Abstract
Filopodia, which are actin-rich finger-like membrane protrusions, have an important role in cell migration and tumor metastasis. Here we identify 13 novel calumenin (Calu) isoforms (Calu 3–15) produced by alternative splicing, and find that Calu-15 promotes filopodia formation and cell migration. Calu-15 shuttles between the nucleus and cytoplasm through interacting with importin α, Ran GTPase, and Crm1. The phosphorylation of the threonine at position 73 (Thr-73) by casein kinase 2 (CK2) is essential for the nuclear import of Calu-15, and either Thr-73 mutation or inhibition of CK2 interrupts its nuclear localization. In the nucleus, Calu-15 increases the transcription of growth differentiation factor-15 (GDF-15), a member of the transforming growth factor-β (TGF-β) superfamily, via binding to its promoter region. Furthermore, Calu-15 induces filopodia formation mediated by GDF-15. Together, we identify that Calu-15, a novel isoform of Calu with phosphorylation-dependent nuclear localization, has a critical role in promoting filopodia formation and cell migration by upregulating the GDF-15 transcription.
Keywords: calumenin-15, isoform, phosphorylation, GDF-15, transcription, filopodia
Filopodia, which are finger-like projections supported by tightly parallel-bundled filamentous actin, are involved in many key physiological and pathological processes,1, 2, 3 whereas abundant filopodia are considered strongly correlated with the enhanced cell migration and tumor metastasis.3, 4, 5 Filopodia contain receptors for various extracellular signals, including cytokines and growth factors, to sense the cell's surroundings during cell migration, then trigger the formation of initial adhesion sites, the recruitment of focal adhesion components, and finally the reorganization of the actin network.3, 6, 7, 8 Transforming growth factor-β (TGF-β), a cytokine, has been reported to induce the expression of paxillin, a focal adhesion complex component, and fascin, an actin filament-bundling protein, which in turn promotes filopodia formation and cell migration.4, 7 Therefore, TGF-β and fascin emerge as potential targets for cancer therapies.4, 9
Growth differentiation factor-15 (GDF-15) is a member of the TGF-β superfamily proteins.10, 11 It is also known as macrophage inhibitory cytokine-1, prostate-derived factor, placental bone morphogenetic protein, non-steroidal anti-inflammatory drug-activated gene-1, and placental TGF-β.10, 11 Similar to other TGF-β superfamily members, GDF-15 is first synthesized as a pro-protein, and then cleaved and secreted in its active mature form.12 It has been reported to have roles in kinds of cellular processes such as cell proliferation, migration, differentiation, and apoptosis.13, 14, 15, 16, 17 GDF-15 has both anti- and pro-tumorigenic functions according to different cell types and different developmental stages in the tumor.13, 17, 18 A high level of GDF-15 in serum is associated with a poor patient survival in colorectal and prostate carcinoma,19, 20 and the level of GDF-15 has also been reported to increase during the transition of colonic lesions to cancer initiation.19 However, there is also evidence that overexpression of GDF-15 induces the apoptosis of breast cancer cells and inhibits the tumorigenicity of LN-Z308 glioblastoma cell line.21, 22 Therefore, to unravel the molecular mechanisms that regulate GDF-15 is the key point for the understanding and treatment of malignant tumor.
Calumenin (Calu) belongs to the CREC protein family, which is composed of Cab45, reticulocalbin-1, reticulocalbin-2 (also known as ERC-55), reticulocalbin-3, and Calu.23, 24 These proteins all contain multiple EF-hand domains and are encoded respectively by five genes.23, 24 Interestingly, most of these genes produce isoforms by alternative mRNA splicing and these isoforms usually have different subcellular localizations and physiological functions.23, 24 The CALU gene has been reported to produce two isoforms, Calu-1 and Calu-2 (also known as crocalbin),25, 26 having equal length (315 amino acids (aa)) with only exons 3 and 4 exchanged.23 Recently, standard Edman degradation assay revealed that both Calu-1 and Calu-2 possess an N-terminal signal peptide (19 aa),27 which leads to their translocations into the ER or Golgi lumen.28 Functionally, Calu-1 and Calu-2 regulate vitamin K-dependent γ-carboxylation activity29 and participate in calcium cycling by interacting with ryanodine receptor-1 and ER Ca2+-ATPase SERCA2.30, 31
Here we set out to determine whether there are other Calu isoforms and identify that the CALU gene produces 13 novel isoforms (named Calu 3–15) by alternative splicing, with only one of them, Calu-15, possessing nuclear localization. Calu-15 shuttles between the nucleus and cytoplasm, and this process is regulated by its phosphorylation. Functionally, Calu-15 increases GDF-15 transcription level in the nucleus, which in turn induces filopodia formation and promotes cell migration.
Results
Identification of Calu isoforms and their subcellular localizations
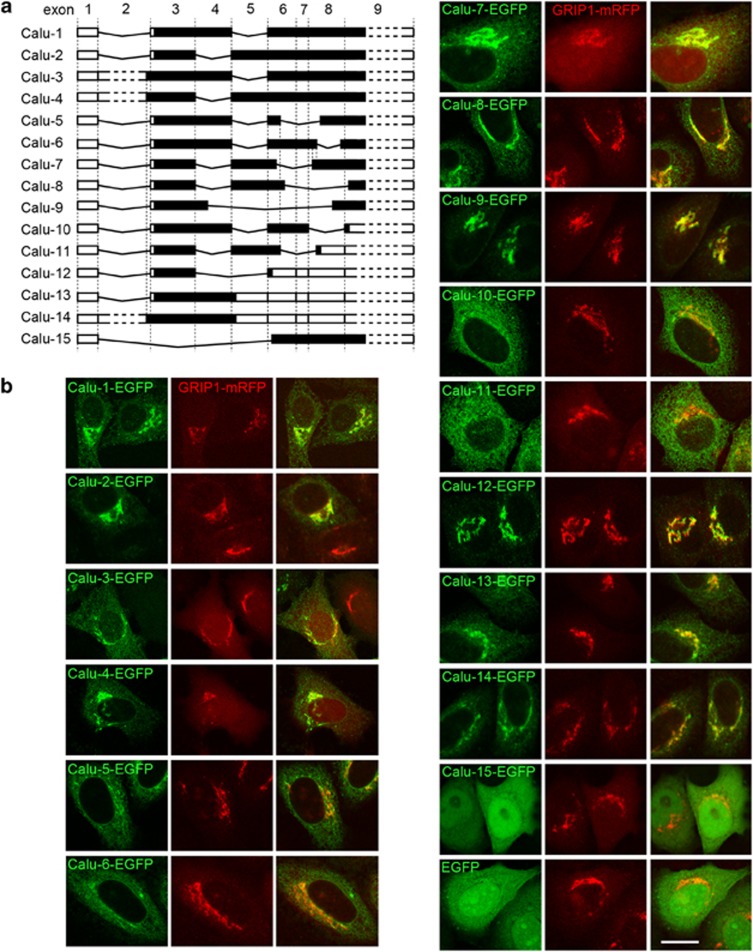
To search for more alternatively spliced isoforms of the CALU gene, we designed a pair of primers localized in the first and last exon (Supplementary Figure S1a), and performed PCR analysis. Several bands were amplified from the cDNA of HeLa cells (Supplementary Figure S1b), and 13 more splicing variants were identified by sequencing. We named these 13 novel variants Calu 3–15 (GenBank accession number: HM002604–HM002616; Figure 1a).
Figure 1.
Fifteen calumenin (Calu) isoforms and their subcellular localizations. (a) Schematic picture shows the exon organization of 15 Calu isoforms encoded by the CALU gene. The number at the top of the picture is the exon number. Black regions represent coding sequence, white regions represent untranslated mRNA sequence. (b) Subcellular localizations of the enhanced green fluorescent protein (EGFP) fusion proteins of Calu isoforms (green). GRIP1-mRFP (red) marks the Golgi apparatus. Bar, 10 μm
Previous reports have shown that Calu-1 and Calu-2 have eight exons with mRNA lengths of ∼3.4 kb.25, 26 When the exon organization of these isoforms was compared, we found that Calu-3 and Calu-4 possessed a novel exon (exon 2 in Figure 1a), leading to an increase of mRNA length to ∼4.2 kb. In addition, Calu-3 and Calu-4 had extra 8 aa at the N-terminus compared with Calu-1 and Calu-2 (Supplementary Figure S1c). As the N-terminal 19 aa of Calu-1 and Calu-2 are the signal peptide,27, 28 we examined whether the extra 8 aa interrupted this signal. The fluorescence assay showed that Calu-3–EGFP and Calu-4–EGFP also colocalized with GRIP1-mRFP, a Golgi apparatus marker, similar to Calu-1–EGFP and Calu-2–EGFP (Figure 1b). Furthermore, EGFP fusion with the N-terminal 27 aa (8+19 aa) of Calu-3 and Calu-4 also colocalized with GRIP1-mRFP (Supplementary Figure S1d). These results suggest that the N-terminal 27 aa of Calu-3 and Calu-4 still function as a signal peptide.
We also determined the subcellular distributions of the other identified isoforms by overexpressing their EGFP fusion proteins in HeLa cells. We found that only Calu-15 lacked the signal peptide and showed nuclear accumulation (Figure 1b). Together, the CALU gene encodes 15 isoforms, and only Calu-15 lacks the N-terminal signal peptide and shows nuclear localization.
Calu-15 shuttles between the nucleus and cytoplasm, mediated by importin-α, Ran GTPase, and Crm1
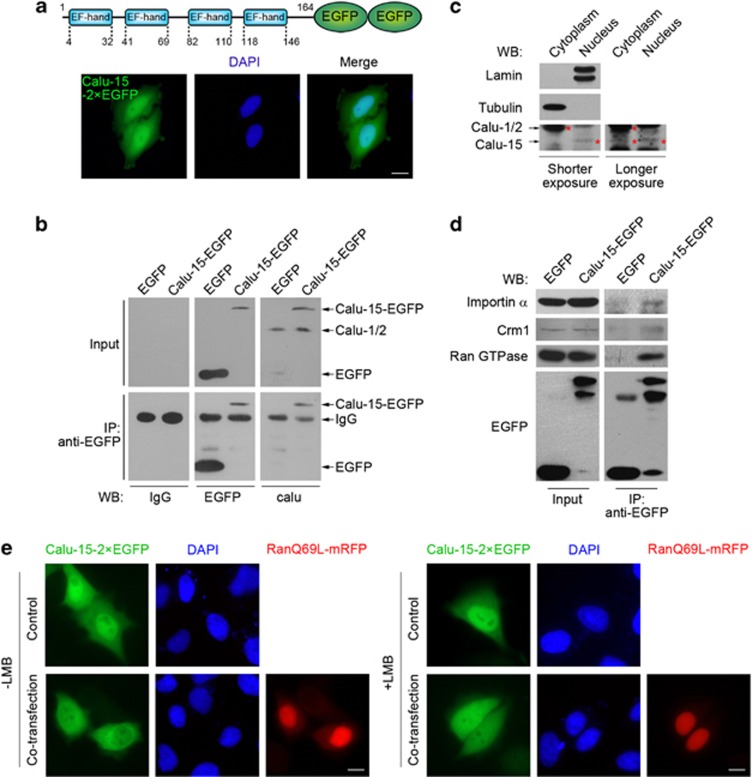
To confirm that Calu-15 shows the nuclear localization, we tagged it with 2 × EGFP and found that Calu-15–2 × EGFP was also localized in the nucleus (Figure 2a). Consistently, β-Gal–EGFP-tagged Calu-15 also localized in the nucleus, whereas the control β-Gal–EGFP did not (Supplementary Figure S2). Thus, Calu-15 is able to drag 2 × EGFP and β-Gal–EGFP into the nucleus.
Figure 2.
Calumenin (Calu)-15 shuttles between the nucleus and the cytoplasm, mediated by importin-α, Ran GTPase, and Crm1. (a) Subcellular localization of Calu-15–2 × enhanced green fluorescent protein (EGFP) fusion protein (green). Nuclear DNA was stained with DAPI (4',6-diamidino-2-phenylindole; blue). Bar, 10 μm. The upper panel shows the domain architecture of Calu-15–2 × EGFP. (b) Immunoprecipitation (IP) with antibody against EGFP were performed in the lysates of HEK293T cells expressing Calu-15–EGFP, followed by immunoblotting (western blotting (WB)) with antibody against Calu-1/2 (calu). Antibody against EGFP (EGFP) as a positive control. IgG as a negative control. (c) Immunoblottings (WB) of endogenous Calu-15 in the cytoplasmic and nuclear fraction from HeLa cells. Asterisks indicate the band of Calu-1/2 or Calu-15. Tubulin and lamin, respectively, labels the cytoplasmic and nuclear fraction. (d) IP with antibody against EGFP was performed in the lysates of HEK293T cells expressing Calu-15–EGFP or EGFP, followed by immunoblotting (WB) with the indicated antibodies. (e) HeLa cells cotransfected with Calu-15–2 × EGFP and RanQ69L-mRFP were treated with (right panels) or without (left panels) leptomycin B (LMB) for 3 h, then fixed for observation. Nuclear DNA was stained with DAPI (blue). Bar, 10 μm
To determine the subcellular localization of endogenous Calu-15, we used the antibody against Calu-1 and Calu-2, which could also recognize the overexpressed Calu-15–EGFP similar to the EGFP antibody (Figure 2b). Using this antibody, we analyzed the nuclear and cytoplasmic fraction proteins extracted from HeLa cells by western blotting, and found that Calu-1 and Calu-2 were in the tubulin-containing cytoplasmic fraction, whereas Calu-15 was mainly detected in the nuclear fraction along with the nuclear marker lamin (Figure 2c).
After confirming that Calu-15 is localized in the nucleus, we next investigated whether it was through the classical pathway to enter the nucleus. We performed immunoprecipitation experiments and found that Calu-15 was associated with importin-α (Figure 2d), a protein participating in the nuclear entry process.32, 33 Besides, immunoprecipitation showed that Calu-15 also interacted with Ran GTPase (Figure 2d), which has been reported to disassociate the importin–cargo complex and be required for the nuclear import process.33, 34 Moreover, the cotransfection with RanQ69L, which is a mutant defective in GTPase activity,35 largely inhibited the nuclear localization of Calu-15 (Figure 2e). Therefore, the nuclear import process of Calu-15 is mediated by importin-α and regulated by Ran GTPase.
Interestingly, Calu-15 also interacted with the exportin protein Crm1 (Figure 2d), which is reported to mediate nucleocytoplasmic transport.33, 36 Therefore, it is reasonable to hypothesize that Calu-15 might be exported from the nucleus through a Crm1-dependent pathway. To test this, we used leptomycin B (LMB), a small molecule that interferes Crm1–cargo binding,37 to suppress the Crm1-dependent nuclear export. In cells overexpressing Calu-15–2 × EGFP due to its high level of nuclear accumulation, LMB treatment could not increase the nuclear accumulation of Calu-15 (Figure 2e). However, in the presence of RanQ69L, LMB treatment obviously induced Calu-15 to be localized in the nucleus (Figure 2e). Thus, Calu-15 can shuttle between the nucleus and cytoplasm, and this is mediated by importin-α, Ran GTPase, and Crm1.
The nuclear localization of Calu-15 depends on phosphorylation at Thr-73 by CK2
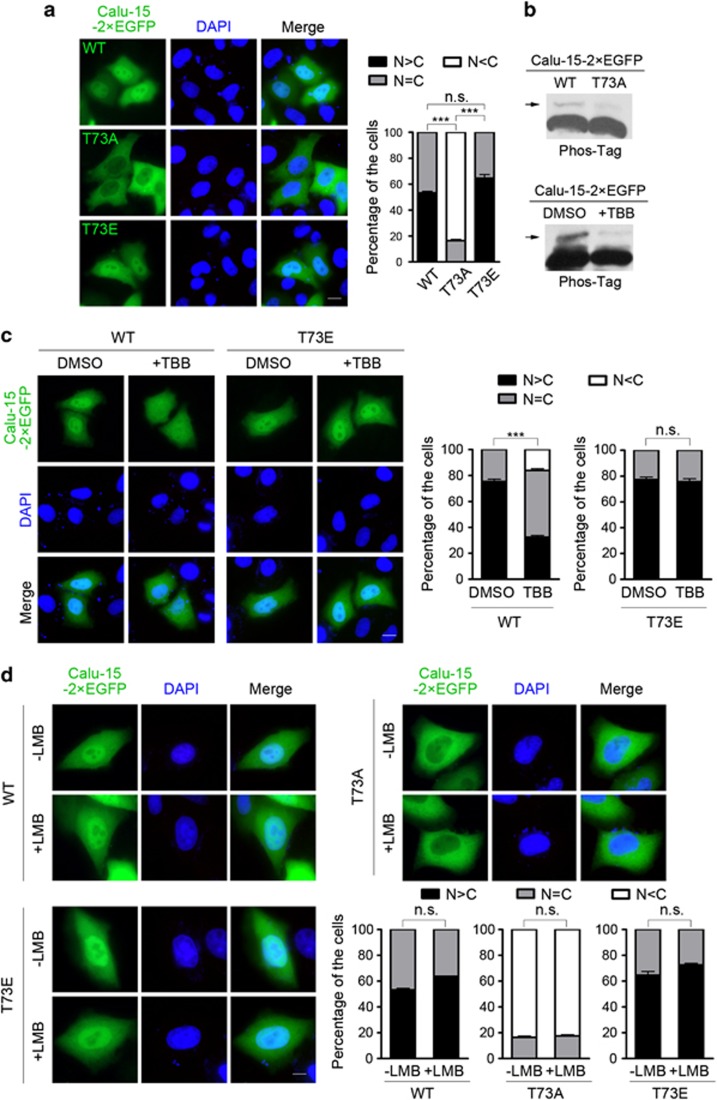
Given that phosphorylation can regulate the nuclear-cytoplasmic shuttling of proteins,38, 39 we analyzed the amino acid sequence of Calu-15 with bioinformatics tools40 and found that the threonine at position 73 (Thr-73) might be phosphorylated by casein kinase 2 (CK2). This finding prompted us to examine whether this site had a role in the nuclear-cytoplasmic shuttling of Calu-15. We mutated this amino acid to alanine (T73A) and glutamic acid (T73E) to mimic the unphosphorylated and phosphorylated state, respectively, and examined their subcellular localizations. To quantify the localization pattern, we classified it into three types: (1) cells with predominantly nuclear localization of EGFP fusion protein (N>C); (2) with both nuclear and cytoplasmic localization (N=C); and (3) with predominantly cytoplasmic localization (N<C). Compared with the wild-type Calu-15 (52.2% of N>C), Calu-15-T73A showed no nuclear localization (0% of N>C), whereas no significant difference was observed between Calu-15-T73E and wild type (Figure 3a, P=1.41 × 10−33 and P=0.10, respectively). Thus, these results suggest that the phosphorylation of Calu-15 at Thr-73 is related to its nuclear localization.
Figure 3.
Casein kinase 2 (CK2) phosphorylates calumenin (Calu)-15 at threonine at position 73 (Thr-73) and facilitates its nuclear localization. (a) The subcellular localization of Calu-15 wild-type (WT), T73A, or T73E. Left panels, representative pictures. Nuclear DNA was stained with DAPI (4',6-diamidino-2-phenylindole; blue). Bar, 10 μm. Right panel, the graph shows the percentage of cells with predominantly nuclear (N>C), both nuclear and cytoplasmic (N=C) or predominantly cytoplasmic (N<C) localization of enhanced green fluorescent protein (EGFP) fusion protein (n=3; >50 cells per experiment). Data are mean±S.E.M.; ***P<0.001; n.s., no significant difference (contingency table test for independence). (b) Lysates of HEK293T cells expressing Calu-15–2 × EGFP WT or T73A mutants (upper panel), and lysates of HEK293T cells expressing Calu-15–2 × EGFP treated with dimethylsulfoxide (DMSO) or 4,5,6,7-tetrabromo-2-azabenzimidazole (TBB; lower panel) were separated by Phos-tag SDS-polyacrylamide gel electrophoresis, then immunoblotted with antibody against EGFP. Arrows indicate the phosphorylated form. (c) HeLa cells transfected with Calu-15–2 × EGFP WT or T73E mutant were treated with DMSO or TBB for 10 h post transfection and then fixed for observation. Left panels, representative pictures. Nuclear DNA was stained with DAPI (blue). Bar, 10 μm. Right panel, the graphs show the percentage of cells with different localization patterns (n=3; >50 cells per experiment). Data are mean±S.E.M.; ***P<0.001; n.s., no significant difference (contingency table test for independence). (d) HeLa cells transfected with Calu-15–2 × EGFP WT, T73A, or T73E were treated with or without leptomycin B (LMB) before fixation and observation. Nuclear DNA was stained with DAPI (blue). Bar, 10 μm. The graphs show the percentage of cells with different localization patterns (n=3; >50 cells per experiment). Data are mean±S.E.M.; n.s., no significant difference (contingency table test for independence)
To confirm that Calu-15 is phosphorylated at Thr-73, we used SDS-PAGE with 50 μM Phos-tag acrylamide, in which phosphorylated bands migrate slower.41 An upshifted phosphorylated band of Calu-15 was detected in wild type, whereas this band almost disappeared in Calu-15-T73A mutant (Figure 3b). Next, to determine whether it is CK2 that phosphorylates the Thr-73 of Calu-15 and affects its nuclear localization, we used 4,5,6,7-tetrabromo-2-azabenzimidazole (TBB) to specifically inhibit CK2 activity42 immediately after the transfection of Calu-15–2 × EGFP. TBB treatment decreased the phosphorylation level of Calu-15 (Figure 3b), and the nuclear localization of Calu-15 was also significantly interrupted (from 75.3% to 32.3% of N>C) after 10 h TBB treatment (Figure 3c, P=5.27 × 10−10). However, TBB treatment did not change the subcellular localization of Calu-15-T73E mutant (Figure 3c, P=0.78), suggesting that CK2 promotes the nuclear localization of Calu-15 through phosphorylation at Thr-73. As the loss of phosphorylation at Thr-73 led to no nuclear localization of Calu-15 (Figure 3a), we then examined whether it was due to the decrease of the nuclear import or an increase of the nuclear export process. The treatment of the nuclear export inhibitor LMB could not increase the nuclear localization of Calu-15-T73A (Figure 3d, P=0.84), suggesting that the reason why the mutant Calu-15–T73A showed no nuclear localization is not the increase of nuclear export. Together, we conclude that the phosphorylation of Calu-15 at Thr-73 by CK2 is indispensable for its nuclear localization.
Calu-15 promotes GDF-15 transcription
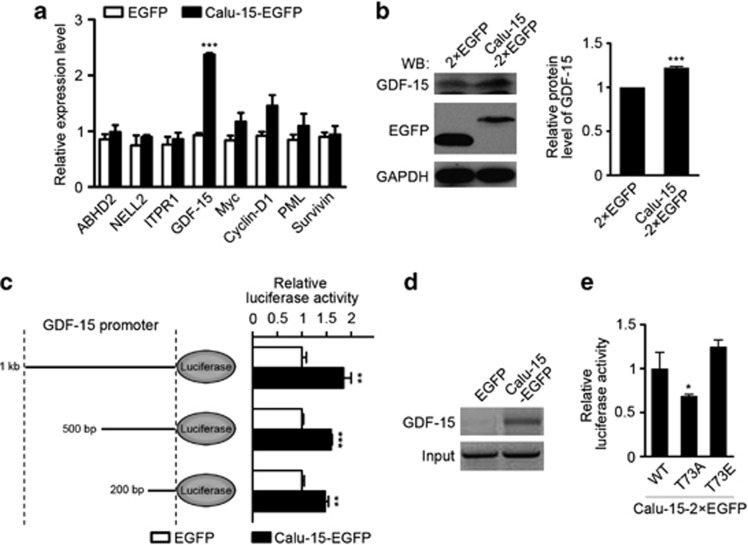
To unravel the physiological function of Calu-15 in the nucleus, we examined the transcription levels of a group of genes (Figure 4a). Quantitative real-time PCR analysis showed that GDF-15 mRNA level was significantly increased in the HeLa cell line stably expressing Calu-15–EGFP (Figure 4a, P=6.85 × 10−6, and Supplementary Figure S3). Correspondingly, GDF-15 protein level was also increased when overexpressing Calu-15–2 × EGFP, which is revealed by western blotting (Figure 4b, P=0.0006).
Figure 4.
Calumenin (Calu)-15 promotes the transcription of growth differentiation factor-15 (GDF-15). (a) Quantitative real-time PCR analysis of the relative mRNA expression levels of the indicated genes in the HeLa cells stably expressing Calu-15–EGFP (n=3). ABHD2, α-/β-hydrolase domain containing protein 2; NELL2, neural epidermal growth factor-like 2; ITPR1, inositol 1,4,5-triphosphate receptor, type 1; PML, promyelocytic leukemia; EGFP, as a control. Data are mean±S.E.M.; ***P<0.001 (unpaired two-tailed Student's t-test). (b) Lysates of HEK293T cells expressing Calu-15–2 × EGFP or 2 × EGFP were immunoblotted (western blotting (WB)) with the indicated antibodies. Right panel, the graph shows the relative protein level of GDF-15 (n=4). Data are mean±S.E.M.; ***P<0.001 (unpaired two-tailed Student's t-test). (c) Analysis of GDF-15 promoter activity by dual-luciferase reporter assay. The left panel shows the schematic structures of plasmids with different lengths of GDF-15 promoter followed by luciferase gene. The right graph indicates the relative luciferase activity in Calu-15–EGFP-overexpressing HeLa cells transfected with the indicated plasmids (n=4). EGFP, as a control. Data are mean±S.E.M.; **P<0.01, ***P<0.001 (unpaired two-tailed Student's t-test). (d) Chromatin immunoprecipitation (ChIP) assay of HeLa cells expressing Calu-15–EGFP or EGFP. (e) Quantification of the GDF-15 promoter activity in HeLa cells expressing Calu-15–2 × EGFP wild type (WT), T73A, or T73E (n=3). Data are mean±S.E.M.; *P<0.05 (unpaired two-tailed Student's t-test)
To investigate whether Calu-15 changes the GDF-15 expression level through interacting with its promoter, we performed dual-luciferase reporter assay and found that under the control of the promoter of GDF-15 (1 kb) the relative luciferase activity was largely increased in Calu-15–EGFP-expressing cells (Figure 4c, P=3.67 × 10–3). Further analysis showed that only a 200-bp region of the GDF-15 promoter was sufficient (Figure 4c, P=1.13 × 10–3). Moreover, ChIP assay confirmed the interaction between Calu-15 and the promoter of GDF-15 (Figure 4d). To further verify our findings, we showed that the non-phosphorylation mimic mutant T73A, with cytoplasmic localization pattern, was insufficient to increase the relative luciferase activity, which was under the control of the 200-bp region of the GDF-15 promoter, whereas both the wild-type and the phosphorylation mimic mutant T73E could (Figure 4e, P=0.0337). Together, these results suggest that nuclear Calu-15 regulates the GDF-15 transcription via binding to its promoter region.
Calu-15 facilitates filopodia formation through a GDF-15-mediated pathway
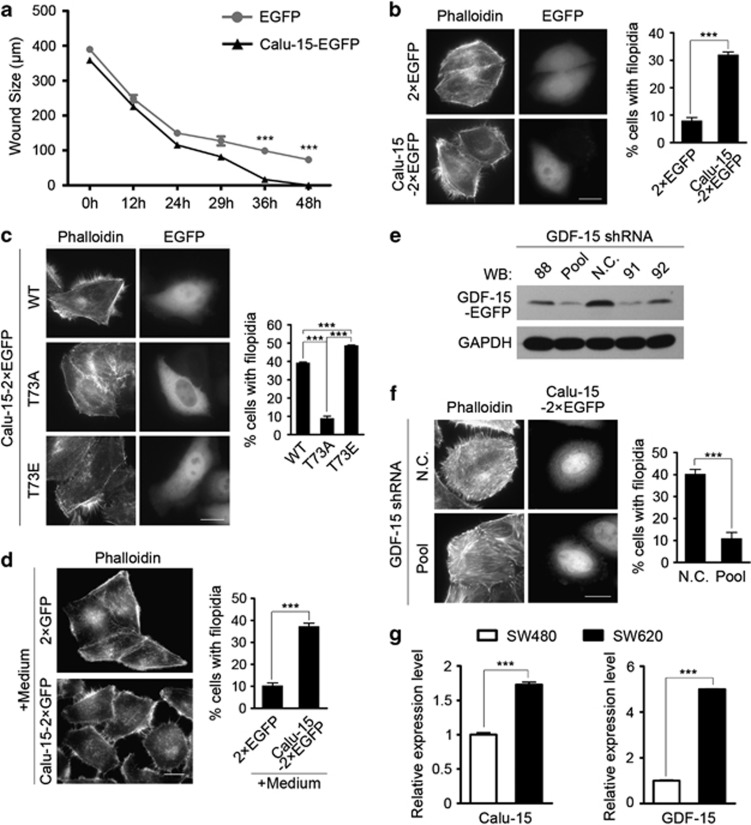
Interestingly, we found that overexpression of Calu-15–EGFP promoted cell migration in the wound-healing assay (Figure 5a and Supplementary Figure S4). Besides, Calu-15 also significantly increased the number and size of filopodia (Figure 5b, P=9.59 × 10–5), which facilitates cell migration through highly active actin rearrangement.13 Accordingly, the phosphorylation mimic mutant T73E also induced filopodia formation, whereas the unphosphorylation mimic mutant T73A failed (Figure 5c, P=1.46 × 10–5, P=5.03 × 10−6 and P=3.84 × 10−5, respectively). All these observations are consistent with previous reports that GDF-15 is involved in the regulation of cell migration.13 Considering that GDF-15 is a cytokine and is secreted into the extracellular space, we then tested the conditioned medium from Calu-15–2 × EGFP-expressing HEK293T cells and the filopodia formation was also induced (Figure 5d, P=1.38 × 10–4). Moreover, we knocked down GDF-15 in Calu-15–2 × EGFP-overexpressing HeLa cells and found that the depletion of GDF-15 inhibited the function of Calu-15 on filopodia formation (Figure 5f, P=9.20 × 10–4). Collectively, these data suggest that Calu-15 can induce filopodia formation through GDF-15, which in turn promotes cell migration.
Figure 5.
Calumenin (Calu)-15 promotes cell migration and facilitates filopodia formation in a growth differentiation factor-15 (GDF-15)-dependent pathway. (a) Wound sizes of HeLa cells expressing Calu-15–EGFP or EGFP at the indicated time points (n=3). Data are mean±S.E.M.; ***P<0.001 (unpaired two-tailed Student's t-test). (b) HeLa cells transfected with Calu-15–2 × EGFP or 2 × EGFP were stained with Atto 565 phalloidin to show the localization and distribution of filamentous (F)-actin. Left panel, representative pictures. Bar, 10 μm. Right panel, the graph shows the percentage of cells with filopodia (n=3; >100 cells per experiment). Data are mean±S.E.M.; ***P<0.001 (unpaired two-tailed Student's t-test). (c) HeLa cells transfected with Calu-15–2 × EGFP wild type (WT), T73A, or T73E were stained with Atto 565 phalloidin. Left panel, representative pictures. Bar, 10 μm. Right panel, the graph shows the percentage of cells with filopodia (n=3; >100 cells per experiment). Data are mean±S.E.M.; ***P<0.001 (unpaired two-tailed Student's t-test). (d) Conditioned medium collected from HEK293T cells transfected with Calu-15–2 × EGFP or 2 × EGFP were added into the cultures of HeLa cells, and then the HeLa cells were stained with Atto 565 phalloidin. Left panel, representative pictures. Bar, 10 μm. Right panel, the graph shows the percentage of cells with filopodia (n=3; >100 cells per experiment). Data are mean±S.E.M.; ***P<0.001 (unpaired two-tailed Student's t-test). (e) At 48 h after GDF-15 shRNA transfection, the protein level of GDF-15–EGFP in HEK293T cells were examined by immunoblotting (western blotting (WB)). N.C., negative control; 88, 91, and 92 indicate GDF-15 shRNA with different target sequences; Pool, a mixture of GDF-15 shRNA 88, 91, and 92. (f) At 48 h after cotransfection with Calu-15–2 × EGFP and GDF-15 shRNA pool (Pool, lower panels) or N.C. shRNA (upper panels), HeLa cells were stained with Atto 565 phalloidin. Left panel, representative pictures. Bar, 10 μm. Right panel, the graph shows the percentage of cells with filopodia (n=3; >100 cells per experiment). Data are mean±S.E.M.; ***P<0.001 (unpaired two-tailed Student's t-test). (g) Quantitative real-time PCR analysis of the relative mRNA expression levels of Calu-15 and GDF-15 in SW480 and SW620 cancer cells (n=5). Data are mean±S.E.M.; ***P<0.001 (unpaired two-tailed Student's t-test). For b, c, d and f, to quantify the number of cells with filopodia, cells were scored positive when presenting at least 10 filopodia ≥4 μm in length
To further confirm the correlation among Calu-15, GDF-15, and tumor metastatic potential, we performed quantitative real-time PCR analysis in colorectal cancer cell lines SW480 and SW620, which were from the same patient. Compared with the SW480 primary clone, the SW620 that originated from the metastatic lymph node shows a higher tumor metastatic potential.43,44 Consistently, both Calu-15 and GDF-15 mRNA levels were significantly increased in SW620 cells (Figure 5g, P=2.25 × 10−7 and P=5.13 × 10−14, respectively), suggesting a significant correlation among Calu-15, GDF-15, and tumor metastatic potential.
Discussion
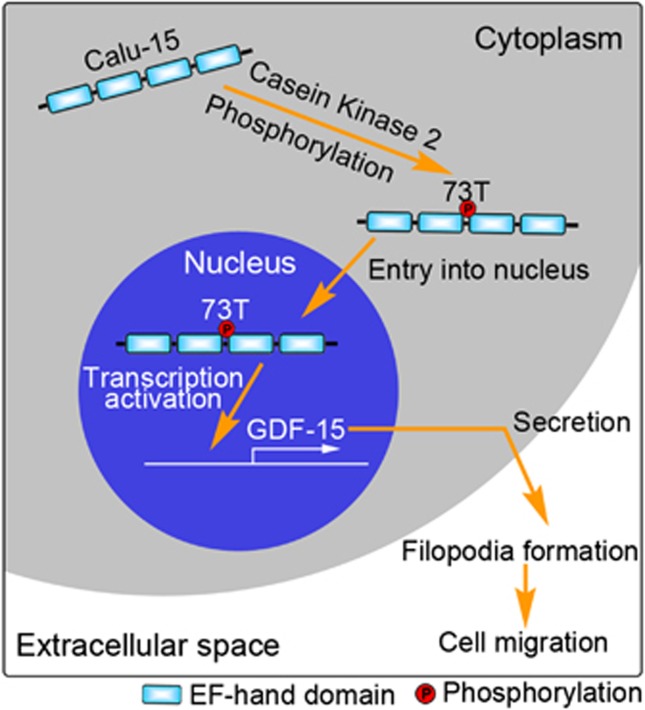
In this study, we found that the CALU gene encodes 13 novel isoforms besides Calu-1 and Calu-2. Among them, only Calu-15 shows nuclear localization. We demonstrate that Calu-15 is phosphorylated at Thr-73 by CK2, and then enters the nucleus to promote the transcription of GDF-15 via binding to its promoter, resulting in enhanced filopodia formation and cell migration (Figure 6). Moreover, the entry of Calu-15 into the nucleus is mediated by importin-α and Ran GTPase, whereas its export from the nucleus is mediated by Crm1.
Figure 6.
Proposed working model of calumenin (Calu)-15. Calu-15 undergoes phosphorylation at threonine at position 73 (Thr-73) by casein kinase 2 (CK2) and then promotes its entry into the nucleus. In the nucleus, Calu-15 can activate the transcription of growth differentiation factor-15 (GDF-15) via binding to its promoter region, and then leads to the formation of filopodia, which in turn promotes cell migration
One gene can produce more than one isoform to function either similarly or differently.23 Our results show that the CALU gene produces 15 isoforms in human cells by alternative splicing, while 13 of them are first identified. Among these 15 isoforms, Calu 1–14 all possess an N-terminal signal peptide and might localize in the lumen of the endomembrane system. However, only Calu-15, lacking the N-terminal 151 aa, shows nuclear accumulation and has novel functions as a transcription regulator. Interestingly, the homologue gene of Drosophila melanogaster, SCF, also encodes a nuclear isoform DmSCF (DNA supercoiling factor) through alternative splicing.45 Therefore, it suggests that the production of a nuclear isoform of the CALU gene is evolutionarily conserved and has a significant physiological function.
In eukaryotic cells, nuclear import receptors such as importin-α and -β recognize the nuclear localization signal (NLS) of the proteins and then form an import complex with them to enter the nucleus,33 whereas exportins such as Crm1 recognize the nuclear export signal (NES) and form an export complex to exit.33, 36 In our experiment, Calu-15 interacts with both importin-α and Crm1. However, we have not identified any classical NLS and NES in Calu-15, suggesting that there may be other proteins to mediate its nuclear-cytoplasmic shuttling, or that Calu-15 contains non-classical NLS and NES. Besides, we show that the phosphorylation of Calu-15 at Thr-73 by CK2 is indispensable for its nuclear localization, which is similar to recent reports that nuclear-cytoplasmic shuttling can be regulated by protein modification such as phosphorylation and dephosphorylation.38, 39 CK2 has been reported to regulate the nuclear export of S6K1 II,46 but in our study it acts as a regulator to facilitate the nuclear import of Calu-15. However, we still do not know whether the phosphorylation at Thr-73 facilitates the binding of Calu-15 directly to importin-α or to some other scaffold proteins. More detailed mechanism needs further investigation.
GDF-15 functions as a cytokine, participating in various cellular processes.13, 14, 15, 16, 17 Meanwhile, it has pleiotropic functions in cancer progression.17 Because of its importance, the transcription level of GDF-15 is under stringent control of various regulatory pathways. Many transcription factors have been reported to regulate its expression, such as p53, early growth response protein 1 (EGR-1), Sp1, nuclear factor-κB, poly (ADP-ribose) polymerase-1, and hypoxia-inducible factor-1α.14, 47, 48, 49, 50, 51 Our results show that Calu-15, as a new GDF-15 regulator, binds to the promoter region of GDF-15, and our preliminary data suggests that other factors might be required for this binding (data not shown). The binding region of Calu-15 to GDF-15 we identified ranges from −168 to +32 bp around the transcription start site, in which it also contains one p53- (at +21 bp), three Sp1- (within the −133 to +41 bp), and two EGR-1- (within the −73 to −51 bp) binding sites.21, 47, 48 Whether Calu-15 functions coordinately with them to promote the expression of GDF-15 remains unclear, however, it is evident that this 200-bp region of GDF-15 promoter is crucial for its transcription. In addition, we do not exclude the possibility that Calu-15 regulates the transcription activities of other genes.
GDF-15 has been reported to promote filopodia formation and metastasis of human prostate cancer cells through the focal adhesion kinase (FAK)-RhoA signaling pathway.13 Here we show that Calu-15 functionally promotes cell migration by facilitating filopodia formation and this function depends on GDF-15. Therefore, future research to investigate whether Calu-15 also promotes filopodia formation through FAK-RhoA signaling pathway could facilitate our understanding of the detailed mechanism. Besides, the function of GDF-15 in tumor progression is multiple but the regulatory mechanism of GDF-15 in cancer cells is still incompletely understood, whereas tumor cells with abundant filopodia are identified to be more malignant and invasive during tumor progression.3, 4, 5 Considering the above two facts, Calu-15, as a novel regulator of GDF-15, might have a pro-tumorigenic role in cancer development and serve as a new target for cancer therapy.
Materials and Methods
Cell lines and culture
HeLa, HEK293T, and SW480 cells were cultured at 37 °C with 5% CO2 in Dulbecco's modified Eagle's medium (GIBCO BRL, Grand Island, NY, USA) containing 10% heat-inactivated fetal bovine serum.52 SW620 cells were cultured at 37 °C without CO2 in Leibovitz's L-15 medium (GIBCO BRL) containing 10% heat-inactivated fetal bovine serum.
RNA isolation and cDNA cloning
Cells were homogenized in TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and the total RNA was isolated as described in Molecular Cloning (third version). Subsequently, total RNA and Superscript III Reverse Transcriptase (Invitrogen) were used to synthesize the first-strand cDNA, according to the manufacturers' instructions. The cDNAs synthesized were then used for Calu cloning and quantitative real-time PCR analysis. All PCR products were delivered for sequencing (Invitrogen).
Vector construction
To gain all the transcripts of Calu isoforms, specific primers were synthesized and isoforms were cloned into the EcoRI and SalI site of the pEGFP-N3 vector (Clontech Laboratories, Mountain View, CA, USA). 2 × EGFP plasmid was constructed by cloning EGFP into the KpnI and SmaI site of the pEGFP-N3 vector. All the other mutants of Calu-15 were cloned into the EcoRI and SalI site of the 2 × EGFP vector. The promoter of GDF-15 was cloned into the pGL3-Basic vector (Promega, Madison, WI, USA).
Antibodies
Mouse anti-calu polyclonal antibody was produced previously.52 Rabbit anti-EGFP polyclonal antibody was produced in our lab and was used in immunoblotting and immunoprecipitation. Rabbit anti-importin-α, rabbit anti-lamin-A and -C polyclonal antibody, and mouse anti-Ran GTPase polyclonal antibody were from Professor CM Zhang (Peking University). Mouse anti-α-tubulin monoclonal antibody (DM1α) was from Sigma (St. Louis, MO, USA). Mouse anti-GAPDH monoclonal antibody (ab8245) was from Abcam (Cambridge, UK). Mouse anti-GFP monoclonal antibody (clone 1E4) was from MBL (Woburn, MA, USA). Rabbit anti-GDF-15 monoclonal antibody (D2A3) was from Cell Signaling Technology (Danvers, MA, USA).
Immunoblotting analysis
Immunoblotting analysis were preformed as described.52 Briefly, the proteins were transferred onto PVDF membranes (Millipore, Billerica, MA, USA) in a semidry transfer cell (Bio-Rad, Hercules, CA, USA) after gel separation by SDS-PAGE. Next, the membranes were blocked in 3–5% milk in TTBS for 30 min and probed with primary antibodies at 4 °C overnight. HRP-conjugated goat anti-mouse or anti-rabbit IgG (Jackson ImmunoResearch Laboratories, West Grove, PA, USA) were used as secondary antibodies. To test the phosphorylation, SDS-PAGE with 50 μM Phos-tag acrylamide AAL-107 (Wako Pure Chemical Industries, Osaka, Japan) was used.
Transfection, RNAi, and immunofluorescence
HeLa cells were transiently transfected by jetPEI (Polyplus, Illkirch, France) at ∼60% confluence following the manufacturer's instructions. At 24 h post transfection, cells were washed in PBS and then fixed with freshly prepared 4% PFA (paraformaldehyde) in PBS for 15 min at 37 °C. Subsequently, cells were permeabilized with 0.1% Triton X-100 in PBS buffer for another 15 min and blocked by 3% BSA in PBS for 30 min. Next, they were incubated with primary and secondary antibodies or Atto 565 phalloidin (Sigma), and observed under a fluorescence microscope (Olympus, Shinjuku, Tokyo, Japan) as previously described.53 For GDF-15 knockdown, cells were transfected with plasmids expressing short hairpin RNA targeting GDF-15 (88: 5′-GCTCCAGACCTATGATGACTT-3′ 91: 5′-CCGGATACTCACGCCAGAAGT-3′ and 92: 5′-CTATGATGACTTGTTAGCCAA-3′ Sigma), then they were lysed for western blotting or fixed for immunofluorescence at 48 h post transfection.
Immunoprecipitation assay
For the immunoprecipitation assay, HEK293T cells transfected with Calu-15–EGFP were lysed in the buffer (20 mM Tris-HCl, 150 mM NaCl, 0.1% NP-40, 1 mM DTT, 0.5 mM EGTA, 10% glycerol, 1 mM MgCl2, pH 7.9) containing a protease inhibitor cocktail (Roche, Basel, Switzerland). Next, the total cell lysates were incubated with anti-EGFP antibody at 4 °C overnight. The protein samples that combined with the Protein A Sepharose (GE Healthcare, Uppsala, Sweden) were collected and subjected to western blotting assay.
Nuclear and cytoplasmic extraction
HeLa cells were first incubated with buffer 1 (20 mM HEPES, 5 mM CH3COOK, 0.5 mM MgCl2, 1 mM DTT, pH 7.8) on ice for 10 min and then homogenized by 25 strokes of a glass Dounce homogenizer. The cytoplasmic fraction was gained by first centrifugation at 4000 r.p.m. for 5 min and second centrifugation of the supernatant at 20 000 × g for 30 min. The fraction remaining from the first centrifugation was incubated with buffer 2 (20 mM HEPES, 5 mM CH3COOK, 0.5 mM MgCl2, 1 mM DTT, 0.4 M NaCl, pH 7.8) at 4 °C for 9 min and then centrifuged at 14 000 r.p.m. for 30 min to gain the nuclear fraction. Nuclear and cytoplasmic fractions were then subjected to western blotting.
Quantitative real-time PCR
The ABI 7300 Detection System (Applied Biosystems, Foster City, CA, USA) was used to perform quantitative real-time PCR using the SYBR Green PCR Master Mix (Applied Biosystems) as previously described.54, 55 The primers used for quantitative real-time PCR were listed in Supplementary Table S1. GAPDH served as a reference control and the 2−ΔΔCT method was used.56
Dual-luciferase reporter assay
Different lengths of the promoter region of GDF-15 were amplified from genomic DNA and cloned into the pGL3-Basic vector. Next, 50 ng of the pGL3-Basic vectors containing different lengths of GDF-15 promoter were transfected into HeLa cells, together with the indicated plasmids and 50 ng of TK-Renilla receptor gene as an internal control. After 24 h, the luciferase activity of total cell lysates was tested by the Dual-Luciferase Reporter Assay System (Promega).
Chromatin immunoprecipitation assay
ChIP assay was performed as described.57 Briefly, cells were cross-linked by adding a final concentration of 1% formaldehyde for 10 min at room temperature and cross-linking was stopped by adding glycine to a final concentration of 125 mM. Cells were then washed with 1 × PBS, resuspended in cell lysis buffer (5 mM Pipes (KOH), 85 mM KCl, 0.5% NP-40, pH 8.0), and centrifuged to get the nuclei. The nuclear fraction was then resuspended in nuclear lysis buffer (50 mM Tris, 10 mM EDTA, 1% SDS, pH 8.1), sonicated, and centrifuged to get the supernatant containing chromatin shorter than 500 bp. The supernatant was diluted five-fold in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris, 167 mM NaCl, pH 8.1) and incubated with anti-EGFP antibody at 4 °C overnight. Protein A Sepharose beads (GE Healthcare) were used to collect the immune complex and then washed consecutively for 3–5 min with 1 ml of each solution: low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris, 150 mM NaCl, pH 8.1), high-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris, 500 mM NaCl, pH 8.1), and LiCl wash buffer (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris, pH 8.0), twice in 1 × TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0). The complexes were eluted by adding elution buffer (1% SDS, 0.1 M NaHCO3). Next, the formaldehyde cross-linking in the elutant was reversed and the DNA was purified by a Gel Extraction Kit (CWBIO, Beijing, China). The purified DNA fraction was then analyzed by PCR using appropriate primers.
Wound-healing assay
HeLa cells stably expressing EGFP or Calu-15–EGFP were planted on coverslips at the same density. A micropipette tip was used to scratch a wound. The detached cells were removed and the remaining cells were cultured at 37 °C. The wound sizes at the same site were recorded as the indicated time.
Acknowledgments
We thank Professor WG Zhu from the Peking University for SW480 cell line, Professor CM Zhang from the Peking University for the rabbit anti-importin-α, and rabbit anti-lamin-A and -C polyclonal antibody and mouse anti-Ran GTPase polyclonal antibody, and Professor IC Bruce for reading the manuscript. This work was supported by the National Natural Science Foundation of China (31271424) and the Major State Basic Research Development Program of China (973 Program; 2010CB833705).
Glossary
- Calu
calumenin
- ChIP
chromatin immunoprecipitation
- CK2
casein kinase 2
- EGR-1
early growth response protein 1
- FAK
focal adhesion kinase
- GDF-15
growth differentiation factor-15
- LMB
leptomycin B
- NLS
nuclear localization signal
- NES
nuclear export signal
- TBB
4,5,6,7-tetrabromo-2-azabenzimidazole
- TGF-β
transforming growth factor-β
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by G Melino
Supplementary Material
References
- Swaney KF, Huang CH, Devreotes PN. Eukaryotic chemotaxis: a network of signaling pathways controls motility, directional sensing, and polarity. Annu Rev Biophys. 2010;39:265–289. doi: 10.1146/annurev.biophys.093008.131228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. The cytoskeleton and cancer. Cancer Metastasis Rev. 2009;28:5–14. doi: 10.1007/s10555-008-9166-3. [DOI] [PubMed] [Google Scholar]
- Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- Sun J, He H, Xiong Y, Lu S, Shen J, Cheng A, et al. Fascin protein is critical for transforming growth factor beta protein-induced invasion and filopodia formation in spindle-shaped tumor cells. J Biol Chem. 2011;286:38865–38875. doi: 10.1074/jbc.M111.270413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignjevic D, Schoumacher M, Gavert N, Janssen KP, Jih G, Lae M, et al. Fascin, a novel target of beta-catenin-TCF signaling, is expressed at the invasive front of human colon cancer. Cancer Res. 2007;67:6844–6853. doi: 10.1158/0008-5472.CAN-07-0929. [DOI] [PubMed] [Google Scholar]
- Lidke DS, Lidke KA, Rieger B, Jovin TM, Arndt-Jovin DJ. Reaching out for signals: filopodia sense EGF and respond by directed retrograde transport of activated receptors. J Cell Biol. 2005;170:619–626. doi: 10.1083/jcb.200503140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Stewart JE, Jr., Bellis SL, Benveniste EN, Ding Q, Tachibana K, et al. TGF-beta1 up-regulates paxillin protein expression in malignant astrocytoma cells: requirement for a fibronectin substrate. Oncogene. 2001;20:7976–7986. doi: 10.1038/sj.onc.1204996. [DOI] [PubMed] [Google Scholar]
- Giancotti FG, Ruoslahti E. Integrin signaling. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- Akhurst RJ, Hata A. Targeting the TGFbeta signalling pathway in disease. Nat Rev Drug Discov. 2012;11:790–811. doi: 10.1038/nrd3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bootcov MR, Bauskin AR, Valenzuela SM, Moore AG, Bansal M, He XY, et al. MIC-1, a novel macrophage inhibitory cytokine, is a divergent member of the TGF-beta superfamily. Proc Natl Acad Sci USA. 1997;94:11514–11519. doi: 10.1073/pnas.94.21.11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottner M, Suter-Crazzolara C, Schober A, Unsicker K. Expression of a novel member of the TGF-beta superfamily, growth/differentiation factor-15/macrophage-inhibiting cytokine-1 (GDF-15/MIC-1) in adult rat tissues. Cell Tissue Res. 1999;297:103–110. doi: 10.1007/s004410051337. [DOI] [PubMed] [Google Scholar]
- Uchida K, Chaudhary LR, Sugimura Y, Adkisson HD, Hruska KA. Proprotein convertases regulate activity of prostate epithelial cell differentiation markers and are modulated in human prostate cancer cells. J Cell Biochem. 2003;88:394–399. doi: 10.1002/jcb.10327. [DOI] [PubMed] [Google Scholar]
- Senapati S, Rachagani S, Chaudhary K, Johansson SL, Singh RK, Batra SK. Overexpression of macrophage inhibitory cytokine-1 induces metastasis of human prostate cancer cells through the FAK-RhoA signaling pathway. Oncogene. 2010;29:1293–1302. doi: 10.1038/onc.2009.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Wang Y, Guan K, Y Sun. PTGF-beta, a type beta transforming growth factor (TGF-beta) superfamily member, is a p53 target gene that inhibits tumor cell growth via TGF-beta signaling pathway. Proc Natl Acad Sci USA. 2000;97:109–114. doi: 10.1073/pnas.97.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roninson IB. Tumor cell senescence in cancer treatment. Cancer Res. 2003;63:2705–2715. [PubMed] [Google Scholar]
- Ichikawa T, Suenaga Y, Koda T, Ozaki T, Nakagawara A. TAp63-dependent induction of growth differentiation factor 15 (GDF15) plays a critical role in the regulation of keratinocyte differentiation. Oncogene. 2008;27:409–420. doi: 10.1038/sj.onc.1210658. [DOI] [PubMed] [Google Scholar]
- Mimeault M, Batra SK. Divergent molecular mechanisms underlying the pleiotropic functions of macrophage inhibitory cytokine-1 in cancer. J Cell Physiol. 2010;224:626–635. doi: 10.1002/jcp.22196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Bauskin AR, Zaunders J, Brown DA, Pankhurst S, Russell PJ, et al. Macrophage inhibitory cytokine 1 reduces cell adhesion and induces apoptosis in prostate cancer cells. Cancer Res. 2003;63:5034–5040. [PubMed] [Google Scholar]
- Brown DA, Ward RL, Buckhaults P, Liu T, Romans KE, Hawkins NJ, et al. MIC-1 serum level and genotype: associations with progress and prognosis of colorectal carcinoma. Clin Cancer Res. 2003;9:2642–2650. [PubMed] [Google Scholar]
- Brown DA, Lindmark F, Stattin P, Balter K, Adami HO, Zheng SL, et al. Macrophage inhibitory cytokine 1: a new prognostic marker in prostate cancer. Clin Cancer Res. 2009;15:6658–6664. doi: 10.1158/1078-0432.CCR-08-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PX, Wong J, Ayed A, Ngo D, Brade AM, Arrowsmith C, et al. Placental transforming growth factor-beta is a downstream mediator of the growth arrest and apoptotic response of tumor cells to DNA damage and p53 overexpression. J Biol Chem. 2000;275:20127–20135. doi: 10.1074/jbc.M909580199. [DOI] [PubMed] [Google Scholar]
- Albertoni M, Shaw PH, Nozaki M, Godard S, Tenan M, Hamou MF, et al. Anoxia induces macrophage inhibitory cytokine-1 (MIC-1) in glioblastoma cells independently of p53 and HIF-1. Oncogene. 2002;21:4212–4219. doi: 10.1038/sj.onc.1205610. [DOI] [PubMed] [Google Scholar]
- Honore B. The rapidly expanding CREC protein family: members, localization, function, and role in disease. Bio Essays. 2009;31:262–277. doi: 10.1002/bies.200800186. [DOI] [PubMed] [Google Scholar]
- Honore B, Vorum H. The CREC family, a novel family of multiple EF-hand, low-affinity Ca(2+)-binding proteins localised to the secretory pathway of mammalian cells. FEBS Lett. 2000;466:11–18. doi: 10.1016/s0014-5793(99)01780-9. [DOI] [PubMed] [Google Scholar]
- Vorum H, Liu X, Madsen P, Rasmussen HH, Honore B. Molecular cloning of a cDNA encoding human calumenin, expression in Escherichia coli and analysis of its Ca2+-binding activity. Biochim Biophys Acta. 1998;1386:121–131. doi: 10.1016/s0167-4838(98)00089-2. [DOI] [PubMed] [Google Scholar]
- Hseu MJ, Yen CH, Tzeng MC. Crocalbin: a new calcium-binding protein that is also a binding protein for crotoxin, a neurotoxic phospholipase A2. FEBS Lett. 1999;445:440–444. doi: 10.1016/s0014-5793(99)00177-5. [DOI] [PubMed] [Google Scholar]
- Tsukumo Y, Tsukahara S, Saito S, Tsuruo T, Tomida A. A novel endoplasmic reticulum export signal: proline at the +2-position from the signal peptide cleavage site. J Biol Chem. 2009;284:27500–27510. doi: 10.1074/jbc.M109.021592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorum H, Hager H, Christensen BM, Nielsen S, Honore B. Human calumenin localizes to the secretory pathway and is secreted to the medium. Exp Cell Res. 1999;248:473–481. doi: 10.1006/excr.1999.4431. [DOI] [PubMed] [Google Scholar]
- Wajih N, Sane DC, Hutson SM, Wallin R. The inhibitory effect of calumenin on the vitamin K-dependent gamma-carboxylation system. Characterization of the system in normal and warfarin-resistant rats. J Biol Chem. 2004;279:25276–25283. doi: 10.1074/jbc.M401645200. [DOI] [PubMed] [Google Scholar]
- Sahoo SK, Kim T, Kang GB, Lee JG, Eom SH, Kim do H. Characterization of calumenin-SERCA2 interaction in mouse cardiac sarcoplasmic reticulum. J Biol Chem. 2009;284:31109–31121. doi: 10.1074/jbc.M109.031989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung DH, Mo SH, DH Kim. Calumenin a multiple EF-hands Ca2+-binding protein, interacts with ryanodine receptor-1 in rabbit skeletal sarcoplasmic reticulum. Biochem Biophys Res Commun. 2006;343:34–42. doi: 10.1016/j.bbrc.2006.02.115. [DOI] [PubMed] [Google Scholar]
- Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- Liu Q, Yu J, Zhuo X, Jiang Q, Zhang C. Pericentrin contains five NESs and an NLS essential for its nucleocytoplasmic trafficking during the cell cycle. Cell Res. 2010;20:948–962. doi: 10.1038/cr.2010.89. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, et al. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma RY, Tong TH, Cheung AM, Tsang AC, Leung WY, Yao KM. Raf/MEK/MAPK signaling stimulates the nuclear translocation and transactivating activity of FOXM1c. J Cell Sci. 2005;118 (Pt 4:795–806. doi: 10.1242/jcs.01657. [DOI] [PubMed] [Google Scholar]
- Lidke DS, Huang F, Post JN, Rieger B, Wilsbacher J, Thomas JL, et al. ERK nuclear translocation is dimerization-independent but controlled by the rate of phosphorylation. J Biol Chem. 2010;285:3092–3102. doi: 10.1074/jbc.M109.064972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Koike T. Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE. Nat Protoc. 2009;4:1513–1521. doi: 10.1038/nprot.2009.154. [DOI] [PubMed] [Google Scholar]
- Sarno S, Reddy H, Meggio F, Ruzzene M, Davies SP, Donella-Deana A, et al. Selectivity of 4,5,6,7-tetrabromobenzotriazole, an ATP site-directed inhibitor of protein kinase CK2 ('casein kinase-2') FEBS Lett. 2001;496:44–48. doi: 10.1016/s0014-5793(01)02404-8. [DOI] [PubMed] [Google Scholar]
- Chen ML, Liang LS, Wang XK. miR-200c inhibits invasion and migration in human colon cancer cells SW480/620 by targeting ZEB1. Clin Exp Metastasis. 2012;29:457–469. doi: 10.1007/s10585-012-9463-7. [DOI] [PubMed] [Google Scholar]
- Yehezkel G, Cohen L, Kliger A, Manor E, Khalaila I. O-linked beta-N-acetylglucosaminylation (O-GlcNAcylation) in primary and metastatic colorectal cancer clones and effect of N-acetyl-beta-D-glucosaminidase silencing on cell phenotype and transcriptome. J Biol Chem. 2012;287:28755–28769. doi: 10.1074/jbc.M112.345546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Aita N, Hayashi S, Okada K, Ohta T, Hirose S. DNA supercoiling factor localizes to puffs on polytene chromosomes in Drosophila melanogaster. Mol Cell Biol. 1998;18:6737–6744. doi: 10.1128/mcb.18.11.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panasyuk G, Nemazanyy I, Zhyvoloup A, Bretner M, Litchfield DW, Filonenko V, et al. Nuclear export of S6K1 II is regulated by protein kinase CK2 phosphorylation at Ser-17. J Biol Chem. 2006;281:31188–31201. doi: 10.1074/jbc.M602618200. [DOI] [PubMed] [Google Scholar]
- Baek SJ, Kim JS, Moore SM, Lee SH, Martinez J, Eling TE. Cyclooxygenase inhibitors induce the expression of the tumor suppressor gene EGR-1, which results in the up-regulation of NAG-1, an antitumorigenic protein. Mol Pharmacol. 2005;67:356–364. doi: 10.1124/mol.104.005108. [DOI] [PubMed] [Google Scholar]
- Baek SJ, Horowitz JM, Eling TE. Molecular cloning and characterization of human nonsteroidal anti-inflammatory drug-activated gene promoter. Basal transcription is mediated by Sp1 and Sp3. J Biol Chem. 2001;276:33384–33392. doi: 10.1074/jbc.M101814200. [DOI] [PubMed] [Google Scholar]
- Shim M, Eling TE. Protein kinase C-dependent regulation of NAG-1/placental bone morphogenic protein/MIC-1 expression in LNCaP prostate carcinoma cells. J Biol Chem. 2005;280:18636–18642. doi: 10.1074/jbc.M414613200. [DOI] [PubMed] [Google Scholar]
- Frizzell KM, Gamble MJ, Berrocal JG, Zhang T, Krishnakumar R, Cen Y, et al. Global analysis of transcriptional regulation by poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in MCF-7 human breast cancer cells. J Biol Chem. 2009;284:33926–33938. doi: 10.1074/jbc.M109.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg AJ, Rankin EB, Chan D, Razorenova O, Fernandez S, Giaccia AJ. Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene expression and tumor growth. Mol Cell Biol. 2010;30:344–353. doi: 10.1128/MCB.00444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Feng H, Zheng P, Shen B, Chen L, Liu L, et al. The intracellular transport and secretion of calumenin-1/2 in living cells. PLoS One. 2012;7:e35344. doi: 10.1371/journal.pone.0035344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Shen B, Zheng P, Feng H, Chen L, Zhang J, et al. Silkworm coatomers and their role in tube expansion of posterior silkgland. PLoS One. 2010;5:e13252. doi: 10.1371/journal.pone.0013252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Teng J, Shen B, Zhang W, Guo Y, Su X, et al. Characterization of kinesin-like proteins in silkworm posterior silk gland cells. Cell Res. 2010;20:713–727. doi: 10.1038/cr.2010.47. [DOI] [PubMed] [Google Scholar]
- Wang Q, Chen L, Shen B, Liu Y, Chen J, Teng J. The tau-like protein in silkworm (Bombyx mori) induces microtubule bundle formation. Front Biosci (Elite Ed) 2012;4:998–1008. doi: 10.2741/E435. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Xie W, Ling T, Zhou Y, Feng W, Zhu Q, Stunnenberg HG, et al. The chromatin remodeling complex NuRD establishes the poised state of rRNA genes characterized by bivalent histone modifications and altered nucleosome positions. Proc Natl Acad Sci USA. 2012;109:8161–8166. doi: 10.1073/pnas.1201262109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.