Abstract
Mutants, called p-, that result from extensive deletions of the 75-kilobase Saccharomyces cerevisiae mitochondrial genome arise at high frequency. The remaining mitochondrial DNA is amplified in the p- cells, often as head-to-tail multimers, producing a cell with the normal amount of mitochondrial DNA. In matings, some of these p- mutants exhibit zygotic hypersuppressiveness, excluding the wild-type mitochondrial genome (p+) from all the diploids that are produced. From a hypersuppressive p- strain, we isolated two mutants with reduced suppressiveness. These mutants, one moderately suppressive and one nonsuppressive, retain only 89 and 70 base pairs, respectively, of the wild-type mitochondrial genome. Their sequences consist of 100% A . T base pairs. Replication of DNA in the mitochondrion, formation and amplification of new deletion genomes, and exhibition of suppressiveness do not require a single G . C base pair.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blanc H., Dujon B. Replicator regions of the yeast mitochondrial DNA responsible for suppressiveness. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3942–3946. doi: 10.1073/pnas.77.7.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach J. R., Li Y. Y., Feldman J., Jayaram M., Abraham J., Nasmyth K. A., Hicks J. B. Localization and sequence analysis of yeast origins of DNA replication. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1165–1173. doi: 10.1101/sqb.1983.047.01.132. [DOI] [PubMed] [Google Scholar]
- Coen D., Deutsch J., Netter P., Petrochilo E., Slonimski P. P. Mitochondrial genetics. I. Methodology and phenomenology. Symp Soc Exp Biol. 1970;24:449–496. [PubMed] [Google Scholar]
- EPHRUSS B., GRANDCHAMP S. ETUDES SUR LA SUPPRESSIVIT'E DES MUTANTS 'A DEFICIENCE RESPIRATOIRE DE LA LEVURE. I. EXISTENCE AU NIVEAU CELLULAIRE DE DIVERS "DEGR'ES DE SUPPRESSIVIT'E". Heredity (Edinb) 1965 Feb;20:1–7. doi: 10.1038/hdy.1965.1. [DOI] [PubMed] [Google Scholar]
- Ephrussi B., Jakob H., Grandchamp S. Etudes Sur La SuppressivitE Des Mutants a Deficience Respiratoire De La Levure. II. Etapes De La Mutation Grande En Petite Provoquee Par Le Facteur Suppressif. Genetics. 1966 Jul;54(1):1–29. doi: 10.1093/genetics/54.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi B., de Margerie-Hottinguer H., Roman H. SUPPRESSIVENESS: A NEW FACTOR IN THE GENETIC DETERMINISM OF THE SYNTHESIS OF RESPIRATORY ENZYMES IN YEAST. Proc Natl Acad Sci U S A. 1955 Dec 15;41(12):1065–1071. doi: 10.1073/pnas.41.12.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard C., Strauss F., Bernardi G. Excision sequences in the mitochondrial genome of yeast. Nature. 1980 Jan 10;283(5743):218–220. doi: 10.1038/283218a0. [DOI] [PubMed] [Google Scholar]
- Goursot R., Mangin M., Bernardi G. Surrogate origins of replication in the mitochondrial genomes of ori-zero petite mutants of yeast. EMBO J. 1982;1(6):705–711. doi: 10.1002/j.1460-2075.1982.tb01234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudspeth M. E., Vincent R. D., Perlman P. S., Shumard D. S., Treisman L. O., Grossman L. I. Expandable var1 gene of yeast mitochondrial DNA: in-frame insertions can explain the strain-specific protein size polymorphisms. Proc Natl Acad Sci U S A. 1984 May;81(10):3148–3152. doi: 10.1073/pnas.81.10.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman B. C., Cramer J. H., Rownd R. H. Properties of a Saccharomyces cerevisiae mtDNA segment conferring high-frequency yeast transformation. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1578–1582. doi: 10.1073/pnas.79.5.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman B. C., Cramer J. H., Rownd R. H. The mitochondrial genome of Saccharomyces cerevisiae contains numerous, densely spaced autonomously replicating sequences. Gene. 1983 Dec;26(2-3):223–230. doi: 10.1016/0378-1119(83)90192-0. [DOI] [PubMed] [Google Scholar]
- Jacquier A., Dujon B. The intron of the mitochondrial 21S rRNA gene: distribution in different yeast species and sequence comparison between Kluyveromyces thermotolerans and Saccharomyces cerevisiae. Mol Gen Genet. 1983;192(3):487–499. doi: 10.1007/BF00392195. [DOI] [PubMed] [Google Scholar]
- Locker J., Lewin A., Rabinowitz M. The structure and organization of mitochondrial DNA from petite yeast. Plasmid. 1979 Apr;2(2):155–181. doi: 10.1016/0147-619x(79)90036-2. [DOI] [PubMed] [Google Scholar]
- Lukins H. B., Tate J. R., Saunders G. W., Linnane A. W. The biogenesis of mitochondria 26. Mitochondrial recombination: the segregation of parental and recombinant mitochondrial genotypes during vegetative division of yeast. Mol Gen Genet. 1973 Jan 18;120(1):17–25. doi: 10.1007/BF00332981. [DOI] [PubMed] [Google Scholar]
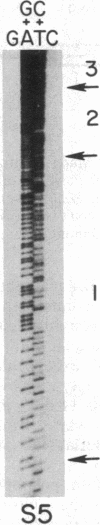
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Moustacchi E. Determination of the degree of suppressivity of Saccharomyces cerevisiae strain RD I A. Biochim Biophys Acta. 1972 Aug 16;277(1):59–60. doi: 10.1016/0005-2787(72)90351-6. [DOI] [PubMed] [Google Scholar]
- Nagley P., Gingold E. B., Lukins H. B., Linnane A. W. Biogenesis of mitochondria. XXV. Studies on the mitochondrial genomes of petite mutants of yeast using ethidium bromide as a probe. J Mol Biol. 1973 Aug 5;78(2):335–350. doi: 10.1016/0022-2836(73)90120-4. [DOI] [PubMed] [Google Scholar]
- Shore D., Baldwin R. L. Energetics of DNA twisting. I. Relation between twist and cyclization probability. J Mol Biol. 1983 Nov 15;170(4):957–981. doi: 10.1016/s0022-2836(83)80198-3. [DOI] [PubMed] [Google Scholar]
- Sor F., Fukuhara H. Unequal excision of complementary strands is involved in the generation of palindromic repetitions of rho- mitochondrial DNA in yeast. Cell. 1983 Feb;32(2):391–396. doi: 10.1016/0092-8674(83)90458-0. [DOI] [PubMed] [Google Scholar]
- Van Kreijl C. F., Bos J. L. The repeating nucleotide sequence in the repetitive mitochondrial DNA from a "low-density" petite mutant of yeast. Nucleic Acids Res. 1977 Jul;4(7):2369–2388. doi: 10.1093/nar/4.7.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zamaroczy M., Faugeron-Fonty G., Bernardi G. Excision sequences in the mitochondrial genome of yeast. Gene. 1983 Mar;21(3):193–202. doi: 10.1016/0378-1119(83)90002-1. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M., Marotta R., Faugeron-Fonty G., Goursot R., Mangin M., Baldacci G., Bernardi G. The origins of replication of the yeast mitochondrial genome and the phenomenon of suppressivity. Nature. 1981 Jul 2;292(5818):75–78. doi: 10.1038/292075a0. [DOI] [PubMed] [Google Scholar]