Abstract
Gene promoter activity can be studied in vivo by molecular imaging methods using reporter gene technology. Transcription of the reporter and the reported genes occurs simultaneously. However, imaging depends on reporter protein translation, stability, and cellular fate that may differ among the various proteins. A double transgenic mouse strain expressing the firefly luciferase (lucF) and fluorescent mPlum protein under the transcriptional control of the thermo-inducible heat-shock protein (Hspa1b) promoter was generated allowing to follow up the reporter proteins by different and complementary in vivo imaging technologies. These mice were used for in vivo imaging by bioluminescence and epi fluorescence reflectance imaging (BLI & FRI) and as a source of embryonic fibroblast (MEF) for in vitro approaches. LucF, mPlum and endogenous Hsp70 mRNAs were transcribed simultaneously. The increase in mRNA was transient, peaking at 3 h and then returning to the basal level about 6 h after the thermal stimulations. The bioluminescent signal was transient and initiated with a 3 h delay versus mRNA expression. The onset of mPlum fluorescence was more delayed, increasing slowly up to 30 h after heat-shock and remaining for several days. This mouse allows for both bioluminescence imaging (BLI) and fluorescence reflectance imaging (FRI) of Hsp70 promoter activation showing an early and transient lucF activity and a retrospective and persistent mPlum fluorescence. This transgenic mouse will allow following the transient local induction of Hsp-70 promoter beyond its induction time-frame and relate into subsequent dynamic biological effects of the heat-shock response.
OCIS codes: (280.1415) Biological sensing and sensors, (280.6780) Temperature
1. Introduction
Molecular imaging enables in vivo visualization of cellular processes in living animals including proteomic, metabolic, cellular biologic and genetic events. Because the molecular and genetic changes precede functional modifications, imaging of genetic processes (so called molecular-genetic imaging) allows for detection of cellular events occurring in the early step of the disease development or in the early response to therapeutic treatments [1,2]. Molecular genetic imaging is also essential for the follow up of gene therapies [3]. Genetic processes are evaluated by assaying reporter gene expression. The reporter proteins induce accumulation of a specific imaging signal that reflects the genetic process. However, each reporter imaging protein exhibits its own maturation process, stability and cellular fate. Short maturation time reduces the delay between transcriptional induction and imaging detection. Stable proteins accumulate in cells and thus facilitate detection in vitro and in vivo. However, stability also limits application in studies that require rapid reporter turnover, including transcriptional induction studies [4]. Furthermore, half-life of reporter protein in vivo is difficult to determine and quite often unavailable in the current literature thus, the choice for the reporter gene should be adapted to the imaging strategy and the biological question.
The heat-shock response is a remarkable example of an inducible and transient transcriptional regulation in eukaryotic organisms subsequent to a stressful event [5]. The most prominent protein of the heat-shock response is a 70 kDa protein called heat-shock protein 70 (HSP70). Promoters for HSP70 (namely Hspa1a and Hspa1b) are known to exhibit low basal level and high induced activities. They are activated by a large number of inducers including stress conditions (heat-shock, heavy metal, arsenic, reactive oxygen species, ...), physiopathological situations (fever, ischemia, hypoxia, cancer, ...) or physiological challenges (embryonic development, hormonal stimulation) [6]. The sharp pattern of response of the Hsp70 promoter has also drawn attention to possible therapeutic use as a molecular tool for gene therapy [7–9]. In order to provide a rapid in vivo readout for induction of Hsp transcription related to the aforementioned research field several transgenic mice have been generated [10] and are currently used for in vivo imaging. Available reporter genes encode for fluorescent proteins (eGFP and mPlum) or for luciferase firefly (lucF) [11–13] and they have been used in vivo to monitor the HSP response to thermal challenges [12–16] or brain ischemia [13]. Different imaging patterns were found in vivo depending on the biological stimulus and the reporter gene [13]. Reporter signal imaging depends on protein translation, stability, and cellular fate. These latter steps add complexity in interpretation of the imaging findings since they may differ among the various reporters and the reported protein. Furthermore, imaging HSP70 is complex since different pathways are involved in the HSP response [17]. Therefore, it is currently unclear whether the various in vivo imaging profiles described in previous works in the available transgenic mice report on the same transcriptional events.
In the present work, a double transgenic mouse strain was generated by crossing a Hspa1b-lucF mice [11] with Hspa1b-mPlum mice [13]. Mouse Embryonic Fibroblasts (MEF) were obtained from transgenic embryos in order to compare the in vitro and in vivo patterns of reporter genes expression in response to a single short thermal stress. The in vitro transcription pattern of Hsp70, lucF and mPlum mRNAs in response to a heat challenge was correlated to both the in vitro protein expression pattern by MEF and the in vivo monitoring of mice by optical imaging.
2. Materials & methods
2.1. Animals
Animal manipulation and husbandry were performed in agreement with the European Commission, directives of the French Research Ministry (Agreement 33-04392 to F.C). Mice were reared at University of Bordeaux transgenic facilities. The transgenic mouse Hspa1b-lucF (+/+) were previously reported [11]. The Hspa1b-mPlum (+/−) transgenic mice were generated from purified DNA containing the mPlum minigene injected into (C57BL/6J x CBA/J) F2 mouse blastocyst. Four female founders that carried the mPlum minigene as deduced by PCR-genotyping were obtained. Three of the founder females produced offspring in crosses with wt C57BL/6J males. Transgenic female offspring were back-crossed to C57BL/6J wild type males for 7 generations. Litters were genotyped in order to identify transgenic male mice to be used in the experiments [13]. Female transgenic Hspa1b-lucF (+/+) mice were crossed with Hspa1b-mPlum (+/−) males. Each pup was PCR-tested for mPlum insertion. Although corresponding to the same gene, the Hsp promoter sequence driving the lucF expression is 800 bp long [11] while the sequence driving the mPlum expression is only 516 bp [13]. Animals were anesthetized with 2% isoflurane (Belamont, Nicholas Piramal Limited, London, GB) in air. Skin samples (about 25 mm2) were removed on anesthetized mice just prior euthanasia.
2.2. Mouse Embryonic Fibroblasts (MEF) culture
MEF were prepared from individual embryos of 14 days post-coitus. Embryos were removed from uterine horns and separated from placenta. Head, limbs and organs were cut away and embryos were washed in PBS. Embryos were minced and incubated in trypsin 0.05% (v/v) EDTA (Invitrogen, Carlsbad, California, USA) during 15 min at 37°C, with gentle shaking. Supernatant was removed and trypsin activity was stopped with addition of 2 mL of DMEM medium (Invitrogen) supplemented with 10% (v/v) fetal bovine serum (FBS, Invitrogen), and 1% (v/v) of a Penicillin, Streptomycin and Amphotericin mixture (PSA, Invitrogen). Cells were pelleted by centrifugation (350 g, 10 min), expanded in DMEM supplemented with 10% FBS, 1% PSA and maintained in culture (37°C, 5% CO2 and humidity atmosphere saturated). Each MEF sample was genotyped from 2.105 cells for both mPlum and lucF insertion.
2.3. In vitro MEF heating
MEF were plated in 4 wells plate at 26,300 cells/cm2 2 days before heating in a water bath. Cell’s medium was replaced by pre-heated medium and plates were incubated at 44°C for 25 min. Thereafter, cells were placed back at 37°C for 0, 3, 6, 24 and 48 h until measurements.
2.4. In vitro luciferase activity measurement
The luciferase enzymatic activity was measured using the Luciferase Assay System (Promega) according to the manufacturer’s instructions. Cells were incubated in cell culture lysis reagent buffer (10 min at room temperature) with shaking and samples (5 µL) were mixed with Luciferase Assay Substrate (LAS, 50 µL). Luminescence was measured 10 s after mixing with a luminometer apparatus (Lumat, Berthold Technologies).
2.5. Flow cytometry
MEF were harvested with trypsin and washed with PBS. More than 10,000 events were captured for each analysis with excitation at 561 nm and emission filter at 660/20 nm (Cy5 filter set). Data were collected on a BD Biosciences LSRFortessa II apparatus (Becton Dickinson, San José, California, USA) and analyzed with FACSDiva software.
2.6. Extraction and quantification of nucleic acid
Genomic DNA was extracted from the mice tail tips and from MEF pellets (2.105 cells) using the Wizard SV Genomic DNA Purification System (Promega). Total RNA was extracted from MEF (3.105 cells) or skin samples using TRIzol® reagent (Invitrogen). Nucleic acids were quantified using a Versafluor apparatus (BIO-RAD, Hercules, California, USA). DNA was quantified using the Fluorescent DNA Quantification kit (BIO-RAD) and RNA was quantified using the Ribogreen RNA quantification kit (Invitrogen, Molecular probes). Total RNA (1 µg) was treated with DNase I (Fermentas, ThermoFisher Scientific, Waltham, Massachusetts, USA) and converted to cDNA with Improm II (Promega) using random hexameric primers according to the manufacturer’s recommendations.
2.7. PCR amplification
PCR amplifications were carried out on a Biometra T-gradient apparatus usually programmed for 39 cycles (30 s at 95°C, 30 s at Tm = 50°C-62°C according to the primers, 1 min per Kb at 72°C) with the following final conditions: 50 µL final volume, 0.2 µM of primers, 0.2 mM of each dNTP, 1.25 unit of Taq DNA polymerase (DreamTaq, Fermentas) in a reaction buffer provided by the manufacturer. DMSO (1% (v/v)) was added for lucF and mPlum amplification. Primers for GAPDH were GAPDH-5′ (5′-ACCACAGTCCATGCCATCAC-3′) and GAPDH-3′ (5′-TCCACCACCCTGTTGCTGTA-3′) used at Tm = 55°C. Primers for lucF were lucF-12D (5′-TCCATTCCATCACGGTTTTGG-3′) and lucF-13R (5′-GCTATGTCTCCAGAATGTAGC-3′) used at Tm = 60°C. Primers for mPlum were mPlum-3D (5′-AGGGCGAGGAGGTCATCAAG-3′) and mPlum-5R (5′-GATGTCGGTCTTGTAGGCGC −3′) used at Tm = 61.8°C.
2.8. Real time PCR
mRNA was quantified with a real-time thermal iQ cycler (BIO-RAD). All sets of primers were assayed for qPCR effectiveness and found to exhibit PCR efficiency greater than 90%. Primers for lucF and mPlum were the same as those used for genotyping. The absence of unwanted products was confirmed by automated melting curve analysis. Each sample was assayed in triplicate. Each mRNA species was quantified respective to acidic ribosomal phosphoprotein P0 (36B4) mRNA abundance (ΔCt) [18]. Primers for 36B4 amplification are 36B4-S (5′-GCTTCATTGTGGGAGCAGAC-3′) and 36B4-AS (5′-CATGGTGTTCTTGCCCATCAG-3′) used at Tm = 56°C or 62.5°C and primers for Hsp70 are Hspa1-1D (5′-CGCCTACTTCAACGACTCTCAG-3′) and Hspa1-4R (5′-GCTTGTTCTGGCTGATGTC-3′) used at Tm = 56°C.
2.9. In vivo heating
Three days before the heating or imaging setup, the posterior part of mice was shaved under sedation with isoflurane (2% in air). In vivo heating was done in a large water bath with automatic temperature regulation, preventing fluctuations in temperature during the experiment. Water temperature was measured with a calibrated thermometer (Luxtron, California, USA). For applying the heat-shock (45°C, 8 min), animals were sedated with isoflurane (2% in air) and their left leg was introduced in the water while the rest of their body was lying on isolation material to prevent heating of the mouse.
2.10. In vivo bioluminescence (BLI) and fluorescence reflectance imaging (FRI)
BLI and FRI were performed sequentially using a NightOWL II LB 983 calibrated system equipped with a NC 100 CCD deep-cooled camera (Berthold Technologies, Bad Wildbad, Germany). Mice, before and after heat-shock, were first injected intraperitoneally with D-luciferin (2.9 mg in 100 µL PBS, Promega, Madison, Wisconsin, USA,) and sedated 7 min later with isoflurane (2%). Bioluminescence images (2 min exposure, 4 × 4 binning) and photographs (100 ms exposure) were taken 8 min after the luciferin injection. A low light emitting standard (Glowell, LUX biotechnology, UK) was placed next to the animal during each image acquisition to provide a constant reference for BLI. Next, fluorescence images (1 sec exposure, 1 × 1 binning) and photographs (100 ms exposure) were obtained. The mPlum protein (excitation and emission maxima: 590 and 649 nm, respectively) was epi-illuminated at 590/20 nm and the fluorescence emission was detected at 680/30 nm. For both BLI and FRI, mice were imaged individually and placed in the center of a constant field of view. Pseudo-color luminescent images representing the spatial distribution of emitted photons were generated using IndiGO 2 software (Berthold Technologies) and superposed to the corresponding photographs. BLI and FRI analyses were done semi-automatically by first placing a small region of interest (ROI) on the heated leg. The mean light intensity (in photons.s−1.mm−2) was measured within this ROI.
2.11. Statistical analysis
Statistical significance (***P<0.01, **P between 0.01 and 0.05, *P between 0.05 and 0.1) was calculated using unpaired, heteroscedastic, one-tailed t test.
3. Results
3.1. Time course of heat-induced mRNA levels for Hsp70, lucF and mPlum in MEF
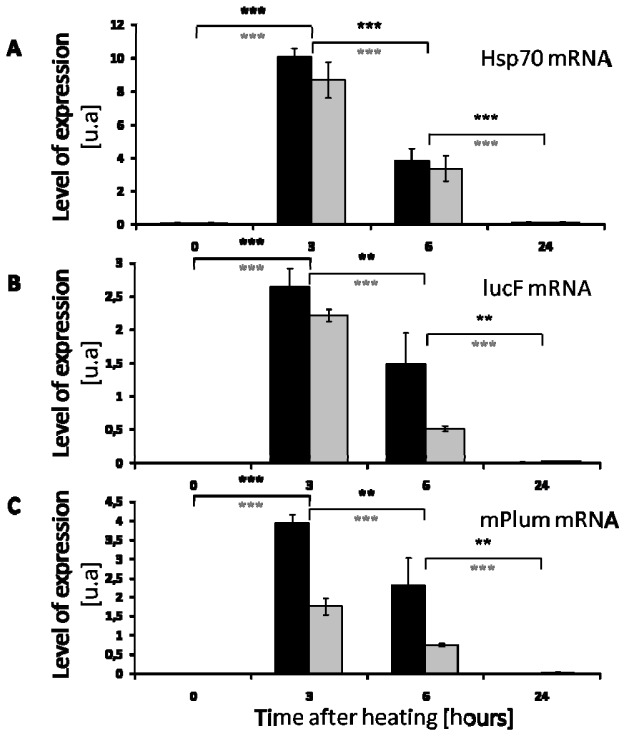
MEF were isolated from transgenic embryos (E.14) obtained by crossing Hspa1b-lucF (+/+) females with Hspa1b-mPlum (+/−) males. Each MEF line originating from a single embryo was genotyped by PCR for lucF and mPlum. Two MEF lines containing both lucF and mPlum transgenes were selected and amplified. MEF were heated at 44°C for 25 min and then incubated at 37°C and 5% CO2 for 3, 6, 24 and 48 h. Non-heated MEF were used as control. HSP70 mRNA refers to the two mRNA transcripts resulting from transcription of both Hspa1a and Hspa1b inducible promoter’s activities. The RT-qPCR setup was unable to discriminate between the 2 transcripts as they are almost identical [19]. The levels of mRNA encoding for HSP70 proteins (Hspa1a and Hspa1b) (Fig. 1(A) ), lucF (Fig. 1(B)) and mPlum (Fig. 1(C)) were determined by RT-qPCR. As shown in Fig. 1, the mRNA levels increased strongly 3 h after heating and then decreased at 6 h. The mRNA levels for HSP70, lucF and mPlum returned to the initial basal levels 24 h after heating. Reporter gene expression was not followed after 48 h because cells in culture rapidly reach confluence in the culture dish and cell splitting activates HSP70 promoters.
Fig. 1.
Time course of Hsp70, lucF and mPlum mRNA expression after heating 2 MEF lines (black bars = D1 and grey bars = E2) from distinct Hspa1b-lucF (+/+) Hspa1b-Plum (+/−) mouse embryos as determined by real time RT-PCR. (A) Hsp70 mRNA, (B) lucF mRNA and (C) mPlum mRNA (mean +/−SD; n = 3).
3.2. Time course of luciferase activity in MEF after heat-shock
The time course of luciferase activity after heat-shock was followed in the 2 MEF cells lines (D1 and E2 population). As shown in Fig. 2 , luciferase activity was detectable 3 h after heating and reached a peak at 6 h. The lucF activity decrease at 24 h and almost disappeared at 48 h.
Fig. 2.

Time course of lucF activity after heating MEF in vitro. Luciferase activity of cell lysates after heating (350,000 cells; 44°C, 25 min) MEF lines (dark = D1 and grey = E2 bars) from Hspa1b-lucF (+/+) Hspa1b-mPlum (+/−) mouse embryos (mean +/−SD; n = 3).
3.3. Time course of mPlum fluorescence in MEF after heat-shock
Heat-induced time-course changes in fluorescence related to mPlum expression in the MEF D1 cell line were followed by flow cytometry (Fig. 3 ). Before heat-shock (time 0), the MEF D1 cell line did not exhibit any fluorescent signal in the above filter settings for the mPlum channel (Fig. 3(B)). After heating, the fluorescent signal increased with time and was maximal at 48 h (Fig. 3(C)). Figure 3(D) shows the time evolution of mPlum fluorescence level in the D1 MEF cell line.
Fig. 3.
Time course of mPlum expression after heating MEF cell line. (A) Representative distribution of cell population (P1). (B) Representative cell distribution before heating the MEF line D1 (150,000 cells, 44°C, 25 min), (P2) is the positive population to the mPlum fluorescence. (C) Representative cell distribution 48 h post heating MEF (150,000 cells, 45°C, 8 min), (P2) is the positive population to the mPlum fluorescence. (D) Evolution of mPlum fluorescence in MEF as determined by flow cytometry after heating (mean +/−SD; n = 3).
3.4. In vivo imaging of transgenic mice after heat-shock
Transgenic mice resulting from crossing Hspa1b-lucF (+/+) females with Hspa1b-mPlum (+/−) males were followed by both BLI and FRI before and after heating of the left posterior leg in a water bath (45°C, 8 min). The right leg was used as non-heated control. As illustrated in Fig. 4(A) , mice exhibited no BLI signal before heating. At 6 h post heating, a strong BLI signal restricted to the heated leg was found. At 30 h and 48 h post heating, BLI signal on the heated leg almost disappeared and no signal was detected at day 7. Fluorescence signal before heating was very weak (ex: 590/20 nm; em: 680/30 nm), and a similar signal was detected in the right and left legs (Fig. 4(B)). After heating, a very low FRI signal was detected at 6 h only on the heated leg, it became very intense at 30 h and 48 h and it was still observed at 7 days. The kinetics of BLI and FRI signal are presented in Fig. 4(C). Based on graphical assessment, half-life of the bioluminescence and fluorescence signals were evaluated to about 20 hours and 45 hours, respectively.
Fig. 4.
Time course of in vivo bioluminescence and fluorescence signals after heat-shock of Hspa1b-lucF (+/+) Hspa1b-mPlum (+/−) mice. Representative images of bioluminescence (A) and fluorescence (B) signals at different time after heating of the left leg (45°C, 8 min) of Hspa1b-lucF (+/+) Hspa1b-mPlum (+/−) mice. The color scales (identical for each image) show light intensity. (C) Time course of BLI (grey squares) and FRI (black circles) signal intensity of the mouse left leg after heating (45°C, 8 min) (mean +/−SD; n = 8 mice).
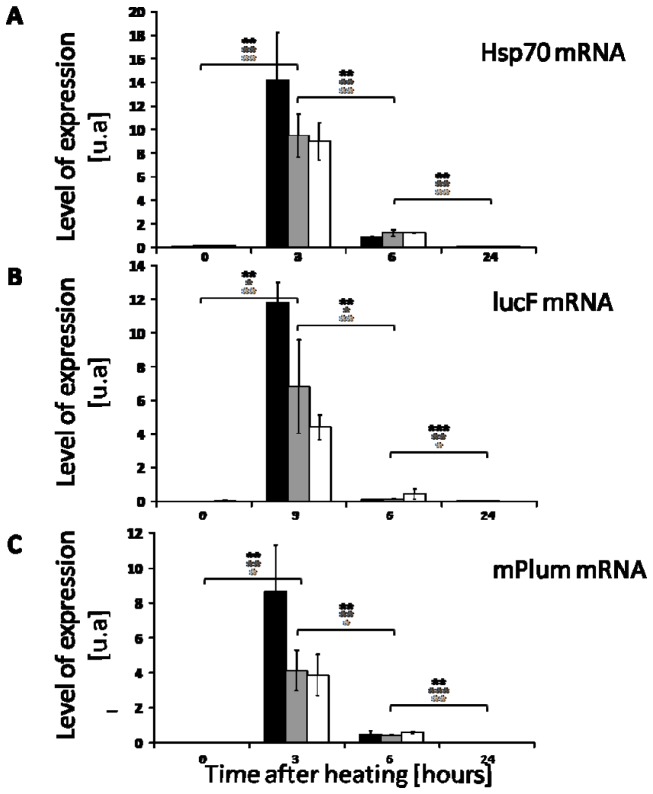
3.5. Time course of heat-induced mRNA levels for Hsp70, lucF and mPlum in skin of transgenic mice
At different times after heat shock, the mRNA levels for Hsp70, lucF and mPlum was determined by RT-qPCR. As shown on Fig. 5 , the mRNA level for the 3 mRNA species strongly increase 3 h after heat shock. It returned to the initial basal levels 24 h after heating.
Fig. 5.
Time course of Hsp70, lucF and mPlum mRNA expression in 3 mice Hspa1b-lucF (+/+) Hspa1b-mPlum (+/−) after heating (45°C, 8 min). Hsp70mRNA (A) lucF mRNA (B) and mPlum mRNA (C) content was determined by real time RT-PCR at different time after heating (mean +/−SD; n = 3).
4. Discussion
In the present paper, we crossed transgenic mice expressing two different optical reporter genes each driven by the inducible Hsp70 promoter (namely Hspa1b). Resulting mice were used for in vivo imaging and their embryos as a source of MEF to be handled in vitro. Mice and MEF exhibited exactly the same genetic background and transgenic modifications. Optical reporter lucF and mPlum were under transcriptional control of the mouse Hspa1b promoter but the molecular constructs were cloned separately and are slightly different. They both contain four tandemly arranged heat-shock elements for the binding of specific heat-shock transcription factors (HSF), an inverted CCAAT box, a TATA box, two binding sites for the common transcription factor Spl, and one binding site for the transcription factor AP2. However, the promoter sequence driving the lucF expression is 800 bp long [11] while the sequence driving the mPlum expression is only 516 bp [13].
The molecular constructs also differ in the sequence coding for the 5′UTR region of each mRNA. Finally, they are certainly inserted at different location inside the mouse genome. In spite of this molecular differences, the expression patterns for lucF and mPlum mRNA as determined both in vitro and in vivo after heat shock, were perfectly synchronic. In vitro, mRNAs peaked 3 h after heat-shock and then decreased to return to the basal rate at 24 h. The kinetics of reporter gene mRNA expression was also identical to the kinetics of endogenous Hsp70 mRNA. In vivo, expression of lucF, mPlum and Hsp70 mRNA is also transient and synchronic. Synchronicity of mRNA expression profiles was expected and consistent with the simultaneous activation of the 4 different promoters (2 reporters and 2 endogenous) by the same pool of nucleus-translocated HSF1 trimer that bind to HSE [20,21]. Thus, in vitro and in vivo, lucF and mPlum mRNA both report for the transient activation of the endogenous Hsp70 promoters after heat shock.
While mRNA expression for the 2 reporter gene is synchronic after heat-shock, the temporal pattern of lucF and mPlum protein detection clearly differ. LucF activity peaked 6 h after heating and returned to the basal level at 48 h both in vitro and in vivo. In contrast, the mPlum fluorescence signal appeared later and remained longer. The mPlum fluorescence in vitro, increase over the 48 h experimental period and in vivo the fluorescence signal peaked between 24 and 30 h then slowly decreased. The most surprising observation is that mPlum fluorescent signal still remains detectable up to 7 days in vivo.
Differences in lucF and mPlum expression patterns would clearly be attributable to divergences in translational and/or post-translational processes and in the fate of the proteins. In future studies, it would be interesting to investigate further whether these effects are associated to distinct subcellular localization of the reporter proteins. After heat-shock, the fluorescence signal of mPlum appeared later than lucF activity. LucF is known as a monomer that does not require any post-translational modification to provide mature enzyme that catalyze luciferin to generate the BLI signal. Regarding to the signal duration, the time course of BLI signal in vivo is consistent a with the short in vitro half-life (about 2 h) reported for lucF [22]. As already reported [12,16], these properties make lucF an extremely responsive reporter for in vivo imaging of the transient activation of the Hsp70 promoter and the transient expression of the heat induced mRNA. The time required for mPlum to acquire fluorescence properties is relatively long due to post-translational modifications and may be responsible for the delay for fluorescence detection after heat shock. The monomeric mPlum protein is very stable and is degraded very slowly. The mPlum half-life has been estimated between 100 and 200 min in vitro [23,24] but it is clearly longer in vivo since mRNA disappeared in one day but fluorescence signal remained up to 7 days after heating. The late growth and the long persistence of the fluorescence signal did not report the sharp transient properties of Hspa1b promoter activation and the mPlum mRNA content, but is the memory of earlier Hsp70 transcriptional activation. The delayed and long-term pattern of FRI signal mPlum-based may be useful to report heat-induced long-term process such as thermotolerance [25], and can be useful to compare Hsp70 induction with its delayed biological consequences in the same individuals and time frame. The FRI signal of mPlum has already been shown to report in vivo the Hsp70 response to brain ischemia while BLI signal was not detectable in the same context [13].
Half-life of proteins is usually determined in vitro, using biochemistry methods but they do not correspond necessarily with the in vivo observations. The fate of the protein in vivo may result from additional events such as for example protein interactions (binding, chaperone effect) or organelle storage that changes the protein half-life. Placing the reporter protein under transcriptional control of the Hsp70 promoter provides a convenient way to estimate the reporter protein’s half-life in vivo after heat-shock.
In conclusion, BLI and FRI provide in vivo, two different profiles of imaging for a unique and transient molecular event. BLI follow up of lucF activity provides early reporting of the transient activation of the Hspa1b promoter, with about a 3 h delay. Expression of mPlum occurred later (30 h) but the fluorescent signal was persistent allowing FRI signal up to 7 days after promoter activation. It thus provides a retrospective imaging of the stress event.
Acknowledgments
This work was supported by the “Conseil Régional d’Aquitaine” and the “Diagnostic Molecular Imaging Project, DIMI, EC-FP6-LSHB-CT-2005-512146”. The authors thank Dr. Pierre Costet (Bordeaux) for his efforts to maintain the double transgenic mice colony and Dr. Vincent Pitard (Bordeaux) for his technical assistance to FACS analysis, Dr. Claire Rome (Grenoble) and Dr. Claire Mazzocco (Bordeaux) for reading the manuscript.
References and links
- 1.Kang J. H., Chung J.-K., “Molecular-genetic imaging based on reporter gene expression,” J. Nucl. Med. 49(2Suppl 2), 164S–179S (2008). 10.2967/jnumed.107.045955 [DOI] [PubMed] [Google Scholar]
- 2.Grenier N., Brader P., “Principles and basic concepts of molecular imaging,” Pediatr. Radiol. 41(2), 144–160 (2011). 10.1007/s00247-010-1835-z [DOI] [PubMed] [Google Scholar]
- 3.Waerzeggers Y., Monfared P., Viel T., Winkeler A., Voges J., Jacobs A. H., “Methods to monitor gene therapy with molecular imaging,” Methods 48(2), 146–160 (2009). 10.1016/j.ymeth.2009.03.007 [DOI] [PubMed] [Google Scholar]
- 4.Li X., Zhao X., Fang Y., Jiang X., Duong T., Fan C., Huang C. C., Kain S. R., “Generation of destabilized green fluorescent protein as a transcription reporter,” J. Biol. Chem. 273(52), 34970–34975 (1998). 10.1074/jbc.273.52.34970 [DOI] [PubMed] [Google Scholar]
- 5.Voellmy R., “Transduction of the stress signal and mechanisms of transcriptional regulation of heat shock/stress protein gene expression in higher eukaryotes,” Crit. Rev. Eukaryot. Gene Expr. 4(4), 357–401 (1994). [PubMed] [Google Scholar]
- 6.Morimoto R. I., “Cells in stress: transcriptional activation of heat shock genes,” Science 259(5100), 1409–1410 (1993). 10.1126/science.8451637 [DOI] [PubMed] [Google Scholar]
- 7.Dreano M., Brochot J., Myers A., Cheng-Meyer C., Rungger D., Voellmy R., Bromley P., “High-level, heat-regulated synthesis of proteins in eukaryotic cells,” Gene 49(1), 1–8 (1986). 10.1016/0378-1119(86)90380-X [DOI] [PubMed] [Google Scholar]
- 8.Vekris A., Maurange C., Moonen C., Mazurier F., De Verneuil H., Canioni P., Voisin P., “Control of transgene expression using local hyperthermia in combination with a heat-sensitive promoter,” J. Gene Med. 2(2), 89–96 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Smith R. C., Machluf M., Bromley P., Atala A., Walsh K., “Spatial and temporal control of transgene expression through ultrasound-mediated induction of the heat shock protein 70B promoter in vivo,” Hum. Gene Ther. 13(6), 697–706 (2002). 10.1089/104303402317322267 [DOI] [PubMed] [Google Scholar]
- 10.Christians E. S., Benjamin I. J., “The stress or heat shock (HS) response: insights from transgenic mouse models,” Methods 35(2), 170–175 (2005). 10.1016/j.ymeth.2004.08.008 [DOI] [PubMed] [Google Scholar]
- 11.Christians E., Campion E., Thompson E. M., Renard J. P., “Expression of the HSP 70.1 gene, a landmark of early zygotic activity in the mouse embryo, is restricted to the first burst of transcription,” Development 121(1), 113–122 (1995). [DOI] [PubMed] [Google Scholar]
- 12.O’Connell-Rodwell C. E., Mackanos M. A., Simanovskii D., Cao Y.-A., Bachmann M. H., Schwettman H. A., Contag C. H., “In vivo analysis of heat-shock-protein-70 induction following pulsed laser irradiation in a transgenic reporter mouse,” J. Biomed. Opt. 13(3), 030501 (2008). 10.1117/1.2904665 [DOI] [PubMed] [Google Scholar]
- 13.de la Rosa X., Santalucía T., Fortin P.-Y., Purroy J., Calvo M., Salas-Perdomo A., Justicia C., Couillaud F., Planas A. M., “In vivo imaging of induction of heat-shock protein-70 gene expression with fluorescence reflectance imaging and intravital confocal microscopy following brain ischaemia in reporter mice,” Eur. J. Nucl. Med. Mol. Imaging 40(3), 426–438 (2013). 10.1007/s00259-012-2277-7 [DOI] [PubMed] [Google Scholar]
- 14.Kruse D. E., Mackanos M. A., O’Connell-Rodwell C. E., Contag C. H., Ferrara K. W., “Short-duration-focused ultrasound stimulation of Hsp70 expression in vivo,” Phys. Med. Biol. 53(13), 3641–3660 (2008). 10.1088/0031-9155/53/13/017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deckers R., Quesson B., Arsaut J., Eimer S., Couillaud F., Moonen C. T. W., “Image-guided, noninvasive, spatiotemporal control of gene expression,” Proc. Natl. Acad. Sci. U.S.A. 106(4), 1175–1180 (2009). 10.1073/pnas.0806936106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deckers R., Debeissat C., Fortin P.-Y., Moonen C. T. W., Couillaud F., “Arrhenius analysis of the relationship between hyperthermia and Hsp70 promoter activation: A comparison between ex vivo and in vivo data,” Int. J. Hyperthermia 28(5), 441–450 (2012). 10.3109/02656736.2012.674620 [DOI] [PubMed] [Google Scholar]
- 17.Velichko A. K., Markova E. N., Petrova N. V., Razin S. V., Kantidze O. L., “Mechanisms of heat shock response in mammals,” Cell. Mol. Life Sci. 70(22), 4229–4241 (2013). 10.1007/s00018-013-1348-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akamine R., Yamamoto T., Watanabe M., Yamazaki N., Kataoka M., Ishikawa M., Ooie T., Baba Y., Shinohara Y., “Usefulness of the 5′ region of the cDNA encoding acidic ribosomal phosphoprotein P0 conserved among rats, mice, and humans as a standard probe for gene expression analysis in different tissues and animal species,” J. Biochem. Biophys. Methods 70(3), 481–486 (2007). 10.1016/j.jbbm.2006.11.008 [DOI] [PubMed] [Google Scholar]
- 19.Günther E., Walter L., “Genetic aspects of the hsp70 multigene family in vertebrates,” Experientia 50(11-12), 987–1001 (1994). 10.1007/BF01923453 [DOI] [PubMed] [Google Scholar]
- 20.Voellmy R., “Feedback regulation of the heat shock response,” Handbook Exp. Pharmacol. 172(172), 43–68 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Powers M. V., Workman P., “Inhibitors of the heat shock response: Biology and pharmacology,” FEBS Lett. 581(19), 3758–3769 (2007). 10.1016/j.febslet.2007.05.040 [DOI] [PubMed] [Google Scholar]
- 22.Thompson J. F., Hayes L. S., Lloyd D. B., “Modulation of firefly luciferase stability and impact on studies of gene regulation,” Gene 103(2), 171–177 (1991). 10.1016/0378-1119(91)90270-L [DOI] [PubMed] [Google Scholar]
- 23.Baird G. S., Zacharias D. A., Tsien R. Y., “Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral,” Proc. Natl. Acad. Sci. U.S.A. 97(22), 11984–11989 (2000). 10.1073/pnas.97.22.11984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L., Jackson W. C., Steinbach P. A., Tsien R. Y., “Evolution of new nonantibody proteins via iterative somatic hypermutation,” Proc. Natl. Acad. Sci. U.S.A. 101(48), 16745–16749 (2004). 10.1073/pnas.0407752101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kregel K. C., “Heat shock proteins: modifying factors in physiological stress responses and acquired thermotolerance,” J. Appl. Physiol. 92(5), 2177–2186 (2002). [DOI] [PubMed] [Google Scholar]