Abstract
LIM-homeodomain (LIM-HD) transcription factors have been extensively studied for their role in the development of the central nervous system. Their function is key to several developmental events like cell proliferation, differentiation and subtype specification. However, their roles in retinal neurogenesis remain largely unknown. Here we report a detailed expression study of LIM-HD transcription factors LHX9 and LHX2, LHX3 and LHX4, and LHX6 in the developing and mature mouse retina using immunohistochemistry and in situ hybridization techniques. We show that LHX9 is expressed during the early stages of development in the retinal ganglion cell layer and the inner nuclear layer. We also show that LHX9 is expressed in a subset of amacrine cells in the adult retina. LHX2 is known to be expressed in retinal progenitor cells during development and in Müller glial cells and a subset of amacrine cells in the adult retina. We found that the LHX2 subset of amacrine cells is not cholinergic and that a very few of LHX2 amacrine cells express calretinin. LHX3 and LHX4 are expressed in a subset of bipolar cells in the adult retina. LHX6 is expressed in cells in the ganglion cell layer and the neuroblast layer starting at embryonic stage 13.5 (E13.5) and continues to be expressed in cells in the ganglion cell layer and inner nuclear layer, postnatally, suggesting its likely expression in amacrine cells or a subset thereof. Taken together, our comprehensive assay of expression patterns of LIM-HD transcription factors during mouse retinal development will help further studies elucidating their biological functions in the differentiation of retinal cell subtypes.
Keywords: LIM-homeodomain, Lhx genes, retinogenesis, retinal development, transcription factors
The mammalian neural retina is comprised of six major neuronal cell types and one glial cell type. Structurally, the retina can be divided into three layers: the outer nuclear layer (ONL) containing cell bodies of rod and cone photoreceptors, the inner nuclear layer (INL) containing cell bodies of horizontal cells, bipolar cells, amacrine cells and Müller glial cells, and the ganglion cell layer (GCL) containing cell bodies of retinal ganglion cells and displaced amacrine cells (Livesey and Cepko 2001; Masland 2001; Hatakeyama and Kageyama 2004). Retinal neurons further display heterogeneity in morphology and functions in visual signal processing and are hence further classified into retinal cell subtypes (Masland 2001). For instance, bipolar cells are classified into rod and cone bipolar cells depending on the photoreceptor type they receive their synaptic input from, and further as ON and OFF bipolar cells based on their polarizing response to light stimulus (Masland 2012). Amacrine cells have also been classified into different categories based on the neurotransmitter type they express (Masland 2012). Amacrine cells can be GABAergic or Glycenergic or as recently discovered, neither (nGnG) (Kay, Voinescu et al. 2011). They can further be classified as cholinergic, tyrosine hydroxylas-expressing or parvalbumin (PV)-expressing amacrine cells, to name a few (Wassle 2004; Bhati, Lee et al. 2008; Voinescu, Kay et al. 2009). Several transmitter markers such as ChAT and GABA mark amacrine cell subtypes and markers such as PKCα and Goα distinguish between classes of bipolar cells (Wassle 2004), but they do not facilitate the labeling of cells undergoing differentiation from a pool of progenitors. This emphasizes the need for developmental biomarkers that can specifically label different subtypes of retinal cells. Several developmental markers that label subtypes of amacrine cells such as Isl1, Lmo4, Bhlhb5 (Feng, Xie et al. 2006; Elshatory, Deng et al. 2007a; Elshatory, Everhart et al. 2007b; Duquette, Zhou et al. 2010) and subtypes of bipolar cells such as Bhlhb4 and Bhlhb5 (Bramblett, Pennesi et al. 2004; Feng, Xie et al. 2006) have been recently identified. While the list of retinal cell subtypes is still growing, molecular markers that can identify and track them developmentally are mostly undiscovered.
LIM-homeodomain (HD) transcription factors are characterized by the presence of two protein binding zinc finger motifs, the LIM domains, located at the N-terminal of a central HD that specifically bind TAAT-containing DNA sequences. Owing to the presence of LIM domains, LIM-HD transcription factors also have the unique ability to form homomeric or heteromeric combinatorial complexes with other transcription factors (Bach 2000, Bhati, Lee et al. 2008, Dawid, Breen et al. 1998). Several proteins belonging to the LIM-HD family of transcription factors have been studied for their roles during the specification and differentiation of several central nervous system neurons (Reviewed in Hobert and Westphal, 2000 and Shirasaki and Pfaff, 2002).
The expression of some LIM-HD factors during retinogenesis has been previously studied. LHX1 (also known as LIM1), is expressed in horizontal cells and directs the migration of developing horizontal cells to their correct laminar position in the inner nuclear layer (Liu et al. 2000; Poche et al. 2007). The expression and function of the ISLET group of LIM-HD factors during retinogenesis has also been extensively studied. While ISL1 is expressed during retinal development and in retinal ganglion cells, cholinergic amacrine cells and ON-bipolar cells in the adult retina (Pan et al., 2008; Elshatory et al., 2007a; Elshatory et al., 2007b), ISL2 is expressed only in post-mitotic retinal ganglion cells (Pak et al. 2004). However, the expression of other LIM-HD factors during retinal development, have not been thoroughly characterized. Here we report the expression patterns of five of the LIM-HD proteins during the development of retina in mice: LHX9 and LHX2 belonging to the APTEROUS group, LHX3 and LHX4 belonging to the LIM-3 group and LHX6 belonging to the LHX6/7 group.
LHX9 and LHX2 belong to the APTEROUS group of LIM-HD transcription factors. LHX9 expression has been described in the mammalian system central nervous system structures such as spinal cord neurons, diencephalon, telencephalic vessels and dorsal mesencephalon (Retaux et al., 1999, Bertuzzi et al., 1999). Recently, LHX9 was also found to be expressed in a subset of hypocretin neurons in the hypothalamus and to have a crucial role in regulating somnolence (Dalal et al., 2013). Further, combinatorial expression of LIM-HD factors LHX9 and LHX2 has been demonstrated during the development of several systems (Nakagawa and O’Leary 2001, Wilson et al. 2008, Abellan et al. 2009, Peukert et al. 2011, Chatterjee et al. 2012).
The expression of LHX2 was first described during mammalian central nervous system development by Bourgouin et al., 1992 and Xu et al., 1993. Subsequently Porter et al., 1997 described the importance of LHX2 during the early development of the forebrain, eye and in erythrocyte development. LHX2 mutants are anophthalmic, display severe malformations of the cerebral cortex and are not viable since they present with defective erythropoiesis leading to severe anemia. Specifically, during eye development, LHX2 expression is noted in the optic vesicle as early as E8.5 and continues to be expressed in neural retinal progenitor cells (Porter et al., 1997). In the absence of LHX2, the specification of the optic vesicle occurs but development is arrested prior to the formation of optic cup (Porter et al., 2007, Yun et al., 2009, Hägglund et al., 2009). It has also been shown to interact with and activate other eye-field specific transcription factors. LHX2 interacts with PAX6, which in turn activates the expression of SIX6 in retinal progenitor cells (Tètreault et al., 2009). LHX2 has also been shown to link several extrinsic and intrinsic factors to co-ordinate multiple patterning events for the formation of the optic cup (Yun et al., 2009). Conditional disruption of Lhx2 shows that the expression of Lhx2 is not only necessary for optic cup formation but is also necessary for differentiation of the neuroretina (Roy et al., 2013) by facilitating a transition in competence states (Gordon et al., 2013).
LHX3 and LHX4 belong to the LIM-3 (Drosophila) group of LIM-HD proteins (Yamashita et al. 1997, Sharma et al. 1998, Hobert and Westphal, 2000). Pioneering work describing the combinatorial regulation of development by LHX3 and LHX4 has been widely studied in two systems: pituitary organogenesis (Sheng et al., 1997) and spinal cord motor neuron subtype development (Tsuchida et al. 1994, Sharma et al. 1998 and Thor et al. 1999).
LHX6 expression was first reported in the first branchial arch and the developing medial ganglionic eminence of the basal forebrain by Grigoriou et al., 1998, suggesting that LHX6 along with its closely related LIM-HD member – LHX7 might have functional roles in development of craniofacial structures and the forebrain. Soon after, Marin et al., 2000 provided further evidence implicating LHX6 along with LHX7 in the development of striatal interneuron subtype. Alifragis et al., 2004 and Liodis et al., 2007 further showed that LHX6 does not regulate the neurotransmitter choice of these interneurons but does control the tangential migration of the cortical interneurons. The role of LHX6 has also been extensively studied in the amygdala (Choi et al. 2005). The expression of LHX6 in the developing retina however has not been reported to date.
RESULTS AND DISCUSSION
Expression of Lhx9 and Lhx2
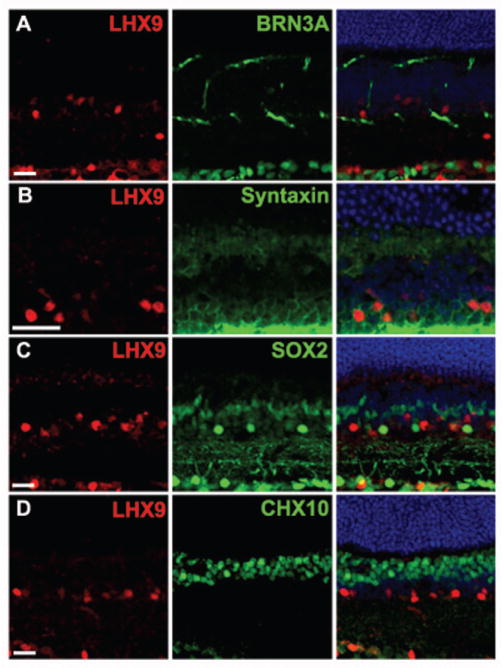
Atkinson-Leadbeater et al., 2009 briefly reported LHX9 expression in INL neurons of developing Xenopus retina. The identity of these cells was not further characterized and the expression of LHX9 in the mammalian retina has not been reported to date. Thus, we first looked at the expression of Lhx9 using in situ hybridization (Fig. 1). Retinogenesis in the mouse begins midway through the gestational period at E10 and continues through the second postnatal week. First, retinal ganglion cells differentiate from retinal progenitors at E11 followed by horizontal cells, cone photoreceptors and amacrine cells. Bipolar cells, rod photoreceptors and Müller glial cells are born in the later half of retinal development (Livesey and Cepko 2001). The expression of Lhx9 mRNA transcript was not detected at E12.5 or earlier (data not shown). At E13.5, a weak Lhx9 signal appeared in the retinal progenitor cells of the neuroblast layer (NBL) (Fig. 1A) and beginning at E15.5, strong expression of Lhx9 was observed in cells in both the GCL and NBL (Fig. 1B). This expression is sustained throughout embryonic development (E17.5: Fig. 1C) and postnatally (P5, P7: Fig. 1D, E). After the inner and outer nuclear layers are specified after P7, the expression of Lhx9 was noted at P15 or later to be present in cells in both the GCL and INL (Fig. 1F). Since amacrine cells and retinal ganglion cells are both present in the GCL and INL, the expression of Lhx9 could be in either of these two cell types or in both. As the onset of Lhx9 expression starts after E13.5, by which point most retinal progenitor cells have already started differentiating into retinal ganglion cells, and there are much fewer displaced retinal ganglion cells in the INL than there are displaced amacrine cells in the GCL, we theorized that the Lhx9 expressing cells are very likely a subset of amacrine cells. We thus characterized this expression in a mature retina (P30) using immunohistochemistry to colabel P30 retinal sections with LHX9 antibody along with retinal cell markers. Colabeling with anti-BRN3A, a marker for retinal ganglion cells (Wang, Mu et al. 2002; Pan, Yang et al. 2005), showed that the LHX9 positive cells in the GCL did not express BRN3A, hence excluding the expression of LHX9 in retinal ganglion cells (Fig. 2A). Colabeling for syntaxin, a pan amacrine cell marker (Inoue and Akagawa 1993), demonstrated that LHX9 is expressed in amacrine cells (Fig. 2B). Colabeling for SOX2 (Fig. 2C), a Müller glial cell marker (Lin, Ouchi et al. 2009) and CHX10 (Fig. 2D), a pan bipolar cell marker (Chen and Cepko 2000), confirmed that LHX9 is not expressed in Müller glial cells or bipolar cells. Taken together, the above results demonstrate that Lhx9 is exclusively expressed in a subset of amacrine cells.
Figure 1.
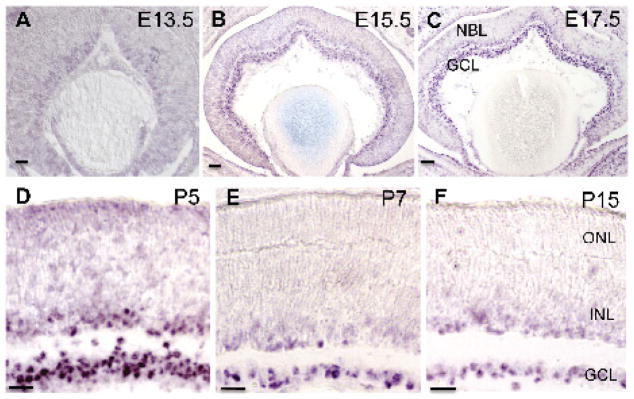
Expression of Lhx9 mRNA transcript in the developing retina. A: Expression of Lhx9 is faint and mostly undetectable at E13.5 in retinal progenitor cells. B: Strong expression of Lhx9 beings at E15.5 in the GCL and developing NBL. C-D: Expression of Lhx9 continues (E17.5, P5) in the GCL and developing NBL. E-F: Expression of Lhx9 is in cells in the GCL and cells in the INL postnatally (P7, P15). All scale bars are 50 μm.
Figure 2.
Characterization of LHX9 expressing cells in the adult retina (P30). A: Co-expression of LHX9 with BRN3A indicates that LHX9 might not be expressed in retinal ganglion cells. B: Co-localization of all LHX9 cells with syntaxin indicates that LHX9 expressing cells are amacrine cells. C: Co-expression of LHX9 with SOX2 does not reveal any LHX9 expressing Müller glial cells. D: Co-expression of LHX9 with CHX10 does not reveal any LHX9 expressing bipolar cells. All scale bars are 50 μm.
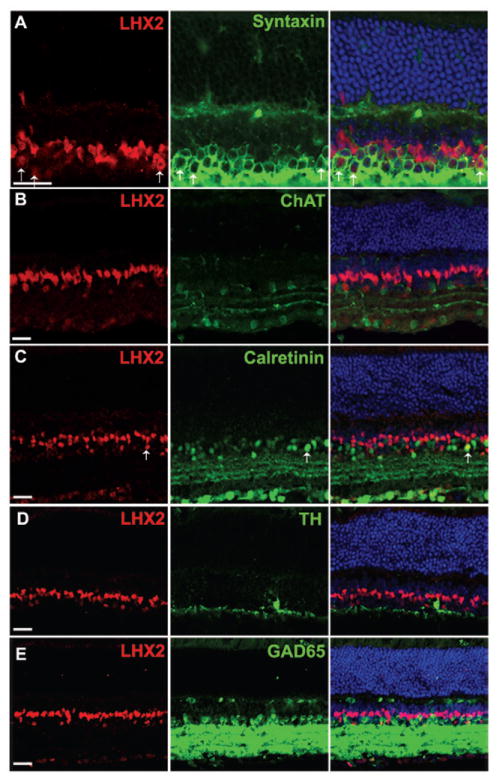
LHX2 expression is found to be confined to the INL of the adult retina by Porter et al., 1997. Recently de Melo et al., 2012 studied the expression of LHX2 in the adult retina. They find that LHX2 is expressed in Müller glial cells and a few amacrine cells in the INL of the mature retina. They also found that LHX2 maintains Müller glial cells in their quiescent state, while a conditional disruption of Lhx2 in Müller glial cells initiated a non-proliferative reactive gliosis. Here, we examined the expression of LHX2 in a subset of amacrine cells in the P30 adult retina (Fig. 3). Colabeling anti-LHX2 with the pan-amacrine cell marker anti-syntaxin showed that all LHX2 cells that were not Müller glial cells, were syntaxin-positive and are thus amacrine cells (Fig. 3A), consistent with the previous report (de Melo et al.). We further looked to see if these amacrine cells were starburst amacrine cells and expressed choline acetyle transferase (ChAT). We found that none of the LHX2-expressing amacrine cells was cholinergic (Fig. 3B). Further, very few amacrine cells co-localized with calretinin (Fig. 3C), which marks a subset of amacrine cells. LHX2 cells also do not co-localize with TH-expressing dopaminergic amacrine cells (Fig. 3D) or with GAD65 (Fig. 3E), which marks a large percentage of GABAergic amacrine cells. Though the possibility that LHX2 expressing amacrine cells are not GABAergic cannot be completely excluded, the above results do strongly suggest in favor of glycinergic amacrine cells – the other major subtype of amacrine cells in the mammalian retina.
Figure 3.
Characterization of LHX2 expressing amacrine cells in the adult retina (P30). A: Co-localization of LHX2 with syntaxin indicates the presence of LHX2 expressing amacrine cells (Arrows). B: Co-expression of LHX2 with ChAT shows that LHX2 expressing amacrine cells are not cholinergic. C: Co-expression of LHX2 with calretinin reveals that few LHX2 expressing amacrine cells co-localize calretinin (Arrow). D: LHX2 expressing amacrine cells do not co-localize with TH expressing dopaminergic amacrine cells. E: LHX2 expressing amacrine cells do not co-localize with GAD65, a GABAergic amacrine cell marker. All scale bars are 50μm.
Expression of Lhx3 and Lhx4
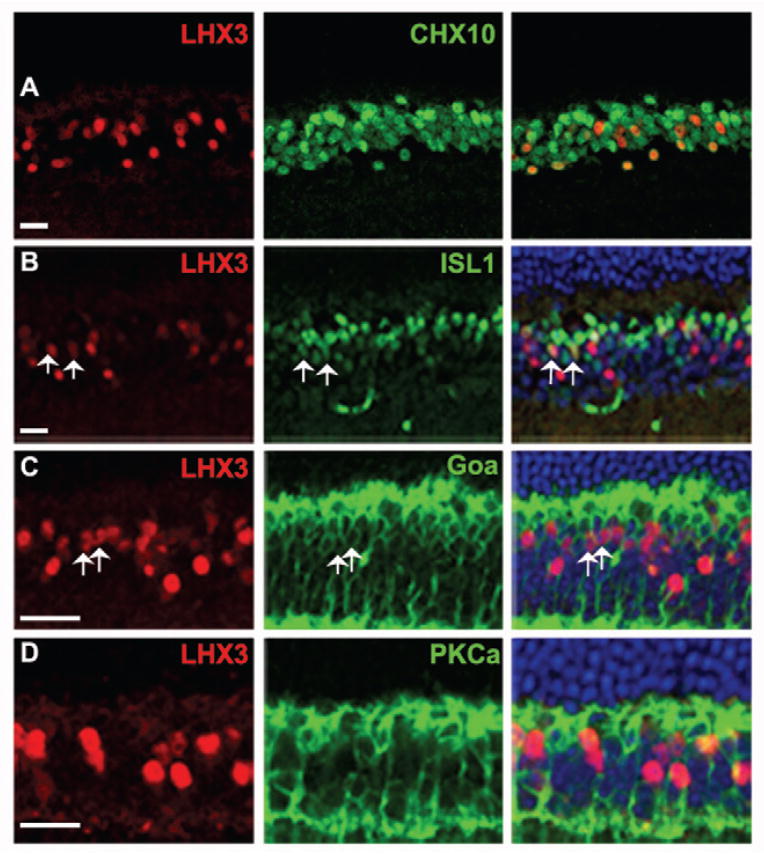
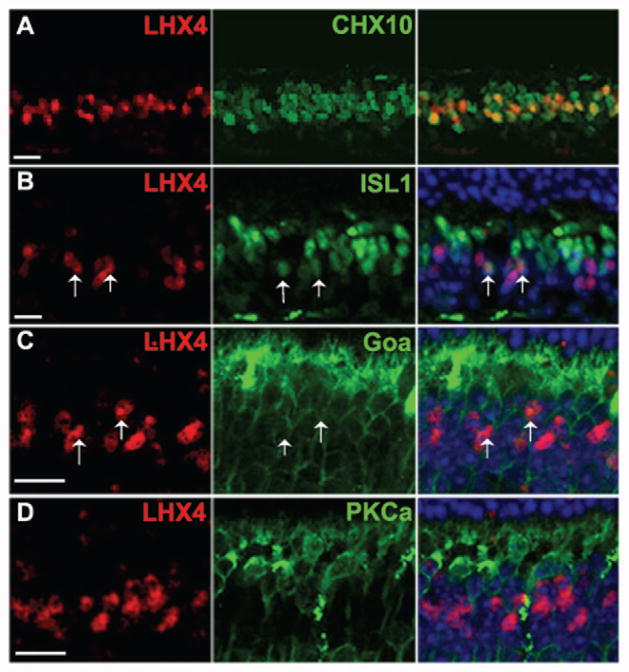
The expression of Lhx3 and Lhx4 has been reported in bipolar cells of the developing and adult mammalian retina (Blackshaw et al., 2004, Elshatory et al., 2007a, Kim et al., 2008) and in the chick retina (Edqvist, Myers et al. 2006). We sought to further characterize the bipolar cell subtype expression in the adult retina by immunohistochemistry (Fig. 4 and 5). We first co-localized LHX3 (Fig. 4A) and LHX4 (Fig. 5A) with CHX10. While 19±2.3% of CHX10 cells expressed LHX3 and 22±1.8% of CHX10 cells expressed LHX4, 100% of LHX3- and LHX4-positive cells were CHX10-positive, confirming that LHX3 and LHX4 are expressed in a subset of bipolar cells. Bipolar cells can be characterized into rod and cone bipolar cells based on the photoreceptor type they receive their input from, and into ON and OFF bipolar cells based on whether they depolarize or hyperpolarize in response to light stimuli respectively. Goα is expressed by ON bipolar cells including rod bipolar cells and ON-cone bipolar cells while PKCα expression marks only rod bipolar cells. Also ISL1 is expressed exclusively in ON bipolar cells with a higher ISL1 expression level in rod bipolar cells and a lower expression in ON-cone bipolar cells. We thus co-expressed LHX3 and LHX4 individually with ISL1, Goα, and PKCα. Both LHX3 and LHX4 co-expressed with few ‘weakly’ ISL1-positive cells, indicating that they are expressed in ON-cone bipolar cells (Fig. 4B, 5B). However there were LHX3 and LHX4 cells that did not co-localize with ISL1-positive cells indicating their expression in OFF-cone bipolar cells as well. While some LHX3- and LHX4-positive cells co-localized with Goα (Fig. 4C, 5C), none co-localized with PKCα (Fig. 4D, 5D). These results taken together indicate that LHX3 and LHX4 are expressed in cone bipolar cells only and might thus serve as useful developmental markers for cone bipolar (ON and OFF) cells.
Figure 4.
Characterization of LHX3 expressing cells in the adult retina (P30). A: Co-localization of LHX3 with CHX10 indicates that all LHX3 expressing cells are bipolar cells. B: Co-expression of LHX3 is seen with “weakly” ISL1 expressing cells (Arrows). C: Co-expression of a few LHX3 cells is seen with Goα expressing bipolar cells (Arrows). D: LHX3 cells do not co-localize with PKCα expressing bipolar cells. All scale bars are 50 μm.
Figure 5.
Characterization of LHX4 expressing cells in the adult retina (P30). A: Co-localization of LHX4 with CHX10 indicates that all LHX4 expressing cells are bipolar cells. B: Co-expression of LHX4 is seen with “weakly” ISL1 expressing cells (Arrows). C: Co-expression of some LHX4 cells is seen with Goα expressing bipolar cells (Arrows). D: LHX4 cells do not co-localize with PKCα expressing bipolar cells. All scale bars are 50 μm.
Expression of Lhx6
The expression of Lhx6 in the developing retina was analyzed by using in situ hybridization (Fig. 6). The onset of Lhx6 expression was not detected at E11.5 (Fig. 6A). It was first seen in the developing GCL at E13.5 (Fig. 6B) and this expression was sustained in the GCL throughout embryonic stages of development and postnatally (Fig. 6C–F). At E17.5, Lhx6 expression was seen in the INL in addition to the GCL (Fig. 6C–F). With similar arguments as that presented for Lhx9 above, we hypothesized that Lhx6 expressing cells are also amacrine cells. However, it could not be conclusively demonstrated to be the case, in the absence of a commercially available good working antibody.
Figure 6.
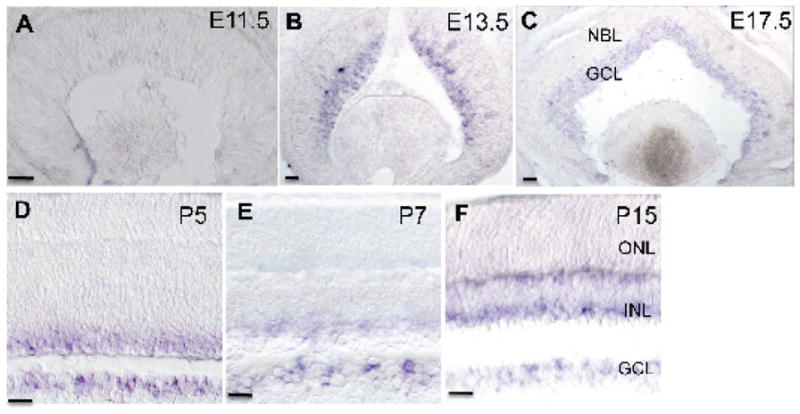
Expression of Lhx6 mRNA transcript in the developing retina. A: Expression of Lhx6 is not detected at E11.5. B: Expression of Lhx6 is seen in retinal progenitor cells starting at E13.5. C-D: Lhx6 expression is detected in cells in the GCL and the developing NBL (E17.5, P5). E-F: Lhx6 expression is detected in cells in the GCL and cells in the INL postnatally (P7, P15). All scale bars are 50 μm.
In summary, LIM-HD transcription factors are broadly expressed and play essential roles in the development and specification of neuronal subtypes in the central nervous system. Currently, their expression and function in the retina are not fully understood. We show here that LHX2 expressing amacrine cells are not cholinergic, dopaminergic or to a large extent GABAergic. Postnatal expression of LHX3 and LHX4 is seen in cone bipolar cells or a subset thereof. We also show that Lhx6 expression begins embryonically in retinal progenitors at E13.5 and continues to be expressed in cells in the ganglion cell layer (GCL) and the outer portion of the inner nuclear layer (INL), postnatally. Lhx9 expression begins embryonically in retinal progenitors after E13.5 and is seen postnatally in a subset of amacrine cells. Using in situ hybridization, we also looked at the expression of Lhx5 and Lhx7 (also known as Lhx8) but were unable to detect any significant expression during the development of the retina (data not shown). Taken together, the expression of LIM-HD transcription factors is for most part restricted to the INL and some in the GCL, but not in the ONL. Our expression study combined with previous studies on expression of LHX1, ISL1 and ISL2 indicates that most retinal cell types/subtypes in the INL can be identified with one or more combinations of LIM-HD factors.
MATERIALS AND METHODS
Animals
Retinas of embryonic and postnatal C57BL/6J mice (The Jackson Laboratory, Stock #000664) were used in this study. The mice were time mated and embryos were designated as E0.5 at noon on the day the vaginal plugs were first observed. The expression studies were performed at least three times, each time with a different embryo or animal and at time points between E11.5 and P30. All animal procedures in this study were in accordance to NIH guidelines and were approved by the University Committee of Animal Resources (UCAR) at the University of Rochester.
In-situ hybridization and Immunohistochemistry
Embryos were dissected in phosphate buffered saline (PBS). Retinas of postnatal mice were enucleated to remove the anterior segment and vitreous. The posterior retinal cups and embryonic heads were fixed in 4% paraformaldehyde for 2 hours for immunohistochemistry or overnight at 4°C for in situ hybridization. Following fixation, samples were washed in PBS and cryoprotected in 20% sucrose. To obtain sections, samples were embedded in OCT medium (Tissue-Tek), stored at −80°C and sectioned at 20 μm. Sections were then processed for non-radioactive digoxygenin-labeled in situ hybridization and immunohistochemical analysis as previously described (Yang, Ding et al. 2003).
The following primary antibodies were used: mouse anti-BRN3A (1:200; Santa Cruz Biotech), rabbit anti-calretinin (1:200; Oncogene); goat anti-ChAT (1:1000; Millipore), sheep anti-CHX10 (1:1000; Exalpha Biologicals); rabbit anti-GAD65 (1:500; Millipore); mouse anti-Goα (1:500; Millipore); mouse anti-ISL1 (1:200; DSHB); goat anti-LHX2 (1:200; Santa Cruz Biotechnology); rabbit anti-LHX3 and anti-LHX4 (1:2500 each; courtesy of Dr. S. Pfaff, The Salk institute, La Jolla, CA); guinea pig anti-LHX9 (1:2000; courtesy of Dr. Jane Dodd, Columbia University, NY), mouse anti-PKCα (1:500; Millipore), rabbit anti-SOX2 (1:500; Chemicon), mouse anti-syntaxin (1:1000, Sigma Aldrich); Alexa secondary antibodies (Molecular probes) were used at a concentration of 1:1000.
Highlights.
We analyzed expression of LIM-HD transcription factors during mouse retinal development
LHX9 is expressed in a subset of amacrine cells in the adult retina.
LHX2 amacrine cells are not cholinergic; few LHX2 cells express calretinin
LHX4 and LHX3 are expressed in a subset of bipolar cells postnatally
LHX6 is expressed in cells in the ganglion cell layer and the inner nuclear layer
Acknowledgments
We thank Dr. Heiner Westphal of National Institute of Health for the Lhx6 in situ hybridization probe. We are also grateful to Dr. Samuel Pfaff of Salk Institute and Dr. Jane Dodd of Columbia University for kindly sharing LHX3 and LHX4, and LHX9 antibodies, respectively. We also thank the other members of the Gan laboratory for helpful discussions and technical assistance. This work was supported by National Institutes of Health Grant 1R21EY023104 and a Research to Prevent Blindness Senior Scientific Investigator Award to L.G., the NYSTEM pre-doctoral training grant NYS DOH C026877 to R.B. as well as by a Research to Prevent Blindness Challenge Grant to the Department of Ophthalmology at the University of Rochester.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
LITERATURE CITED
- Abellan A, Legaz I, et al. Olfactory and amygdalar structures of the chicken ventral pallium based on the combinatorial expression patterns of LIM and other developmental regulatory genes. J Comp Neurol. 2009;516(3):166–186. doi: 10.1002/cne.22102. [DOI] [PubMed] [Google Scholar]
- Alifragis P, Liapi A, et al. Lhx6 regulates the migration of cortical interneurons from the ventral telencephalon but does not specify their GABA phenotype. J Neurosci. 2004;24(24):5643–5648. doi: 10.1523/JNEUROSCI.1245-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson-Leadbeater K, Bertolesi GE, et al. FGF receptor dependent regulation of Lhx9 expression in the developing nervous system. Dev Dyn. 2009;238(2):367–375. doi: 10.1002/dvdy.21850. [DOI] [PubMed] [Google Scholar]
- Bach I. The LIM domain: regulation by association. Mech Dev. 2000;91(1–2):5–17. doi: 10.1016/s0925-4773(99)00314-7. [DOI] [PubMed] [Google Scholar]
- Bertuzzi S, Porter FD, et al. Characterization of Lhx9, a novel LIM/homeobox gene expressed by the pioneer neurons in the mouse cerebral cortex. Mech Dev. 1999;81(1–2):193–198. doi: 10.1016/s0925-4773(98)00233-0. [DOI] [PubMed] [Google Scholar]
- Bhati M, Lee C, et al. Implementing the LIM code: the structural basis for cell type-specific assembly of LIM-homeodomain complexes. EMBO J. 2008;27(14):2018–2029. doi: 10.1038/emboj.2008.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshaw S, Harpavat S, et al. Genomic analysis of mouse retinal development. PLoS Biol. 2004;2(9):E247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgouin C, Lundgren SE, et al. Apterous is a Drosophila LIM domain gene required for the development of a subset of embryonic muscles. Neuron. 1992;9(3):549–561. doi: 10.1016/0896-6273(92)90192-g. [DOI] [PubMed] [Google Scholar]
- Bramblett DE, Pennesi ME, et al. The transcription factor Bhlhb4 is required for rod bipolar cell maturation. Neuron. 2004;43(6):779–793. doi: 10.1016/j.neuron.2004.08.032. [DOI] [PubMed] [Google Scholar]
- Chatterjee M, Li K, et al. Gbx2 regulates thalamocortical axon guidance by modifying the LIM and Robo codes. Development. 2012;139(24):4633–4643. doi: 10.1242/dev.086991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CM, Cepko CL. Expression of Chx10 and Chx10-1 in the developing chicken retina. Mech Dev. 2000;90(2):293–297. doi: 10.1016/s0925-4773(99)00251-8. [DOI] [PubMed] [Google Scholar]
- Choi GB, Dong HW, et al. Lhx6 delineates a pathway mediating innate reproductive behaviors from the amygdala to the hypothalamus. Neuron. 2005;46(4):647–660. doi: 10.1016/j.neuron.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Dalal J, Roh JH, et al. Translational profiling of hypocretin neurons identifies candidate molecules for sleep regulation. Genes Dev. 2013;27(5):565–578. doi: 10.1101/gad.207654.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid IB, Breen JJ, et al. LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 1998;14(4):156–162. doi: 10.1016/s0168-9525(98)01424-3. [DOI] [PubMed] [Google Scholar]
- de Melo J, Miki K, et al. Injury-independent induction of reactive gliosis in retina by loss of function of the LIM homeodomain transcription factor Lhx2. Proc Natl Acad Sci U S A. 2012;109(12):4657–4662. doi: 10.1073/pnas.1107488109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquette PM, Zhou X, et al. Loss of LMO4 in the retina leads to reduction of GABAergic amacrine cells and functional deficits. PLoS One. 2010;5(10):e13232. doi: 10.1371/journal.pone.0013232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edqvist PH, Myers SM, et al. Early identification of retinal subtypes in the developing, pre-laminated chick retina using the transcription factors Prox1, Lim1, Ap2alpha, Pax6, Isl1, Isl2, Lim3 and Chx10. Eur J Histochem. 2006;50(2):147–154. [PubMed] [Google Scholar]
- Elshatory Y, Deng M, et al. Expression of the LIM-homeodomain protein Isl1 in the developing and mature mouse retina. J Comp Neurol. 2007a;503(1):182–197. doi: 10.1002/cne.21390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshatory Y, Everhart D, et al. Islet-1 controls the differentiation of retinal bipolar and cholinergic amacrine cells. J Neurosci. 2007b;27(46):12707–12720. doi: 10.1523/JNEUROSCI.3951-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Xie X, et al. Requirement for Bhlhb5 in the specification of amacrine and cone bipolar subtypes in mouse retina. Development. 2006;133(24):4815–4825. doi: 10.1242/dev.02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon PJ, Yun S, et al. Lhx2 balances progenitor maintenance with neurogenic output and promotes competence state progression in the developing retina. J Neurosci. 2013;33(30):12197–12207. doi: 10.1523/JNEUROSCI.1494-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriou M, Tucker AS, et al. Expression and regulation of Lhx6 and Lhx7, a novel subfamily of LIM homeodomain encoding genes, suggests a role in mammalian head development. Development. 1998;125(11):2063–2074. doi: 10.1242/dev.125.11.2063. [DOI] [PubMed] [Google Scholar]
- Hagglund AC, Dahl L, et al. Lhx2 is required for patterning and expansion of a distinct progenitor cell population committed to eye development. PLoS One. 2011;6(8):e23387. doi: 10.1371/journal.pone.0023387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama J, Kageyama R. Retinal cell fate determination and bHLH factors. Semin Cell Dev Biol. 2004;15(1):83–89. doi: 10.1016/j.semcdb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Hobert O, Westphal H. Functions of LIM-homeobox genes. Trends Genet. 2000;16(2):75–83. doi: 10.1016/s0168-9525(99)01883-1. [DOI] [PubMed] [Google Scholar]
- Inoue A, Akagawa K. Neuron specific expression of a membrane protein, HPC-1: tissue distribution, and cellular and subcellular localization of immunoreactivity and mRNA. Brain Res Mol Brain Res. 1993;19(1–2):121–128. doi: 10.1016/0169-328x(93)90156-j. [DOI] [PubMed] [Google Scholar]
- Kay JN, Voinescu PE, et al. Neurod6 expression defines new retinal amacrine cell subtypes and regulates their fate. Nat Neurosci. 2011;14(8):965–972. doi: 10.1038/nn.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DS, Ross SE, et al. Identification of molecular markers of bipolar cells in the murine retina. J Comp Neurol. 2008;507(5):1795–1810. doi: 10.1002/cne.21639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YP, Ouchi Y, et al. Sox2 plays a role in the induction of amacrine and Muller glial cells in mouse retinal progenitor cells. Invest Ophthalmol Vis Sci. 2009;50(1):68–74. doi: 10.1167/iovs.07-1619. [DOI] [PubMed] [Google Scholar]
- Liodis P, Denaxa M, et al. Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J Neurosci. 2007;27(12):3078–3089. doi: 10.1523/JNEUROSCI.3055-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Wang JH, et al. Specific expression of the LIM/homeodomain protein Lim-1 in horizontal cells during retinogenesis. Dev Dyn. 2000;217(3):320–325. doi: 10.1002/(SICI)1097-0177(200003)217:3<320::AID-DVDY10>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2(2):109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- Marin O, Anderson SA, et al. Origin and molecular specification of striatal interneurons. J Neurosci. 2000;20 (16):6063–6076. doi: 10.1523/JNEUROSCI.20-16-06063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masland RH. The fundamental plan of the retina. Nat Neurosci. 2001a;4(9):877–886. doi: 10.1038/nn0901-877. [DOI] [PubMed] [Google Scholar]
- Masland RH. Neuronal diversity in the retina. Curr Opin Neurobiol. 2001b;11(4):431–436. doi: 10.1016/s0959-4388(00)00230-0. [DOI] [PubMed] [Google Scholar]
- Masland RH. The neuronal organization of the retina. Neuron. 2012a;76(2):266–280. doi: 10.1016/j.neuron.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masland RH. The tasks of amacrine cells. Vis Neurosci. 2012b;29(1):3–9. doi: 10.1017/s0952523811000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa Y, O’Leary DD. Combinatorial expression patterns of LIM-homeodomain and other regulatory genes parcellate developing thalamus. J Neurosci. 2001;21(8):2711–2725. doi: 10.1523/JNEUROSCI.21-08-02711.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak W, Hindges R, et al. Magnitude of binocular vision controlled by islet-2 repression of a genetic program that specifies laterality of retinal axon pathfinding. Cell. 2004;119(4):567–578. doi: 10.1016/j.cell.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Pan L, Yang Z, et al. Functional equivalence of Brn3 POU-domain transcription factors in mouse retinal neurogenesis. Development. 2005;132(4):703–712. doi: 10.1242/dev.01646. [DOI] [PubMed] [Google Scholar]
- Peukert D, Weber S, et al. Lhx2 and Lhx9 determine neuronal differentiation and compartition in the caudal forebrain by regulating Wnt signaling. PLoS Biol. 2011;9(12):e1001218. doi: 10.1371/journal.pbio.1001218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poche RA, Kwan KM, et al. Lim1 is essential for the correct laminar positioning of retinal horizontal cells. J Neurosci. 2007;27(51):14099–14107. doi: 10.1523/JNEUROSCI.4046-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter FD, Drago J, et al. Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development. 1997;124(15):2935–2944. doi: 10.1242/dev.124.15.2935. [DOI] [PubMed] [Google Scholar]
- Retaux S, Rogard M, et al. Lhx9: a novel LIM-homeodomain gene expressed in the developing forebrain. J Neurosci. 1999;19(2):783–793. doi: 10.1523/JNEUROSCI.19-02-00783.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, de Melo J, et al. LHX2 is necessary for the maintenance of optic identity and for the progression of optic morphogenesis. J Neurosci. 2013;33(16):6877–6884. doi: 10.1523/JNEUROSCI.4216-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K, Sheng HZ, et al. LIM homeodomain factors Lhx3 and Lhx4 assign subtype identities for motor neurons. Cell. 1998;95(6):817–828. doi: 10.1016/s0092-8674(00)81704-3. [DOI] [PubMed] [Google Scholar]
- Sheng HZ, Moriyama K, et al. Multistep control of pituitary organogenesis. Science. 1997;278(5344):1809–1812. doi: 10.1126/science.278.5344.1809. [DOI] [PubMed] [Google Scholar]
- Shirasaki R, Pfaff SL. Transcriptional codes and the control of neuronal identity. Annu Rev Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- Tetreault N, Champagne MP, et al. The LIM homeobox transcription factor Lhx2 is required to specify the retina field and synergistically cooperates with Pax6 for Six6 trans-activation. Dev Biol. 2009;327(2):541–550. doi: 10.1016/j.ydbio.2008.12.022. [DOI] [PubMed] [Google Scholar]
- Thor S, Andersson SG, et al. A LIM-homeodomain combinatorial code for motor-neuron pathway selection. Nature. 1999;397(6714):76–80. doi: 10.1038/16275. [DOI] [PubMed] [Google Scholar]
- Tsuchida T, Ensini M, et al. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79(6):957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Voinescu PE, Kay JN, et al. Birthdays of retinal amacrine cell subtypes are systematically related to their molecular identity and soma position. J Comp Neurol. 2009;517(5):737–750. doi: 10.1002/cne.22200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SW, Mu X, et al. Brn3b/Brn3c double knockout mice reveal an unsuspected role for Brn3c in retinal ganglion cell axon outgrowth. Development. 2002;129(2):467–477. doi: 10.1242/dev.129.2.467. [DOI] [PubMed] [Google Scholar]
- Wassle H. Parallel processing in the mammalian retina. Nat Rev Neurosci. 2004;5(10):747–757. doi: 10.1038/nrn1497. [DOI] [PubMed] [Google Scholar]
- Wilson SI, Shafer B, et al. A molecular program for contralateral trajectory: Rig-1 control by LIM homeodomain transcription factors. Neuron. 2008;59(3):413–424. doi: 10.1016/j.neuron.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Xu Y, Baldassare M, et al. LH-2: a LIM/homeodomain gene expressed in developing lymphocytes and neural cells. Proc Natl Acad Sci U S A. 1993;90(1):227–231. doi: 10.1073/pnas.90.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T, Moriyama K, et al. Lhx4, a LIM homeobox gene. Genomics. 1997;44(1):144–146. doi: 10.1006/geno.1997.4852. [DOI] [PubMed] [Google Scholar]
- Yang Z, Ding K, et al. Math5 determines the competence state of retinal ganglion cell progenitors. Dev Biol. 2003;264(1):240–254. doi: 10.1016/j.ydbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Yun S, Saijoh Y, et al. Lhx2 links the intrinsic and extrinsic factors that control optic cup formation. Development. 2009;136(23):3895–3906. doi: 10.1242/dev.041202. [DOI] [PMC free article] [PubMed] [Google Scholar]