Abstract
We describe the properties of a repetitive transposable element isolated from a chromosomal site close to the Adh (alcohol dehydrogenase gene) region of the Hawaiian Drosophila species, D. heteroneura. The cloned element is less than 2 kilobases in length. Although its polytene chromosome sites are constant in an individual, it shows a pattern of in situ hybridization that varies both within and between five species of the D. planitibia subgroup. These species are closely related, having diverged at various times from 0.5 to 5 million years ago. The distribution of the element appears to reflect the evolutionary relationships of the species except that the differences between D. planitibia and D. differens are ambiguous. Evidence of ragged excision of the element is found in one species.
Full text
PDF
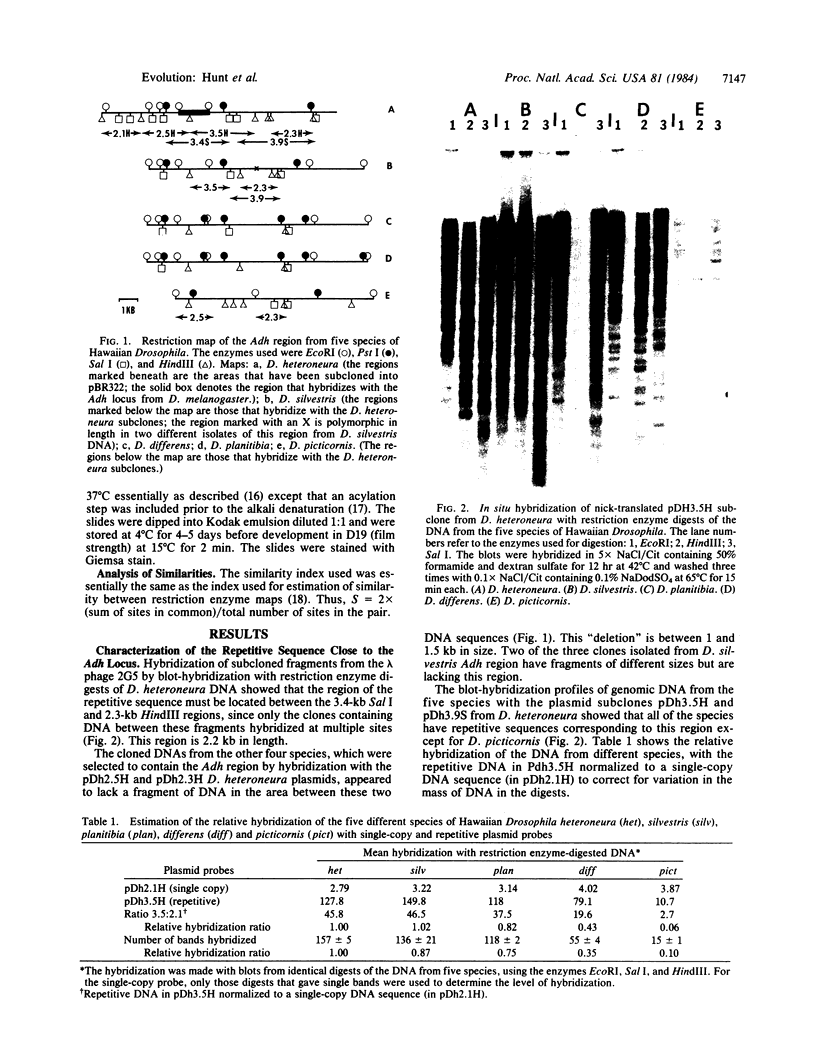

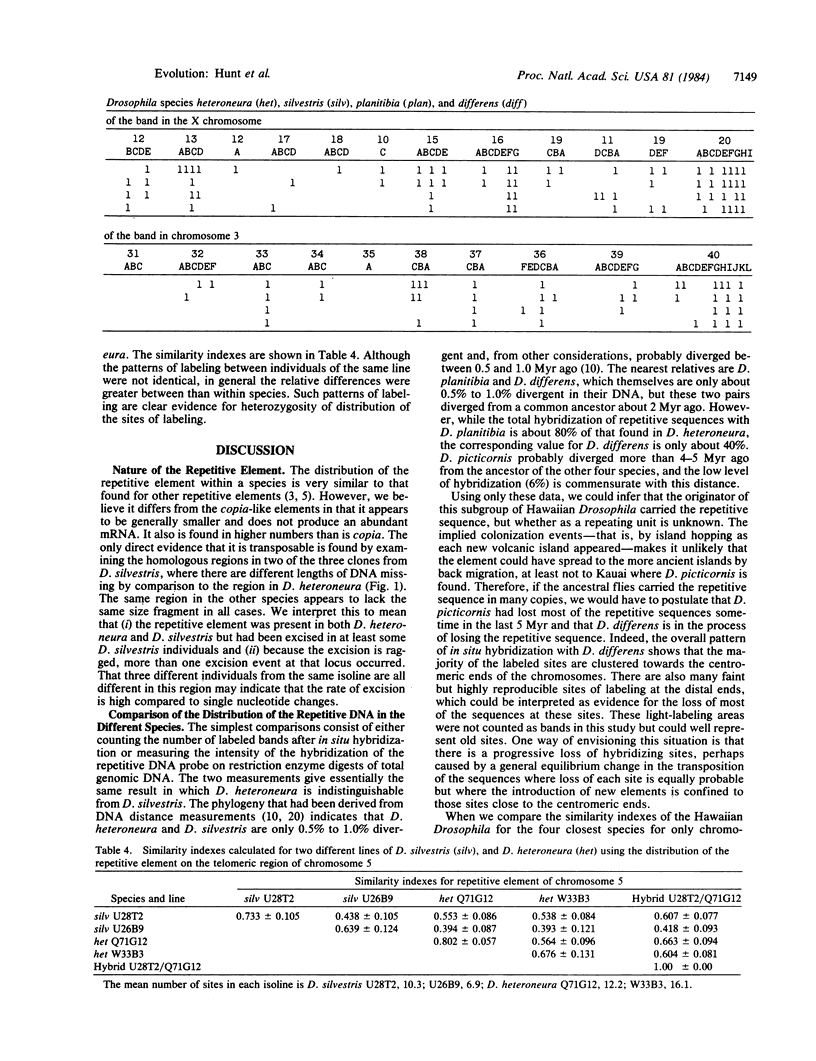

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blin N., Stafford D. W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976 Sep;3(9):2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner J. J., Pardue M. L. Ecdysone-stimulated RNA synthesis in imaginal discs of Drosophila melanogaster. Assay by in situ hybridization. Chromosoma. 1976 Oct 12;58(1):87–99. doi: 10.1007/BF00293443. [DOI] [PubMed] [Google Scholar]
- Dowsett A. P. Closely related species of Drosophila can contain different libraries of middle repetitive DNA sequences. Chromosoma. 1983;88(2):104–108. doi: 10.1007/BF00327329. [DOI] [PubMed] [Google Scholar]
- Dowsett A. P., Young M. W. Differing levels of dispersed repetitive DNA among closely related species of Drosophila. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4570–4574. doi: 10.1073/pnas.79.15.4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan D. J., Rubin G. M., Young M. W., Hogness D. S. Repeated gene families in Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1053–1063. doi: 10.1101/sqb.1978.042.01.106. [DOI] [PubMed] [Google Scholar]
- Flavell A. J., Ish-Horowicz D. Extrachromosomal circular copies of the eukaryotic transposable element copia in cultured Drosophila cells. Nature. 1981 Aug 13;292(5824):591–595. doi: 10.1038/292591a0. [DOI] [PubMed] [Google Scholar]
- Goldberg D. A. Isolation and partial characterization of the Drosophila alcohol dehydrogenase gene. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5794–5798. doi: 10.1073/pnas.77.10.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gvozdev V. A., Belyaeva E. S., Ilyin Y. V., Amosova I. S., Kaidanov L. Z. Selection and transposition of mobile dispersed genes in Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):673–685. doi: 10.1101/sqb.1981.045.01.085. [DOI] [PubMed] [Google Scholar]
- Hayashi S., Gillam I. C., Delaney A. D., Tener G. M. Acetylation of chromosome squashes of Drosophila melanogaster decreases the background in autoradiographs from hybridization with [125I]-labeled RNA. J Histochem Cytochem. 1978 Aug;26(8):677–679. doi: 10.1177/26.8.99471. [DOI] [PubMed] [Google Scholar]
- Hunt J. A., Carson H. L. Evolutionary relationships of four species of hawaiian Drosophila as measured by DNA reassociation. Genetics. 1983 Jun;104(2):353–364. doi: 10.1093/genetics/104.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt J. A., Hall T. J., Britten R. J. Evolutionary distances in Hawaiian Drosophila measured by DNA reassociation. J Mol Evol. 1981;17(6):361–367. doi: 10.1007/BF01734358. [DOI] [PubMed] [Google Scholar]
- Martin G., Wiernasz D., Schedl P. Evolution of Drosophila repetitive-dispersed DNA. J Mol Evol. 1983;19(3-4):203–213. doi: 10.1007/BF02099967. [DOI] [PubMed] [Google Scholar]
- Montgomery E. A., Langley C. H. Transposable Elements in Mendelian Populations. II. Distribution of Three COPIA-like Elements in a Natural Population of DROSOPHILA MELANOGASTER. Genetics. 1983 Jul;104(3):473–483. doi: 10.1093/genetics/104.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Li W. H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A. C., Rubin G. M. Drosophila genome organization: conserved and dynamic aspects. Annu Rev Genet. 1981;15:219–264. doi: 10.1146/annurev.ge.15.120181.001251. [DOI] [PubMed] [Google Scholar]
- Wensink P. C., Finnegan D. J., Donelson J. E., Hogness D. S. A system for mapping DNA sequences in the chromosomes of Drosophila melanogaster. Cell. 1974 Dec;3(4):315–325. doi: 10.1016/0092-8674(74)90045-2. [DOI] [PubMed] [Google Scholar]
- Young M. W. Middle repetitive DNA: a fluid component of the Drosophila genome. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6274–6278. doi: 10.1073/pnas.76.12.6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. W., Schwartz H. E. Nomadic gene families in Drosophila. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):629–640. doi: 10.1101/sqb.1981.045.01.081. [DOI] [PubMed] [Google Scholar]