Abstract
Recent advances in human embryonic stem cell (ESC) and induced pluripotent stem cell (iPSC) biology enable generation of dopaminergic neurons for potential therapy and drug screening. However, our current understanding of molecular and cellular signaling that controls human dopaminergic development and function is limited. Here, we report on a whole genome analysis of gene expression during dopaminergic differentiation of human ESC/iPSC using Illumina bead microarrays. We generated a transcriptome data set containing the expression levels of 28,688 unique transcripts by profiling five lines (three ESC and two iPSC lines) at four stages of differentiation: (1) undifferentiated ESC/iPSC, (2) neural stem cells, (3) dopaminergic precursors, and (4) dopaminergic neurons. This data set provides comprehensive information about genes expressed at each stage of differentiation. Our data indicate that distinct pathways are activated during neural and dopaminergic neuronal differentiation. For example, WNT, sonic hedgehog (SHH), and cAMP signaling pathways were found over-represented in dopaminergic populations by gene enrichment and pathway analysis, and their role was confirmed by perturbation analyses using RNAi (small interfering RNA of SHH and WNT) or small molecule [dibutyryl cyclic AMP (dcAMP)]. In summary, whole genome profiling of dopaminergic differentiation enables systematic analysis of genes/pathways, networks, and cellular/molecular processes that control cell fate decisions. Such analyses will serve as the foundation for better understanding of dopaminergic development, function, and development of future stem cell-based therapies.
Introduction
Dopaminergic neurons form a neurotransmitter system that originates in the substantia nigra pars compacta, ventral tegmental area, and hypothalamus. Several diseases of the central nervous system, such as Parkinson's disease (PD), schizophrenia, and attention deficit hyperactivity disorder, are associated with dysfunctions of the dopamine system. Knowledge of the molecular mechanisms that regulate human dopaminergic development and function may provide better understanding of the causes for these diseases and offer clues for new treatments. The development and function of human dopaminergic neurons remain relatively uncharacterized at the molecular level compared to rodents. Therefore, studies of the underlying molecular mechanisms of the above-mentioned diseases have been largely hampered by the limited availability of human cells at the appropriate stages of dopaminergic development.
The recent advances in human pluripotent stem cell (PSC) biology enable reprogramming of human adult somatic cells from healthy subjects and patients to induced pluripotent stem cells (iPSC) [1,2]. The reprogramming technology and derivation of iPSC followed by their differentiation allow the generation of sufficient numbers of human dopaminergic neurons at different stages of development for both in vitro and in vivo studies. Several groups have reported on the generation of functional dopaminergic neurons from PSC [3–7]. Our laboratory has established a stage-wise protocol for generating midbrain dopaminergic neurons from human embryonic stem cell (ESC) and iPSC lines [4,8]. This process includes four steps and cells at intermediate stages can be cryopreserved. These steps include (1) culture of PSC, (2) derivation of neural stem cells (NSC) from PSC, (3) induction of midbrain type dopaminergic precursors (DA1), and (4) maturation of dopaminergic neurons (DA2) (Fig. 1A). Neurons generated by this process appear to be authentic midbrain dopaminergic neurons as they express midbrain dopaminergic determinants, and can survive and ameliorate behavioral deficits when transplanted into a 6-hydroxydopamine rat PD model [4,8].
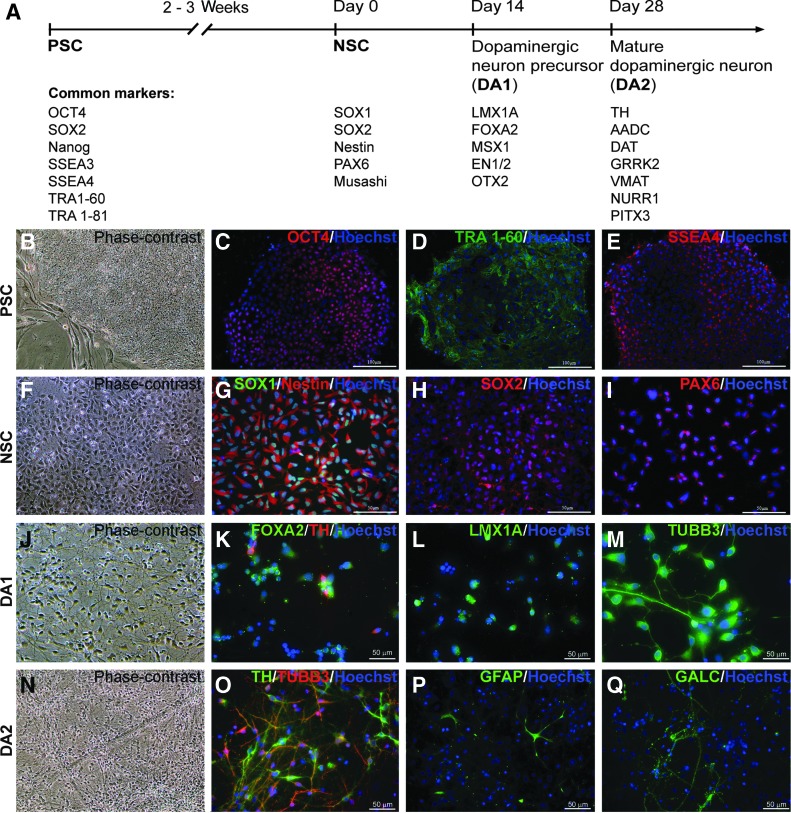
FIG. 1.
Stage-specific dopaminergic differentiation protocol. (A) Schematic depiction of the differentiation protocol with the list of representative markers at the each stage. (B–E) Phase contrast (B) and immunocytochemistry images in pluripotent stem cells (PSCs) with antibodies against OCT4 (C), TRA1–60 (D), and SSEA4 (E). (F–I) Phase contrast (F) and immunocytochemistry images in neural stem cells (NSC) with antibodies against SOX1/Nestin (G), SOX2 (H), and PAX6 (I). (J–M) Phase contrast (J) and immunocytochemistry images in dopaminergic precursors (DA1) with antibodies against FOXA2/TH (K), LMX1A (L), and β-III-tubulin (M). (N–Q) Phase contrast (N) and immunocytochemistry images in mature dopaminergic population (DA2) for neuronal/dopaminergic markers β-III-tubulin/TH (O), astrocyte marker GFAP (P), and oligodendrocyte marker GALC (Q). Blue—DNA. Scale bar as marked.
Based on rodent studies several signaling factors and major pathways in dopaminergic development have been identified, but many aspects of human dopaminergic neuronal differentiation remain elusive. Genome wide transcriptional profiling has emerged as a useful tool for understanding of the development and function of defined cell types. Examining gene expression profiles during dopaminergic neuronal differentiation can confirm known signaling cascades and identify novel genes and pathways that regulate this differentiation process. The knowledge gained from such analyses can be used to establish more efficient protocols for generation of functional (and possibly patient-specific) dopaminergic neurons from PSC for cell replacement therapy, disease modeling, and drug screening. Further, as the in vitro differentiation of PSC into dopaminergic neurons is improved and such neurons begin to be utilized for research, pharmaceutical industry and in clinical applications, robust and reliable methods for their analysis are needed.
In this study we investigated changes in gene expression during dopaminergic differentiation of PSC using Illumina Beadchips that provide coverage of 47,231 transcripts and splice variants across the human transcriptome (www.illumina.com). We have successfully used this methodology earlier with different cell types including NSCs, astrocytes, and oligodendrocytes [9,10]. Here, we report on a data set of global gene expression in five PSC lines (three ESC lines and two iPSC lines) at four stages of dopaminergic differentiation: PSC, NSC, DA1, and DA2. The complete data set is accompanying this article as Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/scd). Analysis of this data set confirmed cell-type specific expression of numerous genes and signaling pathways that play a role in dopaminergic differentiation and/or maturation. This data set provides comprehensive genome profile of dopaminergic differentiation and enables systematic analysis of genes/pathways, networks, and cellular/molecular processes that control cell (dopaminergic) fate decisions. We believe this will be a valuable resource for the stem cell and PD communities.
Materials and Methods
PSC culture
ESC and iPSC lines were maintained on mitomycin C-inactivated mouse embryonic fibroblasts (MEF; Millipore Corporation) in medium comprised of knockout Dulbecco's modified Eagle's medium/Ham's F12 supplemented with 20% knockout serum replacement, 1% nonessential amino acids, 2 mM l-glutamine, 0.1 mM β-mercaptoethanol, 1% penicillin/streptomycin (100 U/100 μg/mL; all from Invitrogen, www.invitrogen.com), and 4 ng/mL of basic fibroblast growth factor (bFGF; Sigma, www.sigmaaldrich.com), or on Geltrex (Invitrogen, http://products.invitrogen.com)-coated dishes in medium conditioned with MEF for 24 h as previously described [3]. Three ESC lines (H1, H9, and H14) (http://grants.nih.gov/stem_cells/registry/current.htm?sort=lna), and two iPSC lines (MMW2 and MR31) [8,11] were used for this study.
Generation of NSC and dopaminergic neurons
NSC lines were derived from ESC or iPSC lines and were cultured in neurobasal medium containing 1×NEAA, 1×l-glutamine (2 mM), 1×B27, and bFGF (10 ng/mL), as previously described [4]. Dopaminergic differentiation was achieved by culturing NSC in medium conditioned on PA6 stromal cells (PA6-CM) for 4 weeks on culture dishes or glass cover slips coated with Poly-l-ornithine (20 μg/mL) and laminin (10 μg/mL) as previously described [12]. To suppress cell death, 2 ng/mL of ascorbic acid was added to PA6-CM as needed.
To test the effect of cAMP signaling on DA differentiation, NSC were cultured for 2 weeks in PA6-CM and 0.2 mM dibutyryl cyclic AMP (dcAMP) was added on day 15 for 5 days (days 15–20). For the last 8 days of differentiation, cells were cultured in PA6-CM without any supplements.
Immunocytochemistry
Immunocytochemistry and staining procedures were performed as described previously [12,13]. Briefly, cells were fixed with 4% paraformaldehyde for 20 min, blocked in buffer containing 10% goat serum, 1% bovine serum albumin (BSA), 0.1% Triton X-100 (all from Sigma) at room temperature for 1 h, followed by incubation with the primary antibody in 8% goat serum, 1% BSA, 0.1% Triton X-100 at 4°C overnight. Appropriately coupled secondary antibodies, Alexa-488 and Alexa-546 (Molecular Probes) were used for primary antibody labeling. All secondary antibodies were tested for cross reactivity and nonspecific immunoreactivity. The following primary antibodies were used: Nestin (611658; BD Transduction Laboratories, 1:500), β-III-tubulin (TUBB3; clone SDL.3D10, T8660; Sigma, 1:1,000), GFAP (Z0334; DakoCytomation, 1:2,000), TH (P40101; Pel-Freeze, 1:500), SOX1 (AB5768; Chemicon, 1:250), SOX2 (MAB4343; Chemicon, 1:250), PAX6 (AB5790; Abcam, 1:500), GALC (MAB342; Millipore, 1:250), FOXA2 (AB40874; Abcam, 1:500), and LMX1A (AB31006; Abcam, 1:500). The quantification of immunoreactive cells in culture was performed by analyzing minimum of 5,000 cells of at least 10 randomly chosen fields derived from three or more independent experiments. The number of Hoechst labeled nuclei on each image was referred as total cell number (100%).
Microarray and quantitative polymerase chain reaction gene expression analysis
Total RNA was prepared using the RNeasy Mini kit according to the manufacturer's instructions (Qiagen). RNA isolated from human ESC, iPSC, NCS, DA1, and DA2 cell populations was hybridized to Illumina Human HT-12 BeadChip v4 (Illumina, Inc.) at the Microarray core facility at the Sanford Burnham Institute for Medical Research (www.sanfordburnham.org/technology/sr/Pages/LaJolla_GenomicsMicroarrayQPCR.aspx). Initial data processing and analyses were performed using the algorithms included with the Illumina BeadStudio software. The background method was used for normalization. The complete data set is available in Supplementary Table S1. For the processed data, the dendrogram was conducted by global array clustering of genes across all the tested samples by using the complete linkage method. Differentially expressed genes were defined as the genes that show at least twofold expression change between any two samples. Unsupervised two-way hierarchical clustering of differentially expressed genes using log2 signal values for each gene across all samples was analyzed with The Institute for Genomic Research (TIGR) Multiexperiments Viewer (MEV) v4.5.1, which used complete linkage and Euclidean distance metric to generate the heatmap. All cell line correlations were a measure of Pearson's coefficient, implemented in R system.
Total RNA (1 μg) was used for the reverse transcription (RT) reactions using SuperScript II First-Strand Synthesis System for RT-PCR (Invitrogen) according to the manufacturer's recommendations. Quantitative polymerase chain reactions (qPCR) were carried out on the ABI 9000HT instrument (Applied Biosystems) using SYBR green kit (Roche) or TaqMan Gene Expression Assay (Applied Biosystems) according to the manufacturers' instructions. PCR reactions were conducted in triplicate for each sample. For microarray validation experiments, samples included H14 ESC, NSC, DA1, and DA2 neurons. For WNT1 and SHH knockdown experiments, samples included NSC and mature 28 days old DA2 neurons. Human TBP and GAPDH were amplified as an internal standard. Reported values were calculated using ΔΔCt method and normalized against endogenous GAPDH. Primer sequences are listed in Supplementary Table S2. RNA Quality assay (RQ1 and RQ2) and genomic DNA contamination assay obtained from Bio-Rad were carried out to ensure the quality and purity of prepared RNA samples for microarray and qPCR analyses.
Gene function and pathway enrichment analysis
The lists of differentially expressed genes from microarray analysis were passed to the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/) for functional analysis. DAVID groups candidate genes that share similar functional annotation and detects statistically significant enrichment of the functional groups. In additional to functional enrichment, differentially expressed genes that shared biological pathways were also identified by using IPA (Ingenuity® Systems, www.ingenuity.com). IPA organizes candidate genes and detects biological pathway enrichment among them by searching known pathways from various sources.
Prioritization of differentially expressed genes
To provide a shortened list of genes that are most likely to be involved in dopaminergic neuronal differentiation processes, the differentially expressed genes were prioritized using Endeavour (www.esat.kuleuven.be/endeavour). Endeavour is a supervised algorithm that, when given a set of causative genes, prioritizes candidate genes by identifying other genes with similar bioinformatic profiles. These profiles are based on literature, function, sequence, protein interaction, gene expression, pathways, and other bioinformatic information. To train the Endeavour algorithm, we used genes known to be involved in lipid metabolism. The differentially expressed genes identified by the microarray were then scored and ranked using this model.
Gene repression using small interfering RNA
Double-stranded, small interfering RNAs (siRNAs) (21-mer) targeting SMO and WNT1 were ordered from Applied Biosystems. The corresponding target mRNA sequences for the siRNAs were as follows: SMO-specific siRNA-1, GCUUUGUGCUCAUUACCUUtt; SMO-specific siRNA-2, GGGACUAUGUGCUAUGUCA; WNT1-specific siRNA-1, CAUCGAAUCCUGCACGUGUtt; WNT1-specific siRNA-2, CCACGAACCUGUUACAGAtt; nontarget siRNA as a negative control, TTCTCCGAACGTGTCACGTtt, and GAPDH-specific siRNA, as a positive control (Invitrogen ID no. s13164, s13165, s14863, s14864, and s5574, respectively).
NSC were enzymatically dissociated into single cell suspensions with accutase (Invitrogen) and 105 cells were plated on individual 24-well geltrex-coated plates and cultured in PA6-CM. After 24 h, NSC were transfected with siRNA following the RNAiMAX lipofectamine transfection protocol (Invitrogen). Briefly, gene-specific siRNA oligomers (20 μM) were diluted in 100 μL Opti-MEM reduced serum medium (Opti-MEM; Invitrogen) and mixed with 3 μL of transfection reagent (RNAiMAX; Invitrogen) pre-diluted in 97 μL Opti-MEM. After 30 min incubation at room temperature, the complexes were added to the cells in a final volume of 1 mL. siRNA transfections were performed every 72 h for 8 days, for a total of three transfections. Control experiments were performed in the same 24-well plate by transfecting cells with GAPDH-specific siRNA (positive control) and scrambled (nontargeting) siRNA (negative control). Other controls included nontransfected cells and cells only exposed to the transfection medium (RNAiMAX lipofectamine only). Cells from representative wells of experimental treatments and controls were collected at the end of the differentiation process (28 days later). The cell lysates were frozen and stored for RNA isolation and qPCR analysis.
Results
Sample characterization prior to global transcriptome analysis
In this study we analyzed the whole genome gene expression of five human PSC lines (three ESC and two iPSC lines) during dopaminergic differentiation using a four-step protocol developed by our laboratory (Fig. 1A) [4,12]. Samples were subdivided into four groups according to their developmental stage (PSC, NSC, dopaminergic precursors—DA1, and dopaminergic neurons—DA2) (Table 1). The first group (PSC—pluripotent stage, Fig. 1B) included three ESC lines (H1, H9, and H14) and two iPSC lines (MMW2 and MR31). Cells at the PSC stage expressed pluripotency markers OCT4 (POU5F1), TRA1–60, and SSEA4 (Fig. 1C–E). The second group included five NSC samples derived from each of the ESC or iPSC lines (Fig. 1F). NSCs can be cultured for long periods of time while uniformly expressing NSC markers SOX1, SOX2, Nestin, and PAX6 (Fig. 1G–I), and without losing the ability to differentiate into neurons and glia (data not shown). Groups 3 and 4 included 10 dopaminergic samples: 5 at the dopaminergic precursor stage (DA1, Fig. 1J) and 5 at the mature dopaminergic neuron stage (DA2, Fig. 1N). We confirmed floor plate identity of dopaminergic precursors (DA1) by staining with FOXA2 and LMX1A, known markers of midbrain floor plate (Fig. 1K–L). The majority of cells at dopaminergic stages were neuronal cells: on average, more than 70% of total cells expressed β-III-tubulin (TUBB3, Fig. 1M) and ∼30% of total cells expressed dopaminergic marker TH (Fig. 1O) at the DA2 stage. A small percentage of cells (<10%) expressed astrocyte marker GFAP (Fig. 1P) and few cells (1%) expressed the oligodendrocyte marker GALC (Fig. 1Q).
Table 1.
List of Samples Used in the Study
| Sample index | Category | No. of genes P<0.05 | No. of genes P<0.01 |
|---|---|---|---|
| ES_H1 | ESC | 23,698 | 20,574 |
| ES_H9 | ESC | 13,426 | 9,863 |
| ES_H14 | ESC | 19,410 | 14,979 |
| iPS_MMW2 | iPSC | 14,202 | 10,626 |
| iPS_MR31 | iPSC | 14,718 | 11,112 |
| NSC_H1 | ES_NSC | 21,078 | 18,815 |
| NSC_H9 | ES_NSC | 13,997 | 10,659 |
| NSC_H14 | ES_NSC | 21,856 | 18,474 |
| NSC_MMW2 | iPS_NSC | 13,756 | 9,966 |
| NSC_MR31 | iPS_NSC | 13,860 | 10,188 |
| DA1_H1 | ES_DA | 21,905 | 18,576 |
| DA1_H9 | ES_DA | 21,518 | 19,393 |
| DA1_H14 | ES_DA | 15,137 | 12,662 |
| DA1_MMW2 | iPS_DA | 14,459 | 10,878 |
| DA1_MR31 | iPS_DA | 14,610 | 10,036 |
| DA2_H1 | ES_DA | 21,645 | 18,612 |
| DA2_H9 | ES_DA | 21,658 | 18,784 |
| DA2_H14 | ES_DA | 15,127 | 12,287 |
| DA2_MMW2 | iPS_DA | 14,054 | 10,173 |
| DA2_MR31 | iPS_DA | 14,573 | 10,087 |
iPSC, induced pluripotent stem cell; ESC, embryonic stem cell; NSC, neural stem cell; DA1, dopaminergic precursors; DA2, dopaminergic neurons.
Array data acquisition and verification
The Illumina BeadArray platform (HumanHT-12 v4 Expression BeadChip) was used to detect the whole genome gene expression in all samples. The microarray contains 47,231 probes representing 28,688 unique transcripts. Microarray results were analyzed by GenomeStudio software; the probe hybridization intensities were normalized after background correction subtraction as previously described [14]. We chose H9, a commonly used ESC line, as the representative line for which data are shown in tables in the article; the data for all five lines at four stages of development are available in Supplementary Table S1.
We performed quality control of our data set, as several criteria must be met prior to biological analyses of the data. First, the expression patterns should be similar for all samples, that is, no wide divergence between different samples should be detected. In general, a successful array should detect between 10,000 and 14,000 genes for each sample, and the intensity distribution should fit Poisson distribution (Fig. 2A). The numbers and percentages of genes detected at different signal intensities in H9 samples are listed in Table 2 (and all other samples in Supplementary Table S3). Overall, the number of detected genes was consistent across the samples, and more than 60% of genes had signal intensity greater than 30. Based on the pattern of gene expression intensities across samples, we chose an arbitrary signal intensity of 30 as a cutoff and restricted our downstream analyses to genes with expression of at least 30 in any one sample. Second, we calculated the pairwise correlation coefficients to determine overall relatedness of samples. The correlation coefficients (R2 value) between technical replicates across arrays should be more than 0.97. Samples at the same differentiation stage should have a correlation coefficient no less than 0.9. However, differences in genetic background, culture conditions, and general cell handling technique could result in lower coefficient of correlation values. The correlation coefficients between different H9 samples were calculated by Pearson's rho and are listed in Table 3 (and among all other samples are reported in Supplementary Table S4). As expected, samples farther apart on developmental scale had lower correlation coefficients than those closer to each other. For example, H9_ESC and H9_DA2 samples had correlation coefficient of 0.75, whereas H9_NSC and H9_DA1 had correlation coefficient of 0.94. These results indicated large enough differences to identify markers that would be able to reliably distinguish these populations.
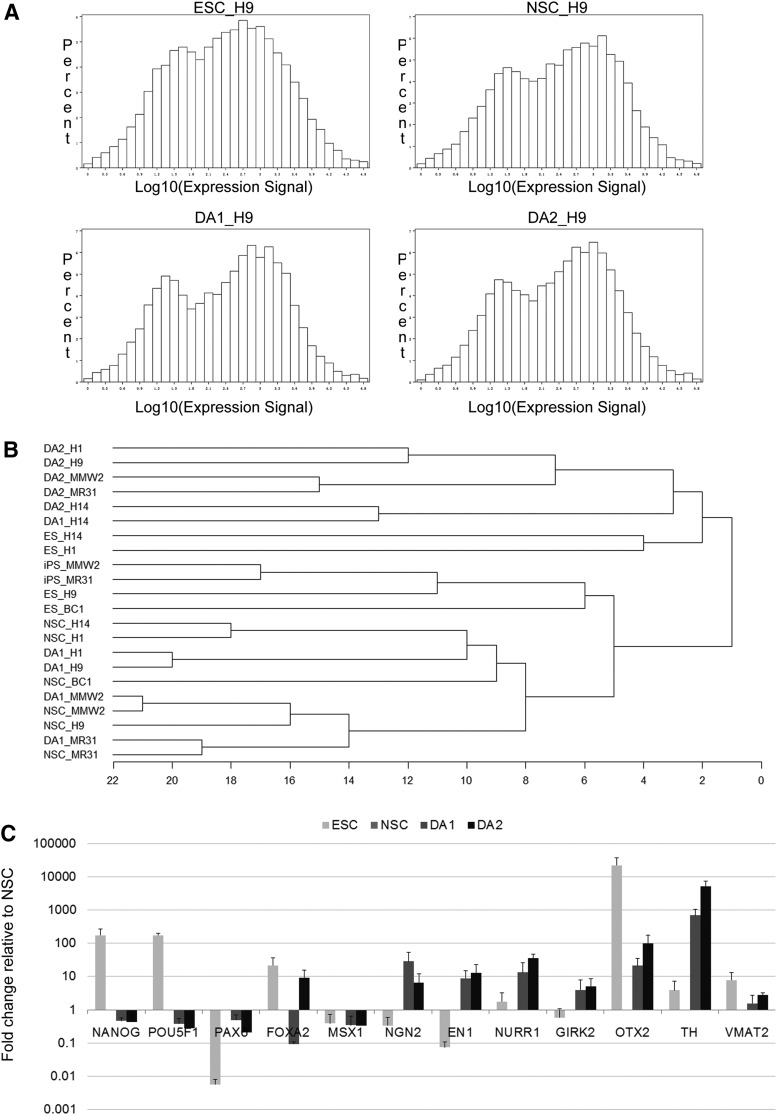
FIG. 2.
Quality control analysis of microarray data. (A) Histogram representation of percentage of genes with different expression intensities for four H9 samples—embryonic stem cell (ESC), NSC, DA1, and DA2. (B) Unsupervised hierarchical clustering of gene expression data for all 20 samples from five PSC lines at four differentiation stages. (C) Quantitative polymerase chain reaction (qPCR) validation of microarray data. GAPDH and TBP served as endogenous standards, and data were normalized against NSC sample. Note the use of logarithmic scale on the y-axis (ie, the x-axis crosses the y-axis at 1, which is also the relative gene expression in NSC).
Table 2.
Number of Genes Detected in Each Sample of the Representative Line (H9) at Different Signal Intensities
| ESC_H9 | NSC_H9 | DA1_H9 | DA2_H9 | |||||
|---|---|---|---|---|---|---|---|---|
| Signal intensity | Number | % | Number | % | Number | % | Number | % |
| >=10,000 | 459 | 3.45 | 411 | 3.09 | 362 | 2.72 | 356 | 2.68 |
| >=5,000 | 508 | 3.82 | 506 | 3.81 | 478 | 3.60 | 428 | 3.22 |
| >=1,000 | 2,404 | 18.09 | 2,761 | 20.78 | 2,719 | 20.47 | 2,555 | 19.23 |
| >=500 | 1,266 | 9.53 | 1,305 | 9.82 | 1,341 | 10.09 | 1,415 | 10.65 |
| >=100 | 2,692 | 20.26 | 2,501 | 18.82 | 2,360 | 17.76 | 2,618 | 19.70 |
| >=30 | 1,734 | 13.05 | 1,659 | 12.49 | 1,423 | 10.71 | 1,506 | 11.34 |
| <30 | 4,223 | 31.79 | 4,143 | 31.18 | 4,603 | 34.65 | 4,408 | 33.18 |
| Total (%) | 13,286 | 100 | 13,286 | 100 | 13,286 | 100 | 13,286 | 100 |
Table 3.
Correlation Coefficients Among Samples of the Representative Line (H9)
| Correlation coefficient | ESC_H9 | NSC_H9 | DA1_H9 | DA2_H9 |
|---|---|---|---|---|
| ESC_H9 | 1 | 0.8973 | 0.84689 | 0.75082 |
| NSC_H9 | 0.8973 | 1 | 0.94072 | 0.86347 |
| DA1_H9 | 0.84689 | 0.94072 | 1 | 0.90779 |
| DA2_H9 | 0.75082 | 0.86347 | 0.90779 | 1 |
We then performed unsupervised one-way hierarchical clustering analysis to group all 20 samples according to the degree of gene expression similarity. The outputs displayed two distribution features: (1) the 20 samples were roughly clustered into four groups; and (2) samples at the far ends of developmental scale clustered separately (ie, PSC were clearly different from DA2), whereas developmentally close stages clustered together (ie, NSC and DA1 samples were intermixed). Thus, the overall gene expression profile in PSC could be clearly discerned from differentiated cells. The gene expression pattern of DA1 neurons was closer to NSC compared with DA2 neurons, but significantly different from PSC (Fig. 2B), reflecting the conclusion gained from the correlation coefficient results.
Overall quality of our microarray results was good and therefore we proceeded with the analysis of gene expression at different developmental stages.
Analysis of known neural and non-neural gene expression
We first examined the levels of expression in several known neural and non-neural genes during dopaminergic differentiation (Table 4). Several genes know to regulate neural induction such as SOX1, SOX2, NES (Nestin), and PAX6, were highly expressed or upregulated in NSC compared with PSC. Genes associated with neurogenesis, including TUBB3, NEUROG2 (NGN2), NEUROD1, NCAM, and DCX were significantly upregulated in DA1 and DA2 populations, whereas glial and nonectodermal lineage genes were largely downregulated under the same culture conditions. For example, the expression intensities of astrocyte marker GFAP and endodermal marker AFP were less than 30 in both NSC and DA populations. These results suggest that the microarrays provide sufficient sensitivity to identify cell lineage differences at various stages of differentiation.
Table 4.
Summary of the Bead Microarray Data for Known Neural and Non-Neural Genes in the Representative Line (H9)
| Gene | ESC_H9 | NSC_H9 | DA1_H9 | DA2_H9 |
|---|---|---|---|---|
| Neural induction | ||||
| SOX1 | 1 | 35 | 120 | 16 |
| SOX2 | 8,858 | 13,000 | 14,679 | 8,544 |
| NES | 3,982 | 9,280 | 10,934 | 7,622 |
| PROM1 | 5,885 | 6,018 | 5,413 | 799 |
| MSI1 | 330 | 1,734 | 2,892 | 1,655 |
| PAX6 | 15 | 3,406 | 5,541 | 1,346 |
| SMO | 880 | 1,667 | 1,430 | 313 |
| GLI1 | 434 | 271 | 21 | 0 |
| TMEM88 | 51 | 655 | 383 | 290 |
| CDH2 | 7,505 | 30,338 | 19,778 | 13,976 |
| FZD9 | 322 | 2,012 | 1,577 | 805 |
| MSX1 | 57 | 187 | 26 | 293 |
| Neurogenesis | ||||
| NEFH | 561 | 326 | 372 | 200 |
| GAD1 | 62 | 303 | 282 | 5,291 |
| CHAT | 164 | 186 | 7 | 30 |
| TH | 25 | 6 | 1 | 373 |
| TUBB3 | 6,276 | 23,727 | 15,455 | 32,618 |
| NCAM1 | 44 | 1,200 | 472 | 1,416 |
| DCX | 203 | 8,253 | 8,170 | 23,663 |
| NEUROD1 | 2 | 86 | 278 | 340 |
| MAP2 | 71 | 1,088 | 697 | 1,180 |
| THY1 | 3,810 | 1,184 | 398 | 774 |
| TUBA1A | 31,582 | 66,973 | 53,403 | 62,057 |
| CORIN | 1 | 1 | 1 | 1 |
| ASCL1 | 30 | 4,115 | 1,386 | 2,839 |
| NEUROG2 | 3 | 360 | 2,126 | 3,336 |
| ITGB1 | 17,356 | 14,524 | 7,859 | 4,283 |
| Gliogenesis | ||||
| GFAP | 1 | 1 | 1 | 1 |
| MAG | 55 | 6 | 28 | 36 |
| CLDN10 | 1,517 | 14 | 46 | 25 |
| SOX9 | 127 | 782 | 834 | 3,366 |
| NFIA | 1 | 1 | 1 | 42 |
| NFIX | 1 | 18 | 1 | 2 |
| SLC1A3 | 115 | 1,001 | 1,462 | 1,549 |
| CD44 | 634 | 140 | 26 | 8 |
| Nonectodermal lineage | ||||
| HAND1 | 373 | 1 | 1 | 1 |
| POU5F1 | 15,860 | 0 | 22 | 27 |
| NANOG | 3,896 | 1 | 1 | 1 |
| AFP | 722 | 1 | 5 | 1 |
| DCN | 15 | 1,461 | 15 | 3 |
| EOMES | 1 | 1 | 1 | 67 |
| WNT5A | 104 | 632 | 1,264 | 288 |
To confirm the microarray gene expression results, we selected 12 transcripts/genes representative for each developmental stage for qPCR validation: EN1, FOXA2, GIRK2 (KCNJ6), MSX1, Nanog, NGN2 (NEUROG2), NURR1 (NR4A2), OCT4 (POU5F1), OTX2, PAX6, TH, and VMAT2 (SLC18A2). We observed that the floor plate marker FOXA2 was not detected by the microarray, and given that it was proposed to be a useful marker of dopaminergic precursors and neurons [15], we included FOXA2 into the panel of genes to be tested by qPCR. Importantly, FOXA2 was detected in TH-positive cells by immunocytochemistry, as mentioned earlier (Fig. 1K). The qPCR results (Fig. 2C) show that pluripotency markers OCT4 and Nanog were downregulated in all differentiated cells (NSC, DA1, and DA2) in comparison with ESC, whereas PAX6, an NSC marker, was expressed at significantly lower levels in ESC and DA2 populations. Pro-neural gene NGN2 was elevated in DA1 and DA2 neurons and had low expression in ESC, whereas floor plate marker FOXA2 was strongly expressed in DA2 neurons, in comparison with NSC. This result indicates that microarrays may not detect all expressed genes. There are several possible reasons, such as poor probe hybridization efficiency and low signal intensity, poor probe coverage, or presence of the new splicing variant in the sample. Finally, midbrain and dopaminergic determinants, such as EN1, GIRK2, NURR1, OTX2, TH, and VMAT2 were all upregulated in DA1 and DA2 populations relative to NSC.
Taken together these results indicate that microarrays have sufficient sensitivity to distinguish between cells at different developmental stages and successfully detected expression of known neural genes.
Expression analysis of known PD-related and mitochondrial genes
One of the goals of this study was to create a data set of multiple PSC lines and their neural differentiated progeny to be used as a standard for future comparisons with other lines, such as, for example, PD patient-specific lines. Several PD-related genes have been shown to localize to mitochondria and/or affect mitochondrial function by modulation of oxidative phosphorylation and energy metabolism. Further, mutations in some PD-related genes may affect production of reactive oxygen species in mitochondria, which appear to contribute to the death of dopaminergic neurons. Therefore, we tested whether microarrays can detect the expression of PD-related and mitochondrial genes in our samples (Supplementary Table S5). Out of 31 PD-related genes 24 were detected (ie, signal intensity was above cut off threshold in at least three out of four samples). The expression of ATP13A2 (PARK9), HIP1R, HTRA2 (PARK13), MAPT, DJ1 (PARK7), PINK1 (PARK6), PARK2, SNCA (PARK1), SNCAIP, STK39, UCHL1 (PARK5), VPS35 (PARK17), and MCR1 was increased, whereas GAK, MED13, and EIF4G1 were decreased during dopaminergic differentiation. The expression of seven genes [BST1, GPNMB, LRRK2 (PARK8), PM20D1, RAB25, GIGYF2 (PARK11), and ADH1C] was below the threshold in at least three samples and these genes were considered undetected.
Among 166 mitochondrial genes, 19 were undetected, 24 were upregulated, and 48 downregulated during dopaminergic differentiation. The rest of the genes (75) had either no change or no clear trend in expression. Nevertheless, it is important to note that the gene expression was detected (above the threshold for at least three out of four samples) for the majority (147 out of 166) of mitochondrial genes by microarray, suggesting that changes in mitochondrial biology can be assessed using a microarray analysis.
Functional and pathway enrichment of differentially expressed genes
To gain insight into the functional profile of genes related to dopaminergic differentiation, we analyzed differentially expressed genes among the four stages using DAVID [16,17]. DAVID is software commonly used to identify enriched gene ontology (GO) terms and reduce large lists of genes into functionally related groups of genes. This analysis is illustrated by using the H9 samples and the data are presented in Table 5.
Table 5.
Gene Ontology-Categories Over-Represented Among the Transcripts Significantly Upregulated During Dopaminergic Differentiation in the Representative Line (H9)
| Annotation cluster | Enrichment score |
|---|---|
| ESC to NSC | |
| Glycoprotein protein | 12.30444 |
| Transmembrane | 7.471824 |
| Extracellular secretory protein | 7.204387 |
| Homobox genes | 7.03388 |
| Positive regulation of cell development and differentiation | 6.070288 |
| G-protein coupled receptor signaling | 5.087306 |
| Cell migration | 4.240437 |
| Neuron development and differentiation | 4.081836 |
| Neurogenesis | 4.022812 |
| Extracellular matrix | 3.225187 |
| Transmission of cell signaling | 3.216997 |
| Embryonic development | 3.119112 |
| Transcription regulation | 3.061987 |
| Embryonic skeletal system development | 2.869361 |
| Plasma membrane | 2.793207 |
| Taste system | 2.777406 |
| C-type lectin | 2.765537 |
| Cell adhesion | 2.387129 |
| Transmission of nerve impulse | 2.326694 |
| NSC to DA1 | |
| Glycoprotein protein | 12.04295144 |
| G-protein coupled receptor | 10.98962735 |
| Transmembrane | 10.83653961 |
| Inflammatory response | 5.012644786 |
| Plasma membrane | 4.181308303 |
| Extracellular secretory protein | 3.2359077 |
| C-type lectin | 2.074264893 |
| NSC to DA2 | |
| Transmembrane protein | 16.7917273 |
| Glycoprotein protein | 13.87956519 |
| Plasma membrane | 8.458944517 |
| Transmission of nerve impulse | 7.415659779 |
| G-protein coupled receptor signaling | 6.069832858 |
| Cell junction | 5.546829305 |
| Extracellular secretory protein | 5.330095148 |
| Sodium transport | 3.491433499 |
| Cation channel activity | 3.36502705 |
| Lipoprotein | 3.341084719 |
| Immunoglobulin | 3.19097774 |
| Inflammatory response | 2.880322901 |
| Homobox genes | 2.844129267 |
| Sarcoplasmic reticulum | 2.485687873 |
| Hormone metabolic process | 2.41357203 |
| Neurotransmitter transport | 2.401570662 |
| Regulation of neurotransmitter transport | 2.350224977 |
| Metal ion-binding | 2.217460564 |
| Membrane fraction | 2.208653179 |
| Cellular ion homeostasis | 2.131879857 |
| Anchored to membrane | 2.111749396 |
| Neurotransmitter receptor activity | 2.09457724 |
We first examined the first step of differentiation process, that is, from the pluripotent stage to the NSC stage. A total of 10,067 transcripts/genes had more than a twofold difference in expression levels and were differentially expressed between ESC and NSC samples. Following the enrichment analysis, upregulated transcripts/genes were segregated into 19 groups (Table 5), including homeobox genes, cell development and differentiation, neuron development and differentiation, and neurogenesis, consistent with the neural induction. Likewise, we examined differentially expressed genes in the following developmental steps. During differentiation of NSC to DA1 stage, seven groups were enriched, mostly related to membrane and extracellular protein functions. Clustering of 7,511 upregulated genes in DA2 sample identified 22 enriched groups, including genes related to transmembrane protein, glycoprotein, G-protein coupled receptor signaling, and cell junction. Further, transmission of nerve impulse, sodium transport, cation channel activity, neurotransmitter transport, and regulation of neurotransmitter transport were GO terms enriched in this sample, consistent with neuron maturation and function. For example, the transmission of nerve impulse group included genes that regulate synaptic transmission and sequential electrical changes across the membrane of the neuron in response to stimulation. A cluster group with an enrichment score of 1.66 that includes genes specific to dopaminergic neurons, such as ligand-gated ion channel activity, GABA receptor activity, postsynaptic cell membrane and GABA-A receptor activity, was also detected, thereby corroborating the sensitivity of array and the validity of the enrichment analysis.
Next, we analyzed genes that are connected with each other in pathways that accomplish particular biological function. We performed pathway analysis of differentially expressed genes using the Ingenuity Systems IPA software (IPA). IPA searches currently available pathway information from KEGG, BioCarta, and GenMapp databases to construct system networks and identify pathway enrichment in our data sets. Using Ingenuity software we found that a large number of pathways were differentially expressed in samples at different developmental stages (Table 6). First, 21 pathways were over-represented in the NSC compared with the PSC. These included axonal guidance signaling, glutamate receptor signaling, G-protein coupled receptor signaling, and WNT/β-catenin signaling, which are consistent with the neural induction. Second, among the networks that were differentially expressed between the NSC and DA1 stages we found multiple immune system pathways, such as communication between innate and adaptive immune cells, role of macrophages, fibroblasts, and endothelial cells in rheumatoid arthritis, autoimmune thyroid disease signaling, graft-versus-host disease signaling, B cell development, role of cytokines in mediating communication between immune cells, and dendritic cell maturation. These findings are in agreement with other reports that previously showed the important role of cytokine signaling during NSC differentiation in vivo [18,19]. Finally, multiple mature neuron pathways were upregulated during the final stage of dopaminergic differentiation (DA1 to DA2). These included GABA and glutamate receptor signaling, dopamine-DARPP32 feedback in cAMP signaling, synaptic long-term potentiation, synaptic long-term depression, and nNOS signaling in neurons, calcium signaling, and cAMP-mediated signaling.
Table 6.
Over-Represented Cellular Pathways Involved in Dopaminergic Neuron Differentiation (Selected Pathways of Interest)
| Ingenuity canonical pathways | −log (P-value) | Ratio |
|---|---|---|
| ESC to NSC | ||
| Axonal guidance signaling | 5E00 | 3E-01 |
| Glutamate receptor signaling | 4.23E00 | 3.91E-01 |
| G-protein coupled receptor signaling | 3.11E00 | 2.88E-01 |
| Neuropathic pain signaling in dorsal horn neurons | 2.77E00 | 3.43E-01 |
| Thyroid hormone metabolism II (via conjugation and/or degradation) | 2.48E00 | 2.5E-01 |
| Heparan sulfate biosynthesis (late stages) | 2.33E00 | 3.28E-01 |
| Role of NFAT in cardiac hypertrophy | 2.09E00 | 2.71E-01 |
| Basal cell carcinoma signaling | 2.07E00 | 3.56E-01 |
| Chondroitin sulfate biosynthesis (late stages) | 2.05E00 | 3.33E-01 |
| Dermatan sulfate biosynthesis (late stages) | 2.03E00 | 3.62E-01 |
| Human ESC pluripotency | 1.95E00 | 2.8E-01 |
| Glioblastoma multiforme signaling | 1.95E00 | 2.87E-01 |
| Uracil degradation II (reductive) | 1.89E00 | 3.33E-01 |
| Thymine degradation | 1.89E00 | 3.33E-01 |
| Antiproliferative role of somatostatin receptor 2 | 1.88E00 | 3.24E-01 |
| Inhibition of angiogenesis by TSP1 | 1.86E00 | 3.59E-01 |
| HER-2 signaling in breast cancer | 1.85E00 | 3.38E-01 |
| Role of NANOG in mammalian ESC pluripotency | 1.74E00 | 3.16E-01 |
| Role of macrophages, fibroblasts, and endothelial cells in rheumatoid arthritis | 1.66E00 | 2.49E-01 |
| Cellular effects of sildenafil (viagra) | 1.65E00 | 2.72E-01 |
| Wnt/β-catenin signaling | 1.61E00 | 2.95E-01 |
| NSC to DA1 | ||
| Communication between innate and adaptive immune cells | 4.4E00 | 2.39E-01 |
| Nicotine degradation II | 3.75E00 | 1.94E-01 |
| Nicotine degradation III | 3.62E00 | 1.98E-01 |
| Role of macrophages, fibroblasts, and endothelial cells in rheumatoid arthritis | 3.09E00 | 1.98E-01 |
| Ethanol degradation II | 2.88E00 | 2.79E-01 |
| Atherosclerosis signaling | 2.82E00 | 2.35E-01 |
| Melatonin degradation I | 2.78E00 | 2.05E-01 |
| Serotonin degradation | 2.78E00 | 2.24E-01 |
| G-protein coupled receptor signaling | 2.64E00 | 1.99E-01 |
| Bupropion degradation | 2.46E00 | 2.12E-01 |
| Macropinocytosis signaling | 2.44E00 | 2.63E-01 |
| Autoimmune thyroid disease signaling | 2.34E00 | 1.97E-01 |
| Acetone degradation I (to methylglyoxal) | 2.33E00 | 2E-01 |
| Graft-versus-host disease signaling | 2.25E00 | 2.6E-01 |
| B cell development | 2.2E00 | 2.73E-01 |
| Noradrenaline and adrenaline degradation | 2.09E00 | 2.12E-01 |
| LPS/IL-1 mediated inhibition of RXR function | 2.08E00 | 1.97E-01 |
| Estrogen biosynthesis | 2.05E00 | 1.91E-01 |
| Dendritic cell maturation | 1.89E00 | 1.79E-01 |
| Role of cytokines in mediating communication between immune cells | 1.88E00 | 2.73E-01 |
| NSC to DA2 | ||
| Neuropathic pain signaling in dorsal horn neurons | 5.76E00 | 3.89E-01 |
| Dopamine-DARPP32 feedback in cAMP signaling | 4.23E00 | 3.01E-01 |
| G-protein coupled receptor signaling | 4.08E00 | 2.73E-01 |
| GABA receptor signaling | 4.07E00 | 3.86E-01 |
| Glutamate receptor signaling | 3.64E00 | 3.48E-01 |
| cAMP-mediated signaling | 3.55E00 | 3.03E-01 |
| Nicotine degradation II | 3.32E00 | 2.23E-01 |
| CREB signaling in neurons | 3.1E00 | 2.67E-01 |
| Nicotine degradation III | 2.94E00 | 2.2E-01 |
| nNOS signaling in neurons | 2.77E00 | 3.65E-01 |
| Synaptic long-term potentiation | 2.72E00 | 2.99E-01 |
| Amyotrophic lateral sclerosis signaling | 2.59E00 | 2.8E-01 |
| Melatonin degradation I | 2.56E00 | 2.41E-01 |
| Calcium signaling | 2.55E00 | 2.46E-01 |
| Synaptic long-term depression | 2.5E00 | 2.7E-01 |
| Natural killer cell signaling | 2.32E00 | 3.02E-01 |
| Reelin signaling in neurons | 2.25E00 | 3.29E-01 |
| Autoimmune thyroid disease signaling | 2.16E00 | 2.3E-01 |
| Corticotropin releasing hormone signaling | 2.06E00 | 2.5E-01 |
| Melatonin signaling | 2.02E00 | 2.95E-01 |
Taken together, these results suggested that microarray analysis is useful in predicting gene function or designing experiments to validate gene function during differentiation.
Prioritization of differentially expressed transcripts/genes
There are several drawbacks to the function or pathway enrichment approach: (1) they are less effective because they use partial information only, (2) they do not provide quantitative measurements for the importance of candidate genes, and (3) they can identify group of genes that respond to differentiation, but are not necessarily related to dopaminergic neuronal differentiation. To circumvent these problems and identify more relevant candidate genes related to dopaminergic neuronal differentiation, we used the Endeavour algorithm. Endeavour integrates a large number of different data sources and generates biologically relevant prioritizations of candidate genes. Table 7 summarizes the prioritized list of 30 differentially expressed genes during dopaminergic neuronal differentiation. As expected, most of the genes are involved in dopaminergic development and function. For example, multiple dopamine receptor genes (DRD2, DRD5, DRD3, and DRD1IP), WNT, and Hedgehog signaling components (WNT6, WNT7B, IHH, BTRC, APC, and WNT9A), dopaminergic specification and maturation genes [TH, NR4A2 (NURR1), and LMX1A], and neuron growth/survival genes (RET—GDNF receptor, STMN2, and NGFR) were ranked among the top 30 genes. Several genes in which mutations are associated with PD, such as SNCA and UCHL1, were also ranked highly. Interestingly, PRKCA, gene encoding protein kinase C alpha, was ranked number 5 and is involved in phosphorylation of microtubule-associated protein tau (MAPT) [20], which is implicated in PD [21]. Another significant result is that both alpha and beta catalytic subunits of cAMP-dependent protein kinase (PRKACA and PRKACB) were ranked 1 and 29, respectively, consistent with the role of cAMP signaling identified by Ingenuity software in this study.
Table 7.
Prioritized List of Differentially Expressed Genes During Dopaminergic Neuron Differentiation in the Representative Line (H9)
| Global prioritization | ||
|---|---|---|
| Gene | Rank | Score rank ratio |
| PRKACA (ENSG00000072062) | 1 | 3.27E-11 |
| BMP4 (ENSG00000125378) | 2 | 1.44E-09 |
| DRD2 (ENSG00000149295) | 3 | 1.49E-09 |
| WNT6 (ENSG00000115596) | 4 | 4.63E-09 |
| PRKCA (ENSG00000154229) | 5 | 4.75E-09 |
| WNT7B (ENSG00000188064) | 6 | 8.50E-09 |
| WNT4 (ENSG00000162552) | 7 | 1.09E-08 |
| RET (ENSG00000165731) | 8 | 2.10E-08 |
| POU5F1|POU5F1P1 (ENSG00000204531) | 9 | 3.31E-08 |
| TH (ENSG00000180176) | 10 | 1.63E-07 |
| STMN2 (ENSG00000104435) | 11 | 2.07E-07 |
| NGFR (ENSG00000064300) | 12 | 2.12E-07 |
| SNCA (ENSG00000145335) | 13 | 3.37E-07 |
| IHH (ENSG00000163501) | 14 | 3.78E-07 |
| UCHL1 (ENSG00000154277) | 15 | 5.01E-07 |
| DRD5 (ENSG00000169676) | 16 | 6.84E-07 |
| BTRC (ENSG00000166167) | 17 | 7.09E-07 |
| NR4A2 (ENSG00000153234) | 18 | 8.51E-07 |
| HOXA9 (ENSG00000078399) | 19 | 0.00000113 |
| DRD3 (ENSG00000151577) | 20 | 0.00000182 |
| GATA2 (ENSG00000179348) | 21 | 0.00000271 |
| LMX1A (ENSG00000162761) | 22 | 0.00000279 |
| POU3F1 (ENSG00000185668) | 23 | 0.00000312 |
| APC (ENSG00000134982) | 24 | 0.00000511 |
| BMP8B (ENSG00000116985) | 25 | 0.00000546 |
| HOXA1 (ENSG00000105991) | 26 | 0.00000563 |
| DRD1IP (ENSG00000130643) | 27 | 0.00000606 |
| CART1 (ENSG00000180318) | 28 | 0.00000689 |
| PRKACB (ENSG00000142875) | 29 | 0.00000867 |
| WNT9A (ENSG00000143816) | 30 | 0.00000896 |
Transcription factor promoter binding sites prediction for differentially expressed genes
Once global changes in gene expression were defined, we utilized the Transcription Element Listening System (TELiS) to identify the transcription control pathways mediating those changes. We found about 196 transcription factor binding motifs (TFBM) in the promoters of differentially expressed genes for each developmental stage (Supplementary Table S6). Notably, the most over-represented TFBM in all differentiation stages is catabolite activator protein (CAP; also known as CRP for cAMP receptor protein). CAP can promote transcription at several sites, affecting the metabolism of sugars and amino acids, transport processes, and protein folding. It also promotes transcription at numerous catabolite-sensitive operons in the presence of cAMP. This finding is consistent with the pathway enrichment analysis in which cAMP-mediated signaling was significantly upregulated. Second, we found that GATA2/3 TFBMs were also over-represented in the promoters of differentially expressed genes, consistent with ranking of GATA2 among top 30 prioritized differentially expressed genes (Table 7). These findings are in agreement with previous reports showing that GATA is a major determinant for guidance of motoneurons and that GATA2/GATA3 are involved in the maintenance of the pool of ventral neuronal progenitors [22,23]. Thus, the TELiS predictions are in accordance with the rest of our findings and previously reported research results.
Functional validations of microarray results
Two signaling pathways known to be involved in dopaminergic development, sonic hedgehog (SHH) and WNT, were identified by the enrichment analysis. Based on previous reports and our microarray results, we hypothesized that manipulation of these pathways at the appropriate developmental stage would affect dopaminergic neuronal differentiation process using our protocol. SHH is a member of the hedgehog family of signaling molecules, which are implicated in numerous developmental events, including growth and patterning of the neural tube [24–28]. SHH is a secreted signaling protein that binds to the receptor smoothened (SMO) and activates signal transduction pathway [24] that promotes differentiation of NSC or neural precursors into immature dopaminergic neurons [12,24,25]. The WNT family of glycoproteins regulates cell proliferation, fate decisions, and cell differentiation [29].
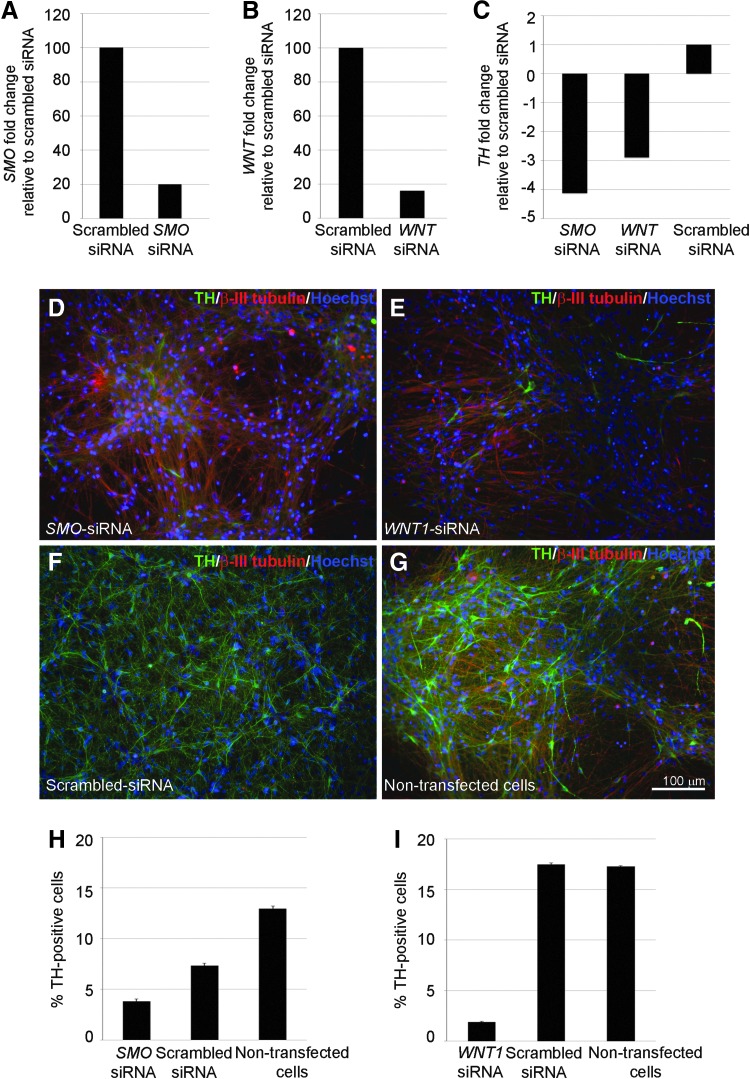
Since SHH and WNT1 signaling pathways are important in neural development, their knockdown would exert multiple effects and might confound the results. We, therefore, transiently silenced the expression of these genes during dopaminergic differentiation in NSC by using siRNA targeting SMO (SHH receptor) and WNT1. To confirm the silencing of SMO and WNT1, we analyzed SMO and WNT1 expression levels in cells transfected with target-specific siRNA versus the scrambled control siRNA by qPCR analysis (Fig. 3). In NSC transfected with SMO-specific siRNA fivefold decrease in SMO expression was determined (Fig. 3A). Similarly, NSC treated with WNT1-specific siRNA showed significantly lower (fivefold) WNT1 mRNA expression in comparison with the controls (Fig. 3B). TH mRNA level was measured in DA2 population by qPCR to quantify the effect of SMO- and WNT1-specific siRNA treatments (Fig. 3C). Both WNT1- and SMO-specific siRNA treatments resulted in significantly lower TH gene expression compared with scrambled siRNA control cells. These observations were confirmed by immunocytochemical analysis of TH-positive neurons at the DA2 stage. NSC transfected with SMO-specific (Fig. 3D) or WNT1-specific (Fig. 3E) siRNA had a lower yield of TH-positive neurons when compared with NSC transfected with scrambled siRNA (Fig. 3F) and to nontransfected cells (Fig. 3G). Quantification of TH-positive cells showed that there was a significant difference between the samples in which SMO or WNT1 were repressed (<5% TH-postive cells) and controls (over 10% TH-positive cells) (Fig. 3H, I).
FIG. 3.
The effect of repression of sonic hedgehog (SHH) and WNT1 signaling pathways on dopaminergic differentiation. (A–C) qPCR analysis of SMO (A), WNT1 (B), and TH (C) gene expression following SMO (A), WNT1 (B), SMO, or WNT1 small interfering RNA (siRNA) treatment. (D–G) Representative TH (green) and β-III-tubulin (red) immunocytochemistry images following SMO (D), WNT1 (E), scrambled siRNA treatment (F), or in nontransfected cells (G). Scale bar as marked. (H, I) Quantification of TH-positive cells following SMO siRNA (H) or WNT1 siRNA (I) treatment.
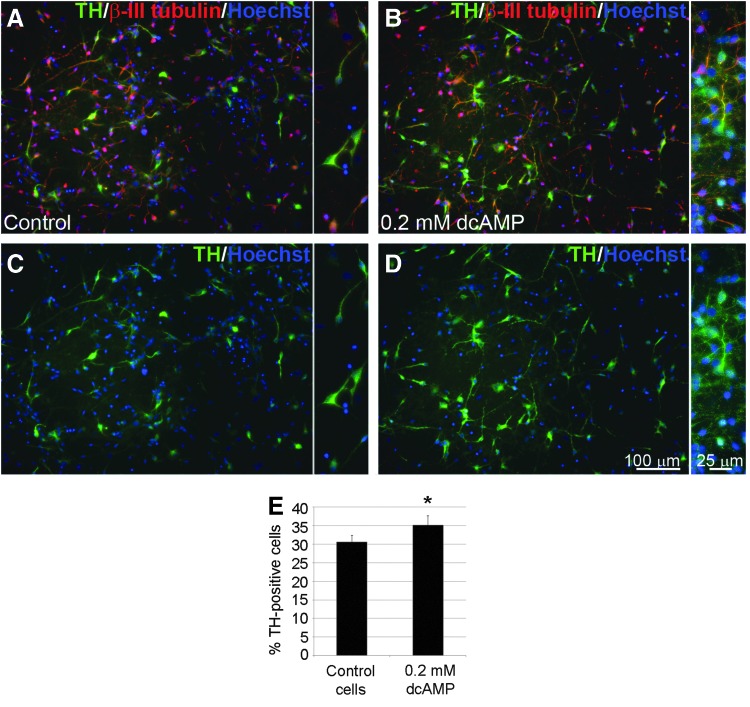
We also identified cAMP signaling as one of the over-represented cellular pathways in the last stage of dopaminergic differentiation (NSC to DA2) by pathway analysis. We hypothesized that the addition of dcAMP in the culture may promote dopaminergic maturation. To test this hypothesis, we added 0.2 mM dcAMP to the culture at the transition from DA1 to DA2 stage (when maturation of dopaminergic precursors to dopaminergic neurons occurs). Immunocytochemistry results showed that dcAMP significantly (two-tail paired t-test P-value 0.0104) increased dopaminergic differentiation (Fig. 4), as ∼5% more TH-positive neurons were observed in cells treated with dcAMP than in controls (Fig. 4E).
FIG. 4.
Dibutyryl cyclic AMP (dcAMP) promotes dopaminergic differentiation. (A–D) Representative TH (green) and β-III-tubulin (red) immunocytochemistry images in control (A, B) and 0.2 mM dcAMP-treated (C, D) cells; inserts—higher magnification. Scale bar as marked. (E) Quantification of TH-positive cells in control and dcAMP-treated samples. The asterisk denotes 0.01<P value <0.05.
Taken together, our pathway perturbation experiments confirm findings of the microarray gene expression analysis and support our hypothesis that the microarrays are robust and reliable methods for detecting similarities and differences in gene expression profiles of multiple cell populations.
Discussion
In this article we generated a whole genome expression data set for five human PSC lines (three ESC lines and two iPSC lines) at four developmental stages: undifferentiated PSC, multipotent NSC, dopaminergic neuron precursors (DA1), and mature dopaminergic neurons (DA2). We believe these data can be used for (1) identification of novel candidate genes and pathways involved in dopaminergic differentiation, (2) future comparisons with other lines, for example, PD patient-derived iPSC and their derivatives for the purposes of understanding the pathological processes and disease modeling, and (3) to identify potential biomarkers that can be used to assess dopaminergic neuron differentiation during a manufacturing process or after transplantation.
We acknowledge that there are limitations in the scope of our data analysis results. First, microarray-based gene expression analyses are constrained by the ability to detect and measure only the amounts of transcripts for which probes exist on the microarray chips. As newer approaches that enable sequencing of every transcript present in the sample, such as RNAseq, are becoming more affordable and widely used, this caveat will likely be eliminated in the future. Second, current differentiation protocols produce a mixed culture of one or more types of neurons and glial cells, and possibly other yet undefined cell types. Thus, all downstream analyses of pooled cells are performed on the mixed cell population instead of pure dopaminergic neuronal population. Development and use of newer technologies can help circumvent this caveat. For example, we have previously used FACS to enrich PSA-NCAM-expressing neural precursors [30], and other researchers have used a floor plate-specific cell surface marker Corin to enrich for mouse dopaminergic neurons [31,32]. Nevertheless, neither of these methods results in isolation of pure population of mature dopaminergic neurons, and thus far cell surface markers that would enable sorting of dopaminergic neurons have yet to be discovered. An alternative approach is to generate reporter lines for dopaminergic neuronal differentiation and use reporter expression for purification or selection of human dopaminergic neurons. To this end, we and other groups have put a significant effort to generate reporter lines using gene targeting tools such as ZFN, TALEN, or CRISPR. In aggregate, while significant technological improvements are paving the road toward broader and more accurate whole-genome expression and epigenetic analyses of dopaminergic neurons in the future, current methodologies enable insightful investigation of gene expression changes during dopaminergic differentiation.
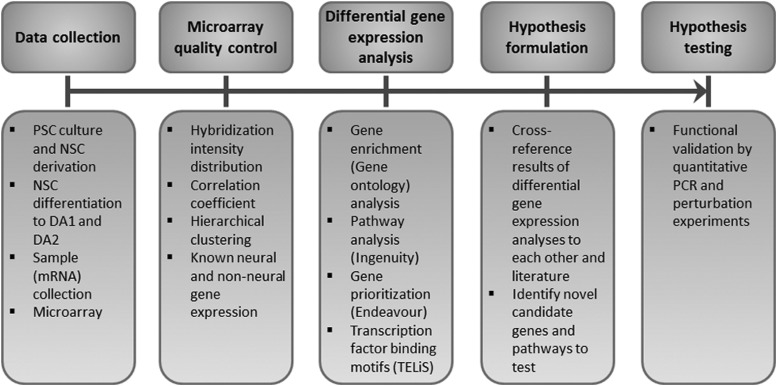
The analytic process is depicted in Figure 5. We first established that the microarrays offered sufficient sensitivity to distinguish among four differentiation stages of dopaminergic differentiation we studied. Quality control of microarray data, including calculation of correlation coefficient (Table 3), hierarchical clustering (Fig. 2B), and analysis of expression of known dopaminergic regulators and non-neural genes (Table 4) confirmed that the microarrays can reliably distinguish between samples at various stages of differentiation. While the overall sensitivity was confirmed, it is important to note that the errors in a large data set are inevitable. For example, the microarray did not detect EN1 or FOXA2, but their expression was detected by qPCR (Fig. 2C) and immunocytochemistry (for FOXA2, Fig. 1K). Further, data on any individual gene should be treated with caution. We therefore limited our analysis to pathways or groups of genes as in our experience this enables better prediction [9,14].
FIG. 5.
The process of microarray data analysis. After mRNA isolation and data collection, we performed a set of quality control steps to ensure the quality of data. In the following steps differentially expressed genes were analyzed and such results were cross-referenced with literature to form hypotheses and validate obtained results.
We next performed functional enrichment of differentially expressed genes, which allowed us to identify GO terms associated with each differentiation step (Table 5). Similarly, pathways analysis using IPA revealed upregulation of signaling pathways in accordance with each developmental stage (Table 6).
There are two major disadvantages in using microarrays for discovering novel candidate genes involved in developmental and pathological processes. First, as already described, microarrays are limited by the quality and the coverage range of the probes used for hybridization with sample RNA/cDNA. Second, microarrays, as all other gene expression studies, are focused on detection of the differences between samples at the transcriptional level. Changes at the post-transcriptional level, however, may be responsible for the pathological processes in many diseases. We addressed some of these issues by validating microarray data using qPCR for a set of genes known to be essential for dopaminergic differentiation (Fig. 2C) and by performing perturbation experiments.
To form hypotheses and design perturbation experiments, we cross referenced our microarray results with the literature. For example, it is well documented that SHH, FGF8, and WNT1 activate transcriptional networks that direct gradual specification and differentiation of dopaminergic neuron progenitors. During the development, midline floor plate secretes SHH, whereas midbrain–hindbrain boundary produces FGF8; these two diffusible growth factors form orthogonal concentration gradients that induce dopaminergic fate in ventral ventricular mesencephalic surface (floor plate) [33]. In addition to SHH and FGF8, WNT1 signaling lateral to the floor plate is needed for midbrain dopaminergic neuronal development and maturation [26]. Among top 30 prioritized differentially expressed genes in our microarray were members of WNT (WNT4, WNT6, BTRC, APC, and WNT9A) and hedgehog (IHH and BTRC) signaling pathways (Table 7). IPA also identified WNT/β-catenin signaling as one of the over-represented cellular pathways during ESC to NSC differentiation (Table 6). To test that in our samples SHH and WNT1 played similar roles as described in vivo and validate microarray results, we repressed SHH receptor smoothened (SMO) and WNT1 at the NSC stage by siRNA. Repression of either of these signaling pathways significantly reduced the percentage of TH-positive dopaminergic neurons (Fig. 3), thereby complementing the microarray findings.
One of the upregulated pathways during NSC to DA2 differentiation step detected by IPA was cAMP-mediated signaling (Table 6). In addition, cAMP-dependent protein kinase (protein kinase A) catalytic subunits A and B (PRKACA and PRKACB) were among top 30 prioritized genes (Table 7), and cAMP receptor protein (CAP) binding site was the most common TFBM among differentially expressed genes (Supplementary Table S6). Therefore, we hypothesized that cAMP may promote dopaminergic differentiation and maturation. Indeed, the addition of dcAMP improved yield of dopaminergic neurons by 5% (Fig. 4). Michel and Agid [34] found that prolonged cAMP signaling specifically promotes development and long-term survival of mesencephalic dopaminergic neurons, and has little or no effect on GABAergic or serotoninergic neurons, which is in agreement with our microarray and perturbation experiment results.
As mentioned, the primary goal of this study was to generate a data set containing whole genome expression profiling during dopaminergic differentiation of multiple PSC lines. We believe this data set can be used as a basis for comparisons with PSC lines and their neural progeny in future studies. More specifically, due to our interest in understanding the molecular events that lead to PD, we investigated the expression of known PD-related genes. Since mutations in some of these genes may contribute to mitochondrial defects and subsequent loss of dopaminergic neurons, we also examined expression of mitochondrial genes. Our data convincingly show that the microarrays can detect the expression of the majority of tested PD related and mitochondrial genes.
In summary, our results indicate that microarray is a suitable methodology for reliable and robust detection of similarities and differences in large number of samples. Microarrays provide a useful platform for initial screening of novel samples and formulating hypotheses for more detailed functional studies.
Supplementary Material
Acknowledgments
We thank all members of our laboratory for many helpful and insightful discussions. This work was supported in part by California Institute for Regenerative Medicine (www.cirm.ca.gov) grants TR-01856, CL1-00501 and TB1-01194, and the NIH CRM common fund initiative (http://commonfund.nih.gov/stemcells/).
Author Disclosure Statement
All authors declare that no competing financial interests exist.
References
- 1.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K. and Yamanaka S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872 [DOI] [PubMed] [Google Scholar]
- 2.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318:1917–1920 [DOI] [PubMed] [Google Scholar]
- 3.Zeng X, Cai J, Chen J, Luo Y, You ZB, Fotter E, Wang Y, Harvey B, Miura T, et al. (2004). Dopaminergic differentiation of human embryonic stem cells. Stem Cells 22:925–940 [DOI] [PubMed] [Google Scholar]
- 4.Swistowski A, Peng J, Han Y, Swistowska AM, Rao MS. and Zeng X. (2009). Xeno-free defined conditions for culture of human embryonic stem cells, neural stem cells and dopaminergic neurons derived from them. PLoS One 4:e6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kriks S, Shim JW, Piao J, Ganat YM, Wakeman DR, Xie Z, Carrillo-Reid L, Auyeung G, Antonacci C, et al. (2011). Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson's disease. Nature 480:547–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M. and Studer L. (2009). Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol 27:275–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang SC, Wernig M, Duncan ID, Brustle O. and Thomson JA. (2001). In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol 19:1129–1133 [DOI] [PubMed] [Google Scholar]
- 8.Swistowski A, Peng J, Liu Q, Mali P, Rao MS, Cheng L. and Zeng X. (2010). Efficient generation of functional dopaminergic neurons from human induced pluripotent stem cells under defined conditions. Stem Cells 28:1893–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo Y, Schwartz C, Shin S, Zeng X, Chen N, Wang Y, Yu X. and Rao MS. (2006). A focused microarray to assess dopaminergic and glial cell differentiation from fetal tissue or embryonic stem cells. Stem Cells 24:865–875 [DOI] [PubMed] [Google Scholar]
- 10.Shaltouki A, Peng J, Liu Q, Rao MS. and Zeng X. (2013). Efficient generation of astrocytes from human pluripotent stem cells in defined conditions. Stem Cells 31:941–952 [DOI] [PubMed] [Google Scholar]
- 11.Chou BK, Mali P, Huang X, Ye Z, Dowey SN, Resar LM, Zou C, Zhang YA, Tong J. and Cheng L. (2011). Efficient human iPS cell derivation by a non-integrating plasmid from blood cells with unique epigenetic and gene expression signatures. Cell Res 21:518–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swistowska AM, da Cruz AB, Han Y, Swistowski A, Liu Y, Shin S, Zhan M, Rao MS. and Zeng X. (2010). Stage-specific role for shh in dopaminergic differentiation of human embryonic stem cells induced by stromal cells. Stem Cells Dev 19:71–82 [DOI] [PubMed] [Google Scholar]
- 13.Zeng X, Chen J, Sanchez JF, Coggiano M, Dillon-Carter O, Petersen J. and Freed WJ. (2003). Stable expression of hrGFP by mouse embryonic stem cells: promoter activity in the undifferentiated state and during dopaminergic neural differentiation. Stem Cells 21:647–653 [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Shin S, Zeng X, Zhan M, Gonzalez R, Mueller FJ, Schwartz CM, Xue H, Li H, et al. (2006). Genome wide profiling of human embryonic stem cells (hESCs), their derivatives and embryonal carcinoma cells to develop base profiles of U.S. Federal government approved hESC lines. BMC Dev Biol 6:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fasano CA, Chambers SM, Lee G, Tomishima MJ. and Studer L. (2010). Efficient derivation of functional floor plate tissue from human embryonic stem cells. Cell Stem Cell 6:336–347 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Huang da W, Sherman BT. and Lempicki RA. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57 [DOI] [PubMed] [Google Scholar]
- 17.Huang da W, Sherman BT. and Lempicki RA. (2009). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong G, Goldshmit Y. and Turnley AM. (2004). Interferon-gamma but not TNF alpha promotes neuronal differentiation and neurite outgrowth of murine adult neural stem cells. Exp Neurol 187:171–177 [DOI] [PubMed] [Google Scholar]
- 19.Boulanger LM. and Shatz CJ. (2004). Immune signalling in neural development, synaptic plasticity and disease. Nat Rev Neurosci 5:521–531 [DOI] [PubMed] [Google Scholar]
- 20.Ekinci FJ. and Shea TB. (1999). Free PKC catalytic subunits (PKM) phosphorylate tau via a pathway distinct from that utilized by intact PKC. Brain Res 850:207–216 [DOI] [PubMed] [Google Scholar]
- 21.Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, et al. (1998). Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 393:702–705 [DOI] [PubMed] [Google Scholar]
- 22.Nardelli J, Thiesson D, Fujiwara Y, Tsai FY. and Orkin SH. (1999). Expression and genetic interaction of transcription factors GATA-2 and GATA-3 during development of the mouse central nervous system. Dev Biol 210:305–321 [DOI] [PubMed] [Google Scholar]
- 23.Zarin AA, Daly AC, Hulsmeier J, Asadzadeh J. and Labrador JP. (2012). A GATA/homeodomain transcriptional code regulates axon guidance through the Unc-5 receptor. Development 139:1798–1805 [DOI] [PubMed] [Google Scholar]
- 24.Hynes M, Ye W, Wang K, Stone D, Murone M, Sauvage F. and Rosenthal A. (2000). The seven-transmembrane receptor smoothened cell-autonomously induces multiple ventral cell types. Nat Neurosci 3:41–46 [DOI] [PubMed] [Google Scholar]
- 25.Parga JA, Rodriguez-Pallares J, Blanco V, Guerra MJ. and Labandeira-Garcia JL. (2008). Different effects of anti-sonic hedgehog antibodies and the hedgehog pathway inhibitor cyclopamine on generation of dopaminergic neurons from neurospheres of mesencephalic precursors. Dev Dyn 237:909–917 [DOI] [PubMed] [Google Scholar]
- 26.Prakash N, Brodski C, Naserke T, Puelles E, Gogoi R, Hall A, Panhuysen M, Echevarria D, Sussel L, et al. (2006). A Wnt1-regulated genetic network controls the identity and fate of midbrain-dopaminergic progenitors in vivo. Development 133:89–98 [DOI] [PubMed] [Google Scholar]
- 27.Abeliovich A. and Hammond R. (2007). Midbrain dopamine neuron differentiation: factors and fates. Dev Biol 304:447–454 [DOI] [PubMed] [Google Scholar]
- 28.Kwon IS, Park RH, Choi JM, Kim SU, Lee YD. and Suh-Kim H. (2006). Differential regulation of tyrosine hydroxylase expression by sonic hedgehog. Neuroreport 17:693–698 [DOI] [PubMed] [Google Scholar]
- 29.Prakash N. and Wurst W. (2007). A Wnt signal regulates stem cell fate and differentiation in vivo. Neurodegener Dis 4:333–338 [DOI] [PubMed] [Google Scholar]
- 30.Freed WJ, Chen J, Backman CM, Schwartz CM, Vazin T, Cai J, Spivak CE, Lupica CR, Rao MS. and Zeng X. (2008). Gene expression profile of neuronal progenitor cells derived from hESCs: activation of chromosome 11p15.5 and comparison to human dopaminergic neurons. PLoS One 3:e1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jonsson ME, Ono Y, Bjorklund A. and Thompson LH. (2009). Identification of transplantable dopamine neuron precursors at different stages of midbrain neurogenesis. Exp Neurol 219:341–354 [DOI] [PubMed] [Google Scholar]
- 32.Ono Y, Nakatani T, Sakamoto Y, Mizuhara E, Minaki Y, Kumai M, Hamaguchi A, Nishimura M, Inoue Y, et al. (2007). Differences in neurogenic potential in floor plate cells along an anteroposterior location: midbrain dopaminergic neurons originate from mesencephalic floor plate cells. Development 134:3213–3225 [DOI] [PubMed] [Google Scholar]
- 33.Ye W, Shimamura K, Rubenstein JL, Hynes MA. and Rosenthal A. (1998). FGF and Shh signals control dopaminergic and serotonergic cell fate in the anterior neural plate. Cell 93:755–766 [DOI] [PubMed] [Google Scholar]
- 34.Michel PP. and Agid Y. (1996). Chronic activation of the cyclic AMP signaling pathway promotes development and long-term survival of mesencephalic dopaminergic neurons. J Neurochem 67:1633–1642 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.