Abstract
Extraocular muscles (EOM) represent a unique muscle group that controls eye movements and originates from head mesoderm, while the more typically studied body and limb muscles are somite-derived. Aiming to investigate myogenic progenitors (satellite cells) in EOM versus limb and diaphragm of adult mice, we have been using flow cytometry in combination with myogenic-specific Cre-loxP lineage marking for cell isolation. While analyzing cells from the EOM of mice that harbor Myf5Cre-driven GFP expression, we identified in addition to the expected GFP+ myogenic cells (presumably satellite cells), a second dominant GFP+ population distinguished as being Sca1+, non-myogenic, and exhibiting a fibro/adipogenic potential. This unexpected population was not only unique to EOM compared to the other muscles but also specific to the Myf5Cre-driven reporter when compared to the MyoDCre driver. Histological studies of periocular tissue preparations demonstrated the presence of Myf5Cre-driven GFP+ cells in connective tissue locations adjacent to the muscle masses, including cells in the vasculature wall. These vasculature-associated GFP+ cells were further identified as mural cells based on the presence of the specific XLacZ4 transgene. Unlike the EOM satellite cells that originate from a Pax3-negative lineage, these non-myogenic Myf5Cre-driven GFP+ cells appear to be related to cells of a Pax3-expressing origin, presumably derived from the neural crest. In all, our lineage tracing based on multiple reporter lines has demonstrated that regardless of common ancestral expression of Myf5, there is a clear distinction between periocular myogenic and non-myogenic cell lineages according to their mutually exclusive antecedence of MyoD and Pax3 gene activity.
Keywords: Extraocular muscles, satellite cells, fibro/adipogenic progenitors, pericytes and vascular smooth muscle cells, Myf5, MyoD, Pax3, XLacZ4, Wnt1, Sca-1
Introduction
Histogenesis of skeletal muscles begins early in embryogenesis where mesenchymal progenitors undergo myogenic determination and the resulting myoblasts fuse to form multinucleated fibers (myofibers). While body and limb muscles are somite-derived, the extraocular muscles (EOM) are of non-somitic origin and develop from the head mesoderm (Buckingham et al., 2003; Noden and Francis-West, 2006). Satellite cells, the myogenic progenitors in postnatal muscles, are thought to be derived from the same embryonic origin as the muscle in which they reside (Gros et al., 2005; Ono et al., 2010; Schienda et al., 2006; Yablonka-Reuveni and Day, 2011). Indeed, in somite-derived muscles, satellite cells show historical and in some muscles current expression of Pax3 (Day et al., 2007; Montarras et al., 2005; Schienda et al., 2006). Differently, in accordance with EOM development from a Pax3-negative lineage, myogenic cells from these muscles have been shown, at least in embryological/early post-natal stages, not to express Pax3 (Harel et al., 2009; Horst et al., 2006).
Despite a distinct lineage origin, EOM development is orchestrated by the same members of the bHLH transcription factor family (MyoD, Myf5, MRF4, myogenin) that are involved in the specification and differentiation of body and limb muscles (Gensch et al., 2008; Kassar-Duchossoy et al., 2004; Noden and Francis-West, 2006; Sambasivan et al., 2009). Cre-loxP lineage tracing demonstrated MyoDCre- and Myf5Cre-driven reporter expression in myofibers of both EOM and somite-derived muscles (Harel et al., 2009; Kanisicak et al., 2009; Kuang et al., 2007). Also, regardless of muscle origin, virtually all satellite cells in adult muscles show expression of MyoDCre-driven reporter (Kanisicak et al., 2009). This reporter expression is thought to reflect ancestral MyoD expression given that quiescent satellite cells of adult muscle do not express MyoD (Kanisicak et al., 2009; Yablonka-Reuveni and Rivera, 1994). Myf5Cre-driven reporter expression in satellite cells has been primarily investigated in limb muscles. In this context, the bulk of satellite cells do express Myf5Cre-driven reporter and endogenous Myf5, although a small subset appeared to be void of ancestral or current Myf5 expression (Beauchamp et al., 2000; Biressi et al., 2013; Day et al., 2010; Gayraud-Morel et al., 2012; Kuang et al., 2007). Whereas Myf5Cre-driven reporter expression has not been studied in EOM satellite cells, such expression is anticipated based on Myf5 expression during EOM development (Sambasivan et al., 2009).
Here, we report on the detection of an unexpected ancestral expression of Myf5 in periocular connective tissue cells of adult mice. This observation was made while searching for an effective means to isolate satellite cells from EOM. We opted to first harvest these small muscles with their associated extensive connective tissues, and then isolate satellite cells from the ‘crude’cell preparation by flow cytometry. Cells were sorted based on a standardized surface antigen signature in combination with a myogenic-specific Cre-loxP reporter. The use of a permanent lineage marker permitted us to further trace the cells in vivo and in culture. Working with mice that harbor Myf5Cre-driven GFP expression, in addition to the expected GFP+ population of satellite cells, we noted a second dominant population of non-myogenic GFP+ cells characterized by a distinctive expression of Sca1 (stem cell antigen-1) and a fibro/adipogenic potential. This additional GFP+ population was not apparent in EOM cells harvested from MyoDCre-driven GFP-expressing mice. Histological analysis of periocular tissue confirmed the presence of Myf5Cre-driven GFP+ cells in connective tissue locations adjacent to the muscle masses, including cells in the vasculature. Furthermore, while the EOM myogenic cells are of a Pax3-negative origin, the non-myogenic Myf5Cre-driven GFP+/Sca1+ cells appear to be related to Pax3-expressing cells and are likely of neural crest origin. In all, our data demonstrate that the periocular Myf5Cre-driven GFP+ myogenic and non-myogenic populations, albeit sharing ancestral expression of Myf5, are derived from different progenitors.
Materials and Methods
Mice
All mice were from colonies we have maintained long-term at the University of Washington and were on enriched or pure C57BL/6 background. Animal care and experimental procedures were approved by the Institutional Animal Care and Use Committee at the University of Washington. The knockin heterozygous Cre males MyoDCre [MyoD1tm2.1(icre)Glh (Kanisicak et al., 2009)], Myf5Cre [Myf5tm3(cre)Sor/J (Tallquist et al., 2000)] and Pax3Cre [Pax3tm1(cre)Joe/J (Engleka et al., 2005)] were crossed with knockin reporter females R26mTmG [Gt(ROSA) 26Sortm4(ACTB-tdTomato,-EGFP)Luo/J (Muzumdar et al., 2007)] to generate adult F1 Cre-loxP experimental animals (typically, 4–9 months old). When indicated, the mural cell reporter mouse XlacZ4 [Tg(Fabp4-lacZ)4Mosh (Klinghoffer et al., 2001; Stuelsatz et al., 2012; Tidhar et al., 2001)] was crossed with male Myf5Cre or Pax3Cre to generate XLacZ4 males on a Cre driver background that were then crossed with females R26mTmG to generate adult F1 double reporter experimental animals.
Tissue isolation
Hindlimb muscles (LIMB;consisting of tibialis anterior, extensor digitorum longus and gastrocnemius) and whole diaphragm (DIA;includes the costal and crural muscles and the central tendon) were harvested according to our routine procedures (Danoviz and Yablonka-Reuveni, 2012; Stuelsatz et al., 2012). For the extraocular tissue we analyzed two types of preparations. One, referred to as “periocular tissue”was used for tissue sectioning, and comprised the EOM (four rectus and two oblique muscles), the retractor bulbi muscle (an ocular accessory muscle, involves in retracting the eye into the orbit forcing the mouse third eyelid across the cornea), the optic nerve and the associated periocular connective tissues, all attached to the eyeball. A second preparation, referred to as “EOM”and used for cell isolation, consisted of EOM that were individually separated from the eyeball. When isolating EOM individually, we harvest the muscles with their surrounding connective tissue without further cleaning, in order to maintain the integrity of those tiny muscles. The EOM are surrounded by an elaborate system of connective tissues that suspend the eyeball within the orbit, permitting a wide range of motion and linking the EOM to each other and to other orbital structures controlling the complex mechanical properties of EOM (Von Noorden and Campos, 2002).
To obtain the aforementioned preparations of “periocular tissue”and of “EOM”, the head was first cleaned from skin and eyelids. Next, a midline cut was done through the sagittal suture of the skull and the parietal bones were broken to expose and remove the brain. The two halves of the skull were then pulled apart but remained connected by the lower jaw and the bottom of the cranium. The nasal cavity was cleaned from the nasal bones and soft tissues, and then the bones of the orbit were broken along their suture lines and removed to expose the eyes. This process breaks the superior oblique muscle into two parts (due to its association with the trochlea), resulting in partial or total absence of this muscle in our preparations. The Harderian gland, surrounding the EOM was then removed, and the optic nerve was severed between the Annulus of Zinn and the optic chiasma to allow for the detachment of the optic nerve from the brain. At this point, the procedure differs depending on the desired preparation. For sectioning of the periocular tissue, the eyes were released from the orbit by cutting the remaining soft tissues and the proximal attachment of the inferior oblique, whereas for cell isolation each of the EOM was dissected from the eyeballs by cutting their proximal and distal attachments.
Harvested tissues were placed in a pre-heated (37°C) DMEM solution and kept at room temperature until ready for processing. The DMEM solution consists of Dulbecco’s modified Eagle’s medium (high glucose, with L-glutamine, 110 mg/l sodium pyruvate, and pyridoxine hydrochloride, Hyclone) supplemented with antibiotics (50 U/ml penicillin and 50 mg/ml streptomycin, Gibco-Invitrogen) and was used throughout the various procedures described below.
Cell isolation
Cells were typically isolated from two mice per muscle group. Harvested tissues were transferred from the DMEM rinse into a digestion solution containing 450 units/ml collagenase type IV, 1 unit/ml dispase II (Worthington) and 2mM CaCl2 in phosphate buffer saline pH 7.2–7.4 (PBS), and minced into fine pieces (with the exception of EOM that do not need to be further reduced in size). Enzymatic dissociation (45 minutes at 37°C) included sample trituration every 15 minutes using a 5-ml polystyrene pipette to breakdown clumps and dissociate single cells. An equal volume of DMEM (supplemented with 10%horse serum, Gibco-Invitrogen) was added at the end of the digestion and samples were filtered through a 70-µm followed by a 40-µm nylon cell strainers (Fisher Scientific). The digestion protocol was adapted from Ieronimakis et al. (2008).
Fluorescence-activated cell sorting (FACS)
Cell suspensions obtained as described above were centrifuged at 1000 × g for 10 minutes and resuspended in a DMEM-10%horse serum solution containing 10 µM Hoechst 33342 (Sigma-Aldrich) and then incubated for 30 min at 37°C. Cells were centrifuged at 400 × g for 10 minutes and resuspended in antibody labeling solution (100 µl per 106 cells). This solution consisted of PBS containing 0.3%of bovine serum albumin (BSA, Omnipur, fraction V;Calbiochem) and a combination of the following fluorescently conjugated monoclonal antibodies: Sca1-APC (clone D7), CD31-PECy7 (clone 390), CD45-PECy7(clone 30-F11), all purchased from eBioscience. Antibodies were diluted at a ratio of 600 ng of antibody per 106 cells for CD31-PECy7 and CD45-PECy7 and 300 ng of antibody per 106 cells for Sca1-APC. Samples were incubated on ice in the dark for 1 hour. Cells were then washed by adding 1 ml of PBS-0.3%BSA, centrifuged at 400 × g, 4oC, for 10 minutes and re-suspended in ice cold PBS-0.3%BSA (300 µl per 106 cells). Samples were then transferred to 5-ml polystyrene tubes equipped with a cell-strainer cap (BD Biosciences), which allows filtering of the samples just before the actual sorts to avoid cell aggregates, and kept on ice from this step on. An Influx Cell Sorter (BD Biosciences) equipped with 350, 488 and 638 nm lasers, was used. All sorted cells were collected within the G0-G1 population based on Hoechst staining eliminating residual cell aggregates or cellular debris. Gates were determined by comparing fluorophore signal intensities between the unstained control and each single antibody/fluorophore control. Data was acquired at 20,000 to 100,000 events per sample and sorted cells were collected in our culture media described below. Subsequent analysis and flow cytometry plots were generated using FlowJo (TreeStar).
Cell culture
Sorted cells were cultured in 24-well culture plates (BD Biosciences) pre-coated with diluted Matrigel (BD Biosciences) using our standard DMEM-based medium containing 20%fetal bovine serum (Gibco-Invitrogen), 10%horse serum (Gibco-Invitrogen), and 1%chicken embryo extract and were incubated at 37°C, 5%CO2 in a humidified tissue culture incubator (Danoviz and Yablonka-Reuveni, 2012; Shefer et al., 2006; Shefer and Yablonka-Reuveni, 2005). Cultures were initiated at a density of 5–10 휐3 cells per well. After the initial plating, growth medium was replaced every 3 days.
Immunohistochemistry
Tissues (held in DMEM following harvesting as described above) were fixed by adding an equal volume of fixative solution (4%paraformaldehyde in 0.1 M sodium phosphate buffer containing 1%sucrose, pH 7.2–7.4, Danoviz and Yablonka-Reuveni, 2012; Shefer and Yablonka-Reuveni, 2005), for 2 hours at room temperature. Tissues were then immersed successively in 10%and 20%sucrose/PBS, each for 30 min, and sunk overnight in 30%sucrose/PBS at 4°C. Tissues were embedded in OCT (Tissue-Tek), rapidly frozen in isopentane cooled with liquid nitrogen and kept in −80oC. Tissues were processed to prepare 10-µm thick transverse cryosections that were placed on Superfrost Plus slides (Fisher Scientific). For direct observation of enhanced GFP (GFP) and tandem dimer Tomato (Tomato) fluorochromes, muscle sections were rinsed with PBS and mounted with Vectashield HardSet Mounting Media containing DAPI (Vector Laboratories). For immunolabeling, muscle sections were first blocked overnight with 2%normal goat serum (Invitrogen), reacted with antibodies, and counterstained with DAPI, following our routine protocol (Danoviz and Yablonka-Reuveni, 2012). The following primary antibodies were used: anti-Sca1 (1:600;Rat IgG2a;clone D7, eBioscience);anti-laminin (1:100;rabbit IgG;AB2034, Chemicon/Millipore);anti-αsmooth muscle actin (1:4000;mouse IgG2a;clone 1A4;Sigma-Aldrich). Secondary antibodies used were all produced in goat and conjugated with AlexaFluor;these included anti-rat IgG2a-488;anti-rabbit IgG-568;anti-mouse IgG2a-568 (1:1000;Invitrogen).
X-gal staining
β-galactosidase (β-gal) activity in cells expressing the lacZ reporter was detected by X-gal staining. Primary cultures were fixed by adding to the culture medium an equal volume of fixative solution (4%paraformaldehyde in 0.1 M sodium phosphate buffer containing 1%sucrose, pH 7.2–7.4, Shefer and Yablonka-Reuveni, 2005) for 10 min at room temperature. Fixed muscle sections (prepared as described above) and cell cultures were rinsed with PBS and incubated in X-gal solution for 4–6 hours at 37°C and rinsed in PBS, as previously described (Day et al., 2010).
Microscopy and imaging
Observations were made with an inverted fluorescent microscope (Eclipse TE2000-S, Nikon). Images were typically acquired using CoolSNAP ES monochrome CCD camera (Photometrics) controlled with MetaVue Imaging System (Universal Imaging Corporation). For acquiring real color images of X-gal staining, images were taken with a Digital Sight DS-Ri1 color camera controlled by NIS-Elements F software (Nikon). Composites of digitized images were prepared and assembled using Adobe Photoshop software.
Results and discussion
Histological overview of periocular tissue preparations
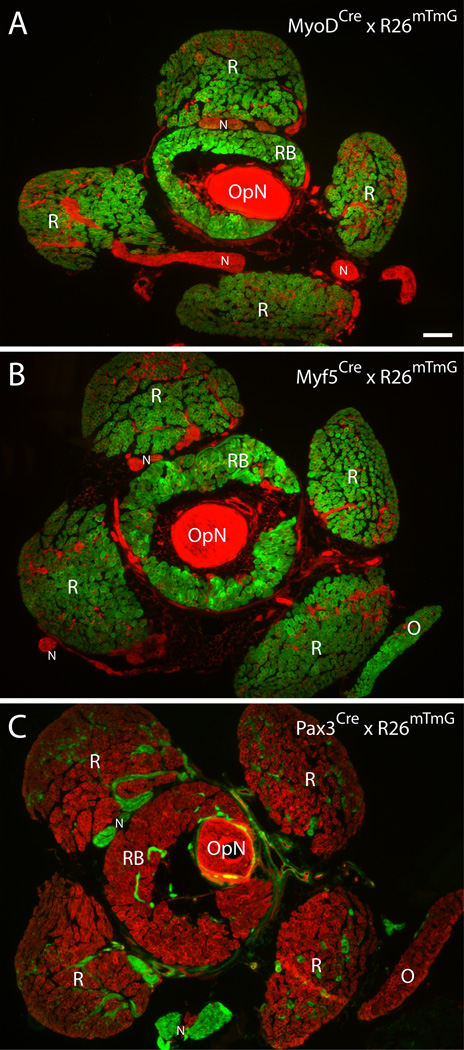
Myf5Cre and MyoDCre mice were crossed with the R26mTmG reporter strain to generate adult mice for cell sorting studies, with the initial aim of isolating satellite cells from different muscle groups. The R26mTmG reporter mouse operates on a dual membrane-localized fluorescent system where all cells express Tomato until Cre-mediated excision of the Tomato coding sequence allows for GFP expression. The two crosses are referred to throughout this study as Myf5Cre × R26mTmG and MyoDCre × R26mTmG. Our histological inspection of these two Cre-loxP reporter lines confirmed a consistent strong GFP expression in myofibers of all muscle groups analyzed. Examples of GFP expression in EOM from these two reporter lines are shown in Fig. 1A and B. The retractor bulbi muscle (RB, Fig. 1) is also included, showing GFP expression, while the optic nerve (OpN, Fig. 1) is negative. Connective tissue and nerve structures within and between muscles can also be observed (Tomato+) in these low magnification images. The specific expression of Myf5Cre- and MyoDCre-driven GFP in myofibers contrasts with the void of GFP reporter expression in myofibers when analyzing EOM from Pax3Cre × R26mTmG mice (Fig. 1C). In fact, Pax3Cre-driven GFP expression is restricted to the connective and nervous tissues (Fig. 1C). Results illustrated in Fig. 1 extend for the first time to adult EOM, the findings previously reported for the fetal stage (E16.5) where EOM depicted Myf5Cre-driven reporter expression, but not Pax3Cre-driven expression (Harel et al., 2009). Additionally, although the current detection of MyoDCre driven expression in adult EOM was anticipated based on the original studies on the persistent expression of MyoDCre-driven reporter in all muscle groups of embryonic and adult mice, these studies did not include direct demonstration pertaining to muscle masses of EOM (Kanisicak et al., 2009; Yamamoto et al., 2009).
Fig. 1. Histological overview of periocular tissue preparations from MyoDCre, Myf5Cre and Pax3Cre reporter lines.
Cross sections were obtained from periocular preparations isolated from (A) MyoDCre × R26mTmG, (B) Myf5Cre × R26mTmG and (C) Pax3Cre × R26mTmG mice. (A–C) Images shown are from cross sections at the level of the optic nerve, immediately posterior to the eye. OpN, optic nerveN, peripheral nerveR, rectus muscleO, inferior oblique muscleRB, retractor bulbi muscle. Scale bar, 200µm. For a schematic overview of the mouse eye with attached rectus, oblique and retractor bulbi muscles refer to LifeMap (2013).
Flow cytometry analyses of Cre-driven GFP+ cells in muscle tissues from Myf5Cre × R26mTmG and MyoDCre × R26mTmG mice
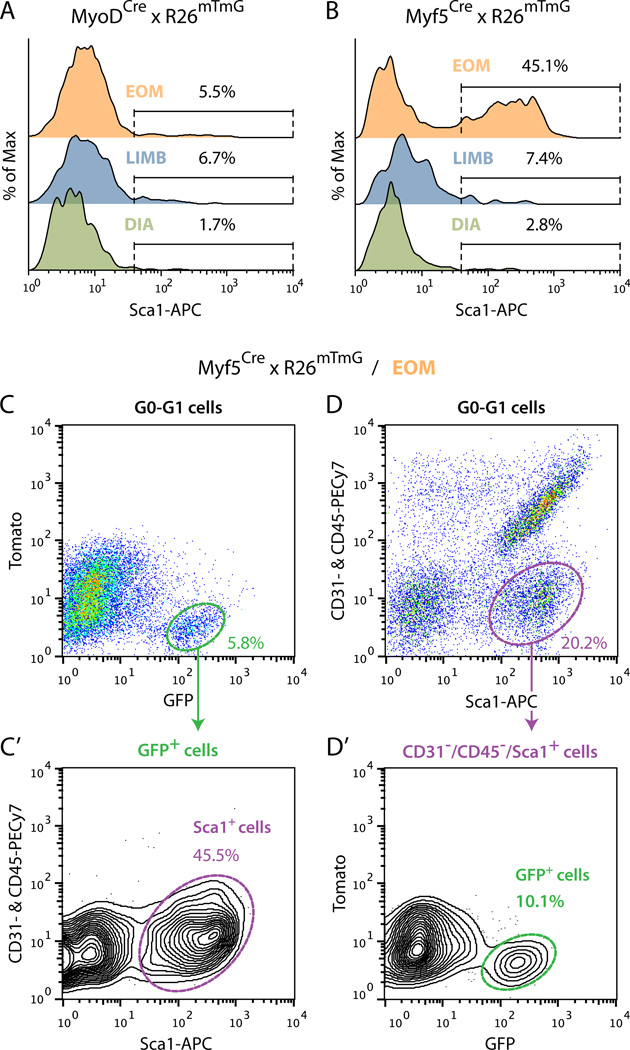
Previous reports with limb muscles demonstrated that the bulk of satellite cells could be isolated from “crude”cell preparations (i.e., released from skeletal muscle by enzymatic dissociation) based on triple negative selection for CD31, CD45 and Sca1 and positive selection for CD34 and α7-integrin cell surface antigens (Ieronimakis et al., 2010; Sacco et al., 2008). Furthermore, additional studies performed in the context of limb muscle revealed a rare subpopulation of Sca1+ satellite cells (Mitchell et al., 2005; Tanaka et al., 2009). The number of such Sca1+ myogenic cells increased in regenerating muscle, possibly related to satellite cell proliferation (Kafadar et al., 2009). Hence, the latter studies have demonstrated that Sca1+ cells should also be considered when developing sort approaches for isolating satellite cells. Here, we reasoned that Myf5Cre- or MyoDCre-driven GFP reporter expression would provide an alternative means to satellite cell enrichment. To validate this approach, we investigated the distribution of Cre-driven GFP+ cells (from Myf5Cre × R26mTmG and MyoDCre × R26mTmG mice) into Sca1− and Sca1+ populations after gating-out endothelial (CD31+) and hematopoietic (CD45+) cells. Using the MyoDCre × R26mTmG line, for each of the muscle group examined (i.e. EOM, LIMB and DIA) the GFP+ cells were distributed into a dominant Sca1− population and a rare Sca1+ population (Fig. 2A). The myogenic nature of these two MyoDCre-driven GFP+ populations was then established in culture (data not shown), likely indicating a satellite cell origin for both populations. When performing the same flow cytometric analyses with the Myf5Cre line, the GFP+ cells from LIMB and DIA muscles were also comprised of a dominant Sca1− and a residual Sca1+ populations (Fig. 2B) whose myogenic nature was also confirmed in culture (data not shown). Surprisingly, EOM preparations from this Myf5Cre × R26mTmG reporter line distinctively showed an abundance of GFP+/Sca1+ cells, amounting to ~45%of GFP+ cells, along with the anticipated GFP+/Sca1− cells.
Fig. 2. Flow cytometric profiles of Cre-driven GFP+ cells isolated from muscle tissues of (A) MyoDCre and (B–D’) Myf5Cre reporter lines.
(A) and (B) Representative FACS overlay histograms analyzing Sca1 staining (X-axis) within GFP+ cells isolated from EOM, LIMB (hindlimb muscles: tibialis anterior, extensor digitorum longus and gastrocnemius) and DIA (diaphragm) of (A) MyoDCre × R26mTmG and (B) Myf5Cre × R26mTmG mice. All populations were analyzed from G0-G1 cells after depletion of CD31+ and CD45+ cells. In each histogram the distribution of GFP+ cells (Y-axis) is depicted in a normalized fashion as %of Max based on the number of cells in each bin divided by the number of cells in the bin that contains the largest number of cells. (C–D’) Detailed flow cytometric characterization of Myf5Cre-driven GFP+ cells isolated from EOM. (C) and (C’) Representative FACS plots showing (C) GFP staining among all G0-G1 cells and subsequent gating of GFP+ cells, and (C’) the distribution of the GFP+ population according to staining for CD31 and CD45 (Y-axis) versus Sca1 (X-axis). (D, D’) Representative FACS plots showing (D) the distribution of G0-G1 cells according to staining for CD31 and CD45 (Y-axis) versus Sca1 (X-axis) and subsequent gating of CD31−/CD45−/Sca1+ cells and (D’) the distribution of the CD31−/CD45−/Sca1+ cells according to GFP staining. In all plots, %values indicate the frequency of the highlighted population out of the total parent population analyzed as indicated at the top of each panel. Histograms and plots are all from one representative experiment of at least 3 independent experiments, which showed no significant differences.
Fig. 2C-D’demonstrates a more detailed flow cytometry analysis of the EOM Myf5Cre-driven GFP+ cells. These GFP+ cells represent 5.8%of total G0-G1 cells and comprise two dominant Sca1+ and Sca1− populations (amounting to 45.5%and 53.6%, of total GFP+ cells respectively), with no notable contribution of CD31+ or CD45+ cells (Fig. 2C-C’). Therefore, CD31+ and CD45+ cells have been routinely excluded throughout our analyses of GFP±/Sca1± populations from the Cre reporter lines used in the context of this manuscript. Noticeably, the GFP+/Sca1+ population in the Myf5Cre reporter line (2.6%of total G0-G1 cells, Fig. 2C’) was distinctively more abundant than in the MyoDCre reporter (0.1%of total G0-G1 cells data not shown). In contrast, cells carrying the classical satellite cell signature (GFP+/Sca1−) were found at a similar frequency in the two Cre-driven lines (~3%of total G0-G1 cells). These results indicate that the GFP+/Sca1+ population in the EOM preparation from the Myf5 driver reflects a second GFP+ population beyond the satellite cells detected with the MyoDCre driver. Analysis of the Myf5Cre reporter line depicted in Fig. 2D and D’further shows that the GFP+/Sca1+ cells contribute an appreciable proportion (10%) of the parent Sca1+ population.
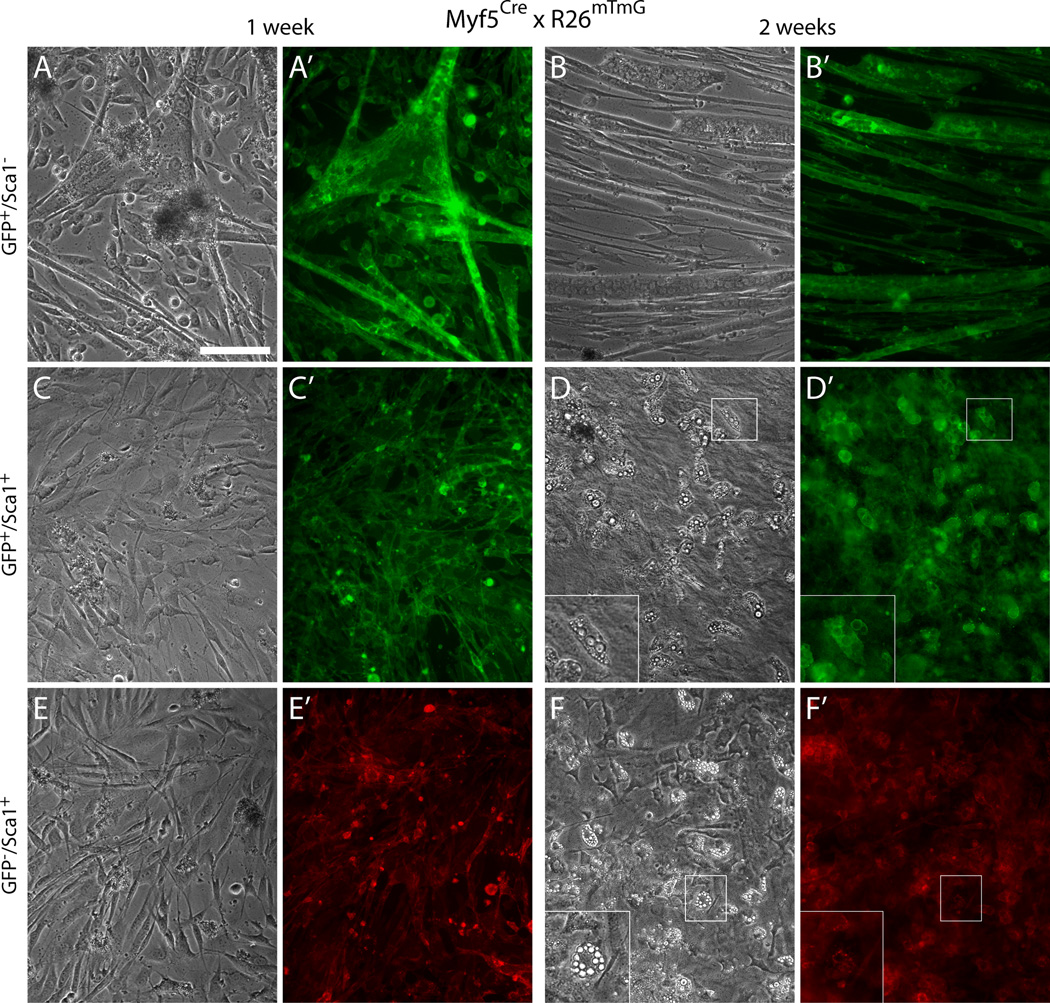
Our cell culture analysis further demonstrates that the two GFP+ populations detected in the Myf5Cre-driven reporter are distinct according to the cell types they give rise to in primary culture (Fig. 3). As anticipated, the GFP+/Sca1− population gave rise entirely to myogenic cells when analyzed in primary culture (Fig. 3A-B’), and are presumably all derived from satellite cells. In contrast, the GFP+/Sca1+ population was composed almost entirely of non-myogenic cells (Fig. 3C-D’). Only rare myotubes were observed in primary cultures from this GFP+/Sca1+ population. The presence of a trace of myogenic cells in this population is in agreement with our detection of trace GFP+/Sca1+ myogenic cells across all muscles analyzed, isolated either from Myf5Cre or MyoDCre (data not shown), that is most likely related to the aforementioned detection of rare Sca1+ satellite cells. Moreover, we never observed mixed myogenic and non-myogenic growth in the clonal patches that developed from low-density cultures of the Sca1+/GFP+ population, attesting that the myogenic and non-myogenic progeny were indeed derived from different progenitors.
Fig. 3. Sca1±/GFP+ cells isolated from EOM preparations of the Myf5Cre reporter line distinctively give rise to myogenic and non-myogenic progeny, respectively.
Representative images of day 7 and day 14 cultures of (A–B’) GFP+/Sca1− cells, (C–D’) GFP+/Sca1+ cells, and (E–F’) GFP−/Sca1+ cells, isolated by FACS as shown in Fig. 2C–D’. All populations were sorted from G0-G1 cells after depletion of CD31+ and CD45+ cells. Inserts in the lower left corner of (D) and (F) represent higher magnification views of the regions delineated by a white box in each corresponding panel, depicting adipogenic cells that develop spontaneously in the cultures of the non-myogenic cells with time. Scale bar, 100µm.
Notably, the GFP+/Sca1+ non-myogenic population isolated from the Myf5 reporter line exhibited a propensity for spontaneous development of adipocytes under our standard culture conditions (Fig. 3D and D’). The GFP+/Sca1+ cells displayed the same morphology, growth pattern, and adipogenic propensity as the remaining Sca1+ cells that were not expressing GFP (Fig. 3E-F’). Adipogenic differentiation, defined by the emergence of cells containing multivacuolar lipid droplets (Gregoire et al., 1998; Mersmann et al., 1975; Napolitano, 1963; Shefer et al., 2004), was exclusively observed within the two Sca1+ populations (GFP− and GFP+), while absent from the myogenic cultures developed from GFP+/Sca1− cells (see higher-zoom views in Fig. 3D and F). These adipogenic cells were first observed at a very low number by the first week and increased over time representing ~10%of the cells by day 14 in culture (Fig. 3D-D’and F-F’). By antigen signature and spontaneous adipogenic potential, these GFP+/Sca1+ and GFP−/Sca1+ populations appear to be related to the reported fibro/adipocyte progenitors, previously discussed in the context of limb and masseter muscles (Joe et al., 2010; Lemos et al., 2012).
Detection of Myf5Cre-driven GFP+ cells in connective tissues of periocular preparations
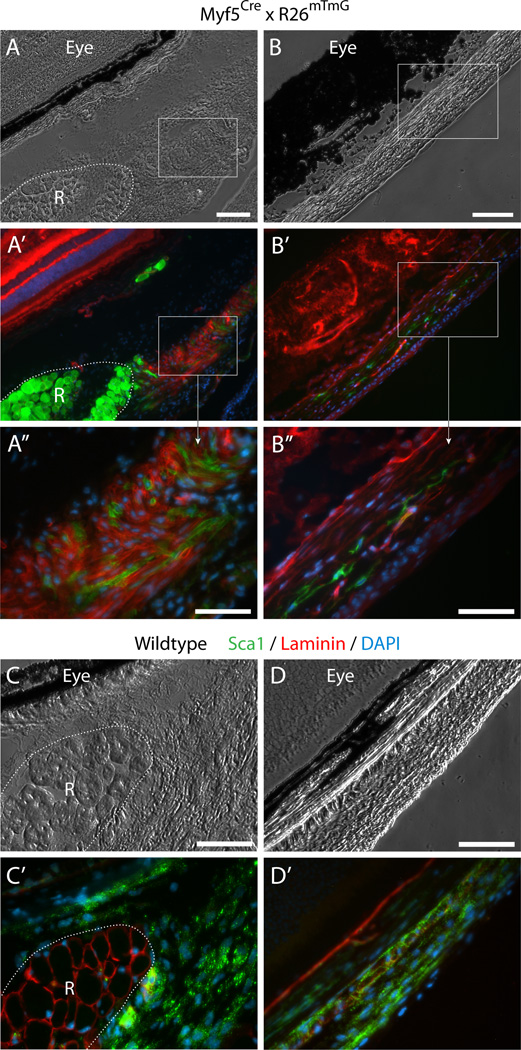
The detection of a non-myogenic Myf5Cre-driven GFP+ population in EOM when analyzing isolated cells, prompted us to investigate further the in-vivo localization of GFP+ cells. To this end, cross sections of periocular tissue (comprising the EOM, the retractor bulbi muscle, the optic nerve and the associated periocular connective tissues, still attached to the eyeballs) were thoroughly analyzed for GFP expression paying attention also to the rich connective tissue surrounding the EOM (se details in the “Materials and methods”section). Indeed, in addition to the expected strong GFP expression in skeletal muscle masses (Fig. 4A’, 5A’and 5B’R and RB denote rectus and retractor bulbi muscles, respectively), we identified several distinct structures within the extensive connective tissues showing Myf5Cre-driven GFP expression, as described next.
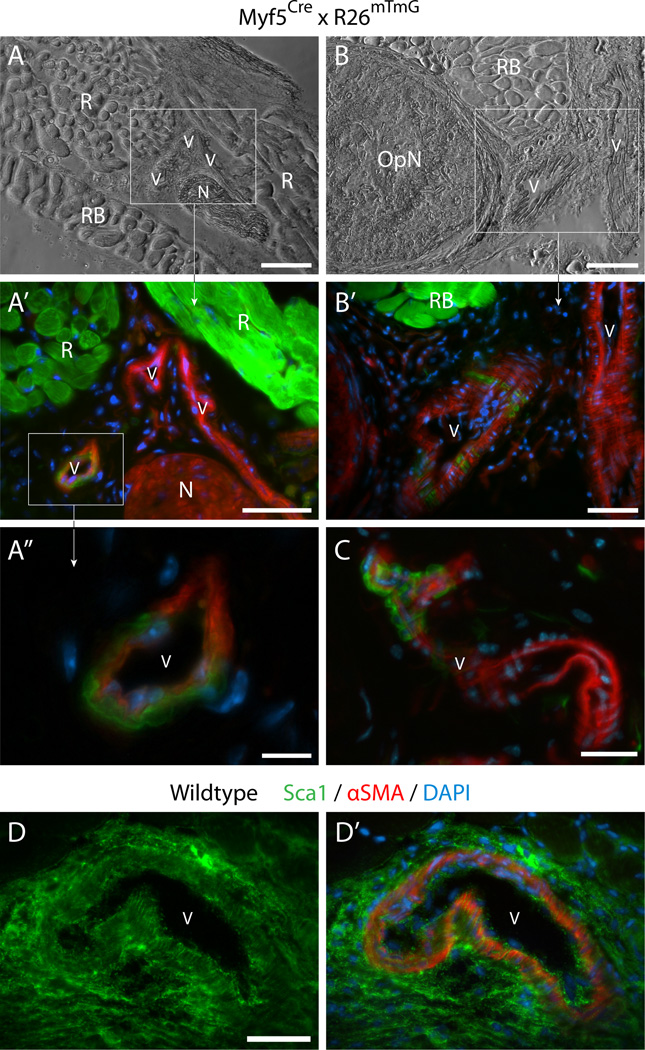
Fig. 4. Histological detection of Myf5Cre-driven GFP+ cells in connective tissues of periocular preparations.
(A–B”) Cross sections from Myf5Cre × R26mTmG mice analyzed for GFP and Tomato fluorescence. (C–D’) Cross sections from wildtype mice labeled by double-immunofluorescence with anti-Sca1 and anti-lamininSca1 marked cells within the periocular connective tissues and laminin identified the basal lamina of individual myofibers, delineating the rectus muscle. Unlike the straightforward Sca1 immunostaining of wildtype tissue, Sca1 immunolabeling of tissue sections from the Myf5Cre × R26mTmG reporter line required using the far-red or the blue channels, but these did not provide effective detection. First, as the red fluorescence bleeds into the far-red channel, and as both the Tomato and Sca1 signals are localized to the cytoplasmic membrane, determination of Sca1 staining in far-red is unreliable. Second the use of a number of secondary antibodies conjugated with different types of blue fluorophores yielded only extremely faint label with a low signal-to-noise ratio. Images shown in the figure are from cross sections at the level of the eyeball, either (A-A”) and (C-C’) posterior, or (B-B”) and (D-D’) anterior to the attachment of the EOM to the eye. R, rectus muscle. In all images DAPI staining is shown in blue. Scale bars in (A) and (B), 100µm, and in (A”), (B”), (C) and (D), 50µm.
First, GFP+ cells were detected within the connective tissue directly surrounding the muscle (Fig. 4A-A”) and within the fascia bulbi and sclera, enveloping and separating the eye from the EOM and other orbital structures (Fig. 4B-B”). These GFP+ cells that clearly reside outside the muscle masses, are likely to be a source of the non-myogenic GFP+ sorted cells. In this case, given that our FACS and ex-vivo analyses showed that the non-myogenic Myf5Cre-driven GFP+ cells were exclusively Sca1+ while the myogenic ones were Sca1− (Fig. 3A-D’), the GFP+ cells residing in the connective tissues would be anticipated to exhibit Sca1 signal. Sca1 immunolabeling of periocular preparations from the dual GFP/Tomato Myf5Cre × R26mTmG reporter line required using the far-red or the blue channels, which in our hands did not provide reliable detection (as detailed in Fig. 4 legend). Nevertheless, immunostaining of control periocular preparations from wildtype mice revealed extensive Sca1 expression throughout the connective tissue structures identified as containing the Myf5Cre-driven GFP expressing cells (Fig. 4C-D’).
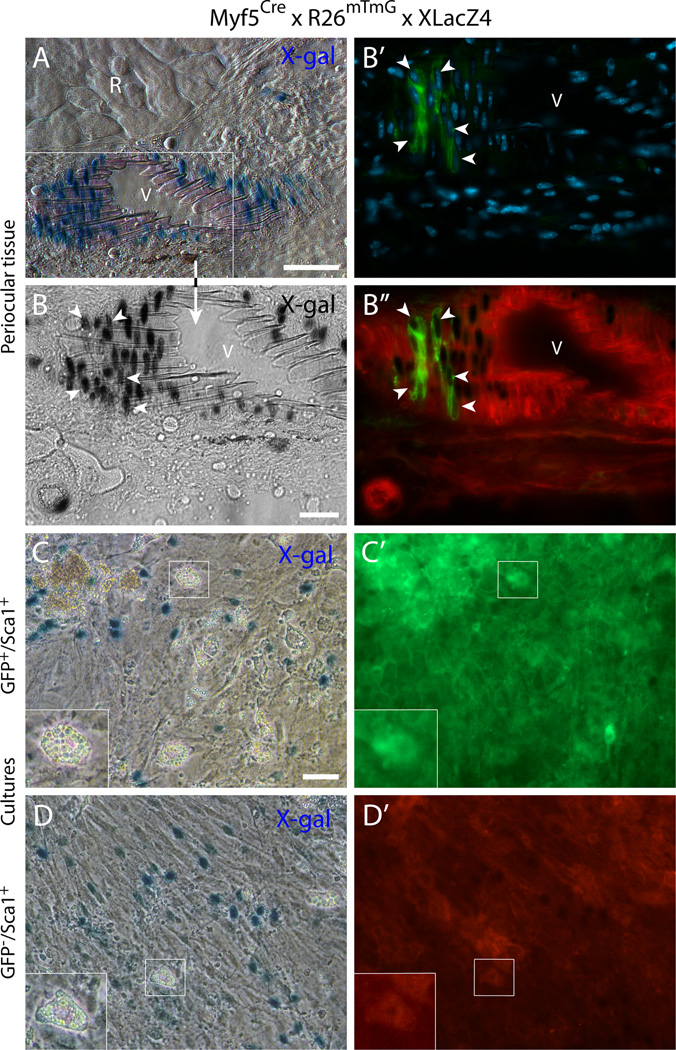
Second, a distinct contribution of Myf5Cre-driven GFP+ cells in non-muscle locations was also revealed in perivascular sites of some major blood vessels within the periocular tissue. Several examples are presented in Fig. 5A-C along with the strong GFP signal in the muscle masses (Fig. 5A-A’and B-B’R and RB identify regions of rectus and retractor bulbi muscles, respectively). Interestingly, images in Fig. 5A and A’depict three blood vessels side by side, with one of them harboring Myf5Cre-driven GFP+ cells. The perivascular locations in which the Myf5Cre-driven GFP+ cells were identified, were shown to express Sca1 in cross sections from control wildtype mice (Fig. 5D and D’). The perivascular Myf5Cre-driven GFP+ cells appeared residing in the anatomical position of smooth muscle cells, as revealed in the high magnification images (Fig. 5A”, B’and C). The possibility that the perivascular GFP+ cells could indeed represent vascular smooth muscle was investigated further using the double reporter mouse, Myf5Cre × R26mTmG × XLacZ4. In the XLacZ4 reporter, nuclear β-gal expression distinctly identifies mural cells (vascular smooth muscle and pericytes) in skeletal muscle and associated connective tissue of adult mice in all muscle groups we have examined (Stuelsatz et al., 2012). In the current EOM study, the vascular-associated Myf5Cre-driven GFP+ cells were clearly localized in the same region as the XLacZ4+ mural cells. The detection of cells co-expressing both GFP and XLacZ4 has further validated the smooth muscle phenotype of these Myf5Cre-driven GFP+ cells (Fig. 6A-B”). When sorting GFP+ cells from the Myf5Cre × R26mTmG × XLacZ4 double reporter mouse into Sca1+ and Sca1− populations (as in Fig. 2), only the GFP+/Sca1+ population depicted XlacZ4+ cells when analyzed in cell culture (Fig. 6C and C’). Furthermore, in agreement with the presence of GFP− (Tomato+) cells in the presumptive smooth muscle layer (Fig. 6A-B”), the sorted GFP−/Sca1+ population also contained XLacZ4+ cells following culturing (Fig. 6D and D’).
Fig. 5. Histological detection of Myf5Cre-driven GFP+ cells in the vasculature of periocular preparations.
(A-C’) Cross sections from Myf5Cre × R26mTmG mice at the level of the optic nerve, immediately posterior to the eye. OpN, optic nerveN, peripheral nerveR, rectus muscleRB, retractor bulbi muscle; V, blood vessel. (D) and (D’) A Cross section from a wildtype mouse labeled by double-immunofluorescence with anti-Sca1 and anti-αSMA, (D) shows Sca1 staining aloneSca1 marked both the smooth muscle cells in the region closer to lumen, coinciding with α-smooth muscle actin (αSMA) staining (Skalli et al., 1986), and the perivascular cells in the more external adventitial region. In all images DAPI staining is shown in blue. Scale bars in (A) and (B), 100µm, in (A’), (B’), and (D), 50µm, in (C), 25µm, and in (A”), 10µm.
Fig. 6. Histological and cell culture analyses of the double reporter mouse Myf5Cre × R26mTmG × XLacZ4 establish a connection between the vascular-associated (Myf5Cre-driven) GFP+ cells and the mural cell marker XLacZ4.
(A-B”) Cross-section from periocular preparation at the level of the optic nerve, immediately posterior to the eye, depicting (A) X-gal+- cells at the wall of a blood vessel close to a rectus muscle and (B-B”) a higher magnification view of the same blood vessel, identifying DAPI-stained cells at the vessel wall that co-express nuclear β-gal and GFP (white arrowheads). R, rectus muscleV, blood vessel. (C-D’) Representative images of day 14 cultures of (C-C’) GFP+/Sca1+ cells and (D-D’) GFP/Sca1+ cells after staining with X-gal. The two populations, isolated from EOM preparations, were sorted by FACS from G0-G1 cells after depletion of CD31+ and CD45+ cells. Inserts in the lower left corner of (C) and (D) represent higher magnification views of the regions delineated by a white box in each corresponding panel, depicting adipogenic cells that develop spontaneously in these cultures with time. Scale bars in (A) and (C), 50µm, and in (B), 25µm.
Overall, these histological studies revealed the presence of an appreciable population of Myf5Cre-driven GFP+ cells distributed between the connective tissues surrounding the EOM and the eye, and the perivascular compartment. These histologically-detected GFP+ cells show, in common with the isolated non-myogenic GFP+ cells, the expression of the Sca1 cell surface antigen and the XLacZ4 transgene. This indicates that the GFP+/Sca1± cells detected in our EOM preparations by flow cytometry are plausibly derived from the GFP+ cells observed in our histological studies of periocular preparations. Nevertheless, we recognize that the histological preparations may contain additional cells beyond what is included in the EOM preparations used in cell sorting. It is also possible that the histological analyses (Fig. 4 and Fig. 5) did not provide a complete inventory of all contributing sources of GFP+ cells. For example, the high microvascular density within EOM masses (Kjellgren et al., 2004) makes it desirable to consider the presence of Myf5Cre-driven GFP+ cells also in the interstitium between myofibers. However, the strong GFP fluorescent signal associated with the myofibers limits the ability to perform such analyses, even when employing high magnification.
Myf5Cre-driven GFP expression in non-myogenic periocular sites does not reflect current expression of Myf5
We used the Myf5nLacZ mouse to investigate current Myf5 gene expression in the harvested EOM tissue. In this Myf5nLacZ knockin line, one of the Myf5 alleles was modified to direct nuclear LacZ expression (Tajbakhsh et al., 1996), resulting in β-gal protein expression in the nuclei of satellite cells (Beauchamp et al., 2000). We have used this reporter mouse extensively to analyze Myf5 expression in satellite cells in isolated myofibers, cross sections and whole muscles (Day et al., 2007; Day et al., 2010; Stuelsatz et al., 2012). Using these three approaches, we only found My5nLacZ expression associated with myofibers in EOM while the vasculature and other connective tissue sites were negative (data not shown).
Do the non-myogenic Myf5Cre-driven GFP+ cells share a common origin with satellite cells?
The expression of Myf5Cre-driven GFP in non-myogenic cells of the EOM tissue could have been initiated during development or postnatal life due to one of the following possibilities:
(i) Transition of myogenic cells into the non-myogenic lineage. This possibility contrasts with the clear demonstration that cells that have expressed MyoD during embryogenesis are essentially all committed to the myogenic lineage (Gerhart et al., 2007; Kanisicak et al., 2009). Whereas satellite cells uniformly demonstrate ancestral expression of MyoD (Kanisicak et al., 2009), adipogenic cells that have been occasionally observed in satellite cell cultures and initially considered to represent non-myogenic derivative of satellite cells (Asakura et al., 2001; Shefer et al., 2004), were proven negative for MyoD-driven reporter, hence, not of satellite cell origin (Starkey et al., 2011). Along this vein, our own flow cytometry studies with the MyoDCre driver line, as briefly detailed in the current study, have shown that all cells expressing this reporter were exclusively myogenic. A recent study nonetheless concluded based on Pax7 and Myf5 lineage marking of satellite cells, that these myogenic cells could give rise to brown adipocytes, albeit at a very low frequency (Yin et al., 2013). Thus, it is possible that a trace of myogenic-related cells, capable of giving rise to adipogenic progenitors, does not go through a MyoDCre-driven reporter expression, and therefore are only detected by Pax7Cre or Myf5Cre drivers. However, the abundance of non-myogenic Myf5Cre driven GFP+ cells identified in the current EOM study would require that a large population of myogenic cells had not gone through MyoD expression, a possibility for which there is no precedence.
(ii) Production of two separate lineages, myogenic and non-myogenic, from a common Myf5-expressing progenitor prior to the acquisition of the complete panel of skeletal muscle progenitor features. Indeed, Myf5Cre-driven reporter, while primarily expressed in the myogenic lineage when analyzed in the context of limb muscles (Kuang et al., 2007), has been also detected in a sub-set of brown adipose tissues and such expression has been linked to a transition of somitic progenitors already expressing some myogenic markers to a non-myogenic fate (Seale et al., 2008).
(iii) The non-myogenic and myogenic populations are not related and derived from two independent progenitors. Indeed, Myf5 expression has been noted outside the myogenic compartment, in the dermomyotome and the sclerotome, as well as in the developing brain and in the neural tube (Daubas et al., 2000; Gensch et al., 2008; Kiefer and Hauschka, 2001; Tajbakhsh and Buckingham, 1995). This has raised uncertainties about the significance of Myf5 expression as an exclusive marker for myogenic lineage derivatives. This hypothesis of a distinctive origin of the two Myf5Cre-driven GFP+ populations is strongly supported by our studies detailed next in which we used the Pax3Cre driver line (Fig. 7 and Fig. 8). While EOMs myogenic cells do not show ancestral expression of Pax3, the periocular connective tissues are known to be contributed by the neural crest (Creuzet et al., 2005; Gage et al., 2005; Noden, 1983), a Pax3-expressing lineage (Epstein et al., 1991; Goulding et al., 1991; Li et al., 2000). Here, we reasoned that examining the Pax3 status of the non-myogenic sites/cells identified earlier with the Myf5Cre driver would permit distinguishing the latter non-myogenic cells from the EOM myogenic lineage.
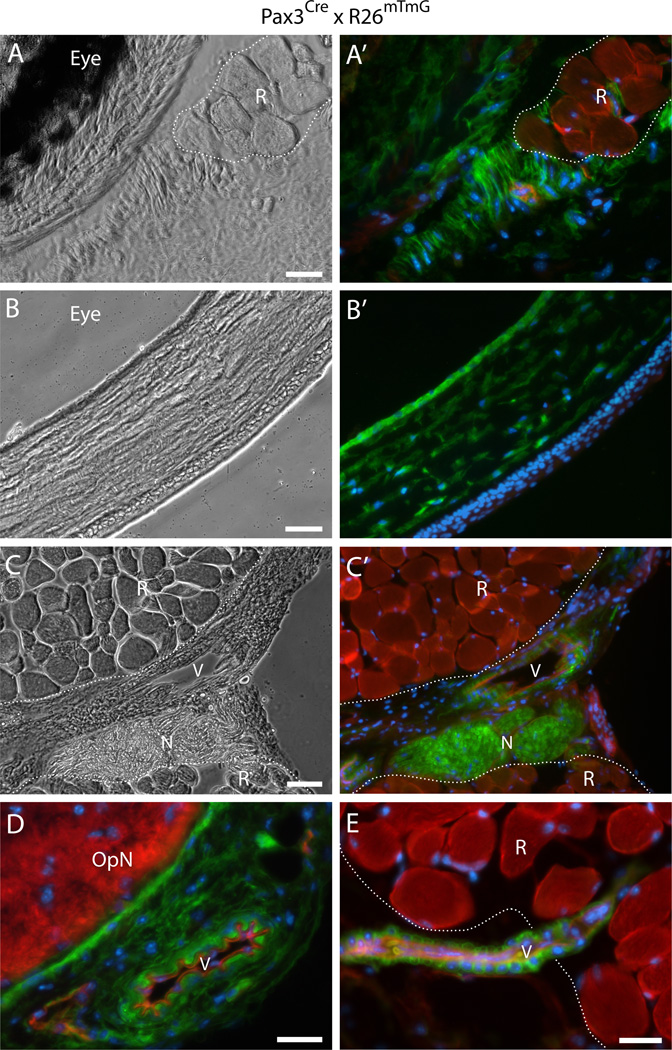
Fig. 7. Histological detection of Pax3Cre-driven GFP+ cells in connective tissues and vasculature of periocular preparations.
(A–E) Cross sections from Pax3Cre × R26mTmG mice at the level of the eye, either (A-A’) posterior, or (B-B’) anterior to the attachment of the EOM to the sclera, or (C-E) at the level of the optic nerve, immediately posterior to the eye. OpN, optic nerveN, peripheral nerveR, rectus muscleV, blood vessel. Noticeably, the connective tissues which originate from the neural crest (as discussed in the “Results”section) and most nervous tissues appeared to be from Pax3 lineage origin (GFP+), with the exception of the optic nerve (seen in D and D’) which derived from the neural ectoderm that form the optic stalk (Evans and Gage, 2005; Pei and Rhodin, 1970; Schwarz et al., 2000). In all images, DAPI staining is shown in blue. Scale bars in (A), (D) and (E), 25µm, in (B) and (C), 50µm.
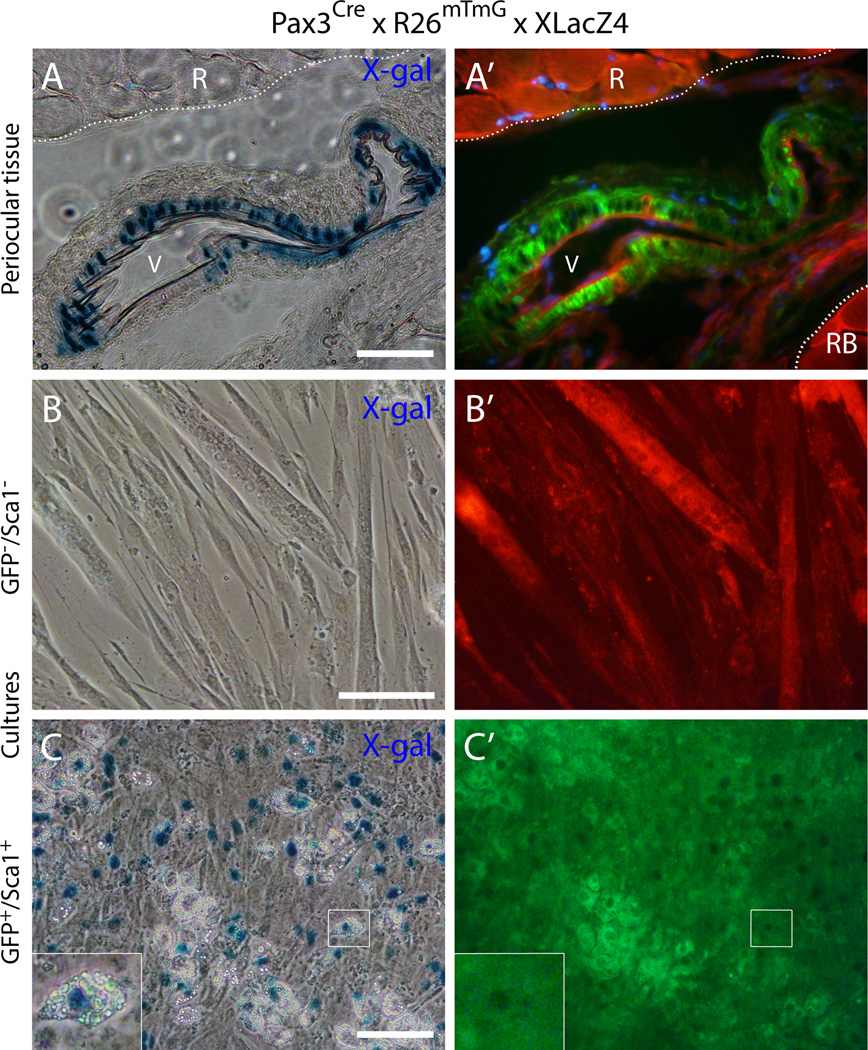
Fig. 8. Histological and cell culture analyses of periocular and EOM preparations from the double reporter mouse Pax3Cre × R26mTmG × XLacZ4.
(A) and (A’) Cross-section from periocular preparation at the level of the optic nerve, immediately posterior to the eye, depicts a blood vessel close to a rectus muscle identifying GFP+ cells at the vessel wall that co-express nuclear β-gal. R, rectus muscleRB, retractor bulbi muscleV, blood vessel. DAPI staining is shown in blue in (A’). (B-C’) Representative images of day 14 cultures of (B-B’) GFP−/Sca1− cells and (C-C’) GFP+/Sca1+ cells after staining with X-gal. Cells were isolated from EOM preparations by FACS and the specified populations were isolated from G0-G1 cells depleted of CD31+ and CD45+ cells. Insert in the lower left corner of (C) represents a higher magnification view of the region delineated by a white box in each corresponding panel, depicting an adipogenic cell expressing the XLacZ4 transgene. Scale bars in (A), 50µm, and in (B) and (C), 100µm.
According to a detailed histological analysis throughout the axial length of periocular preparations, we concluded that all connective tissue structures identified earlier as containing Myf5Cre-driven GFP+ cells (Fig. 5 and Fig. 6) were from a Pax3 lineage origin based on Pax3Cre-driven GFP expression as shown in Fig. 7. These included the extensive connective tissue structures that surround the eye and the muscles (Fig. 7A-B’) and perivascular cells (Fig. 7C-E). Furthermore, by crossing the Pax3Cre × R26mTmG and the mural cell reporter (XLacZ4) lines, we are able to show that the vascular-associated Pax3Cre-driven GFP+ cells are bona fide smooth muscle (i.e., XLacZ4+, Fig. 8A and A’), in accord with the Myf5Cre-driven GFP+ vascular-associated cells (Fig. 6A-B”). EOM preparations from the Pax3Cre × R26mTmG × XLacZ4 double reporter mouse processed by flow cytometry and analyzed in cell culture (in the same fashion as in Fig. 2C-D’and 3), do demonstrate distinctive lineage origin for the myogenic progenitors (GFP−, i.e. Pax3-negative lineage) and the cells that harbor a fibro/adipogenic potential (GFP+, i.e. Pax3-positive lineage) (Fig. 8B-C’). Overall our sort data show that Pax3-driven GFP+ cells represent 25–35%of total G0-G1 cells and are equally distributed between Sca1+ and Sca1− populations, with the GFP+/Sca1+ cells accounting for 60–70%of all the Sca1+ cells. Myogenic progenitors were clearly evident in the GFP−/Sca1− population that demonstrated obvious myotubes (Fig. 8B and B’). Fibro/adipogenic cells were solely detected in the GFP+/Sca1+ population, which also gave rise to numerous XLacZ4+ cells (Fig. 8C and C’). Some of the XLacZ4+ cells spontaneously differentiated into adipocytes, suggesting a connection with mural cells for at least some of the adipocytes developing from this population. Importantly, the EOM fibro/adipogenic progenitors while distributed into GFP+/Sca1+ and GFP−/Sca1+ populations in the Myf5Cre reporter line, are all confined to a single population, GFP+/Sca1+, in the Pax3Cre driver, confirming a Pax3 lineage origin for the non-myogenic Myf5Cre-driven GFP+ cells. The two additional populations resulting from our flow cytometry scheme (i.e. GFP+/Sca1− and GFP−/Sca1+for a similar sort approach see Fig. 2C-D’) grew very poorly in our culture conditions when sorting EOM cells from the Pax3Cre × R26mTmG reporter line mouse and have not been studied further.
Altogether, our FACS and culture studies of EOM cells from the Myf5Cre and Pax3Cre reporter lines indicate that the non-myogenic Myf5Cre-driven GFP+ cells represent a subset of the broader Sca1+ population distinguished by the Pax3Cre-driven reporter expression. These results accord with our histological studies showing that Sca1 and Pax3Cre-driven GFP expressions encompassed the sites containing Myf5Cre-driven GFP+ cells identified within the periocular connective tissues. Collectively, the absence of non-myogenic progeny in the MyoDCre-driven GFP+ population and the phenotypic/functional overlap between the non-myogenic Myf5Cre- and Pax3Cre- expressing cells, support a developmental origin for the non-myogenic Myf5Cre-driven GFP+ cells that is distinct from the EOM myogenic lineage. We are further proposing that these non-myogenic Myf5Cre-driven GFP+ cells are derived from a neural crest origin, similarly to the periocular connective tissues in which they are contained. In the context of the head tissues, Pax3 expression, has indeed, been exclusively reported in the migratory neural crest cells and notably found absent from the head mesoderm (Hacker and Guthrie, 1998; Mootoosamy and Dietrich, 2002). Our pilot histological analyses of periocular preparations from the Wnt1-Cre driven reporter (Fig. S1), an independent neural crest lineage marker (Danielian et al., 1998; Echelard et al., 1994; Jiang et al., 2000), indeed show that Wnt1-Cre driven reporter expression is evident in the connective tissues identified as harboring Myf5Cre-expressing cells (Fig. 4 and Fig. 5) and recapitulates the pattern observed with the Pax3Cre driver (Fig. 7).
Whereas the previously reported subsets of adipose tissues harboring ancestral Myf5 expression could potentially be attributed to a myogenic-related origin (Sanchez-Gurmaches and Guertin, 2013; Seale et al., 2008; Yin et al., 2013), the current study presents an alternative mechanism for the establishment of such adipogenic cells from neural crest. This path to establishing adipogenic cells may prevail in structures rich in neural crest contribution as the periocular tissue (current study) and as it has been shown for other cephalic locations (Billon et al., 2007; Lemos et al., 2012). This alternative mechanism should also be considered in limb and body tissues as derivative of neural crest cells have been found in various adult tissues and it is further held that some embryonic neural crest cells persist throughout adult development as progenitor cells (Crane and Trainor, 2006; Le Lievre and Le Douarin, 1975; Teng and Labosky, 2006). Indeed, recent large-scale genomics studies and in vivo lineage tracing approaches have indicated that individual fat depots located in various sites of the body can differ in their developmental origin (reviewed in Billon and Dani, 2012). Our unpublished work on fibro/adipogenic cells in limb muscles demonstrated that those cells were not of Myf5 nor Pax3 origin, as also indicated by an independent study of limb muscle (Liu et al., 2012). Differently, here we have provided evidence for the existence of fibro/adipogenic progenitors in EOM tissue and revealed their unique lineage origin where all are derived from Pax3-expressing cells while some also presenting a Myf5-expressing origin. This ancestral Myf5 expression however, does not connote a myogenic lineage origin.
Conclusions
The present study is the first to investigate Myf5Cre-driven reporter expression in adult EOM and report a dominant population that is non-myogenic and harbors the Myf5-driven reporter, in addition to the anticipated myogenic population that represents bona fide satellite cells. Our flow cytometry data show that this unexpected population represents nearly 50%of all Myf5Cre-driven GFP+ cells and 10%of the broader Sca1+ (CD31−/CD45−) population. The non-myogenic GFP+/Sca1+ and GFP−/Sca1+ cells from the Myf5Cre reporter mouse essentially represent sub-populations of the GFP+/Sca1+ cells from the Pax3Cre reporter mouse, all demonstrating characteristics of fibro/adipocyte progenitors with spontaneous development of adipocytes in cell culture. Our Histological studies have further indicated that at least some of the non-myogenic Myf5Cre-driven GFP+ cells are located in the connective tissues surrounding the eye and EOM and in perivascular sites reflecting mural cells (XLacZ4+). Whether such an ancestral expression of Myf5 also means antecedence of Myf5 activity is unknown. It is possible that the complexity of the regulatory elements of the Myf5 gene (Buchberger et al., 2003; Francetic and Li, 2011; Hadchouel et al., 2003; Summerbell et al., 2000) leads to increased propensity of this gene to be expressed ectopically during early development and that unwarranted Myf5 protein expression (and function) is ensured by post-transcriptional regulation (Crist et al., 2012). It remains, however, possible that Myf5 also regulates yet an unidentified cascade in fibro/adipogenic lineages. In all, our lineage tracing based on multiple reporter lines has demonstrated that despite common ancestral expression of Myf5, there is a clear distinction between periocular myogenic and non-myogenic cell lineages according to their mutually exclusive antecedence of MyoD and Pax3 expression.
Supplementary Material
Acknowledgments
We are grateful to Donna Prunkard and Dr. Peter Rabinovitch for their valuable assistance with cell sorting (performed at the core facility of the University of Washington Nathan Shock Center of Excellence). This work was supported by grants to Zipora Yablonka-Reuveni from the National Institutes of Health (AG021566AG035377AR057794) and the Muscular Dystrophy Association (135908).
References
- Asakura A, Komaki M, Rudnicki M. Muscle satellite cells are multipotential stem cells that exhibit myogenic, osteogenic, and adipogenic differentiation. Differentiation. 2001;68:245–253. doi: 10.1046/j.1432-0436.2001.680412.x. [DOI] [PubMed] [Google Scholar]
- Beauchamp JR, Heslop L, Yu DS, Tajbakhsh S, Kelly RG, Wernig A, Buckingham ME, Partridge TA, Zammit PS. Expression of CD34 and Myf5 defines the majority of quiescent adult skeletal muscle satellite cells. J. Cell Biol. 2000;151:1221–1234. doi: 10.1083/jcb.151.6.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billon N, Dani C. Developmental origins of the adipocyte lineage: new insights from genetics and genomics studies. Stem Cell Rev. 2012;8:55–66. doi: 10.1007/s12015-011-9242-x. [DOI] [PubMed] [Google Scholar]
- Billon N, Iannarelli P, Monteiro MC, Glavieux-Pardanaud C, Richardson WD, Kessaris N, Dani C, Dupin E. The generation of adipocytes by the neural crest. Development. 2007;134:2283–2292. doi: 10.1242/dev.002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biressi S, Bjornson CR, Carlig PM, Nishijo K, Keller C, Rando TA. Myf5 expression during fetal myogenesis defines the developmental progenitors of adult satellite cells. Dev. Biol. 2013 Apr 29; doi: 10.1016/j.ydbio.2013.04.021. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A, Nomokonova N, Arnold HH. Myf5 expression in somites and limb buds of mouse embryos is controlled by two distinct distal enhancer activities. Development. 2003;130:3297–3307. doi: 10.1242/dev.00557. [DOI] [PubMed] [Google Scholar]
- Buckingham M, Bajard L, Chang T, Daubas P, Hadchouel J, Meilhac S, Montarras D, Rocancourt D, Relaix F. The formation of skeletal muscle: from somite to limb. J. Anat. 2003;202:59–68. doi: 10.1046/j.1469-7580.2003.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane JF, Trainor PA. Neural crest stem and progenitor cells. Annu. Rev. Cell Dev. Biol. 2006;22:267–286. doi: 10.1146/annurev.cellbio.22.010305.103814. [DOI] [PubMed] [Google Scholar]
- Creuzet S, Vincent C, Couly G. Neural crest derivatives in ocular and periocular structures. Int. J. Dev. Biol. 2005;49:161–171. doi: 10.1387/ijdb.041937sc. [DOI] [PubMed] [Google Scholar]
- Crist CG, Montarras D, Buckingham M. Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell. 2012;11:118–126. doi: 10.1016/j.stem.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Danoviz ME, Yablonka-Reuveni Z. Skeletal muscle satellite cells: background and methods for isolation and analysis in a primary culture system. Methods Mol. Biol. 2012;798:21–52. doi: 10.1007/978-1-61779-343-1_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubas P, Tajbakhsh S, Hadchouel J, Primig M, Buckingham M. Myf5 is a novel early axonal marker in the mouse brain and is subjected to post-transcriptional regulation in neurons. Development. 2000;127:319–331. doi: 10.1242/dev.127.2.319. [DOI] [PubMed] [Google Scholar]
- Day K, Shefer G, Richardson JB, Enikolopov G, Yablonka-Reuveni Z. Nestin-GFP reporter expression defines the quiescent state of skeletal muscle satellite cells. Dev. Biol. 2007;304:246–59. doi: 10.1016/j.ydbio.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day K, Shefer G, Shearer A, Yablonka-Reuveni Z. The depletion of skeletal muscle satellite cells with age is concomitant with reduced capacity of single progenitors to produce reserve progeny. Dev. Biol. 2010;340:330–343. doi: 10.1016/j.ydbio.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echelard Y, Vassileva G, McMahon AP. Cis-acting regulatory sequences governing Wnt-1 expression in the developing mouse CNS. Development. 1994;120:2213–2224. doi: 10.1242/dev.120.8.2213. [DOI] [PubMed] [Google Scholar]
- Engleka KA, Gitler AD, Zhang M, Zhou DD, High FA, Epstein JA. Insertion of Cre into the Pax3 locus creates a new allele of Splotch and identifies unexpected Pax3 derivatives. Dev. Biol. 2005;280:396–406. doi: 10.1016/j.ydbio.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Epstein DJ, Vekemans M, Gros P. Splotch (Sp2H), a mutation affecting development of the mouse neural tube, shows a deletion within the paired homeodomain of Pax-3. Cell. 1991;67:767–774. doi: 10.1016/0092-8674(91)90071-6. [DOI] [PubMed] [Google Scholar]
- Evans AL, Gage PJ. Expression of the homeobox gene Pitx2 in neural crest is required for optic stalk and ocular anterior segment development. Hum. Mol. Genet. 2005;14:3347–3359. doi: 10.1093/hmg/ddi365. [DOI] [PubMed] [Google Scholar]
- Francetic T, Li Q. Skeletal myogenesis and Myf5 activation. Transcription. 2011;2:109–114. doi: 10.4161/trns.2.3.15829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage PJ, Rhoades W, Prucka SK, Hjalt T. Fate maps of neural crest and mesoderm in the mammalian eye. Invest. Ophthalmol. Vis. Sci. 2005;46:4200–4208. doi: 10.1167/iovs.05-0691. [DOI] [PubMed] [Google Scholar]
- Gayraud-Morel B, Chretien F, Jory A, Sambasivan R, Negroni E, Flamant P, Soubigou G, Coppee JY, Di Santo J, Cumano A, Mouly V, Tajbakhsh S. Myf5 haploinsufficiency reveals distinct cell fate potentials for adult skeletal muscle stem cells. J. Cell Sci. 2012;125:1738–1749. doi: 10.1242/jcs.097006. [DOI] [PubMed] [Google Scholar]
- Gensch N, Borchardt T, Schneider A, Riethmacher D, Braun T. Different autonomous myogenic cell populations revealed by ablation of Myf5-expressing cells during mouse embryogenesis. Development. 2008;135:1597–1604. doi: 10.1242/dev.019331. [DOI] [PubMed] [Google Scholar]
- Gerhart J, Neely C, Elder J, Pfautz J, Perlman J, Narciso L, Linask KK, Knudsen K, George-Weinstein M. Cells that express MyoD mRNA in the epiblast are stably committed to the skeletal muscle lineage. J. Cell Biol. 2007;178:649–660. doi: 10.1083/jcb.200703060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulding MD, Chalepakis G, Deutsch U, Erselius JR, Gruss P. Pax-3, a novel murine DNA binding protein expressed during early neurogenesis. EMBO J. 1991;10:1135–1147. doi: 10.1002/j.1460-2075.1991.tb08054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire FM, Smas CM, Sul HS. Understanding adipocyte differentiation. Physiol. Rev. 1998;78:783–809. doi: 10.1152/physrev.1998.78.3.783. [DOI] [PubMed] [Google Scholar]
- Gros J, Manceau M, Thome V, Marcelle C. A common somitic origin for embryonic muscle progenitors and satellite cells. Nature. 2005;435:954–958. doi: 10.1038/nature03572. [DOI] [PubMed] [Google Scholar]
- Hacker A, Guthrie S. A distinct developmental programme for the cranial paraxial mesoderm in the chick embryo. Development. 1998;125:3461–3472. doi: 10.1242/dev.125.17.3461. [DOI] [PubMed] [Google Scholar]
- Hadchouel J, Carvajal JJ, Daubas P, Bajard L, Chang T, Rocancourt D, Cox D, Summerbell D, Tajbakhsh S, Rigby PW, Buckingham M. Analysis of a key regulatory region upstream of the Myf5 gene reveals multiple phases of myogenesis, orchestrated at each site by a combination of elements dispersed throughout the locus. Development. 2003;130:3415–3426. doi: 10.1242/dev.00552. [DOI] [PubMed] [Google Scholar]
- Harel I, Nathan E, Tirosh-Finkel L, Zigdon H, Guimaraes-Camboa N, Evans SM, Tzahor E. Distinct origins and genetic programs of head muscle satellite cells. Dev. Cell. 2009;16:822–832. doi: 10.1016/j.devcel.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst D, Ustanina S, Sergi C, Mikuz G, Juergens H, Braun T, Vorobyov E. Comparative expression analysis of Pax3 and Pax7 during mouse myogenesis. Int. J. Dev. Biol. 2006;50:47–54. doi: 10.1387/ijdb.052111dh. [DOI] [PubMed] [Google Scholar]
- Ieronimakis N, Balasundaram G, Rainey S, Srirangam K, Yablonka-Reuveni Z, Reyes M. Absence of CD34 on murine skeletal muscle satellite cells marks a reversible state of activation during acute injury. PLoS ONE. 2010;5:e10920. doi: 10.1371/journal.pone.0010920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieronimakis N, Balasundaram G, Reyes M. Direct isolation, culture and transplant of mouse skeletal muscle derived endothelial cells with angiogenic potential. PLoS ONE. 2008;3:e0001753. doi: 10.1371/journal.pone.0001753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- Joe AW, Yi L, Natarajan A, Le Grand F, So L, Wang J, Rudnicki MA, Rossi FM. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat. Cell Biol. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafadar KA, Yi L, Ahmad Y, So L, Rossi F, Pavlath GK. Sca-1 expression is required for efficient remodeling of the extracellular matrix during skeletal muscle regeneration. Dev. Biol. 2009;326:47–59. doi: 10.1016/j.ydbio.2008.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanisicak O, Mendez JJ, Yamamoto S, Yamamoto M, Goldhamer DJ. Progenitors of skeletal muscle satellite cells express the muscle determination gene, MyoD. Dev. Biol. 2009;332:131–141. doi: 10.1016/j.ydbio.2009.05.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassar-Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V, Tajbakhsh S. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature. 2004;431:466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- Kiefer JC, Hauschka SD. Myf-5 is transiently expressed in nonmuscle mesoderm and exhibits dynamic regional changes within the presegmented mesoderm and somites I-IV. Dev. Biol. 2001;232:77–90. doi: 10.1006/dbio.2000.0114. [DOI] [PubMed] [Google Scholar]
- Kjellgren D, Thornell LE, Virtanen I, Pedrosa-Domellof F. Laminin isoforms in human extraocular muscles. Invest. Ophthalmol. Vis. Sci. 2004;45:4233–4239. doi: 10.1167/iovs.04-0456. [DOI] [PubMed] [Google Scholar]
- Klinghoffer RA, Mueting-Nelsen PF, Faerman A, Shani M, Soriano P. The two PDGF receptors maintain conserved signaling in vivo despite divergent embryological functions. Mol. Cell. 2001;7:343–354. doi: 10.1016/s1097-2765(01)00182-4. [DOI] [PubMed] [Google Scholar]
- Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Lievre CS, Le Douarin NM. Mesenchymal derivatives of the neural crest: analysis of chimaeric quail and chick embryos. J. Embryol. Exp. Morphol. 1975;34:125–154. [PubMed] [Google Scholar]
- Lemos DR, Paylor B, Chang C, Sampaio A, Underhill TM, Rossi FM. Functionally convergent white adipogenic progenitors of different lineages participate in a diffused system supporting tissue regeneration. Stem Cells. 2012;30:1152–1162. doi: 10.1002/stem.1082. [DOI] [PubMed] [Google Scholar]
- Li J, Chen F, Epstein JA. Neural crest expression of Cre recombinase directed by the proximal Pax3 promoter in transgenic mice. Genesis. 2000;26:162–164. doi: 10.1002/(sici)1526-968x(200002)26:2<162::aid-gene21>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- LifeMap. LifeMap Sciences, Inc. 2013 http://discovery.lifemapsc.com/library/images/extraocular-skeletal-muscle-anatomy. [Google Scholar]
- Liu W, Liu Y, Lai X, Kuang S. Intramuscular adipose is derived from a non-Pax3 lineage and required for efficient regeneration of skeletal muscles. Dev. Biol. 2012;361:27–38. doi: 10.1016/j.ydbio.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersmann HJ, Goodman JR, Brown LJ. Development of swine adipose tissue: morphology and chemical composition. J. Lipid Res. 1975;16:269–279. [PubMed] [Google Scholar]
- Mitchell PO, Mills T, O’Connor RS, Kline ER, Graubert T, Dzierzak E, Pavlath GK. Sca-1 negatively regulates proliferation and differentiation of muscle cells. Dev. Biol. 2005;283:240–252. doi: 10.1016/j.ydbio.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Montarras D, Morgan J, Collins C, Relaix F, Zaffran S, Cumano A, Partridge T, Buckingham M. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309:2064–2067. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- Mootoosamy RC, Dietrich S. Distinct regulatory cascades for head and trunk myogenesis. Development. 2002;129:573–583. doi: 10.1242/dev.129.3.573. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Napolitano L. The differentiation of white adipose cells. An electron microscope study. J. Cell Biol. 1963;18:663–679. doi: 10.1083/jcb.18.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noden DM. The embryonic origins of avian cephalic and cervical muscles and associated connective tissues. Am. J. Anat. 1983;168:257–276. doi: 10.1002/aja.1001680302. [DOI] [PubMed] [Google Scholar]
- Noden DM, Francis-West P. The differentiation and morphogenesis of craniofacial muscles. Dev. Dyn. 2006;235:1194–1218. doi: 10.1002/dvdy.20697. [DOI] [PubMed] [Google Scholar]
- Ono Y, Boldrin L, Knopp P, Morgan JE, Zammit PS. Muscle satellite cells are a functionally heterogeneous population in both somite-derived and branchiomeric muscles. Dev. Biol. 2010;337:29–41. doi: 10.1016/j.ydbio.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei YF, Rhodin JA. The prenatal development of the mouse eye. Anat. Rec. 1970;168:105–125. doi: 10.1002/ar.1091680109. [DOI] [PubMed] [Google Scholar]
- Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456:502–506. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivan R, Gayraud-Morel B, Dumas G, Cimper C, Paisant S, Kelly RG, Tajbakhsh S. Distinct regulatory cascades govern extraocular and pharyngeal arch muscle progenitor cell fates. Dev. Cell. 2009;16:810–821. doi: 10.1016/j.devcel.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Sanchez-Gurmaches J, Guertin DA. Adipocyte lineages: Tracing back the origins of fat. Biochim. Biophys. Acta. 2013 Jun 4; doi: 10.1016/j.bbadis.2013.05.027. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schienda J, Engleka KA, Jun S, Hansen MS, Epstein JA, Tabin CJ, Kunkel LM, Kardon G. Somitic origin of limb muscle satellite and side population cells. Proc. Natl. Acad. Sci. USA. 2006;103:945–950. doi: 10.1073/pnas.0510164103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz M, Cecconi F, Bernier G, Andrejewski N, Kammandel B, Wagner M, Gruss P. Spatial specification of mammalian eye territories by reciprocal transcriptional repression of Pax2 and Pax6. Development. 2000;127:4325–4334. doi: 10.1242/dev.127.20.4325. [DOI] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, Tempst P, Rudnicki MA, Beier DR, Spiegelman BM. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shefer G, Van de Mark DP, Richardson JB, Yablonka-Reuveni Z. Satellite-cell pool size does matter: defining the myogenic potency of aging skeletal muscle. Dev. Biol. 2006;294:50–66. doi: 10.1016/j.ydbio.2006.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shefer G, Wleklinski-Lee M, Yablonka-Reuveni Z. Skeletal muscle satellite cells can spontaneously enter an alternative mesenchymal pathway. J. Cell Sci. 2004;117:5393–5404. doi: 10.1242/jcs.01419. [DOI] [PubMed] [Google Scholar]
- Shefer G, Yablonka-Reuveni Z. Isolation and culture of skeletal muscle myofibers as a means to analyze satellite cells. Methods Mol. Biol. 2005;290:281–304. doi: 10.1385/1-59259-838-2:281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalli O, Ropraz P, Trzeciak A, Benzonana G, Gillessen D, Gabbiani G. A monoclonal antibody against alpha-smooth muscle actin: a new probe for smooth muscle differentiation. J. Cell Biol. 1986;103:2787–2796. doi: 10.1083/jcb.103.6.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkey JD, Yamamoto M, Yamamoto S, Goldhamer DJ. Skeletal muscle satellite cells are committed to myogenesis and do not spontaneously adopt nonmyogenic fates. J. Histochem. Cytochem. 2011;59:33–46. doi: 10.1369/jhc.2010.956995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuelsatz P, Keire P, Almuly R, Yablonka-Reuveni Z. A contemporary atlas of the mouse diaphragm: myogenicity, vascularity, and the pax3 connection. J. Histochem. Cytochem. 2012;60:638–657. doi: 10.1369/0022155412452417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerbell D, Ashby PR, Coutelle O, Cox D, Yee S, Rigby PW. The expression of Myf5 in the developing mouse embryo is controlled by discrete and dispersed enhancers specific for particular populations of skeletal muscle precursors. Development. 2000;127:3745–3757. doi: 10.1242/dev.127.17.3745. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S, Bober E, Babinet C, Pournin S, Arnold H, Buckingham M. Gene targeting the myf-5 locus with nlacZ reveals expression of this myogenic factor in mature skeletal muscle fibres as well as early embryonic muscle. Dev. Dyn. 1996;206:291–300. doi: 10.1002/(SICI)1097-0177(199607)206:3<291::AID-AJA6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S, Buckingham ME. Lineage restriction of the myogenic conversion factor myf-5 in the brain. Development. 1995;121:4077–4083. doi: 10.1242/dev.121.12.4077. [DOI] [PubMed] [Google Scholar]
- Tallquist MD, Weismann KE, Hellstrom M, Soriano P. Early myotome specification regulates PDGFA expression and axial skeleton development. Development. 2000;127:5059–5070. doi: 10.1242/dev.127.23.5059. [DOI] [PubMed] [Google Scholar]
- Tanaka KK, Hall JK, Troy AA, Cornelison DD, Majka SM, Olwin BB. Syndecan-4-expressing muscle progenitor cells in the SP engraft as satellite cells during muscle regeneration. Cell Stem Cell. 2009;4:217–225. doi: 10.1016/j.stem.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng L, Labosky PA. Neural crest stem cells. Adv. Exp. Med. Biol. 2006;589:206–212. doi: 10.1007/978-0-387-46954-6_13. [DOI] [PubMed] [Google Scholar]
- Tidhar A, Reichenstein M, Cohen D, Faerman A, Copeland NG, Gilbert DJ, Jenkins NA, Shani M. A novel transgenic marker for migrating limb muscle precursors and for vascular smooth muscle cells. Dev. Dyn. 2001;220:60–73. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1089>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Von Noorden GK, Campos EC. Summary of the Gross Anatomy of the Extraocular Muscles. Binocular Vision and Ocular Motility : Theory and Management of Strabismus. Mosby, St. Louis, MO. 2002:38–51. [Google Scholar]
- Yablonka-Reuveni Z, Day K. Skeletal muscle stem cells in the spotlight: the satellite cell. In: Cohen I, Gaudette G, editors. Regenerating the Heart: Stem Cells and the Cardiovascular System. Humana Press: Springer; 2011. pp. 173–200. Chapter 11. [Google Scholar]
- Yablonka-Reuveni Z, Rivera AJ. Temporal expression of regulatory and structural muscle proteins during myogenesis of satellite cells on isolated adult rat fibers. Dev. Biol. 1994;164:588–603. doi: 10.1006/dbio.1994.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Shook NA, Kanisicak O, Yamamoto S, Wosczyna MN, Camp JR, Goldhamer DJ. A multifunctional reporter mouse line for Cre- and FLP-dependent lineage analysis. Genesis. 2009;47:107–114. doi: 10.1002/dvg.20474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Pasut A, Soleimani VD, Bentzinger CF, Antoun G, Thorn S, Seale P, Fernando P, van Ijcken W, Grosveld F, Dekemp RA, Boushel R, Harper ME, Rudnicki MA. MicroRNA-133 controls brown adipose determination in skeletal muscle satellite cells by targeting Prdm16. Cell Metab. 2013;17:210–224. doi: 10.1016/j.cmet.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.