.2009.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizink AC, Levalahti E, Korhonen T, Dick DM, Pulkkinen L, Rose RJ, Kaprio J. Tobacco, cannabis, and other illicit drug use among Finnish adolescent twins: Causal relationship or correlated liabilities? J Stud Alcohol Drugs. 2010;71(1):5–14. doi: 10.15288/jsad.2010.71.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ittiwut C, Yang BZ, Kranzler HR, Anton RF, Hirunsatit R, Weiss RD, Covault J, Farrer LA, Gelernter J. GABRG1 and GABRA2 Variation Associated with Alcohol Dependence in African Americans. Alcohol Clin Exp Res. 2011 doi: 10.1111/j.1530-0277.2011.01637.x. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen CT, Wang J, Lanktree MB, Cao H, McIntyre AD, Ban MR, Martins RA, Kennedy BA, Hassell RG, Visser ME, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 2010;42(8):684–687. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson CE, Roehrs T, Schuh K, Warbasse L. The effects of cocaine on mood and sleep in cocaine-dependent males. Exp Clin Psychopharmacol. 1999;7(4):338–346. doi: 10.1037//1064-1297.7.4.338. [DOI] [PubMed] [Google Scholar]
- Kaabi B, Elston RC. New multivariate test for linkage, with application to pleiotropy: Fuzzy Haseman-Elston. Genet Epidemiol. 2003;24(4):253–264. doi: 10.1002/gepi.10234. [DOI] [PubMed] [Google Scholar]
- Kaabi B, Gelernter J, Woods SW, Goddard A, Page GP, Elston RC. Genome scan for loci predisposing to anxiety disorders using a novel multivariate approach: Strong evidence for a chromosome 4 risk locus. Am J Hum Genet. 2006;78(4):543–553. doi: 10.1086/501072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman L, Rousseeuw PJ. Finding groups in data: An introduction to cluster analysis. New York: John Wiley & Sons, Inc; 1990. [Google Scholar]
- Kendler KS, Jacobson KC, Prescott CA, Neale MC. Specificity of genetic and environmental risk factors for use and abuse/dependence of cannabis, cocaine, hallucinogens, sedatives, stimulants, and opiates in male twins. Am J Psychiatry. 2003;160(4):687–695. doi: 10.1176/appi.ajp.160.4.687. [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ. Linkage thresholds for two-stage genome scans. Am J Hum Genet. 1998;62(4):994–997. doi: 10.1086/301792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11(3):241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Lappalainen J, Krupitsky E, Remizov M, Pchelina S, Taraskina A, Zvartau E, Somberg LK, Covault J, Kranzler HR, Krystal JH, et al. Association between alcoholism and gamma-amino butyric acid alpha2 receptor subtype in a Russian population. Alcohol Clin Exp Res. 2005;29(4):493–498. doi: 10.1097/01.alc.0000158938.97464.90. [DOI] [PubMed] [Google Scholar]
- Lei SF, Yang TL, Tan LJ, Chen XD, Guo Y, Guo YF, Zhang L, Liu XG, Yan H, Pan F, et al. Genome-wide association scan for stature in Chinese: Evidence for ethnic specific loci. Hum Genet. 2009;125(1):1–9. doi: 10.1007/s00439-008-0590-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshner AI. Addiction is a brain disease, and it matters. Science. 1997;278(5335):45–47. doi: 10.1126/science.278.5335.45. [DOI] [PubMed] [Google Scholar]
- Lewis JP, Palmer ND, Hicks PJ, Sale MM, Langefeld CD, Freedman BI, Divers J, Bowden DW. Association analysis in african americans of European-derived type 2 diabetes single nucleotide polymorphisms from whole-genome association studies. Diabetes. 2008;57(8):2220–2225. doi: 10.2337/db07-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD, Ma JZ, Payne TJ, Lou XY, Zhang D, Dupont RT, Elston RC. Genome-wide linkage scan for nicotine dependence in European Americans and its converging results with African Americans in the Mid-South Tobacco Family sample. Mol Psychiatry. 2008;13(4):407–416. doi: 10.1038/sj.mp.4002038. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Lappalainen J, Yang BZ, Gelernter J. ADH4 gene variation is associated with alcohol dependence and drug dependence in European Americans: Results from HWD tests and case-control association studies. Neuropsychopharmacology. 2006;31(5):1085–1095. doi: 10.1038/sj.npp.1300925. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Wang S, Schork NJ, Gelernter J. Multiple ADH genes modulate risk for drug dependence in both African- and European-Americans. Hum Mol Genet. 2007;16(4):380–390. doi: 10.1093/hmg/ddl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthar SS, Rounsaville BJ. Substance misuse and comorbid psychopathology in a high-risk group: A study of siblings of cocaine misusers. Int J Addict. 1993;28(5):415–434. doi: 10.3109/10826089309039639. [DOI] [PubMed] [Google Scholar]
- Mangin B, Thoquet P, Grimsley N. Pleiotropic QTL analysis. Biometrics. 1998;54:88–99. [Google Scholar]
- McClung CA, Sidiropoulou K, Vitaterna M, Takahashi JS, White FJ, Cooper DC, Nestler EJ. Regulation of dopaminergic transmission and cocaine reward by the Clock gene. Proc Natl Acad Sci USA. 2005;102(26):9377–9381. doi: 10.1073/pnas.0503584102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern MP, Wrisley BR, Drake RE. Relapse of substance use disorder and its prevention among persons with co-occurring disorders. Psychiatr Serv. 2005;56(10):1270–1273. doi: 10.1176/appi.ps.56.10.1270. [DOI] [PubMed] [Google Scholar]
- McLellan AT, Lewis DC, O’Brien CP, Kleber HD. Drug dependence, a chronic medical illness: Implications for treatment, insurance, and outcomes evaluation. JAMA. 2000;284(13):1689–1695. doi: 10.1001/jama.284.13.1689. [DOI] [PubMed] [Google Scholar]
- McPeek MS, Sun L. Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet. 2000;66(3):1076–1094. doi: 10.1086/302800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merikangas KR, Stolar M, Stevens DE, Goulet J, Preisig MA, Fenton B, Zhang H, O’Malley SS, Rounsaville BJ. Familial transmission of substance use disorders. Arch Gen Psychiatry. 1998;55(11):973–979. doi: 10.1001/archpsyc.55.11.973. [DOI] [PubMed] [Google Scholar]
- Metten P, Crabbe JC. Common genetic determinants of severity of acute withdrawal from ethanol, pentobarbital and diazepam in inbred mice. Behav Pharmacol. 1994;5(4 and 5):533–547. doi: 10.1097/00008877-199408000-00014. [DOI] [PubMed] [Google Scholar]
- Metten P, Crabbe JC. Genetic determinants of severity of acute withdrawal from diazepam in mice: Commonality with ethanol and pentobarbital. Pharmacol Biochem Behav. 1999;63(3):473–479. doi: 10.1016/s0091-3057(99)00017-9. [DOI] [PubMed] [Google Scholar]
- Mirin SM, Weiss RD, Griffin ML, Michael JL. Psychopathology in drug abusers and their families. Compr Psychiatry. 1991;32(1):36–51. doi: 10.1016/0010-440x(91)90068-n. [DOI] [PubMed] [Google Scholar]
- Myles S, Davison D, Barrett J, Stoneking M, Timpson N. Worldwide population differentiation at disease-associated SNPs. BMC Med Genomics. 2008;1:22. doi: 10.1186/1755-8794-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE. PedCheck: A program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63(1):259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panhuysen CI, Kranzler HR, Yu Y, Weiss RD, Brady K, Poling J, Farrer LA, Gelernter J. Confirmation and generalization of an alcohol-dependence locus on chromosome 10q. Neuropsychopharmacology. 2010;35(6):1325–1332. doi: 10.1038/npp.2010.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickens RW, Svikis DS, McGue M, LaBuda MC. Common genetic mechanisms in alcohol, drug, and mental disorder comorbidity. Drug Alcohol Depend. 1995;39(2):129–138. doi: 10.1016/0376-8716(95)01151-n. [DOI] [PubMed] [Google Scholar]
- Pickens RW, Svikis DS, McGue M, Lykken DT, Heston LL, Clayton PJ. Heterogeneity in the inheritance of alcoholism. A study of male and female twins. Arch Gen Psychiatry. 1991;48(1):19–28. doi: 10.1001/archpsyc.1991.01810250021002. [DOI] [PubMed] [Google Scholar]
- Pierucci-Lagha A, Gelernter J, Chan G, Arias A, Cubells JF, Farrer L, Kranzler HR. Reliability of DSM-IV diagnostic criteria using the semi-structured assessment for drug dependence and alcoholism (SSADDA) Drug Alcohol Depend. 2007;91(1):85–90. doi: 10.1016/j.drugalcdep.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, Farrer L, Kranzler HR. Diagnostic reliability of the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA) Drug Alcohol Depend. 2005;80(3):303–312. doi: 10.1016/j.drugalcdep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbeck TR, DeMichele A, Tran TV, Panossian S, Bunin GR, Troxel AB, Strom BL. Hormone-dependent effects of FGFR2 and MAP3K1 in breast cancer susceptibility in a population-based sample of post-menopausal African-American and European-American women. Carcinogenesis. 2009;30(2):269–274. doi: 10.1093/carcin/bgn247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rewal M, Jurd R, Gill TM, He DY, Ron D, Janak PH. Alpha4-containing GABAA receptors in the nucleus accumbens mediate moderate intake of alcohol. J Neurosci. 2009;29(2):543–549. doi: 10.1523/JNEUROSCI.3199-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounsaville BJ, Anton SF, Carroll K, Budde D, Prusoff BA, Gawin F. Psychiatric diagnoses of treatment-seeking cocaine abusers. Arch Gen Psychiatry. 1991;48(1):43–51. doi: 10.1001/archpsyc.1991.01810250045005. [DOI] [PubMed] [Google Scholar]
- Rousseeuw PJ. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math. 1987;20:53–65. [Google Scholar]
- SAMSHA. Results from the 2008 National Survey on Drug Use and Health: National Findings. Rockville, MD: Office of Applied Studies NSDUH Series H-36; 2009. [Google Scholar]
- Sawcer S, Jones HB, Judge D, Visser F, Compston A, Goodfellow PN, Clayton D. Empirical genomewide significance levels established by whole genome simulations. Genet Epidemiol. 1997;14(3):223–229. doi: 10.1002/(SICI)1098-2272(1997)14:3<223::AID-GEPI1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Schierenbeck T, Riemann D, Berger M, Hornyak M. Effect of illicit recreational drugs upon sleep: Cocaine, ecstasy and marijuana. Sleep Med Rev. 2008;12(5):381–389. doi: 10.1016/j.smrv.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Sham PC, Purcell S, Cherny SS, Abecasis GR. Powerful regression-based quantitative-trait linkage analysis of general pedigrees. Am J Hum Genet. 2002;71(2):238–253. doi: 10.1086/341560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherva R, Kranzler HR, Yu Y, Logue MW, Poling J, Arias AJ, Anton RF, Oslin D, Farrer LA, Gelernter J. Variation in nicotinic acetylcholine receptor genes is associated with multiple substance dependence phenotypes. Neuropsychopharmacology. 2010;35(9):1921–1931. doi: 10.1038/npp.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinson FS, Grant BF, Dawson DA, Ruan WJ, Huang B, Saha T. Comorbidity between DSM-IV alcohol and specific drug use disorders in the United States: Results from the National Epidemiologic Survey on Alcohol and Related Conditions. Drug Alcohol Depend. 2005;80(1):105–116. doi: 10.1016/j.drugalcdep.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, Goldberg J, True W, Lin N, Toomey R, Eaves L. Co-occurrence of abuse of different drugs in men: The role of drug-specific and shared vulnerabilities. Arch Gen Psychiatry. 1998;55(11):967–972. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320(5875):539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Yang BZ, Kranzler HR, Zhao H, Gruen JR, Luo X, Gelernter J. Haplotypic variants in DRD2, ANKK1, TTC12, and NCAM1 are associated with comorbid alcohol and drug dependence. Alcohol Clin Exp Res. 2008;32(12):2117–2127. doi: 10.1111/j.1530-0277.2008.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]