Abstract
Branched peptides containing histidines and lysines (HK) have been shown to be effective carriers for DNA and siRNA. We anticipate that elucidation of the binding mechanism of HK with siRNA will provide greater insight into the self-assembly and delivery of the HK:siRNA polyplex. Non-covalent bonds between histidine residues and nucleic acids may enhance the stability of siRNA polyplexes. We first compared the polyplex biophysical properties of a branched HK with those of branched asparagines-lysine peptide (NK). Consistent with siRNA silencing experiments, gel electrophoresis demonstrated that the HK siRNA polyplex maintained its integrity with prolonged incubation in serum, whereas siRNA in complex with NK was degraded in a time-dependent manner. Isothermal titration calorimetry of various peptides binding to siRNA at pH 7.3 showed that branched polylysine, interacted with siRNA was initially endothermic, whereas branched HK exhibited an exothermic reaction at initial binding. The exothermic interaction indicates formation of non-ionic bonds between histidines and siRNA; purely electrostatic interaction is entropy-driven and endothermic. To investigate the type of non-ionic bond, we studied the protonation state of imidazole rings of a selectively 15N labeled branched HK by heteronuclear single quantum coherence NMR. The peak of Nδ1-H tautomers of imidazole shifted downfield (in the direction of deprotonation) by 0.5 to 1.0 ppm with addition of siRNA, providing direct evidence that histidines formed hydrogen bonds with siRNA at physiological pH. These results establish that histidine-rich peptides form hydrogen bonds with siRNA, thereby enhancing the stability and biological activity of the polyplex in vitro and in vivo.
1. Introduction
Strategies involving small interference RNA (siRNA) provide the potential to manipulate gene expression and facilitate control of diseases. One of the major hurdles in using siRNA or microRNA as therapeutic agents has been the lack of an efficient, stable, and safe delivery vehicle. A means to prevent the degradation of nucleic acids by nucleases in the serum is to condense the nucleic acids with polycations. By binding to the phosphate backbone of nucleic acids through electrostatic interactions, polycationic carriers neutralize their charges and form polyplexes. Compact forms of polyplexes significantly reduce the exposure of nucleic acids to serum, augment their half-life, and enable their greater penetration within the target tissue. Consequently, polycation carriers such as liposomes, synthetic polymers, and cyclodextrins have shown considerable promise in delivering siRNA or DNA plasmids. Although several promising non-viral polycation agents for gene delivery have been developed and tested in animal models and human investigational trials [1–3], no siRNA carrier has been approved by the FDA for systemic delivery.
To optimize delivery of siRNA by non-viral carriers, it is essential to understand the binding mechanisms and biophysical properties between the carrier and siRNA. Correlating the factors that influence the compactness and stability of polyplexes with biological activity may help development of more effective carriers of siRNA. Fluorescent molecules such as ethidium bromide, acridine orange, Hoechst dye, and other fluorochromes have been used to study the binding affinity of polycations and siRNA or DNA plasmids, but the binding exclusion assays with these dyes only examine binding affinity indirectly [4]. Furthermore, free fluorescent molecules may interact with carriers and result in artifacts. Isothermal titration calorimetry (ITC) has become an important technique for studying thermodynamic behavior of protein-protein or protein-nucleic acid binding. Quantitative analysis of thermodynamic parameters provides greater insight into the binding mechanism of carrier and nucleic acids [5, 6]. In addition to ITC, nuclear magnetic resonance (NMR) spectroscopy can directly detect molecular interactions in biological solutions [7], thereby providing a powerful and versatile technique for assessing peptide siRNA binding at the atomic level. For example, 15N chemical shifts of the imidazole ring have been studied to elucidate the hydrogen bond interactions in serine protease [8, 9], triosephosphate isomerase [10], and a membrane component of phosphotransferase system [11].
Our lab is particularly interested in histidine-lysine (HK) peptide carriers (Fig. 1) of siRNAs, which have effectively inhibited Raf-1 oncogene in tumor xenografts and reduced the tumor growth by 60% [12, 13]. Initially, in several laboratories including ours it was thought that formation of the HK polyplexes depended primarily on the electrostatic interaction of positively charged lysines with negatively charged phosphates and that the role of histidine was to buffer endosomes and aid release of nucleic acids [14–18]. In contrast to poly-L-lysine (PLL) complexes, HK peptide in complex with plasmids were resistant to enzymatic degradation and maintained high transfection efficiency [19]. As a result, we hypothesized that histidines may play an important role in increased stability of polyplexes through non-ionic interactions, in addition to its role in buffering of endosomes.
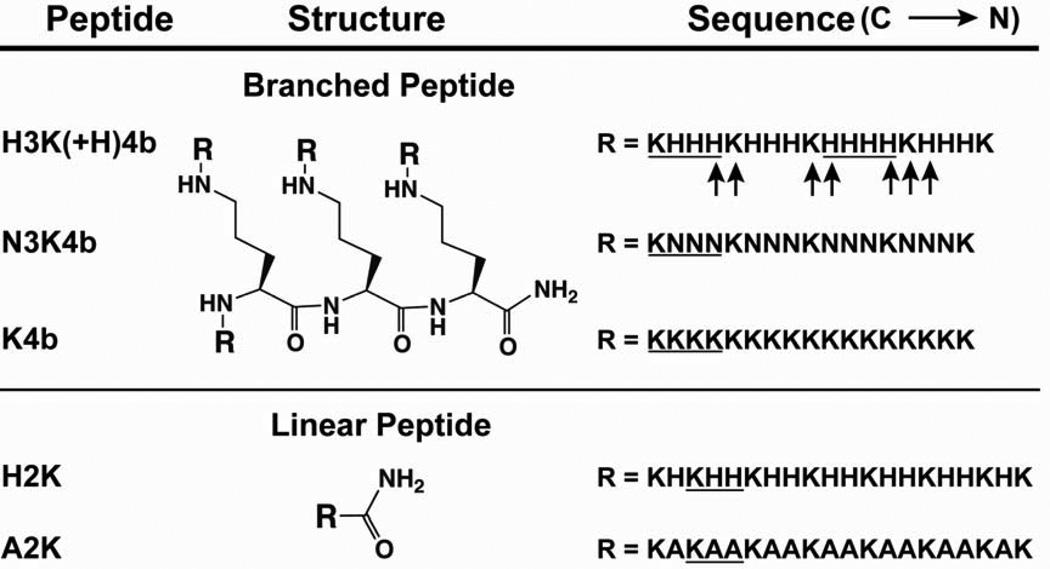
Fig. 1. Schematic structures and sequences of branched and linear peptides.
Branched peptides are composed of a three-lysine core, with four branches attached at the four amino groups (N-terminal) of the core. The 20-mer linear H2K peptides have a sequence similar to a branch of H2K4b. The predominant repeating groups are underline. Labeled histidines (U-15N3) and lysines (U-13C6 and U-15N2) are represented by arrows.
To explore types of non-covalent interactions other than Coulombic forces, a number of studies have examined binding behaviors between polymers and DNA (and/or RNA) in solutions with a range of salt concentrations by using ITC [20–22]. On the basis of the counterion condensation theory [23, 24], experiments at a range of salt condition allow the contributions of non-electrostatic interactions to the binding free energy to be determined. For example, Lohman’s group has shown that a salt-dependent equilibrium constant is thermodynamically comparable for peptides containing lysine or arginine with the same net charge. Nonetheless, compared with oligolysines, oligoarginines have more favorable enthalpic contributions to binding free energy, suggesting hydrogen bond formation between arginines and nucleic acids [21]. Prevette and colleagues modeled the ITC data and calculated that the contribution to the free energy from non-electrostatic interaction of carbohydrates and DNA binding accounts for more than 60% of total free energy [22]. Moreover, in a conformational study using circular dichroism, poly-L-histidine was shown to form complexes with DNA at elevated sodium chloride concentrations (2 M) [25], suggesting that non-ionic interactions may have a role in the complex formation. More recently, investigators have used molecular dynamics to probe hydrogen bond formation and potential hydrogen bond donors/acceptors of carriers and DNA [26, 27]. Although hydrogen bond formation between polymers and nucleic acids has been suggested by both salt-dependent thermodynamics and molecular modeling, direct evidence is lacking.
The aim of this study was to investigate the role of the histidine component of HK peptides in binding to siRNA. We proposed that stable formation of the HK siRNA polyplex, its resistance to serum, and its enhanced biological activity were the consequence of both electrostatic and hydrogen-bond interactions. After synthesizing branched peptides that contained varying amounts of histidines, we determined the influence of serum on the stability and gene silencing activity of these peptides in complex with siRNA. The thermodynamic profiles of siRNA binding to branched or linear peptides with different histidine contents were then analyzed by isothermal titration calorimetry. To explore the presence of histidine-mediated hydrogen bonds, protonation of free and siRNA-bound imidazoles was characterized using heteronuclear single quantum coherence (HSQC) NMR at pH 7.3 and 5.0.
2. Materials and Methods
2.1. Cell line
Human malignant MDA-MB-435 cells were cultivated in Dulbecco’s minimal essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 20 mM glutamine. Stable MDA-MB-435 cells expressing firefly luciferase were obtained by electroporation with a linearized pCpG-Luc plasmid and selection of a high expressing clone in the presence of genetecin (G418). The pCpG-Luc vector was constructed by ligating the luciferase gene (digested from pMOD-Luc plasmid, InvivoGen, San Diego, CA) into the multiple cloning sites of pCpG-mcs (InvivoGen) as previously described [28].
2.2. siRNA
The following siRNA sequences targeting luciferase were used: sense, 5’-CUG-CAC-AAG-GCC-AUG-AAG-A-dTdT-3’; antisense, 5’-UCU-UCA-UGG-CCU-UGU-GCA-G-dTdT-3’ (Thermo Scientific, Dharmacon Division, Lafayette, CO). For duplex annealing, each siRNA was maintained in siRNA buffer at room temperature for 30 min (Dharmacon).
2.3. Peptides
The four-branched H3K(+H)4b, N3K4b, and K4b peptides with predominant repeating groups -HHHK-, -NNNK- and -KKKK-, respectively, as well as the linear H2K (-HHK-) and A2K (-AAK-) peptides (Fig. 1) were synthesized on a Rainin Voyager synthesizer (PTI, Tucson, AZ) by the biopolymer core facility at the University of Maryland Baltimore, as previously described [29].
2.4. Peptide siRNA polyplex preparation
The peptides H3K(+H)4b, and N3K4b were mixed with equal volumes (37.5 µl) of siRNA (10 µg, 20 µM) and maintained at room temperature for 30 min. The resulting polyplexes were tested for stability in serum and/or silencing activity. The peptide/siRNA molar ratios (3/1 for H3K(+H)4b and 6/1 for N3K4b) were chosen based on the amounts needed for complete electrophoretic retardation of the polyplexes in the absence of serum [30].
2.5. Stability of peptide siRNA polyplexes in serum
The stability of peptide siRNA polyplexes was determined with gel electrophoresis. After the peptide siRNA polyplexes formed at room temperature for 30 minutes, the resulting polyplexes (7.5 µl) were mixed with FBS (7.5 µl) and incubated at 37°C for 0, 1, 4, 6, and 24 h. Each sample, containing 1 µg of siRNA (total volume 15 µl) was then analyzed by gel electrophoresis (3% agarose) 100V, 30 min. Naked siRNA (lanes 1), freshly formed polyplexes in the absence of serum (lanes 2), and 50% FBS (lanes 7) were also prepared as controls.
2.6. Gene silencing by peptide siRNA polyplexes incubated with serum
Luciferase-expressing MDA-MB-435 cells were plated on 24-well plates (0.5 ml of DMEM, 10% serum) at a density of 3×104 cells per well 24 h before transfection. The cells were then treated with H3K(+H)4b or N3K4b luciferase siRNA (1 µg siRNA/well) polyplexes that had been previously incubated in 50% FBS for 0, 4, or 24 h. Control (untreated) cells received 15 µl of DMEM/FBS (50%). Forty-eight hours after the addition of polyplexes, the cells were incubated with 200 µl of lysis buffer (Promega, Madison, WI) followed by centrifugation at 13,000 g for 5 min. After luciferase substrate (Promega) was added to the cell lysates, luciferase activity was measured by a Turner TD 20/20 luminometer (Promega).
2.7. Isothermal titration calorimetry
Peptides, and siRNA were dissolved in phosphate buffer (10 mM, pH 7.3). H3K(+H)4b was dissolved in 10 mM 2-(N-morpholino)ethanesulfonic acid (MES) buffer pH 5.0 and 6.0 for pH dependence experiments. The thermodynamic profile of nucleic acid binding with HK peptides was obtained at 37°C with use of V P-ITC (MicroCal Inc., Northampton, MA). The sample cell was filled with siRNA (1 µM, 1.45 ml), and the syringe contained peptide solution (25 µM, 5 µl per injection). Each injection (5 µl, a total of 56 injections) was carried out for 10 sec at 360 sec intervals, except the titration at pH 5 (2.5 ul/injection, 180 sec intervals, and a total of 117 injections). Heat of dilution was determined by titration of peptides into a buffer without siRNA. Molar enthalpy and final figures were generated by Origin 5.0 (OriginLab Corporation, Northampton, MA).
2.8. NMR
Heteronuclear Single Quantum Coherence (HSQC) NMR spectra were recorded at 23°C on a 600 MHz Bruker Avance III spectrometer (Bruker BioSpin, Billerica, MA) equipped with a cryoprobe TXI probe, and analyzed using NMRpipe [31] and Sparky (UCSF). The H3K(+H)4b peptide was selectively labeled with isotope. To each terminal branch sequence KHHHKHHHKHHHHKHHHK, the underlined histidines were labeled with U-15N3, and lysines were double labeled with U-13C6 and U-15N2 (Cambridge Isotopes, Cambridge, MA). Sample buffer was either 10 mM phosphate buffer (pH 7.3; with or without 100 mM NaCl) or 10 mM MES buffer, pH 5.0, containing 5% D2O. For analysis of 15N relaxation data in the presence of siRNA, H3K(+H)4b/siRNA ratios of 10:1, 6:1, and 4:1 were selected. To determine the pKa of each histidine signal, HSQC spectra were obtained with 0.1 mM labeled H3K(+H)4b at pH values of 3.9, 4.4, 4.7, 5.1, 5.5, 5.9, 6.2, 6.5, 6.8, 7.2, and 7.9 in 10 mM phosphate-citrate buffer. This experiment specifically examined the amide proton on the imidazole ring. A pH curve was developed with the proton and nitrogen chemical shifts at different pH values. The pKa values of histidines were calculated by non-linear least squares analysis with the use of Henderson-Hasselbalch equation [32] in Matlab.
3. Results
3.1. Stability of polyplexes in serum
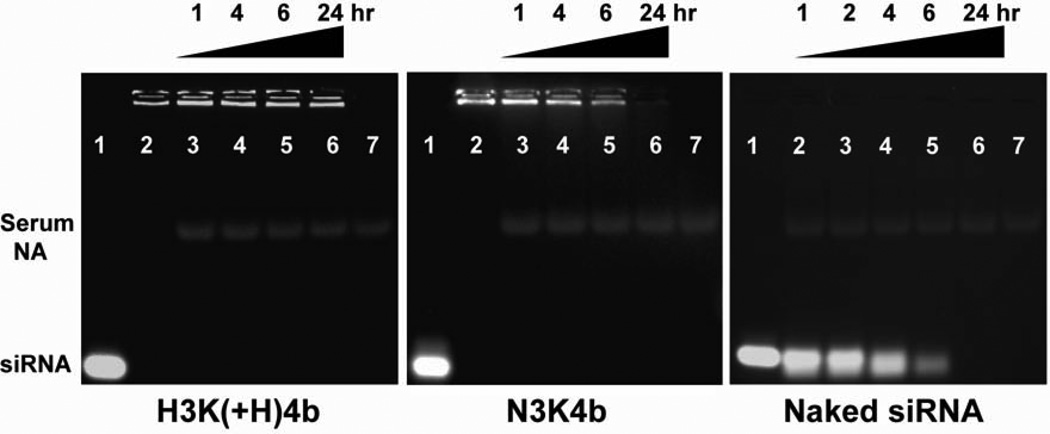
We first studied the stability of peptide siRNA polyplexes in serum (50%) by gel electrophoresis. Polyplexes are retarded in the well, and upon degradation, the siRNA can be released or destroyed. A four-branched HK, H3K(+H)4b (Fig. 1) was compared with a four-branched asparagine-lysine-rich peptide (N3K4b); degradation of naked siRNA in the presence of serum was used as a control. The peptide/siRNA molar ratios (H3K(+H)4b, 3/1; N3K4b, 6/1) used in this study were selected because they provided complete retardation of siRNA (saturated binding). The self-assembled polyplexes were incubated in FBS (50%) for 0, 1, 4, 6, and 24 h at 37°C, and then analyzed by gel electrophoresis. To determine the band intensities and integrity of the polyplexes, the gels were pre-stained with ethidium bromide. As seen in Fig. 2, H3K(+H)4b siRNA polyplexes maintained their integrity for up to 24 h in the medium containing serum. In contrast, the siRNA of N3K4b polyplexes was degraded in a time-dependent manner, with kinetics of degradation comparable to that of naked siRNA alone in serum. This indicates that asparagine-lysine (NK) polyplexes were much more sensitive to serum exposure than were HK polyplexes.
Fig. 2. HK polyplexes showed resistance to dissociation and degradation by serum.
siRNA in complex with H3K(+H)4b or N3K4b was incubated with 50% FBS at 37°C for the indicated times. Lanes are as follows: naked siRNA (1), polyplexes without serum (2), polyplexes with 1 h of serum incubation (3), 4 h (4), 6 h (5), 24 h (6), and 50% FBS without siRNA (7). For naked siRNA in the absence of peptide carriers, lanes 2 to 6 were the siRNA maintained in serum for 1, 2, 4, 6, and 24 h, respectively. Serum NA, serum nucleic acids.
3.2. Silencing by serum-incubated polyplexes
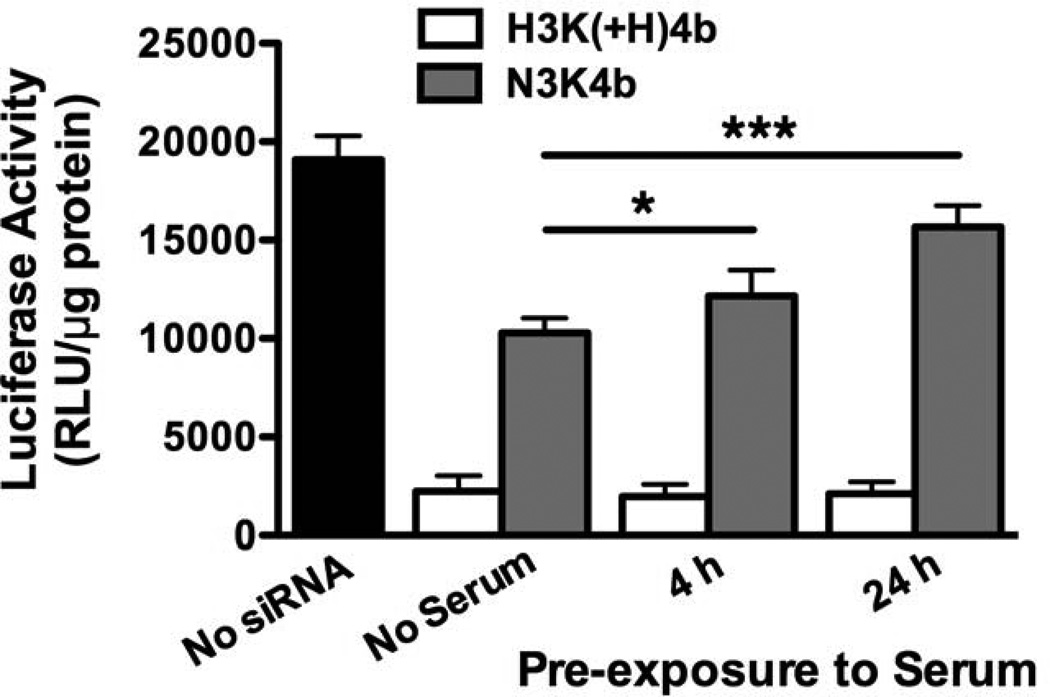
We hypothesized that the stability of complexes would significantly affect the ability of peptide siRNA polyplexes to silence target genes. Polyplexes pre-incubated in serum for 4 or 24 h were examined for their ability to silence luciferase expressed by MDA-MB-435 cells. In the absence of serum, H3K(+H)4b and N3K4b in complex with Luc siRNA inhibited luciferase expression by 90% and 50%, respectively (Fig. 3). The silencing activity of H3K(+H)4b siRNA polyplexes was maintained even after 24 h of serum exposure, whereas silencing activity of serum incubated N3K4b polyplexes was markedly reduced by 20% and 60% in 4 h and 24 h, respectively. Thus, biological activity of the polyplexes appears to correlate with their stability in serum, and HK polyplex is much more stable and effective than NK polyplex.
Fig. 3. H3K(+H)4b polyplexes incubated with serum maintained high transfection efficiency.
Prior to the transfection of MDA-MB-435 cells that stably express luciferase, H3K(+H)4b and N3K4b siRNA polyplexes were maintained at 37°C with serum (50%) for 0, 4, or 24 h. While silencing of luciferase expression with H3K(+H)4b polyplexes was not affected by serum, silencing with N3K4b polyplexes was markedly reduced by incubation with serum. *, P < 0.05; ***, P < 0.001.
3.3. Binding of siRNA to branched peptides
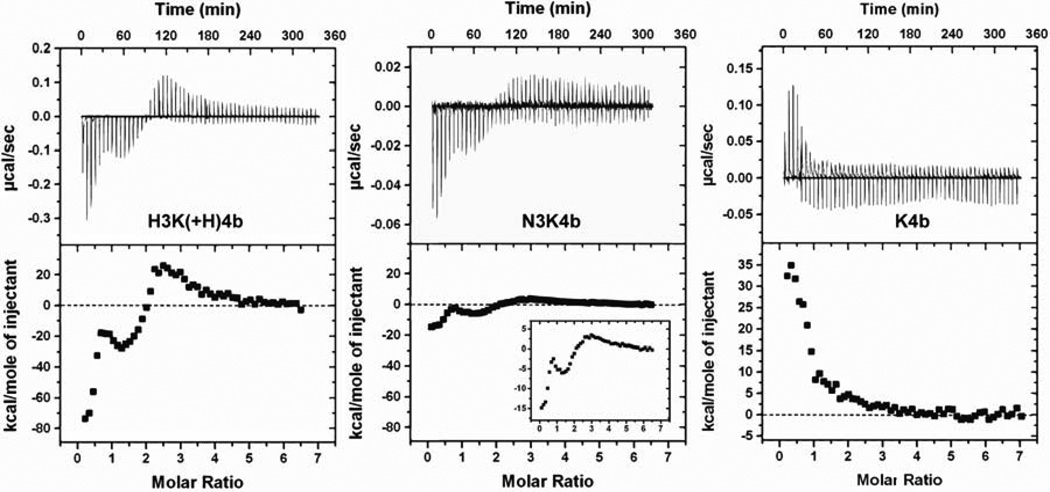
To study the different binding behaviors that presumably underlie the differences in stability of polyplex against nuclease in serum, we investigated peptide-siRNA binding using isothermal titration calorimetry (ITC). The addition of H3K(+H)4b to siRNA (Fig. 4) resulted in three peaks in the integrated enthalpy binding profile. The initial interaction showed an exothermic reaction, with ΔH ~ −80 kcal/mol of injectant. At this stage, with siRNA was in large excess, the interaction likely represents bimolecular binding of HK and siRNA with minimal formation of polyplexes. The absence of siRNA polyplexes as detected by dynamic light scattering (DLS) at ratios corresponded to this initial phase further validates this hypothesis. This initial enthalpy-driven binding of HK peptides to siRNA suggests the importance of non-ionic interactions, such as hydrogen bonding, stacking, or van der Waals interaction [33].
Fig. 4. Isothermal titration calorimetry of peptide siRNA interaction.
Representative isothermal titration calorimetry (ITC) raw data and integrated heat release for H3K(+H)4b, N3K4b and K4b peptides (0.12 molar ratio/injection) in 10 mM phosphate buffer, pH 7.0. Compared H3K(+H)4b and N3K4b with K4b, histidine- and asparagine-containing peptides show exothermic binding at peptide siRNA molar ratio 0 to 2.5 in which K4b is an endothermic reaction.
The initial phase was followed by two more transitions at peptide siRNA molar ratios of 1.5 and 2.5: the second phase showed ΔH ~ −20 kcal/mol and the third was endothermic. The transition from exothermic to endothermic binding was likely due to the particle aggregation induced by charge neutralization of the siRNA phosphate backbone with HK peptides [34]. The dependence of counterions upon aggregation provides an entropic driving force for the interaction. That is, the endothermic binding enthalpy was essentially entropy-driven due to displacement of water molecules and counterions, from electrostatic interaction (e.g., K4b with siRNA) [34, 35]. The aggregation was demonstrated by detection of polyplexes by DLS at peptide/siRNA molar ratios of 0.75 and 2.0, giving average particle size of 247 and 1419 nm, respectively. In contrast to binding of HK with siRNA, the initial binding of the branched K4b with siRNA showed an endothermic reaction (ΔH +35 kcal/mole of injectant), indicative of entropy-driven electrostatic binding based on prior studies [34].
Interestingly, asparagine-containing N3K4b:siRNA interaction showed an enthalpy profile similar to that of H3K(+H)4b but at a reduced scale. Since the imidazole groups of histidines and amide groups of the asparagines can both function as hydrogen bond donors and acceptors, the formation of hydrogen bonds may play a major role in the exothermic binding at a peptide:siRNA molar ratio of 0.1 to 1.0. Moreover, the exothermicity for H3K(+H)4b binding with siRNA was 5 times greater than that for N3K4b. This suggests that, H3K(+H)4b may have greater capacity to form hydrogen bonds with siRNA at physiological pH than N3K4b, thereby decreasing the sensitivity of the HK polyplex to serum (see Discussion below).
3.4. Binding of siRNA to linear peptides
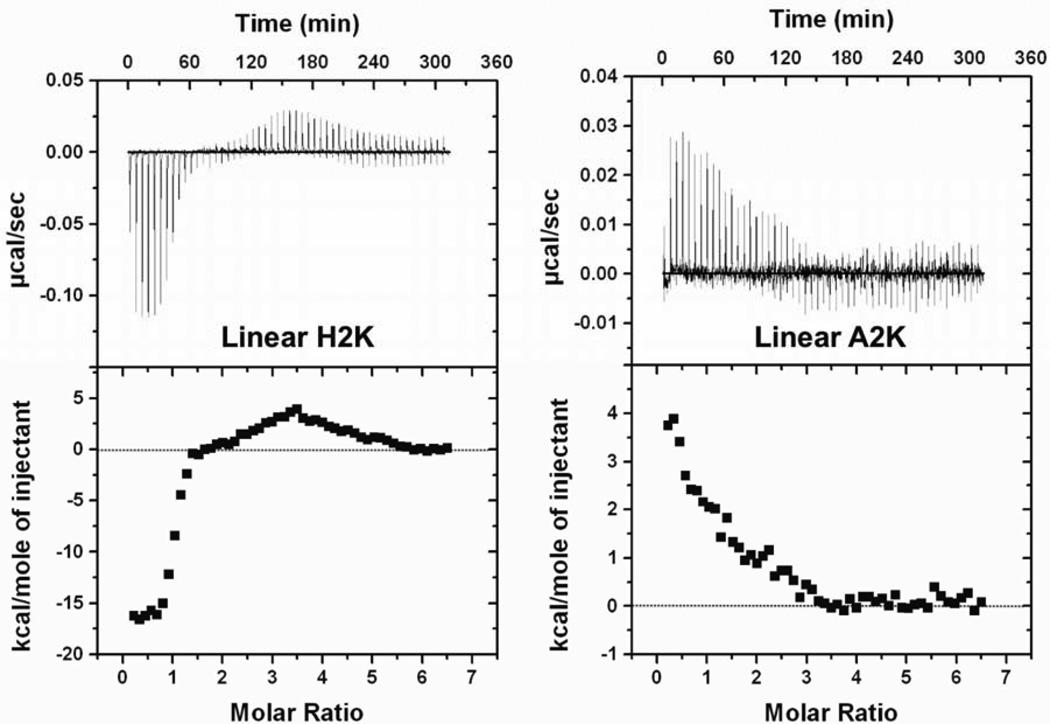
The effect of HK structures on thermodynamic profiles was then investigated. We synthesized two linear peptides (20-mer): histidine-lysine-rich H2K and A2K, in which histidines were replaced with alanines. ITC of linear and branched HKs with siRNA were assessed to distinguish sequence-related results from the two types of polyplexes. Similar to H3K(+H)4b, linear H2K showed negative enthalpy change at low molar ratios, followed by a transition to an endothermic region (Fig. 5). The exothermicity of binding of H2K to siRNA was approximately one-fourth that of H3K(+H)4b, which contained four times more histidine. In contrast, when the histidines were replaced with alanines, A2K showed endothermic binding with siRNA similar to that of K4b. Thus, the effects of linear HK peptides parallel those of branched peptides, validating the hypothesis that both ionic and non-ionic interactions participated in HK siRNA binding.
Fig. 5. Linear peptides exhibit ITC profiles similar to the branched peptides.
ITC profiles for linear peptides H2K and A2K (25 µM, 0.12 molar ratio/injection) titrated into siRNA (1 µM) were studied. Similar to branched peptides, histidine-containing H2K show exothermic binding at molar ratio 0 to 1.5. In contrast, when histidines were replaced with alanines, A2K behaved similarly to K4b: only an endothermic reaction is observed.
3.5. Influence of pH on binding
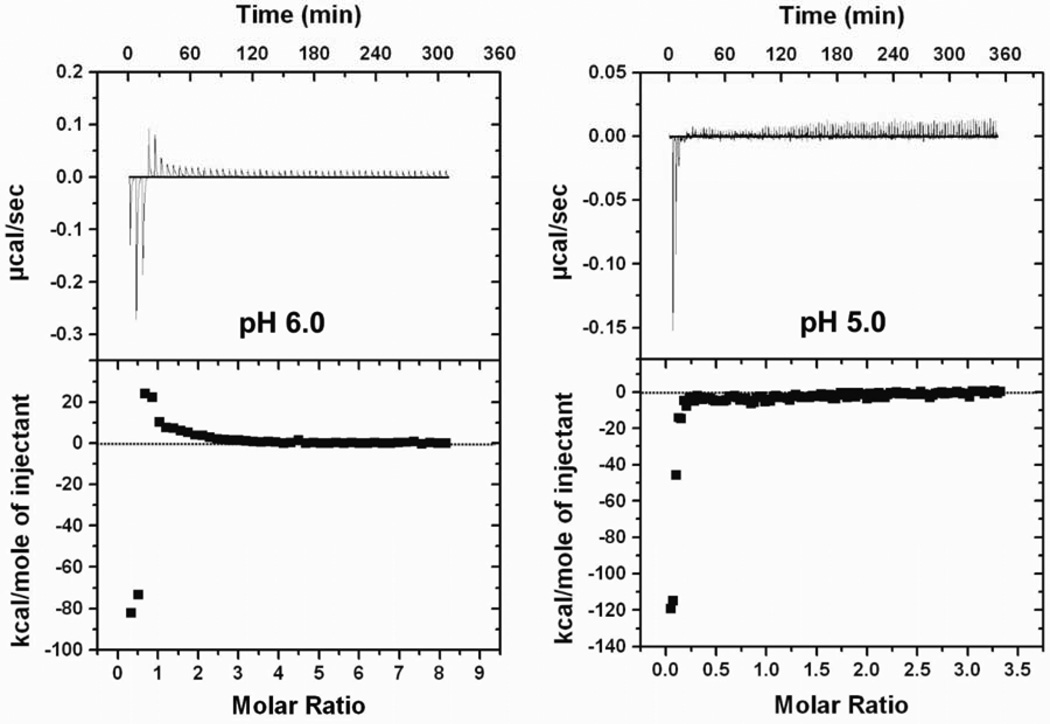
At a pH lower than the pKa of imidazole groups, protonation of histidine residues increased. The decreased molar ratio at saturation binding indicated that the charge density per HK peptide was greatly increased at lower pH and negatively charged phosphate groups of siRNA were saturated at a lower peptide/siRNA molar ratio (Fig. 6). Notably, with decreasing pH, the exothermic reaction was enhanced (pH 6.0, −90; pH 5.0, −120 kcal/mol) while the endothermic reaction was diminished. Protonation of histidines potentially increases the number of hydrogen bond donors and thus increases the likelihood of hydrogen bond formation. Nevertheless, the fully charged HK peptides may also avoid aggregation due to excess positive charges with electrostatic repulsion, which attenuated the rearrangement of small complexes formed in the first phase of binding. Furthermore, the thermodynamic profile of fully protonated H3K(+H)4b was in marked contrast with that of K4b (Fig. 4), indicating that the HK polyplexes formed at low pH were stabilized by non-electrostatic interactions.
Fig. 6. pH dependence of measured enthalpy of H3K(+H)4b siRNA binding.
Representative ITC data for H3K(+H)4b addition to siRNA in 10 mM MES buffer, pH 6.0 and 5.0, are shown. With decreased pH and increased positive charge density of H3K(+H)4b, the amplitude of exothermic reaction was enhanced, while endothermic reaction was diminished.
3.6. NMR spectroscopy
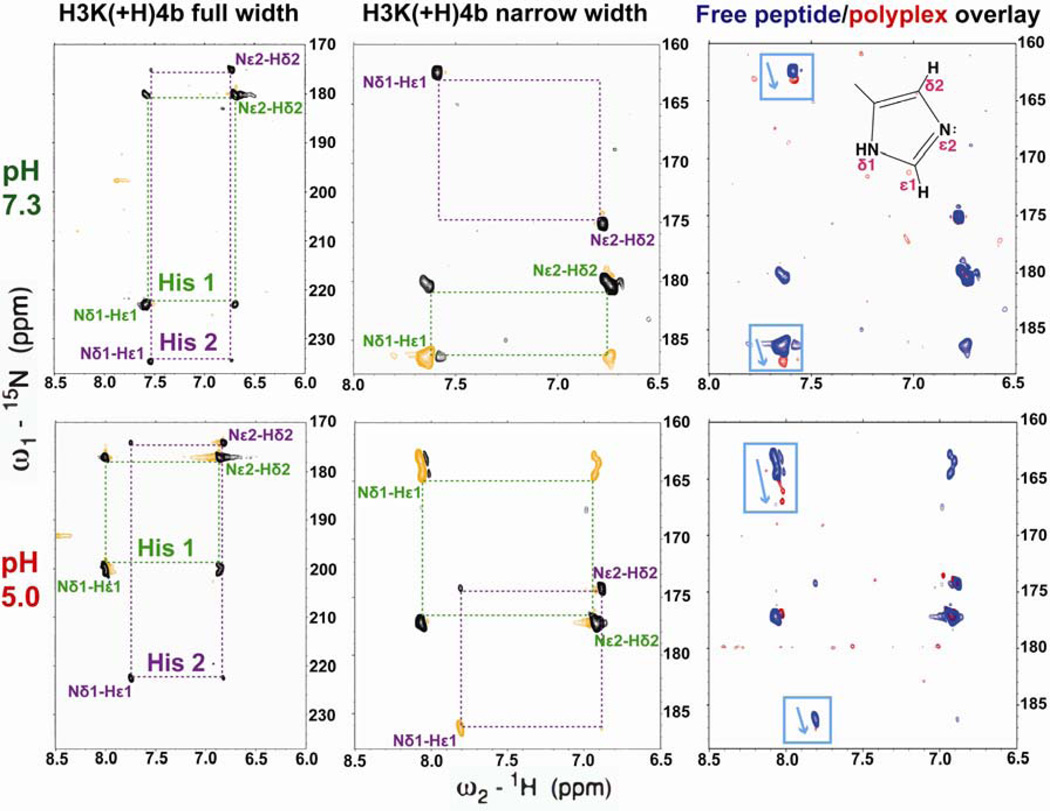
To determine the type of non-ionic interaction that results in an enthalpy-driven binding of HK peptide with siRNA, we studied the protonation state of selectively 15N-labeled histidines directly within H3K(+H)4b by using 1H-15N heteronuclear single quantum coherence (HSQC) NMR (Fig. 7). It has been shown that 15N chemical shifts of the imidazole ring are sensitive to hydrogen bond formation [8, 9, 11, 36]. We recorded 1H-15N HSQC spectra of free HK peptides to measure J-coupling between two carbon-attached, non-exchangeable protons (Hε1 and Hδ2) and two nitrogens of the imidazole ring (Nδ1 and Nε2) at pH values from 3.9 to 7.9 (Fig. S1). The three-bond 3JNδ1Hδ2 coupling results in a relatively weak cross-peak in HSQC spectra [37] whose intensities are diagnostic for histidine tautomer populations. The chemical shifts of histidine nitrogens are classified with three possible protonation types: deprotonated, type-β nitrogen, 249.5 ppm; protonated, type-α nitrogen, 167.5 ppm; charged and protonated, type-α+ nitrogen, 176.5 ppm [8, 11, 37]. At pH 7.3, the nitrogen protonation states identified two predominant tautomeric states of histidines among four labeled histidines. The histidines that were more highly protonated were designated His 1, the other histidine signals His 2.
Fig. 7. Protonated nitrogen shifted toward the direction of deprotonation.
Representative 1H-15N spectra (72 and 36 ppm, 15N width) of H3K(+H)4b (blue) at pH 7.3 (upper panel) and 5.0 (lower panel). Two imidazole ring sets of cross peaks were observed (His 1 and 2). siRNA was added at a peptide/siRNA molar ratio 4:1 (red, right panel). Upon siRNA binding, both Nδ1-H tautomers shifted downfield by 0.5 ppm at pH 7.3. The peak shift upon addition of siRNA demonstrates hydrogen bond formation. The complex-induced peak shift was increased to 2.0 ppm when the pH was lowered to 5.0.
The pH dependence of Nε2-Hδ2 cross-peaks was then determined to calculate the pKa of each histidine residue using Henderson-Hasselbalch equation. A pH titration of the 1H and 15N chemical shifts of Nε2-Hδ2 was performed to obtain pKa of each histidine signal, showing that His 1 and His 2 have pKa values of 6.3 and 5.3–5.8, respectively (Fig. S1). Notably, His 1 differentiated into three individual cross peaks at low pH. Because of its repetitive sequence, however, we were not able to assign residue types to individual histidine residues. Assuming the extent of proton transfer is linearly correlated to the difference of Nδ1 and Nε2 chemical shift, the tautomeric equilibrium of histidines can be quantified [8, 11]. At pH 7.3, the protonation level of Nδ1 of His 1 and His 2 were 25% and 13.5%, and while at pH 5.0, the protonation level of Nδ1 of His 1 and His 2 were increased to 35% and 20%, respectively.
A titration of H3K(+H)4b by siRNA was also similarly monitored by 1H-15N HSQC spectra to determine whether peptide siRNA binding altered the tautomeric forms of His residues. To increase the resolution and efficiency of signal acquisition, siRNA titration experiments were performed with a narrow sweep width (160 to 190 ppm, Fig. 7, middle panel). Compared with the normal full-width spectrum, Nδ1-H signals of His 1 and 2 aliased by 36 ppm (negative phase, orange) and 72 ppm (positive phase, black) in the 15N dimension at pH 7.3, respectively. Upon the addition of siRNA, Nδ1-H of both histidines shifted downfield (towards the direction of deprotonation [9], (Fig. S1)) while the predominant Nε2-H remained unchanged. His 1 and 2 shifted by 0.5 to 1 ppm, respectively, at a peptide:siRNA molar ratio 4:1. Moreover, this peak shift was similar to that in the buffer containing 100 mM NaCl (Fig. S2) demonstrating its nonionic in nature. The siRNA titration experiment showed that as the peptide:siRNA molar ratio decreased to 2:1 (data not shown), cross-peaks were broadened due to polyplex formation (Fig. S2). The Nδ1 peak shifts induced by siRNA binding were enhanced by two fold (1–2.5 ppm) at pH 5.0. The perturbations of Nδ1 chemical shift of HK:siRNA polyplexes were positively correlated with the numbers of hydrogen bonds. Thus, the result of HSQC is comparable to the thermodynamic behavior, which showed increased heat release as the pH was decreased.
4. Discussion
The binding mechanism and interaction between cationic polymers and siRNA are likely to have a critical role in the stability and delivery efficacy of the polyplexes. The goals of this work were to identify the contributions of purely ionic vs. non-electrostatic interactions, to characterize hydrogen bonding patterns in the siRNA and peptides, and to identify the molecular origin of the enhanced stability and biological activity of the HK peptides.
Polyplexes in which ionic interactions are dominant can be sensitive to physiological salt concentration and charged blood components such as albumin, thus reducing the ability to silence target genes in vitro and in vivo. Serum destabilization of cationic liposome:plasmid DNA complexes may be inhibited with a high positive/negative charge ratio [38], but excess polymers can be problematic for siRNA delivery. For example, our laboratory has demonstrated that higher HK to siRNA ratios are associated with poorly formed or small polyplexes that were less effective in gene silencing in vivo [30]. Moreover, an increased charge density of polycations may damage cell membranes, which results in cytotoxicity [39]. Nevertheless, the increased charge density of an ionizable polyplex upon protonation within acidic endosomes may lead to unpacking of the polyplexes with increased delivery of siRNA to the cytosol.
In addition to electrostatic interactions, a number of studies have reported that non-ionic interactions including hydrogen bonding may contribute significantly to binding of cationic carriers and DNA plasmids [22, 40]. Hydrogen bond formation enhances the stability of polyplexes without the use of excess cationic polymers, thereby reducing cytotoxicity induced by excess positive charges. Allen et al. have developed a hydroxyl-containing polymer in which intermediate binding and putative hydrogen bonding with DNA resulted in higher transfection [41]. Hydrogen bond formation also plays a pivotal role in maintaining the condensed state of the polyplex in combination with electrostatic interactions, particularly when exposed to the disruptive effects of serum [42]. Despite the importance of hydrogen bonds in the stability of polyplexes, prior studies with histidine-containing polyplexes have focused on the role of histidine in buffering endosomes and not on its ability to stabilize polyplexes. In this study, we investigated the potential binding and intermolecular forces between HK peptides and siRNA by gel electrophoresis analysis, ITC, and NMR. Together, the data indicate that hydrogen bonds formed between histidine residues and nucleic acids enhance the stability, silencing activity, and transfection efficacy of polyplexes.
Isothermal titration calorimetry is a sensitive technique that has been used to determine the predominant factors critical for binding interactions. Previous studies have suggested that charge-charge interaction of polycations and nucleic acids is often accompanied by the depletion of counterions and the disruption of the hydration layer, greatly increasing entropy and driving an enthalpically unfavorable reaction [34, 43]. In our data, exclusively entropy-driven binding was observed with the titration of K4b or A2K with siRNA, demonstrating that lysines interact primarily with negatively charged phosphate backbone of nucleic acids. In contrast, peptides containing histidines, H3K(+H)4b or H2K, release a significant amount of heat in the initial phase of binding to siRNA. This initial binding event probably represents a monomeric interaction between siRNA and HK peptide with minimal polyplex formation. In contrast to the ionic interaction between K4b and siRNA, exothermic HK:siRNA binding appears to be stabilized primarily by non-ionic interactions [21]. Furthermore, these non-ionic interactions were shown to be hydrogen bonds, as determined by diagnostic changes in NMR spectra under conditions that minimized polyplex formation.
The transition from the exothermic to the endothermic phase of binding, as described in Results, was correlated with the aggregation of polyplexes. The endothermicity is presumably due to strain energy or steric conflicts upon aggregation. Similar thermodynamic results have been obtained for polyethylenimine binding to nucleic acids [5, 6]. Polyplex formation is likely to involve multiple interactions, including ionic interaction, hydrogen bonding, π-π stacking, and/or hydrophobic interaction. Interestingly, H3K(+H)4b and N3K4b shared similar binding patterns in which peptide siRNA interaction was initially an exothermic reaction, followed by two transitions. Considering that both histidines and asparagines have amine groups compatible with formation of hydrogen bonds, it is anticipated that hydrogen bonding, rather than stacking, is the major source of exothermicity.
Several studies have shown that the substitution of histidines with asparagines does not interrupt the binding and biological activity of proteins [44, 45]. However, the initial exothermicity of H3K(+H)4b binding to siRNA is 5 times greater than that of N3K4b (−80 vs. −15 kcal/mole, Fig. 4) binding to siRNA. The thermodynamic difference can be correlated with the serum stability of the polyplex determined by gel electrophoresis. The enthalpy of H3K(+H)4b siRNA initial binding is pH-dependent. At pH 5.0, 6.0, and 7.3, the first exothermic peaks were −120, −90, and −80 kcal/mole, respectively. The results of HSQC also showed that Nδ1-H peaks of both histidines had a greater change in chemical shift at pH 5.0 than at 7.3. Therefore, the exothermic binding that we assign to hydrogen bonding by histidines is enhanced by protonation to give HisH+, and protonation of histidine primarily enhanced the interactions of Nδ1-H protons with siRNA. Furthermore, the hydrogen bond length of N-H donors of imidazole rings is generally shorter than N-H of amide groups on asparagines because of neighboring electron-withdrawing effects [46]. Thus, in addition to increased protonation level of Nδ1-H and charge potential, delocalization of electrons on the imidazole ring may also be responsible for the enhanced exothermic level, when siRNA binding of HK is compared with NK.
Because unprotonated histidines serve as both hydrogen bond donors and acceptors, hydrogen bonds may not necessarily form between histidines and nucleic acids. To determine the orientation of the hydrogen bond, we increased the protonation of histidines by lowering the pH to disrupt potential hydrogen bond interfaces of histidine-histidine (his-his) or histidine-lysine (his-lys) between HK peptides. If his-his or his-lys hydrogen bonding was dominant, protonation of histidines would decrease the number of hydrogen bond acceptors and therefore reduce hydrogen bonding and the exothermic level. The enhanced exothermic interaction at lower pH (Fig. 6), however, provided evidence that hydrogen bond pairs were predominantly at the interface of H3K(+H)4b and siRNA. Importantly, pH-dependent thermodynamic patterns correlated with the chemical shift perturbation in HSQC. Moreover, the addition of siRNA to H3K(+H)4b gave rise to greater shift of Nδ1 peaks at lower pH as assessed with HSQC. Whereas multiple lines of evidence indicate that hydrogen-bonds have an important role in augmenting the binding of HK with siRNA and stabilizing the polyplex, the present studies do not support improved siRNA protection of siRNA by formation of his-his hydrogen bonds.
The ITC data also shows that branched HK peptides have multiple binding stages upon titration into siRNA. As previously described, the transition from exothermic to endothermic reaction is attributed to aggregation induced by the charge neutralization. Nonetheless, the origin of the double peaks in the exothermic phase remained unclear. By replacing the branched HK with linear peptides or replacing the siRNA with DNA oligonucleotides (Fig. S3), double exothermic peaks either regressed to one peak or became less significant. Therefore, we speculate that the complicated binding patterns are correlated with charge heterogeneity in the branched structure of peptides and/or the response of the siRNA duplex. Compared with DNA, dsRNA has narrow, deep major grooves and wide, shallow minor grooves, which differ in solvent accessibility. The second stage of binding may be initiated when hydrogen bond donors and acceptors buried in the major groove may be exposed to branched HK peptides once the binding sites in the minor groove are saturated or the A-form structure is altered by initial binding event.
The HSQC spectroscopy established the involvement of histidines as potential hydrogen bond donors. Although the current technique does not allow us to identify the exact hydrogen bonds involving building blocks on siRNA, a study has shown that phosphate oxygens, hydroxyl groups of riboses, and oxygens or nitrogens of nucleobases of nucleic acids provide potential hydrogen bonds acceptors to the side chain of histidines [47]. In a DNA- and RNA- binding protein-nucleic acids X-ray crystal structure database [47], the most common hydrogen bond acceptors are the phosphate oxygens [41]. When we replaced the siRNA with a 19-mer DNA with similar sequence, however, the thermodynamic profile of H3K(+H)4b and DNA binding showed reduced exothermicity ( initially −40 kcal/mol) and binding affinity. Even though H3K(+H)4b does not bind specifically to double stranded RNA, the energetic difference between RNA and DNA recognition suggests that, in addition to phosphates, minor grooves and/or 2’-hydroxyl groups also form hydrogen bonds [48, 49]. In addition to DNA, a 2’-fluoro-modified siRNA was used to compare with unmodified siRNA binding to H3K(+H)4b to determine the role of 2’-hydroxyl groups. The binding patterns of 2’-fluoro-modified siRNA was indistinguishable from DNA duplex (Fig. S3), suggesting that hydrogen bond formation between H3K(+H)4b and the 2’-hydroxyl groups of unmodified siRNA. We speculate that the polyplex is stabilized by hydrogen bond networks that occupy potential binding sites of ribonucleases or competing binding proteins.
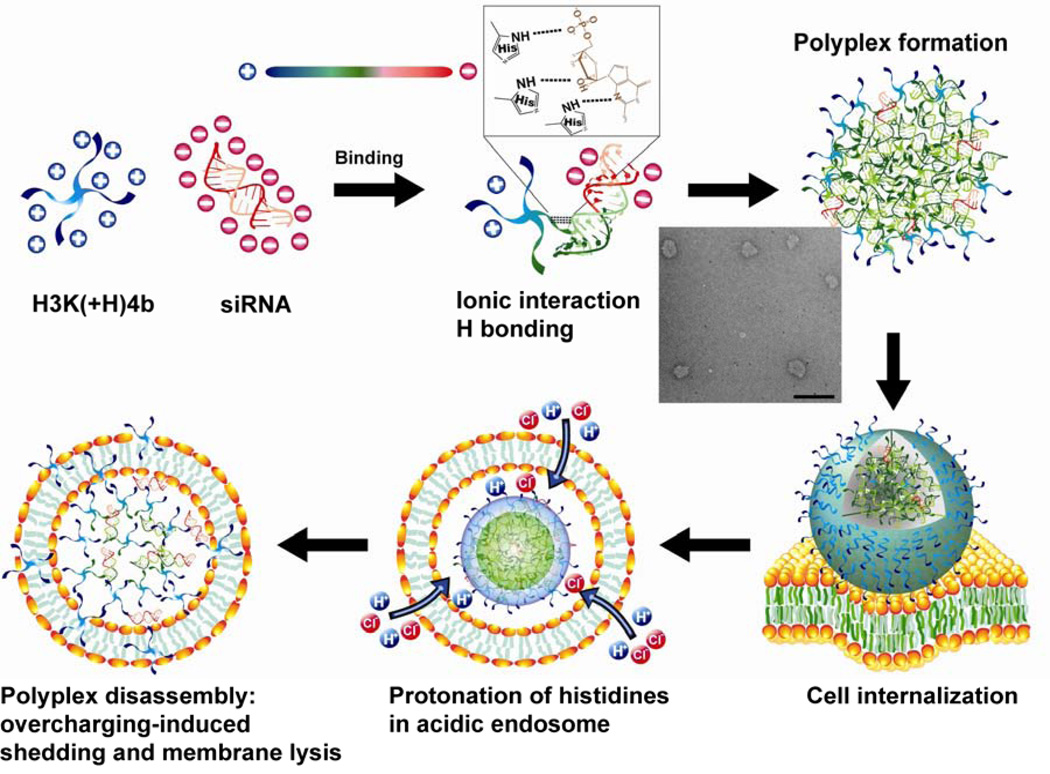
Both the ITC and HSQC data indicated that the overall contribution of hydrogen bonding increases at a pH below 7.3 (physiological), which leads us to explore how the siRNA escapes from polyplexes and endosomes? With the pKa of histidines 6.3 or lower, we speculate that electrostatic repulsion between the histidines upon protonation in the acidic endosomes plays the dominant role in disrupting and/or unpacking the polyplexes (Fig. 8). Furthermore, various charged proteins in the cytosol interacting with the loosely-packed HK polyplex may have a complementary role in promoting decomposition of polyplexes. Unpacking of the polyplex could occur with further polyplex disruption into smaller and less dense smaller polyplexes, with monomeric HK siRNA units as part of the continuum. Upon unpacking of the polyplex, regardless of the extent of disruption, the protonated HK peptides would likely interact with negatively charged endosomal membranes, which would thus act similarly to detergents to aid in the escape of siRNA. This model is consistent with the data provided in this manuscript, as well as previous observations from our laboratory and others [50, 51]. Moreover, if the electrostatic repulsion in the acidic endosomes disrupts polyplexes to the extent of releasing “free” peptide, the secondary structure of H3K(+H)4b may enhance the membrane lytic activity [52]. Accordingly, we propose a model in which the complexation of H3K(+H)4b:siRNA involves electrostatic interaction and hydrogen bonding, whereas decomplexation of polyplexes is induced by overcharging, disruption of the polyplex, and interaction of the peptide with the endosomal membranes (Fig. 8).
Fig. 8.
Model of HK and siRNA binding illustrating compactness in serum and release (or partial release) in endosomes. The colors blue, green, and red represent positive, neutral, and negative electrostatic potential, respectively. Hydrogen bonds are shown as dashed lines. Scale bar in the TEM figure, 500 nm.
5. Conclusion
Characterization of polyplex stability is considered one of the most important properties to understand for more effective carriers to be developed. Our study demonstrates that non-ionic interactions between HK peptides and siRNA is essential for increased stability of HK polyplexes in serum compared to polylysine polyplexes. In particular, spectral shifts in the NMR established that the histidine component of HK formed hydrogen bonds with siRNA. Thus, in addition to the role of buffering endosomes by histidines, their formation of hydrogen-bonds with siRNA is likely a key reason why histidine-containing polyplexes are effective transfection and silencing agents. Moreover, the distinguishing features derived from structural and biophysical studies with HK peptides may be shared by other hydrogen-forming peptides and synthetic polymers, even those without imidazole groups. A fundamental understanding of vector-cargo interactions may establish evidence-based approaches to assess the potential of carriers for siRNA delivery.
Supplementary Material
Acknowledgments
The authors thank P. Talalay for careful reading and invaluable editing assistance of this manuscript. We also thank D. Weber and P. Wilder for the assistance with ITC; W. Decatur for the assistance with siRNA 3D structure; and J. Marino for his comments on the interpretation of NMR spectra. This work was supported by the National Institutes of Health (R01-CA136938).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Appendix A. Supplementary material
Supplementary material associated with this article can be found at
Contributor Information
Szu-Ting Chou, Email: stchou@umd.edu.
Kellie Hom, Email: hom@nmr.umaryland.edu.
Daoning Zhang, Email: zdn3023@umd.edu.
Qixin Leng, Email: QLeng@som.umaryland.edu.
Lucas J. Tricoli, Email: ltricoli@umd.edu.
Jason M. Hustedt, Email: jhustedt@umd.edu.
Amy Lee, Email: amylee16@umd.edu.
Michael J. Shapiro, Email: Michael.J.Shapiro@pfizer.com.
Joonil Seog, Email: jseog@umd.edu.
Jason D. Kahn, Email: jdkahn@umd.edu.
A. James Mixson, Email: amixson@umaryland.edu.
References
- 1.Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brower V. RNA interference advances to early-stage clinical trials. J Natl Cancer Inst. 2010;102:1459–1461. doi: 10.1093/jnci/djq405. [DOI] [PubMed] [Google Scholar]
- 3.Strumberg D, Schultheis B, Traugott U, Vank C, Santel A, Keil O, et al. Phase I clinical development of Atu027, a siRNA formulation targeting PKN3 in patients with advanced solid tumors. Int J Clin Pharmacol Ther. 2012;50:76–78. doi: 10.5414/cpp50076. [DOI] [PubMed] [Google Scholar]
- 4.Basu HS, Schwietert HC, Feuerstein BG, Marton LJ. Effects of variation in the structure of spermine on the association with DNA and the induction of DNA conformational changes. Biochem J. 1990;269:329–334. doi: 10.1042/bj2690329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Utsuno K, Uludag H. Thermodynamics of polyethylenimine-DNA binding and DNA condensation. Biophys J. 2010;99:201–207. doi: 10.1016/j.bpj.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng M, Pavan GM, Neeb M, Schaper AK, Danani A, Klebe G, et al. Targeting the blind spot of polycationic nanocarrier-based siRNA delivery. ACS Nano. 2012;6:9447–9454. doi: 10.1021/nn301966r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuiderweg ER. Mapping protein-protein interactions in solution by NMR spectroscopy. Biochemistry. 2002;41:1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
- 8.Bachovchin WW. 15N NMR spectroscopy of hydrogen-bonding interactions in the active site of serine proteases: evidence for a moving histidine mechanism. Biochemistry. 1986;25:7751–7759. doi: 10.1021/bi00371a070. [DOI] [PubMed] [Google Scholar]
- 9.Farr-Jones S, Wong WYL, Gutheil WG, Bachovchin WW. Direct observation of the tautomeric forms of histidine in nitrogen-15 NMR spectra at low temperatures. Comments on intramolecular hydrogen bonding and on tautomeric equilibrium constants. J Am Chem Soc. 1993;115:6813–6819. [Google Scholar]
- 10.Lodi PJ, Knowles JR. Neutral imidazole is the electrophile in the reaction catalyzed by triosephosphate isomerase: structural origins and catalytic implications. Biochemistry. 1991;30:6948–6956. doi: 10.1021/bi00242a020. [DOI] [PubMed] [Google Scholar]
- 11.Van Dijk AA, Scheek RM, Dijkstra K, Wolters GK, Robillard GT. Characterization of the protonation and hydrogen bonding state of the histidine residues in IIAmtl, a domain of the phosphoenolpyruvate-dependent mannitol-specific transport protein. Biochemistry. 1992;31:9063–9072. doi: 10.1021/bi00152a050. [DOI] [PubMed] [Google Scholar]
- 12.Leng Q, Mixson AJ. Small interfering RNA targeting Raf-1 inhibits tumor growth in vitro and in vivo. Cancer Gene Ther. 2005;12:682–690. doi: 10.1038/sj.cgt.7700831. [DOI] [PubMed] [Google Scholar]
- 13.Leng Q, Scaria P, Zhu J, Ambulos N, Campbell P, Mixson AJ. Highly branched HK peptides are effective carriers of siRNA. J Gene Med. 2005;7:977–986. doi: 10.1002/jgm.748. [DOI] [PubMed] [Google Scholar]
- 14.Putnam D, Gentry CA, Pack DW, Langer R. Polymer-based gene delivery with low cytotoxicity by a unique balance of side-chain termini. Proc Natl Acad Sci U S A. 2001;98:1200–1205. doi: 10.1073/pnas.031577698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kichler A, Mason AJ, Bechinger B. Cationic amphipathic histidine-rich peptides for gene delivery. Biochim Biophys Acta. 2006;1758:301–307. doi: 10.1016/j.bbamem.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Midoux P, Monsigny M. Efficient gene transfer by histidylated polylysine/pDNA complexes. Bioconjug Chem. 1999;10:406–411. doi: 10.1021/bc9801070. [DOI] [PubMed] [Google Scholar]
- 17.Hatefi A, Megeed Z, Ghandehari H. Recombinant polymer-protein fusion: a promising approach towards efficient and targeted gene delivery. J Gene Med. 2006;8:468–476. doi: 10.1002/jgm.872. [DOI] [PubMed] [Google Scholar]
- 18.Chen QR, Zhang L, Stass SA, Mixson AJ. Branched co-polymers of histidine and lysine are efficient carriers of plasmids. Nucleic Acids Res. 2001;29:1334–1340. doi: 10.1093/nar/29.6.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Ambulos N, Mixson AJ. DNA delivery to cells in culture using peptides. Methods Mol Biol. 2004;245:33–52. doi: 10.1385/1-59259-649-5:33. [DOI] [PubMed] [Google Scholar]
- 20.Lohman TM, deHaseth PL, Record MT., Jr Pentalysine-deoxyribonucleic acid interactions: a model for the general effects of ion concentrations on the interactions of proteins with nucleic acids. Biochemistry. 1980;19:3522–3530. doi: 10.1021/bi00556a017. [DOI] [PubMed] [Google Scholar]
- 21.Mascotti DP, Lohman TM. Thermodynamics of oligoarginines binding to RNA and DNA. Biochemistry. 1997;36:7272–7279. doi: 10.1021/bi970272n. [DOI] [PubMed] [Google Scholar]
- 22.Prevette LE, Kodger TE, Reineke TM, Lynch ML. Deciphering the role of hydrogen bonding in enhancing pDNA-polycation interactions. Langmuir. 2007;23:9773–9784. doi: 10.1021/la7009995. [DOI] [PubMed] [Google Scholar]
- 23.Manning GS. Limiting laws and counterion condensation in polyelectrolyte solutions I. colligative properties. J Chem Phys. 1969;51:59. [Google Scholar]
- 24.Manning GS. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q Rev Biophys. 1978;11:179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- 25.Burckhardt G, Zimmer C, Luck G. Conformation and reactivity of DNA in the complex with proteins. IV. Circular dichroism of poly-L-histidine model complexes with DNA polymers and specificity of the interaction. Nucleic Acids Res. 1976;3:561–580. doi: 10.1093/nar/3.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim T, Afonin KA, Viard M, Koyfman AY, Sparks S, Heldman E, et al. In silico, in vitro, and in vivo studies indicate the potential use of bolaamphiphiles for therapeutic siRNAs delivery. Mol Ther Nucleic Acids. 2013;2:e80. doi: 10.1038/mtna.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karatasos K, Posocco P, Laurini E, Pricl S. Poly(amidoamine)-based dendrimer/siRNA complexation studied by computer simulations: effects of pH and generation on dendrimer structure and siRNA binding. Macromol Biosci. 2012;12:225–240. doi: 10.1002/mabi.201100276. [DOI] [PubMed] [Google Scholar]
- 28.Leng Q, Scaria P, Ioffe OB, Woodle M, Mixson AJ. A branched histidine/lysine peptide, H2K4b, in complex with plasmids encoding antitumor proteins inhibits tumor xenografts. J Gene Med. 2006;8:1407–1415. doi: 10.1002/jgm.982. [DOI] [PubMed] [Google Scholar]
- 29.Leng Q, Mixson AJ. Modified branched peptides with a histidine-rich tail enhance in vitro gene transfection. Nucleic Acids Res. 2005;33:e40. doi: 10.1093/nar/gni040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chou ST, Leng Q, Scaria P, Woodle M, Mixson AJ. Selective modification of HK peptides enhances siRNA silencing of tumor targets in vivo. Cancer Gene Ther. 2011;18:707–716. doi: 10.1038/cgt.2011.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 32.Markley JL. Observation of histidine residues in proteins by nuclear magnetic resonance spectroscopy. Acc Chem Res. 1975;8:11. [Google Scholar]
- 33.Ross PD, Subramanian S. Thermodynamics of protein association reactions: forces contributing to stability. Biochemistry. 1981;20:3096–3102. doi: 10.1021/bi00514a017. [DOI] [PubMed] [Google Scholar]
- 34.Matulis D, Rouzina I, Bloomfield VA. Thermodynamics of DNA binding and condensation: isothermal titration calorimetry and electrostatic mechanism. J Mol Biol. 2000;296:1053–1063. doi: 10.1006/jmbi.1999.3470. [DOI] [PubMed] [Google Scholar]
- 35.Mascotti DP, Lohman TM. Thermodynamics of single-stranded RNA binding to oligolysines containing tryptophan. Biochemistry. 1992;31:8932–8946. doi: 10.1021/bi00152a033. [DOI] [PubMed] [Google Scholar]
- 36.Roberts JD, Chun Y, Flanagan C, Birdseye TR. A nitrogen-15 nuclear magnetic resonance study of the acid-base and tautomeric equilibriums of 4-substituted imidazoles and its relevance to the catalytic mechanism of .alpha.-lytic protease. J Am Chem Soc. 1982;115:3945–3949. [Google Scholar]
- 37.Pelton JG, Torchia DA, Meadow ND, Roseman S. Tautomeric states of the active-site histidines of phosphorylated and unphosphorylated IIIGlc, a signal-transducing protein from Escherichia coli, using two-dimensional heteronuclear NMR techniques. Protein Sci. 1993;2:543–558. doi: 10.1002/pro.5560020406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang JP, Huang L. Overcoming the inhibitory effect of serum on lipofection by increasing the charge ratio of cationic liposome to DNA. Gene Ther. 1997;4:950–960. doi: 10.1038/sj.gt.3300485. [DOI] [PubMed] [Google Scholar]
- 39.Fischer D, Li Y, Ahlemeyer B, Krieglstein J, Kissel T. In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24:1121–1131. doi: 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- 40.Ma PL, Lavertu M, Winnik FM, Buschmann MD. New insights into chitosan-DNA interactions using isothermal titration microcalorimetry. Biomacromolecules. 2009;10:1490–1499. doi: 10.1021/bm900097s. [DOI] [PubMed] [Google Scholar]
- 41.Allen MH, Green MD, Getaneh HK, Miller KM, Long TE. Tailoring charge density and hydrogen bonding of imidazolium copolymers for efficient gene delivery. Biomacromolecules. 2011;12:2243–2250. doi: 10.1021/bm2003303. [DOI] [PubMed] [Google Scholar]
- 42.Buyens K, Meyer M, Wagner E, Demeester J, De Smedt SC, Sanders NN. Monitoring the disassembly of siRNA polyplexes in serum is crucial for predicting their biological efficacy. J Control Release. 2010;141:38–41. doi: 10.1016/j.jconrel.2009.08.026. [DOI] [PubMed] [Google Scholar]
- 43.Korolev N, Berezhnoy NV, Eom KD, Tam JP, Nordenskiold L. A universal description for the experimental behavior of salt-(in)dependent oligocation-induced DNA condensation. Nucleic Acids Res. 2012;40:2808–2821. doi: 10.1093/nar/gks214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Narayan K, Chou CL, Kim A, Hartman IZ, Dalai S, Khoruzhenko S, et al. HLA-DM targets the hydrogen bond between the histidine at position beta81 and peptide to dissociate HLA-DR-peptide complexes. Nat Immunol. 2007;8:92–100. doi: 10.1038/ni1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lowe DM, Fersht AR, Wilkinson AJ, Carter P, Winter G. Probing histidine-substrate interactions in tyrosyl-tRNA synthetase using asparagine and glutamine replacements. Biochemistry. 1985;24:5106–5109. doi: 10.1021/bi00340a022. [DOI] [PubMed] [Google Scholar]
- 46.Steiner T. The hydrogen bond in the solid state. Angew Chem Int Ed Engl. 2002;41:49–76. doi: 10.1002/1521-3773(20020104)41:1<48::aid-anie48>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 47.Chen Y, Kortemme T, Robertson T, Baker D, Varani G. A new hydrogen-bonding potential for the design of protein-RNA interactions predicts specific contacts and discriminates decoys. Nucleic Acids Res. 2004;32:5147–5162. doi: 10.1093/nar/gkh785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bevilacqua PC, Cech TR. Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry. 1996;35:9983–9994. doi: 10.1021/bi9607259. [DOI] [PubMed] [Google Scholar]
- 49.Schmedt C, Green SR, Manche L, Taylor DR, Ma Y, Mathews MB. Functional characterization of the RNA-binding domain and motif of the double-stranded RNA-dependent protein kinase DAI (PKR) J Mol Biol. 1995;249:29–44. doi: 10.1006/jmbi.1995.0278. [DOI] [PubMed] [Google Scholar]
- 50.Rehman ZU, Hoekstra D, Zuhorn IS. Mechanism of polyplex- and lipoplex-mediated delivery of nucleic acids: real-time visualization of transient membrane destabilization without endosomal lysis. ACS Nano. 2013;7:3767–3777. doi: 10.1021/nn3049494. [DOI] [PubMed] [Google Scholar]
- 51.Chen QR, Zhang L, Luther PW, Mixson AJ. Optimal transfection with the HK polymer depends on its degree of branching and the pH of endocytic vesicles. Nucleic Acids Res. 2002;30:1338–1345. doi: 10.1093/nar/30.6.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Medina-Kauwe LK, Xie J, Hamm-Alvarez S. Intracellular trafficking of nonviral vectors. Gene Ther. 2005;12:1734–1751. doi: 10.1038/sj.gt.3302592. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.