Abstract
Adult stem cells reside in hypoxic niches and embryonic stem cells (ESCs) are derived from a low oxygen environment. However, it is not clear whether hypoxia is critical for stem cell fate since for example human ESCs (hESCs) are able to self-renew in atmospheric oxygen concentrations as well. We now show that hypoxia can govern cell fate decision since hypoxia alone can revert hESC- or iPSC-derived differentiated cells back to a stem cell-like state, as evidenced by re-activation of an Oct4-promoter reporter. Hypoxia-induced “de-differentiated” cells also mimic hESCs in their morphology, long-term self-renewal capacity, genome wide mRNA and miRNA profiles, Oct4 promoter methylation state, cell surface markers TRA1-60 and SSEA4 expression and capacity to form teratomas. These data demonstrate that hypoxia can influence cell fate decisions and could elucidate hypoxic niche function.
Keywords: hESC, hypoxia, stem cell fate, plasticity
Introduction
Many adult stem cells reside in a hypoxic niche and it is proposed that this low oxygen level in niche regulates stem cell fate. For example long term hematopoietic stem cells (LT-HSC) are found in hypoxic osteoblastic niches [1]. Similarly, inner cell mass and epiblast stem cells (EpiSCs) reside in a hypoxic environment and hypoxia has been shown to be beneficial for the embryonic stem cells (ESCs) or EpiSCs derived from these stages [2-4]. However, it is not clear what the mode of function of hypoxia is in the stem cell niche.
Hypoxia has been shown to promote an undifferentiated state in various stem cell populations [5-10]. Hypoxia Inducible Factors (HIFs) directly regulate the expression of transcription factors implicated in stem cell self-renewal and multipotency and induce hESC signature in cancer cells [11-13]. Hypoxia also enhances reprogramming of fibroblasts into induced pluripotent stem cells (iPSCs; [14, 15]. Some adult stem cells, for example HSC, have highly glycolytic metabolism. One of the key regulators for this process is HIF1α [16]. Similarly, EpiSCs and human ESCs (hESCs) are highly glycolytic, while ESCs are bivalent changing from mitochondrial to glycolytic energy production in demand [17]. HIF1α also regulates this metabolic and developmental transition between pluripotent stages [17]. These findings are in accordance with the importance of hypoxia and HIF-pathway in stem cell fate. However, ESCs can self-renew in normoxia as well and some hypoxic experimental paradigms do not result in a consistent increase in self-renewal suggesting that hypoxia is not essential for maintenance of ESCs [18, 19].
We now show that hypoxia can drive committed cells back to an earlier stage. These hypoxia-induced cells are pluripotent as evidenced by their stem cell marker expression and teratoma forming property. These data are a step toward addressing the function of hypoxia on niche biology. The continuous push toward stem cell fate by the hypoxic niche environment may be essential since stem cells are metastable and have a natural tendency to drift towards differentiation.
Materials and Methods
Tissue culture and hypoxia induction
The H1 (WA-01) and H7 (WA-07) hESC lines were obtained from Wicell Research Institute (Madison, WI, USA). The RUES2 hESC line was obtained from the Rockefeller University (New York, NY, USA). OSLN6 iPSCs were generated from human foreskin fibroblasts [20]. hESCs and iPSCs were cultured as previously described [11, 21]. Briefly, the cells were cultured on a feeder layer of irradiated primary mouse embryonic fibroblasts (MEF) in DMEM/F-12 media supplemented with 20% serum replacer, 1 mM sodium pyruvate, 0.1 mM non-essential amino acids, 50 U/ml penicillin, 50 μg/ml streptomycin, 0.1 mM β-mercapto-ethanol (Sigma-Aldrich, St. Louis, MO), and 4 ng/ml basic fibroblast growth factor (bFGF). Prior to the experiments, the cells were transferred to growth factor reduced Matrigel (Becton Dickinson, Moutainview, CA) in MEF conditioned media (CM). Dispase was used to passage hESC and iPSC colonies. All reagents are from Invitrogen (Carlsbad, CA) unless otherwise specified.
For hypoxic conditions, cells were cultured in multi-gas incubators (Sanyo, San Diego, CA). Nitrogen gas was supplied to the chambers in order to induce a controlled reduced percentage of oxygen. To reduce fluctuation in oxygen level after media changes, either the media was pre-equilibrated under hypoxia or a closed hypoxia workstation (In Vivo2 200 hypoxic station, Ruskinn Technologies, Bridgend, UK) was used. However, some fluctuation can't be avoided in the procedure used in these studies.
For “normoxic” conditions, cells were cultured in incubators containing 5% CO2 and atmospheric concentration of O2, resulting in approximately 20% O2. While 20% is not a “normal” oxygen tension for hESCs in vivo, we will use that nomenclature for the cell culture conditions throughout this paper.
Differentiation of hESCs and “de-differentiation” experiment
Serum forced undirected differentiation of hESCs and iPSCs was induced with the following media formulation: DMEM + Glutamax supplemented with 20% fetal bovine serum (FBS, ES qualified, Invitrogen), penicillin and streptomycin for 4 to 14 days. Similar de-differentiation efficiency was observed when the cells were differentiated up to two weeks. However, the cells differentiated passed two weeks were not de-differentiating with hypoxia alone in the present experimental conditions. The time to reach differentiated cellular morphology varied depending on the cell line and the density they were first plated. The differentiated cells were passaged with Dispase, replated on Matrigel with conditioned media and cultured under either hypoxia (2% O2) or 20% O2. In some cases, plates of differentiated cells were directly transferred in hypoxic chambers or kept at 20% O2. First hESC-like colonies appeared in cells cultured in hypoxia around 5-7 days. When cells became too confluent, they were passaged with Dispase and replated on Matrigel. After two weeks, colonies were picked and plated on mouse embryonic fibroblasts (MEF) and the “de-differentiated” cell line generated was cultured at 20% O2. The differentiated cells cultured at 20% O2 showed a higher percentage of dead cells after passage compared to cells cultured in hypoxia. In order to avoid fluctuations in hypoxia level, in the first three experiments cells differentiated from H1 were passaged in a hypoxic chamber. In later experiments, cells differentiated from hESCs and iPSCs were passaged in a normoxic tissue culture hood with a media equilibrated to hypoxia for a few hours prior to handling. In both cases, between 3 to 5 colonies were obtained in the first week under hypoxia on a plate where 4×105 cells were originally seeded.
Sodium butyrate treatment
In order to assess the effect of HDAC activity in the “de-differentiation” process, the HDAC inhibitor sodium butyrate (1mM, Sigma) was added to the conditioned media when 6-day differentiated H7 cells were transferred to hypoxia (2% O2). Media with or without sodium butyrate was changed every other day.
Stable transfection of hESCs with Oct4-GFP reporter construct, cell sorting and environmental scope analysis
A 3.9kb OCT4 promoter region-GFP fusion construct was linearized using Apal I restriction enzyme and transfected into cells using lipofectamine 2000 as previously described [22]. Since the vector also contained a neomycin resistance gene regulated by an SV40 promoter, cells were treated for two weeks with 200μg/ml of G418 to select for those that stably integrated and expressed the transgene. GFP expression was assessed in H1 cells by fluorescence microscopy (Leica) as well as flow cytometry (FACS Canto, BD).
4 day differentiated Oct4-GFP hESCs were harvested by trypsinization, washed, and resuspended in hESC media for cell sorting. Fluorescence-activated cell sorting was performed using a FACS Aria flow cytometer (Becton-Dickinson) based on green fluorescence intensity. An equal number of GFP negative cells (4×105 cells) were plated in high (20%) or low (2%) oxygen on 35mm Matrigel-coated plates in presence of conditioned media.
After 4 days of serum-induced differentiation, H1 Oct4-GFP cells were cultured in hESC medium within an environmental imaging apparatus (Zeiss) and maintained in hypoxia (2% O2). Bright field and fluorescence images were taken every 3 hours.
“Traffic light” system
“Traffic light” H7 cells ([23], Fig.4A-B) growing on Matrigel were differentiated using 20% serum without CM or FGF. After two days of serum forced differentiation the colonies had dispersed to single flat cells. After differentiation for two days these cells were infected with the CK7-CRE lentivirus (∼3,500 lentiviral particles per cell) in presence of Polybrene (4μg/ml) [23]. Pictures of 6-day differentiated cells were taken with a fluorescence microscope (Leica). Cells were then cultured in hESC media under either normoxia (20%O2) or hypoxia (2%O2) and additional pictures were taken to monitor the appearance of green colonies. In some conditions (in particular infection of hESCs as single cells in suspension with high virus titer) some GFP expression could be detected immediately in hESCs [23]. However, the infection conditions in the data shown here used lower virus titer on pre-plated cells. In order to rule out the possibility of leakiness of the GFP from the construct in our system, undifferentiated “traffic light” H7 cells were infected with the CK7-CRE lentivirus 4 days prior to analysis. No obvious leakiness was observed in these conditions.
Figure 4. Cells expressing a differentiation marker “de-differentiate” to hESC-like colonies in hypoxia.
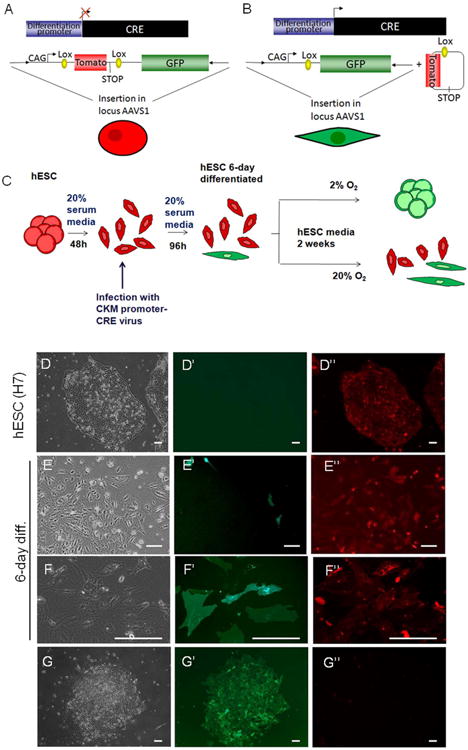
(A-B) Schematic representation of the “traffic light” system in undifferentiated hESCs (A) and differentiated cells (B, see Materials and Methods for details). (C) Flowchart of the experiment. Traffic light H7 cells expressing Tomato were differentiated during 6 days using 20% serum without conditioned media or FGF. At day 2 of differentiation, cells were infected with a lentivirus containing CK7-CRE. Cells were then cultured in hESC media at either 20%O2 or 2%O2 for 2 weeks. (D-G) Bright field, GFP and Tomato fluorescence channel images of undifferentiated H7 cells (D), 6-day differentiated H7 cells (E-F) and H7 cells “de-differentiated” during 15 days in hypoxia (G). Bars represent 100μm.
Retinal progenitor induction
Hypoxia “de-differentiated” cells were differentiated into retinal progenitors as previously described [24]. Briefly, cells were aggregated in six-well ultra-low attachment plate (VWR) to form embryoid bodies (EB) in media containing DMEM/F-12, 10% serum replacer, B-27 supplement (Invitrogen), 1 ng/ml mouse noggin (R & D Systems, Minneapolis, MN), 1 ng/ml human recombinant Dkk-1 (R & D Systems) and 5 ng/ml human recombinant insulin-like growth factor-1 (IGF-1) (R & D Systems). After 3 days, EB were plated onto Matrigel-coated plates and cultured in the presence of DMEM/F-12, B-27 supplement, N-2 Supplement (Invitrogen), 10 ng/ml mouse noggin, 10 ng/ml human recombinant Dkk-1, 10 ng/ml human recombinant IGF-1, and 5 ng/ml bFGF. The media was changed every 2–3 days.
Retinal progenitor marker expression was analyzed by either qPCR anaylsis for PAX6, LHX2 and SIX3 (primer sequences in Suppl.Table5) or immunostaining for TUJ1, PAX6, NESTIN and SOX9. The following antibodies were used: mouse anti-TUJ-1 (Covance, Austin, TX), mouse anti-PAX6 (DHSB, Iowa City, IA), rabbit anti-SOX9 (Abcam, Cambridge, MA), mouse anti-NESTIN (gift from Dr. Eugene Major, NIH, Bethesda, MD). Secondary antibody stainings were done using the corresponding Alexa Fluor 633 fluorescent-tagged antibodies (Molecular Probes, Invitrogen).
Teratoma formation
Hypoxia “de-differentiated” cells were cultured on either conditioned medium or TeSR2 medium (StemCell Technologies, Vancouver, BC, Canada) on Matrigel-coated plates or in hESC medium on a feeder layer. Cells were detached from culture dishes with dispase and pooled. About 4 × 106 cells were resuspended in Matrigel supplemented with a cocktail of prosurvival factors [25], and injected into the femoral muscle of SCID-Beige mice (Charles River, Wilmington, MA). Mice were kept under biosafety containment level 2. Palpable tumor masses developed in approximately 5 weeks. The tumor bearing mice were sacrificed, tumor tissue was fixed in 10% formalin (Richard-Allan Scientific, Kalamazoo, MI) for 24h and stored in 70% ethanol until paraffin imbedding. Five μm sections of the tumor were stained with hematoxylin and eosin using standard protocols. Sections were assessed by a boarded veterinary pathologist.
Gene expression (mRNA and miRNA) analyses
mRNA microarray analysis was performed as described previously [26]. Gene expression data analysis was done with the Rosetta Resolver gene expression analysis software (version 7.1 Rosetta Biosoftware, Seattle, WA). The miRNA levels were determined using Agilent miRNA microarrays. A total of 100 ng of total RNA per profile was labeled, hybridized, and scanned by Agilent Technologies personnel following the manufacturer's protocol (Agilent P/N G4170-90010). Briefly, samples were dephosphorylated and end-labeled using T4 RNA ligase and 3′-cyanine, 5′-cytidine bisphosphate (pCp-Cy3). Labeled material was combined with blocking and hybridization buffer, denatured, and hybridized for 20 hr at 55°C in Agilent SureHyb cartridges. Samples were washed using the manufacturer's protocol and scanned using Agilent XDR scanning (dual-scan at 100% PMT and 5% PMT). Agilent Feature Extraction v9.5.3.1 software was used to acquire and process the array images. MAGE-XML data were uploaded into Rosetta Resolver and analyzed using custom MatLab tools.
mRNA and miRNA qRT-PCR
Total RNA was extracted using TriZol (Invitrogen) and treated with DNase (Ambion, Austin, TX). RNA abundance was determined using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE). After a reverse transcription reaction using the Omniscript RT kit (Qiagen, Valencia, CA), OCT4, SOX2, NANOG, LHX-2, PAX-6, SIX-3, BMP4 and HAND1 were analyzed by qPCR with SybrGreen master mix (Applied Biosystems, Carlsbad, CA), while CGB and FN1 were analyzed by qPCR using the TaqMan assay (Applied Biosystems) on an Applied Biosystems 7300 Real-time PCR cycler. All data were normalized to b-ACTIN transcript levels. The gene primers used are listed in Suppl. Table5. TaqMan miRNA assays (Applied Biosystems) were used for miRNA qPCR according to the manufacturer's protocol using RNU66 snoRNA as a loading control.
Oxygen consumption rate (OCR) and Extra-cellular acidification rate (ECAR) measurements using SeaHorse Cellular Flux assays
hESCs H7 grown in normoxia or in hypoxia (2%O2) for 2 weeks, 6-day differentiated H7 cells, cells “de-differentiated” from 6-day differentiated H7 cells, RUES2 hESCs and 14-day differentiated RUES2 cells were seeded the day prior to the experiment onto Matrigel- or Gelatin-pre-treated SeaHorse plates at 2–4 × 105 cells per XF96 well. Differentiated cells were cultured in DMEM medium complemented with 20% fetal bovine serum (FBS) and all other cells in MEF-conditioned medium. Culture media were exchanged for base medium 1 h before the assay: unbuffered DMEM (Sigma D5030) supplemented sodium pyruvate (Gibco, 1mM) and with 25mM glucose (for MitoStress assay) or 2mM glutamine (for glucose stress assay). The OCR and ECAR values were further normalized to the number of cells present in each well, quantified by Hoechst staining (HO33342; Sigma-Aldrich) as measured using fluorescence at 355 nm excitation and 460 nm emission. Changes in OCR in response to the uncoupler carbonyl cyanide p-triuoromethoxyphenylhydrazone, (FCCP, 1μM) injection in the MitoStress assay was defined as the maximal change after the chemical addition compared with the uncoupled or basal OCR. Changes in ECAR in response to Glucose (2.5mM) injection in the glucose stress assay was defined as the maximal change after glucose addition compared to basal ECAR.
Immunofluorescence analysis of Tra-1-60 and SSEA-4 protein expression
hESCs and “de-differentiated” cells were grown on Matrigel-treated chamber slides (Nalge Nunc Intl., Rochester, NY). Cells were fixed with 4% paraformaldehyde (PFA) and stained for SSEA-4 or TRA-1-60 (1:200, Chemicon, Temecula, CA). The secondary antibody Alexa-568 goat anti-mouse (1:1000, Molecular Probes) was used. Cells were further treated with DAPI and visualized using confocal microscopy. Incubation with the secondary antibody alone was used as a negative control.
Bisulfite sequencing analysis
To identify the methylation status of CpG islands in the Oct-4 gene promoter, a published method [27] was followed. Briefly, conversion of unmethylated cytosines to uracil of purified genomic DNA was carried out as described in EZ DNA Methylation-Gold Kit (ZYMO, Orange, CA). About 400 ng genomic DNA was bisulfate treated in each reaction and 100ng of the treated DNA was used for each PCR reaction. Primers used to amplify a genomic DNA fragment in the human OCT4 promoter were: methyl-Oct4F: 5′-ATTTGTTTTTTGGGTAGTTAAAGGT-3′, and methyl-Oct4F: 5′-CCAACTATCTTCATCTTAATAACATCC-3′, with the following conditions (Zymo Taq DNA Polymerase, Orange, CA): initial denaturation for 10 min at 95°C; 38 cycles of 95°C for 30 seconds, 50°C for 30 seconds, 72°C for 1 min; and followed by 72°C for 10 min. The PCR products were cloned into TOPO PCR cloning vector (Invitrogen, Carlsbad, CA), amplified and sequenced.
Results
Hypoxia reverts committed cells back to the hESC state
To determine whether hypoxia is involved in changing cell fate, we tested if hypoxia can revert early committed cells back to the hESC state, using three pluripotent cell lines. Two hESC lines (H1 and H7) and one induced pluripotent stem cell (iPSC) line (OSLN6, [20]) were forced to differentiate in 20% serum without conditioned media and FGF 4, 5 or 6 days, respectively ([20]; Fig.1A-B). These committed cells were then cultured under hypoxia (2% O2) or atmospheric oxygen (20% O2) in conditions that support stem cell growth (Fig.1A). In all three cell lines, after one to two weeks in hypoxia, colonies were identified with undifferentiated hESC morphology (Fig.1B, “hypoxia-de-differentiated”), whereas cells cultured at 20% O2 continued to differentiate (Suppl.S1A). To test the extent of differentiation that still supports hypoxia driven “de-differentiation”, we differentiated hESCs and tested their “de-differentiation” capacity at different time points. Cells up to, but not beyond two weeks of differentiation were able to revert back to Oct4-GFP positive, stem cell like colonies in hypoxia (Fig.1C). We analyzed the methylation status of the Oct4 promoter by bisulfate sequencing during the course of differentiation and showed that Oct4 promoter was efficiently methylated upon differentiation (4, 6, 11, 14 days, Fig.1D, Suppl.S2). Furthermore, a change in the metabolic profile from glycolytic hESC profile to a more differentiated cell profile with a higher oxygen consumption rate (OCR) and lower Extra-cellular acidification rate (ECAR) was observed during two week differentiation (Fig.1E-F, [17]). Altogether these results indicate that the cells used were committed to a differentiation process. 4day or 6day differentiation paradigms were used in the rest of the manuscript.
Figure 1. Hypoxia, but not atmospheric oxygen concentrations, allows differentiated cells to “de-differentiate” to hESC-like colonies.
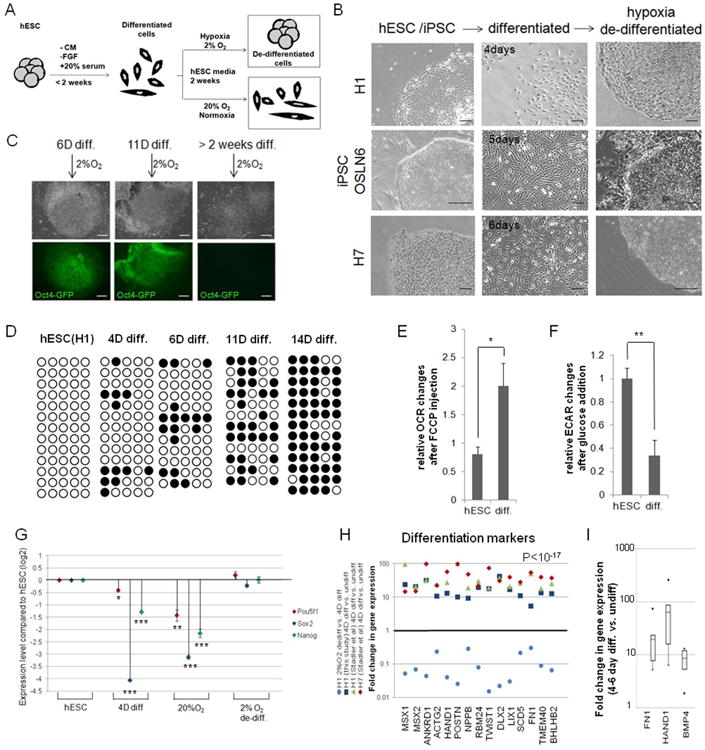
(A) Flowchart of the experiment. hESCs were differentiated (<2weeks, 20%O2) using 20% serum without conditioned media or FGF, then cultured in normoxia (20%O2) or hypoxia (2%O2) for 2 weeks. “De-differentiated” colonies were observed only in hypoxia. (B) Bright field images of hESC (H1 or H7) and iPSC (OSLN6) colonies, differentiated cells for the specified number of days (4, 5 and 6, respectively) and cells “de-differentiated” to hESC-like colonies by hypoxia. Images were taken on a Zeiss microscope and were representative of at least 3 independent experiments. Bars represent 100μm. (C) hESC(H1) differentiated for 6 or 11 days, but not over 2 weeks, are still able to “de-differentiate” in hypoxia, as evidenced by OCT4-promoter activation (Oct4-GFP positive colonies). Bars represent 200μm. (D) Methylation analysis of OCT4 promoter region in hESCs(H1) and after 4, 6, 11 and 14 days of differentiation. White circles represent unmethylated; black circles methylated cytosine-phosphate-guanosine (CpG). (E-F) Differentiated cells lose their hESC metabolic signature. Changes of oxygen consumption rate (OCR, E) and extra-cellular acidification rate (ECAR, F) were measured in SeaHorse Extracellular Flux assay after addition of FCCP or glucose, respectively (hESC=RUES2, 14-day diff.). (G) Stem cell marker expression during the “de-differentiation” process. mRNA expression of OCT4 (POU5F1), SOX2 and NANOG was analyzed by RT-qPCR (hESC=H1, 4-day diff., 14-day in 20%O2 or 2%O2). (H-I) Induction of differentiation markers after serum-induced differentiation. Expression of differentiation markers was monitored by microarray analysis in this study and in a study from Stadler et al. [20] in undifferentiated and 4-day differentiated hESC lines, as well as hypoxia-“de-differentiated” cells (H). FN1, HAND1 and BMP4 mRNA upregulation during 4-6 day differentiation of H1 cells was validated by RT-qPCR. Represented are 4 replicates of 4-day differentiated and 2 replicates of 6-day differentiated samples pooled together (I). Error bars show standard error of the mean (SEM). *, P< 0.05; **, P< 0.01 and ***, P<0.001.
We further detected a dramatic up-regulation of early differentiation markers and stem cell marker down-regulation during 4day and 6day differentiation (Fig.1G-I, Suppl.S1, Suppl.Table1). These changes in gene expression continue in 20% O2, but not in 2%O2. In hypoxia stem cell marker expression is up-regulated and the differentiation markers are repressed, showing molecular indications of “de-differentiation” (Fig. 1G,H).
To test whether hypoxia could affect the growth rate of hESCs, colony diameter and cell cycle profile (H1) and BrDU incorporation (H7) were monitored when cells were cultured at 20% O2 or 2% O2. Under the conditions used, no obvious growth rate differences were observed (Suppl.S3), suggesting that the hESC-like colonies observed in hypoxia, but not at 20% O2, were not a product of an abnormally rapid cell division in low oxygen.
Hypoxic “de-differentiated” cells express hESC markers
Previous studies have revealed that HIFs can regulate the reprogramming factors, OCT4, NANOG, SOX2 and miR-302 in cancer cells [11, 12]. The mRNA levels of these master regulators of stem cells decreased after four days of serum forced differentiation [20, 21, 28] and were further reduced after two weeks in 20% O2 in hESC media (Fig.1G, Suppl.S1B-E). In contrast, these stem cell markers were highly up-regulated reaching the hESC expression levels in the colonies formed during 10-14 day “de-differentiation” in 2% O2 in hESC media (Fig.1G, 2A; Suppl.S1B-C). Accordingly, the OCT4 promoter was highly unmethylated in hypoxia “de-differentiated” cells, comparable to the levels observed in hESCs (Fig.2B, Fig.1D). We also tested the mitochondrial activity of the hypoxia “de-differentiated” cells by analyzing the OCR increase after FCCP addition. OCR of the hypoxia “de-differentiated” cells was significantly reduced compared to 6-day differentiated cells, comparable to the level of OCR observed in hESCs, indicating that the hypoxia “de-differentiated cells” depicted hESC-like glycolytic metabolism (Fig.2C; hESC(H7) [17]). When in primed state, the maximum OCR of hESCs does not change whether cultured in hypoxia (2%O2) or normoxia (20%O2) (Fig.2C; H7).
Figure 2. Hypoxic “de-differentiated” cells express hESC markers.
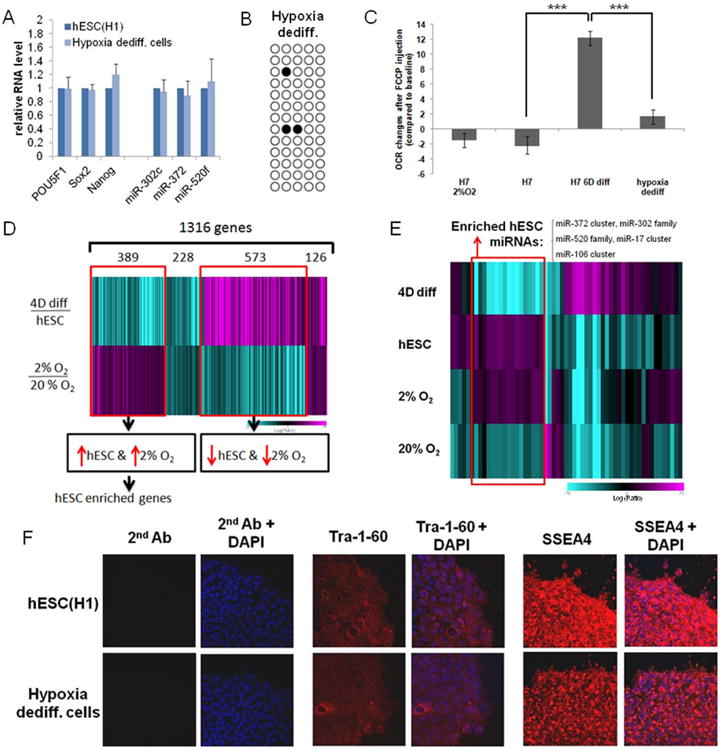
(A) Gene expression in “de-differentiated” cells is similar to hESCs. qPCR analysis for expression of stem cell markers OCT4, SOX2, NANOG, miR-302c, miR-372 and miR-520f in H1 parental cells and H1 hypoxia-“de-differentiated” cells. Error bars show standard error of the mean (SEM) for 3 separate experiments. (B) Methylation analysis of OCT4 promoter region in H1 hypoxia- “de-differentiated” cells. White circles represent unmethylated, black circles methylated cytosine-phosphate-guanisine (CpG). (C) Metabolic profile of “de-differentiated” cells. hESCs and “de-differentiated” cells have lower mitochondrial respiration activity than serum-induced differentiated cells. Changes of oxygen consumption rate (OCR) were measured in SeaHorse Extracellular Flux assay after addition of FCCP compared to basal OCR (hESC H7, 6-day differentiated H7 cells and “de-differentiated” cells (parent=H7) cultured in normoxia, and H7 (2%O2) cultured in hypoxia). (D) mRNA profiling of H1 “de-differentiated” cells. Shown is a heatmap of significantly differentially expressed genes upon differentiation (4 days, top panel) and between cells grown under 2% O2 or 20% O2 (bottom panel). An inverse relationship was expected in this comparison of genes that were up-regulated in H1 and in hypoxia. The color bar represents log10 expression ratio -0.7 (teal) to +0.7 (magenta): a common signature of 1316 genes is seen (P<0.01 in both experiments). (E) miRNA profiling of H1 hypoxia “de-differentiated” cells. Shown is a heatmap representation of miRNA expression changes in samples compared to a “visual pool” as baseline. The “visual pool” is a pool of all 4 experiments. The color bar represents log10 expression ratio -0.7 (teal) to +0.7 (magenta). P<0.01 in at least 1 experiment. (F) Immunofluorescence microscopy of H1 and H1 hypoxia-“de-differentiated” cells indicates the presence of the stem cell surface markers SSEA-4 and Tra-1-60. Magnification 20×.
We further analyzed the genome wide expression profiles of the “de-differentiated”, hESC-like colonies formed during the two week incubation in hypoxia and found them to be similar to hESC expression profiles (Fig.2D,E, Suppl.S4A). The microarray expression analysis revealed an overlap of between hESC and hypoxia “de-differentiated” cells. Among 1316 significantly changed genes, 389 genes were up-regulated in both hESC and hypoxia-induced “de-differentiated” cells, including OCT4, SOX2 and NANOG targets, as well as other hESC enriched genes as revealed by independent GO-analysis (Fig.2D; Suppl.Table2).
We also performed microarray analysis to profile the miRNA signatures (Fig.2E) since subtle miRNA changes have been used as indicators of different stem cell stage [20, 28, 30]. We found that the set of up-regulated miRNAs in hypoxia “de-differentiated” cells was highly similar to those enriched in hESCs (H1) [20]. This set contained miR-302 family, miR-372 and miR-515 family members (Fig.2E, open box; Suppl.Table3). We also analyzed the data for common miRNA signatures (P<0.01) in H1 relative to 4-day differentiated cultures [20], and in 20% O2 relative to 2% O2 for 2 weeks. The datasets identified 64 miRNAs that were significantly up-regulated in H1 cells and “de-differentiated” cells (Suppl.S4B). As a control, we showed that the hypoxia-induced miRNA miR-210 was over-expressed in cells cultured under hypoxia (2% O2) but not cultured at 20% O2 (Suppl.S4B). We also examined the expression of 45 hESC-enriched miRNAs [20] and found a significant overlap between undifferentiated H1 and cells “de-differentiated” for two weeks under hypoxia (Suppl.S4C).
The hypoxia-induced stem cell-like colonies possessed a similar morphology and self-renewing capacity in normoxia or in hypoxia as hESCs, and expressed hESC stem cell markers OCT4, SOX2, NANOG, miR-302c, miR-372and miR-520f, and hESC surface markers Tra-1-60 and SSEA4 (Fig.2A,F; Suppl.S1, S4).
Collectively, the results of miRNA and mRNA profiling and validation, OCT4 promoter methylation analysis, metabolic profiling and the cell surface markers show that, when cultured two weeks in hypoxia in stem cell permissive conditions, early committed cells can adopt an expression signature and functional metabolism characteristic of undifferentiated hESCs.
Hypoxic “de-differentiated” cells arise from already committed cells
To further analyze the process, we transformed the H1 hESC line with an Oct4-GFP-construct, and obtained a GFP-positive stable H1-line ([22]; H1 Oct4-GFP; Fig.3A left panel). As expected, the Oct4 promoter driven GFP expression was highly reduced after serum-forced differentiation (Fig.3A middle panel; [20]). Those committed, Oct4-GFP negative cells were isolated by FACS sorting and cultured at either 2% O2 or 20% O2 (Fig.3B). Only in hypoxia (2% O2) were the GFP-negative cells able to “de-differentiate” to Oct4-GFP positive hESC-like colonies (Fig.3B). We also followed the hypoxia-induced process under an environmental microscope. Interestingly, the first appearance of Oct4-driven GFP in “de-differentiating” cells was observed 4-5 days after initiation of hypoxia treatment using live cell imaging (Fig.3C). After two weeks in hypoxia, we obtained green colonies expressing Oct4-GFP. Colonies were then picked and cultured on irradiated MEF feeders (Fig.3A right panel). Altogether, these results show that early committed Oct4-GFP negative cells are able to re-express Oct4-GFP when cultured under hypoxia.
Figure 3. Oct4-GFP negative cells are able to “de-differentiate” in hypoxia to GFP positive cells.
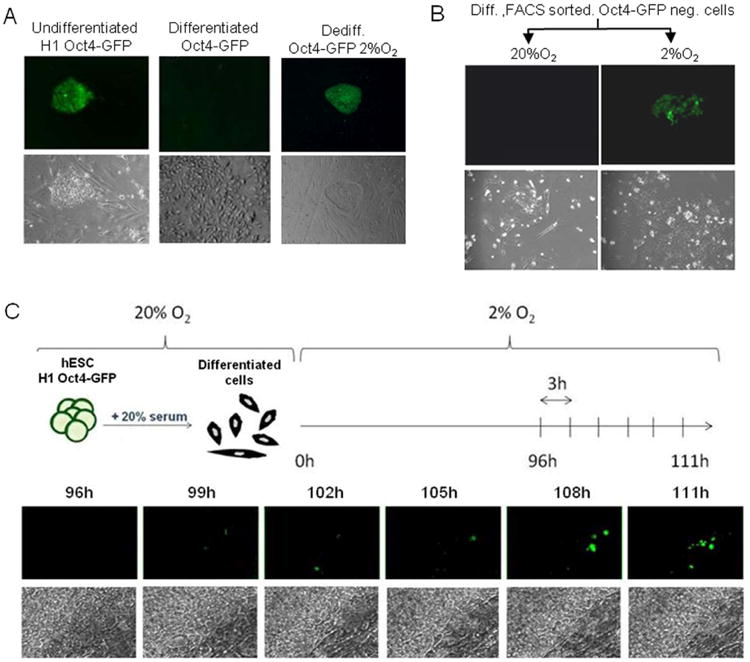
(A) OCT4 expression is lost after 4-6 days of differentiation and re-expressed in hypoxia “de-differentiated” cells. Representative bright field and green fluorescence channel images of H1 Oct4-GFP, H1 Oct4-GFP differentiated for 4-6 days with 20% serum and hypoxia-“de-differentiated” cells. Depending on the experiment and the density of the cells, it takes 4 to 6 days for the majority of the cells to lose GFP expression. Magnification 5×. (B) GFP negative cells were isolated by FACS sorting after H1 Oct4-GFP cells were forced to differentiate for 4 days in 20% serum. GFP negative cells were able to form green colonies when cultured under hypoxia (2% O2) but not at 20% O2 (day 7). Images are representative of two independent biological experiments. Magnification 5×. (C) Visualization of the “de-differentiation” process under an environmental microscope in hypoxic conditions. Following 4 days of serum-induced differentiation, H1 Oct4-GFP cells were cultured within an incubation chamber automated for live imaging under 2% O2. Bright field and fluorescence images were taken every 3 hours. Represented here are images taken between 4 and 5 days of culture in hypoxia. Magnification 10×.
To mark the early committed cells, we used a hESC line (H7) engineered to stably express a “floxed” reporter integrated into the AAVS1 locus using zinc finger nuclease and CK7-CRE [23]. The AAVS1 locus is located in a site with an open chromatin structure, avoiding transgene silencing [31, 32]. The construct used is a dual-fluorescence reporter containing the Tomato and GFP genes under the control of a strong CAG promoter and separated by a STOP codon (“traffic light system”; [23, 33]). The Tomato gene is flanked by loxP recombination sites and can be excised along with the STOP codon when CRE is expressed (Fig.4A). When CRE, driven by an enhancer of the muscle creatine kinase promoter (CK7;[34]) is added by lentivirus, only cells that differentiate can lose Tomato and gain GFP expression due to a CRE dependent excision event. After the excision, GFP is constitutively expressed driven by the CAG promoter activity (Fig.4B). This marking system allowed us to follow a hESC derived, committed cell's capacity to revert back to an undifferentiated state in hypoxia (Fig.4C). We first confirmed that undifferentiated H7 cells expressed only Tomato with our settings (red fluorescence, Fig.4A, 4D, Suppl.S5A-B). We then initiated the differentiation protocol and after two days, when the cells had lost their stem cell like colony morphology, the CRE-recombinase was induced (Fig.4C). After 6 days of serum-induced differentiation, around 1-2% of H7 traffic light cells expressed GFP, indicating differentiation (Fig. 4B, 4E-F). When cultured under hypoxia (2%O2), about 0.025% of the cells expressing GFP formed colonies (Fig.4G, Suppl.S5C-E) while the cells cultured at 20% O2 continued differentiating. These results show that differentiated hESCs as determined by acquisition of GFP signal, can be pushed back to an undifferentiated stage when grown under hypoxic conditions (Fig.4G) and that this effect is not due to cells that escaped differentiation initially.
Hypoxia can push committed cells back to a pluripotent state
We further tested whether hypoxia induced hESC-like cells were pluripotent and had the capacity to differentiate. The hypoxia induced Oct4-GFP positive hESC-like cells lost their capacity to express Oct4-GFP after 4-6 day serum-forced differentiation, comparable to the normal hESCs (Suppl.S6A). Accordingly, after 10 days in serum-forced differentiation, the Oct4 promoter was highly methylated (10D diff., 60% CpG methylation, Fig.5A) compared to hypoxia-“de-differentiated” cells (4.6% CpG methylation, Fig.2B). Differentiation capacity was also indicated by the fact that stem cell markers were down-regulated while differentiation markers were up-regulated as analyzed by qPCR (Fig. 5B).
Figure 5. Hypoxia-“de-differentiated” cells are pluripotent.
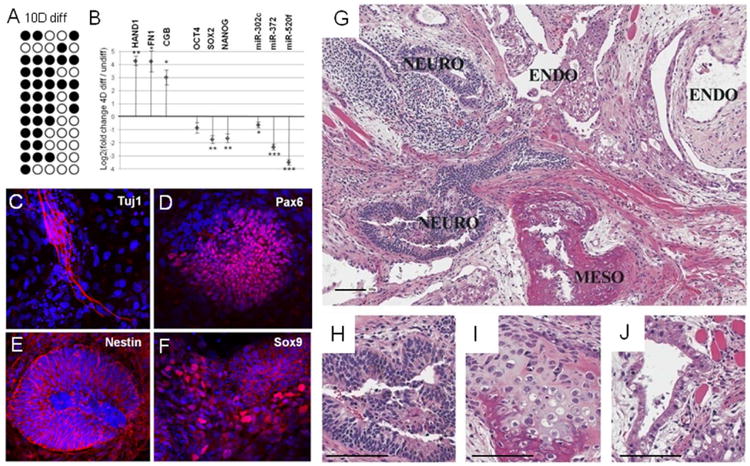
(A) Methylation analysis of OCT4 promoter region in H1 de-differentiated cells differentiated for 10 days with serum. White circles represent unmethylated; black circles methylated cytosine-phosphate-guanosine (CpG). (B) qPCR analysis of differentiation markers (HAND1, FN1 and CGB) and stem cell markers (OCT4, SOX2, NANOG, miR-302c, miR-372 and miR-520f) in 4-day differentiated Oct4-GFP “de-differentiated” cells compared to Oct4-GFP “de-differentiated” cells. Error bars show SEM for 4 separate experiments. *, P< 0.05; **, P< 0.01 and ***, P<0.001. (C-F) Representative pictures of immunostaining of retinal progenitor markers PAX6, TUJ1, NESTIN and SOX9 in H1 “de-differentiated” cells after 3 weeks of retinal induction. (G-J) Hematoxylin and eosin stained sections of teratoma obtained after injection of H1 “de-differentiated” cells in immunodeficient mice. Differentiated tissues from all 3 germ layers are apparent. Survey view (10×) a well-differentiated teratoma is shown in G. Ectoderm-derived included neural tissue with rosette formation and occasional pigmented neuroectodermal regions (Neuro); mesoderm-derived included bone and cartilage (Meso); Endoderm-derived structures included dilated ducts or glands lined by ciliated to attenuated cuboidal epithelium (Endo). Higher magnification (20×) of differentiated tissues: pigmented neuroectoderm (H); cartilage and bone (I); gland lined by ciliated epithelium (J). Bars represent 100μm.
To test whether the hypoxia “de-differentiated” cells were pluripotent, we analyzed whether they were able to induce retinal progenitor differentiation and teratoma formation. Both immunochemistry and qPCR analysis of retinal progenitor markers showed that hypoxia “de-differentiated” cells were able to differentiate to neuronal cells (Fig.5C-F, Suppl.S6B; [24]). Furthermore, teratoma analysis revealed that hypoxia “de-differentiated” cells were capable of differentiating to all three germ layers, endoderm (gland), ectoderm (pigmented neuroectoderm) and mesoderm (cartilage and bone) (Fig.5G-J).
Hypoxia driven “de-differentiation” requires HDAC activity
To reveal the potential mechanism for hypoxia-induced “de-differentiation”, we analyzed the expression profiles and identified highly significant HIF1α or HIF2α target signature up-regulation (P<9×10-35 and 9.91×10-7, respectively). Among the 515 genes significantly up-regulated in the hypoxia “de-differentiated” cells, 18.6% were HIF target genes (11.3% HIF1α targets and 7.3% HIF2α targets). In addition, a significant enrichment (50%, P< 6.12-7) of HIF1α target genes was observed in the top 15% of genes up-regulated in 2% O2 but not in the 20% O2 treatment (Fig.6A, Suppl.Table4). These data indicate that the process instigated in 2% O2 directly stimulated HIF target genes. Most of these HIF1α targets (Fig.6A, Suppl.Table4) are involved in up-regulation of glycolytic metabolism, supporting the recent finding that the primed pluripotent state is highly glycolytic and the push toward an undifferentiated state might be driven by metabolic changes [17, 29].
Figure 6. HIFs and HDAC activity are involved in hypoxia-induced “de-differentiation”.
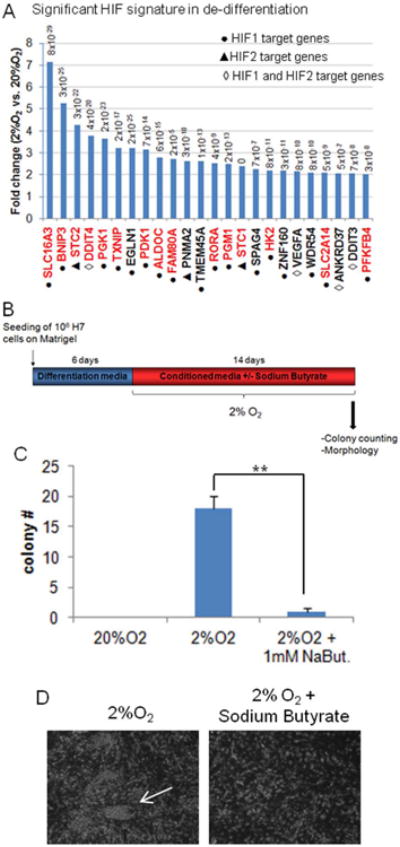
(A) HIF target genes are enriched in hypoxia “de-differentiated” samples. Graph represents the most significantly up-regulated HIF1α and HIF2α target genes in hypoxia-“dedifferentiated” cells (microarray analysis). Shown in red are the HIF target genes involved in regulation of metabolic pathways. P-value from microarray for each gene is indicated. (B) Experimental scheme of HDAC inhibitor sodium butyrate treatment (1mM) during the “de-differentiation” assay. 1×106 H7 cells were originally seeded and differentiated during 6 days prior to transfer into hypoxia in hESC media with or without 1mM sodium butyrate. (C) Quantification of hESC-like colonies formed under normoxia (20%O2) or hypoxia (2%O2) with or without sodium butyrate (NaBut.) treatment (1mM). Colonies were counted 14 days after transfer to hypoxia. (D) Bright field images of “de-differentiation process” with or without sodium butyrate treatment (1mM). The arrow indicates a hESC-like colony.
In order to determine whether epigenetic modifications could play a role in hypoxic-generation of hESC-like colonies, we inhibited histone deacetylase (HDAC) activity with sodium butyrate. This HDAC inhibitor blocked the hypoxia-induced “de-differentiation” since the number of colonies observed was significantly reduced (Fig.6B-D). However HDAC inhibitor did not affect significantly the survival of the differentiated cells.
Discussion
We challenged the controversial notion of hypoxia requirement in stem cell fate by testing whether hypoxia is involved specifically in the re-acquisition of stem cell properties. To do so, we set up a sufficiency assay and showed that hypoxia, but not atmospheric concentrations of oxygen, can push committed cells back to a stem cell state. The hypoxia “de-differentiated” cells were generated from differentiated cells as assessed with differentiation marker profiles and re-activation of OCT4 promoter analyzed by an Oct4-GFP construct and promoter methylation analysis. These hypoxia-induced stem-like cells also mimic hESCs by their colony morphology and long-term self-renewal capacity. Furthermore, as seen before with hESCs, after initial “de-differentiation”, the hypoxia-induced cells were able to self-renew both in hypoxia and in normoxia. Genome wide mRNA and miRNA analysis revealed a significant similarity between the hypoxia-induced stem-like cells and hESCs. In addition, the characteristic cell surface markers, TRA-1-60 and SSEA4, were expressed in hypoxia-induced stem-cell like colonies. Most significantly, the hypoxia-induced stem-like cells showed a pluripotent differentiation capacity; serum-forced early differentiation was efficient, neuronal photoreceptor precursors were observed and the cells were able to generate teratomas in mice. Hence early committed cells can be reverted back to a hESC-like state by hypoxia alone (Fig.7A). In the conditions we have used hypoxia alone is not potent to reprogram adult or fetal fibroblasts. However hypoxia does increase their reprogramming efficiency if other reprogramming factors are included to the process [14, 15]. The population of pluripotent cells in culture is very heterogeneous and has a high degree of plasticity [35, 36, 37]. Recent works in multipotent hematopoietic cells propose that the switch from precursor state to differentiated state is not a bistable system but rather that “metastable” intermediate states exist [38-40]. It is therefore possible that early committed cells derived from hESCs are in a reversible stage where they express markers of differentiation but are still capable of re-entry into pluripotency if the conditions are favorable. In this paper, we show that hypoxia can influence fate determination and push “metastable” committed cells toward a more undifferentiated state (Fig.7).
Figure 7. Model and hypothesis.
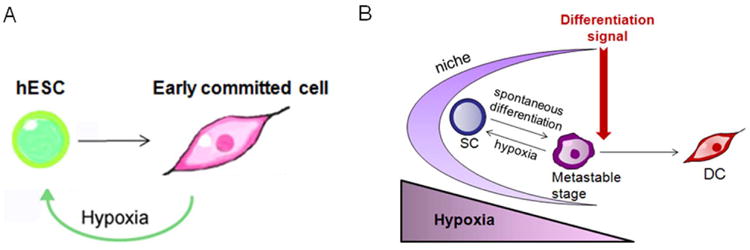
(A) Early committed hESCs can re-acquire pluripotency in hypoxia (2% O2). (B) Hypoxic stem cell niche hypothesis. Stem cells (SC) are believed to reside in a hypoxic niche that maintains their self-renewal capacity. A low level of oxygen might work as a rheostat when stem cells drift towards spontaneous differentiation. Only a strong differentiation signal could induce the cells to differentiate (DC). The hypoxic niche could also facilitate the de-differentiation process observed previously in vivo.
At a concentration of 2% oxygen, HIFs are stabilized and induce expression of genes involved in numerous pathways [41]. We and others have recently shown that HIFs can induce hESC marker expression in cancer cells and HIF1α can modify the fate of the cells by changing their metabolic pathways towards glycolytic metabolism [11-13, 17, 29]. Both HIF1α and HIF2α target genes were found up-regulated in “de-differentiating” cells. Interestingly, those cells highly express HIF1α target genes involved in the regulation of the glycolytic pathway and mitochondrial respiration (including lactate transporter MCT4, PGK1, PDK1, aldolase C, hexokinase 2, PFKFB4, phosphoglucomutase 1, glucose transporter SLC2A14 and miR-210; Fig.6A, Suppl.Table4, Suppl.S4B). It is therefore plausible that HIFs and their downstream targets are responsible for the cell fate push toward pluripotency under hypoxic conditions. Furthermore, the HIF-induced metabolic changes might be the leading force in this push. Previous studies have suggested that HIF is crucial for some adult stem cell fates (reviewed in [42]). In addition, HIF has been shown to play a role in ESC pluripotency [9], and HIF modulator, FM19G11, regulates stem cell differentiation status [43]. It will be important to define whether hypoxia is involved in vivo in the initiation of the stem cell program or de-differentiation processes observed in vivo [44, 45, 46]. If so, HIF activity would be expected to be critical in the induced pluripotency reprogramming process. In support of this, recent results have shown that hypoxia and HIF1α are advantageous for iPSC induction [14, 15].
We have shown that the promoter of OCT4 is reactivated and its methylation status is reverted in hypoxic “de-differentiated” cultures. Chromatin changes and activation of OCT4 promoter can be regulated by transcription factors such as FoxO1 and HIF2α [12, 47] as well as transcriptional modulators like Cited2 [48]. A complex network of epigenetic modifications can control pluripotency [49]. In particular, histone acetylation/deacetylation plays a key role in hESC maintenance and differentiation. It has been shown previously that HIF1α can directly interact with histone deacetylases (HDACs) [50] and hypoxia induces HDAC activity [50]. Moreover, HDAC inhibitors phenocopy HIFβ deletion in trophoblast stem cells, showing that the interplay between HIFs and HDACs is crucial during development [52]. We found that sodium butyrate, a broad HDAC inhibitor, blocks the hypoxia-induced “de-differentiation” process. It is therefore possible, that the “de-differentiation” process requires HDAC activity through HIF. This result provides a beginning of a mechanistic insight of hypoxia in “de-differentiation” process. Further investigation will be needed to fully uncover the role of HIFs and HDACs as well as their interaction in this context.
Many adult stem cells, reside in and interplay with a niche that regulates the balance between stem cell self-renewal and differentiation [53, 54. 55]. It is thought that the hypoxic signaling microenvironment of the niche maintains stem cell self-renewal [10], however the detailed mechanism is not understood. Based on the data presented in this paper, it is plausible that HIF activation due to low oxygen levels in the niche works as a “de-differentiation” rheostat. If the stem cells drift inappropriately towards differentiation, the cells, while in a “metastable” state, could be driven back to a more naïve state by hypoxia. Only when strong differentiation signals are present would hypoxia signaling lose its capacity to de-differentiate the cell (Fig.7B). This hypothesis could explain how a pool of stem cells is maintained in an elusive niche environment and how a lost stem cell can be replaced by a de-differentiating cell [44, 45]. In cancer, hypoxia is shown to induce a hESC signature that correlates with tumor aggressiveness [11]. Hypoxia may therefore promote a stem cell state both in normal and cancer cells.
Supplementary Material
Supplemental Figure S1: (A) Flowchart of the experiment and bright field images of undifferentiated H1 cells, 4day differentiated H1 cells and cells grown at either 20% O2 or 2%O2 during 2 weeks. Images were taken on a Leica microscope and were representative of 5 independents experiments. Bars represent 100μm. (B-C) Stem cell marker expression during the “de-differentiation” process. mRNA expression of OCT4 (POU5F1), SOX2 and NANOG was analyzed by qPCR in undifferentiated H1 cells, 4-day differentiated H1 cells and cells grown at either 20% O2 or 2%O2 for 7, 10 or 14 days. The table shows the mean and range of two experiments (C). (D) mRNA expression of OCT4 (POU5F1), SOX2 and NANOG was analyzed by qPCR in hESC(H1) and 4-day or 6-day differentiated H1 cells. (E) Expression of microRNA miR-302c, miR-372 and miR-520f was analyzed by qPCR in undifferentiated H1 cells and 4-day differentiated H1 cells. Error bars show standard error of the mean (SEM) for 3 separate experiments. *, P< 0.05; **, P< 0.01 and ***, P<0.001.
Supplemental Figure S2: Kinetics of Oct4 promoter inactivation by methylation upon differentiation of H1: Graphic representation of the methylation status of OCT4 promoter region in H1 at various serum-induced differentiation time points. The y axis show the percentage of methylated CpG.
Supplemental Figure S3: Hypoxia does not affect the cell growth of hESCs: (A) Measure of colony diameter of H1 cells and H1 Oct4-GFP cells cultured at either 20% O2 or 2% O2. Cells were passaged when indicated. (B) BrDU incorporation analysis of hESCs(H7) cultured at either 20% O2 or 2% O2 for 8 days. (C) Cell cycle analysis of hESCs H1 cultured at either 20% O2 or 2%O2 for 3 days.
Supplemental Figure S4: mRNA and microRNA profiling in “de-differentiation” experiments. (A) Hypoxia treatment on differentiated H1 induces a mRNA profile similar to undifferentiated H1 hESCs. Microarray data for H1 hESCs, 4-day differentiated H1 cells and cells cultured 2 weeks under hypoxia (2%O2) as depicted by scatter plot. The data are plotted as the log10 ratio (4D diff./Hypoxia) versus the log10 ratio (4D diff./Undiff.) for each gene. (B) Clustering of the hypoxia and differentiation experiments in H1 cells using common miRNA signature (P<0.01 in both experiments). (C) Clustering of 45 selected hESC specific miRNAs.
Supplemental Figure S5: “traffic light” H7 cells: (A-B) Bright field, GFP and Tomato fluorescence channel images of undifferentiated traffic light H7 cells. Pictures represented in A were taken 4 days after infection without the CK7-CRE virus, while pictures represented in B show undifferentiated traffic light cells H7 cells with CRE virus infection. (C-D) Bright field and green florescent channel images of traffic light H7 cells “de-differentiated” during 7 or 10 days in hypoxia. (E) Bright field, GFP and Tomato fluorescence channel images of traffic light H7 cells cultured during 15 days in hypoxia after the 6-day differentiation process. Some cells resume a hESC-like colony morphology (“de-differentiated” cells, high magnification of the colony presented in Fig.4G). Bars represent 100μm.
Supplemental Figure S6: H1 Hypoxia-“de-differentiated” cells are able to differentiate: (A) Bright field and fluorescence microscopy images of hypoxia-“de-differentiated” Oct4-GFP cells and 4-6 day serum-induced differentiated Oct4-GFP “de-differentiated” cells. (B) RT-qPCR analysis of retinal stem cell markers (PAX6, LHX2 and SIX3) in hypoxia-“de-differentiated” cells after one week of retinal induction. Results from 2 independent experiments are shown.
Supplemental Table 1: list of top 65 target mRNAs up-regulated in 4day-differentiated cells compared to undifferentiated hESCs: Presented are the fold changes and p-values of the top 65 mRNAs up-regulated after 4 days of serum-induced differentiation in H7 cells (Stadler et al study) and H1 cells (this study and Stadler et al study). Fold change of those mRNAs between 4 day differentiated H1 cells and cells “de-differentiated” for 2 weeks in hypoxia are also shown. In red are indicated the differentiation markers up-regulated in 4day-differentiated cells vs. undifferentiated hESC lines in both this study and Stadler et al. study. Those markers are presented in Fig.1G.
Supplemental Table 2: list of mRNA in H1 “de-differentiation” experiment: List of genes presented in Figure 2D (significantly differentially expressed genes upon differentiation and between cells grown under 2% O2 or 20% O2). Sets with similar gene signature are indicted for genes found in cluster 1 and 3.
Supplemental Table 3: list of microRNA in H1 “de-differentiated” experiment: List of 57 microRNAs presented in Figure 2E (P<0.01 in at least 1 experiment).
Supplemental Table 4: list of HIF target mRNAs up-regulated in 2% O2 “de-differentiated” cells compared to cells cultured for 2 weeks in normoxia: Lists of HIF1 and HIF2 target genes [11] found up-regulated in cells cultured during 2 weeks under 2% O2 compared to cells cultured during 2 weeks under normoxia. GO signature for each of those genes is indicated. Top 24 genes are presented in Figure 6A.
Supplemental Table 5: Sequences of primers used for RT-qPCR
Acknowledgments
We thank members of the Ruohola-Baker lab for helpful discussions. We thank the Rosetta Gene Expression Laboratory and Dr. Cleary for help in design and processing microarray experiments. We thank Jeff Boyd (ISCRM Flow Cytometry Core) for help with the FACS analysis, Christopher Cavanaugh and Jennifer Hesson (ISRCM Stem Cell Core) with mouse injections, Dr. Treuting (Histology and Imaging Core) with teratoma analysis, and Amy Wang, Tim Dosey and Claudia Maria Pinna for technical help. We thank Jay Gantz, Nathan Palpant, Dr. Murry and Dr. Laflamme for providing the “traffic light” H7 cells and the CK7-CRE virus. We thank Dr. Cui for providing the Oct4-GFP construct and Dr. Major for providing NESTIN antibody. This work was supported by Fred Hutchinson Cancer Research Center Pilot grant and NIH grants R01GM083867-01 and R01GM097372-01 NIGMS for HRB and 1P01GM081619-01 NIGMS and the Institute for Stem Cell and Regenerative Medicine (ISCRM) at the University of Washington for CW and HRB.
Footnotes
Julie Mathieu: Conception and design, Collection and assembly of data, Data analysis and interpretation, Manuscript writing, Final approval of manuscript
Zhan Zhang: Data analysis and interpretation
Angelique Nelson: Collection of data
Deepak A. Lamba: Collection of data, Data analysis and interpretation
Thomas A. Reh: Provision of study material
Carol Ware: Conception and design, Provision of study material
Hannele Ruohola-Baker: Conception and design, Financial support, Data analysis and interpretation, Manuscript writing, Final approval of manuscript
Disclosure of Potential Conflicts of interest: The authors indicate no potential conflicts of interest
References
- 1.Simsek T, Kocabas F, Zheng J, et al. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell. 2010;7(3):380–390. doi: 10.1016/j.stem.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 3.Millman JR, Tan JH, Colton CK. The effects of low oxygen on self-renewal and differentiation of embryonic stem cells. Current Opinion in Organ Transplantation. 2009;14(6):694–700. doi: 10.1097/MOT.0b013e3283329d53. [DOI] [PubMed] [Google Scholar]
- 4.Lengner CJ, Gimelbrant AA, Erwin JA, et al. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell. 2010;141(5):872–883. doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 5.Lin Q, Lee YJ, Yun Z. Differentiation arrest by hypoxia. The Journal of Biological Chemistry. 2006;281(41):30678–30683. doi: 10.1074/jbc.C600120200. [DOI] [PubMed] [Google Scholar]
- 6.Seidel S, Garvalov BK, Wirta V, et al. A hypoxic niche regulates glioblastoma stem cells through hypoxia inducible factor 2 alpha. Brain: a Journal of Neurology. 2010;133(Pt 4):983–995. doi: 10.1093/brain/awq042. [DOI] [PubMed] [Google Scholar]
- 7.Ezashi T, Das P, Roberts RM. Low O2 tensions and the prevention of differentiation of hES cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(13):4783–4788. doi: 10.1073/pnas.0501283102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forsyth NR, Musio A, Vezzoni P, et al. Physiologic oxygen enhances human embryonic stem cell clonal recovery and reduces chromosomal abnormalities. Cloning and Stem Cells. 2006;8(1):16–23. doi: 10.1089/clo.2006.8.16. [DOI] [PubMed] [Google Scholar]
- 9.Forristal CE, Wright KL, Hanley NA, et al. Hypoxia inducible factors regulate pluripotency and proliferation in human embryonic stem cells cultured at reduced oxygen tensions. Reproduction. 2010;139(1):85–97. doi: 10.1530/REP-09-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohyeldin A, Garzon-Muvdi T, Quinones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell. 2010;7(2):150–161. doi: 10.1016/j.stem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Mathieu J, Zhang Z, Zhou W, et al. HIF induces human embryonic stem cell markers in cancer cells. Cancer Research. 2011;71(13):4640–4652. doi: 10.1158/0008-5472.CAN-10-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Covello KL, Kehler J, Yu H, et al. HIF-2alpha regulates Oct-4: effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes & Development. 2006;20(5):557–570. doi: 10.1101/gad.1399906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J, Orkin SH. Embryonic stem cell-specific signatures in cancer: insights into genomic regulatory networks and implications for medicine. Genome Medicine. 2011;3(11):75. doi: 10.1186/gm291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshida Y, Takahashi K, Okita K, et al. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell. 2009;5(3):237–241. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Mathieu J, Zhou W, Sperber H, et al. Hypoxia Inducible Factors in reprogramming. under revision [Google Scholar]
- 16.Takubo K, Goda N, Yamada W, et al. Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell. 2010;7(3):391–402. doi: 10.1016/j.stem.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 17.Zhou W, Choi M, Margineantu D, et al. HIF1alpha induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. The EMBO Journal. 2012;31(9):2103–2116. doi: 10.1038/emboj.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeong CH, Lee HJ, Cha JH, et al. Hypoxia-inducible factor-1 alpha inhibits self-renewal of mouse embryonic stem cells in Vitro via negative regulation of the leukemia inhibitory factor-STAT3 pathway. The Journal of Biological Chemistry. 2007;282(18):13672–13679. doi: 10.1074/jbc.M700534200. [DOI] [PubMed] [Google Scholar]
- 19.Chen HF, Kuo HC, Chen W, et al. A reduced oxygen tension (5%) is not beneficial for maintaining human embryonic stem cells in the undifferentiated state with short splitting intervals. Hum Reprod. 2009;24(1):71–80. doi: 10.1093/humrep/den345. [DOI] [PubMed] [Google Scholar]
- 20.Stadler B, Ivanovska I, Mehta K, et al. Characterization of microRNAs involved in embryonic stem cell states. Stem Cells and Development. 2010;19(7):935–950. doi: 10.1089/scd.2009.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ware CB, Nelson AM, Blau CA. A comparison of NIH-approved human ESC lines. Stem Cells. 2006;24(12):2677–2684. doi: 10.1634/stemcells.2005-0452. [DOI] [PubMed] [Google Scholar]
- 22.Gerrard L, Zhao D, Clark AJ, et al. Stably transfected human embryonic stem cell clones express OCT4-specific green fluorescent protein and maintain self-renewal and pluripotency. Stem Cells. 2005;23(1):124–133. doi: 10.1634/stemcells.2004-0102. [DOI] [PubMed] [Google Scholar]
- 23.Gantz JA, Palpant NJ, Welikson RE, et al. Targeted genomic integration of a selectable floxed dual fluorescence reporter in human embryonic stem cells. PLOS One. 2012;7(10):e46971. doi: 10.1371/journal.pone.0046971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamba DA, Karl MO, Ware CB, et al. Efficient generation of retinal progenitor cells from human embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(34):12769–12774. doi: 10.1073/pnas.0601990103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laflamme MA, Chen KY, Naumova AV, et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nature Biotechnology. 2007;25(9):1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 26.Jackson AL, Bartz SR, Schelter J, et al. Expression profiling reveals off-target gene regulation by RNAi. Nature Biotechnology. 2003;21(6):635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 27.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 28.Qi J, Yu JY, Shcherbata HR, et al. microRNAs regulate human embryonic stem cell division. Cell Cycle. 2009;8(22):3729–3741. doi: 10.4161/cc.8.22.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Folmes CD, Nelson TJ, Martinez-Fernandez A, et al. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metabolism. 2011;14(2):264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463(7281):621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hockemeyer D, Soldner F, Beard C, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nature Biotechnology. 2009;27(9):851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith JR, Maguire S, Davis LA, et al. Robust, persistent transgene expression in human embryonic stem cells is achieved with AAVS1-targeted integration. Stem Cells. 2008;26(2):496–504. doi: 10.1634/stemcells.2007-0039. [DOI] [PubMed] [Google Scholar]
- 33.Muzumdar MD, Tasic B, Miyamichi K, et al. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45(9):593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 34.Salva MZ, Himeda CL, Tai PW, et al. Design of tissue-specific regulatory cassettes for high-level rAAV-mediated expression in skeletal and cardiac muscle. Molecular Therapy: the Journal of the American Society of Gene Therapy. 2007;15(2):320–329. doi: 10.1038/sj.mt.6300027. [DOI] [PubMed] [Google Scholar]
- 35.Enver T, Pera M, Peterson C, et al. Stem cell states, fates, and the rules of attraction. Cell Stem Cell. 2009;4(5):387–397. doi: 10.1016/j.stem.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 36.Bhatia S, Pilquil C, Roth-Albin I, et al. Demarcation of Stable Subpopulations within the Pluripotent hESC Compartment. PLOS One. 2013;8:e57276. doi: 10.1371/journal.pone.0057276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marks H, Kalkan T, Menafra R, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang S, Guo YP, May G, et al. Bifurcation dynamics in lineage-commitment in bipotent progenitor cells. Developmental Biology. 2007;305(2):695–713. doi: 10.1016/j.ydbio.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 39.Chang HH, Oh PY, Ingber DE, et al. Multistable and multistep dynamics in neutrophil differentiation. BMC Cell Biology. 2006;7:11. doi: 10.1186/1471-2121-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang HH, Hemberg M, Barahona M, et al. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453(7194):544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou W, Dosey TL, Biechele T, et al. Assessment of hypoxia inducible factor levels in cancer cell lines upon hypoxic induction using a novel reporter construct. PLOS One. 2011;6(11):e27460. doi: 10.1371/journal.pone.0027460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon MC, Keith B. The role of oxygen availability in embryonic development and stem cell function. Nature Reviews Molecular Cell Biology. 2008;9(4):285–296. doi: 10.1038/nrm2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moreno-Manzano V, Rodriguez-Jimenez FJ, Acena-Bonilla JL, et al. FM19G11, a new hypoxia-inducible factor (HIF) modulator, affects stem cell differentiation status. The Journal of Biological Chemistry. 2010;285(2):1333–1342. doi: 10.1074/jbc.M109.008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barroca V, Lassalle B, Coureuil M, et al. Mouse differentiating spermatogonia can generate germinal stem cells in vivo. Nature Cell Biology. 2009;11:190–196. doi: 10.1038/ncb1826. [DOI] [PubMed] [Google Scholar]
- 45.Sheng XR, Brawley CM, Matunis EL. Dedifferentiating spermatogonia outcompete somatic stem cells for niche occupancy in the Drosophila testis. Cell Stem Cell. 2009;5:191–203. doi: 10.1016/j.stem.2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 2007;449:473–477. doi: 10.1038/nature06159. [DOI] [PubMed] [Google Scholar]
- 47.Zhang X, Yalcin S, Lee DF, et al. FOXO1 is an essential regulator of pluripotency in human embryonic stem cells. Nature Cell Biology. 2011;13:1092–1099. doi: 10.1038/ncb2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Q, Ramirez-Bergeron DL, Dunwoodie SL, et al. Cited2 gene controls pluripotency and cardiomyocyte differentiation of murine embryonic stem cells through Oct4 gene. The Journal of Biological Chemistry. 2012;287:29088–29100. doi: 10.1074/jbc.M112.378034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tollervey JR, Lunyak VV. Epigenetics: judge, jury and executioner of stem cell fate. Epigenetics: Official Journal of the DNA Methylation Society. 2012;7:823–840. doi: 10.4161/epi.21141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kato H, Tamamizu-Kato S, Shibasaki F. Histone deacetylase 7 associates with hypoxia-inducible factor 1alpha and increases transcriptional activity. The Journal of Biological Chemistry. 2004;279:41966–41974. doi: 10.1074/jbc.M406320200. [DOI] [PubMed] [Google Scholar]
- 51.Mie Lee Y, Kim SH, Kim HS, et al. Inhibition of hypoxia-induced angiogenesis by FK228, a specific histone deacetylase inhibitor, via suppression of HIF-1alpha activity. Biochemical and Biophysical Research Communications. 2003;300:241–246. doi: 10.1016/s0006-291x(02)02787-0. [DOI] [PubMed] [Google Scholar]
- 52.Maltepe E, Krampitz GW, Okazaki KM, et al. Hypoxia-inducible factor-dependent histone deacetylase activity determines stem cell fate in the placenta. Development. 2005;132:3393–3403. doi: 10.1242/dev.01923. [DOI] [PubMed] [Google Scholar]
- 53.Voog J, Jones DL. Stem cells and the niche: a dynamic duo. Cell Stem Cell. 2010;6(2):103–115. doi: 10.1016/j.stem.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spradling A, Drummond-Barbosa D, Kai T. Stem cells find their niche. Nature. 2001;414(6859):98–104. doi: 10.1038/35102160. [DOI] [PubMed] [Google Scholar]
- 55.Ward EJ, Shcherbata HR, Reynolds SH, et al. Stem cells signal to the niche through the Notch pathway in the Drosophila ovary. Current Biology. 2006;16:2352–2358. doi: 10.1016/j.cub.2006.10.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1: (A) Flowchart of the experiment and bright field images of undifferentiated H1 cells, 4day differentiated H1 cells and cells grown at either 20% O2 or 2%O2 during 2 weeks. Images were taken on a Leica microscope and were representative of 5 independents experiments. Bars represent 100μm. (B-C) Stem cell marker expression during the “de-differentiation” process. mRNA expression of OCT4 (POU5F1), SOX2 and NANOG was analyzed by qPCR in undifferentiated H1 cells, 4-day differentiated H1 cells and cells grown at either 20% O2 or 2%O2 for 7, 10 or 14 days. The table shows the mean and range of two experiments (C). (D) mRNA expression of OCT4 (POU5F1), SOX2 and NANOG was analyzed by qPCR in hESC(H1) and 4-day or 6-day differentiated H1 cells. (E) Expression of microRNA miR-302c, miR-372 and miR-520f was analyzed by qPCR in undifferentiated H1 cells and 4-day differentiated H1 cells. Error bars show standard error of the mean (SEM) for 3 separate experiments. *, P< 0.05; **, P< 0.01 and ***, P<0.001.
Supplemental Figure S2: Kinetics of Oct4 promoter inactivation by methylation upon differentiation of H1: Graphic representation of the methylation status of OCT4 promoter region in H1 at various serum-induced differentiation time points. The y axis show the percentage of methylated CpG.
Supplemental Figure S3: Hypoxia does not affect the cell growth of hESCs: (A) Measure of colony diameter of H1 cells and H1 Oct4-GFP cells cultured at either 20% O2 or 2% O2. Cells were passaged when indicated. (B) BrDU incorporation analysis of hESCs(H7) cultured at either 20% O2 or 2% O2 for 8 days. (C) Cell cycle analysis of hESCs H1 cultured at either 20% O2 or 2%O2 for 3 days.
Supplemental Figure S4: mRNA and microRNA profiling in “de-differentiation” experiments. (A) Hypoxia treatment on differentiated H1 induces a mRNA profile similar to undifferentiated H1 hESCs. Microarray data for H1 hESCs, 4-day differentiated H1 cells and cells cultured 2 weeks under hypoxia (2%O2) as depicted by scatter plot. The data are plotted as the log10 ratio (4D diff./Hypoxia) versus the log10 ratio (4D diff./Undiff.) for each gene. (B) Clustering of the hypoxia and differentiation experiments in H1 cells using common miRNA signature (P<0.01 in both experiments). (C) Clustering of 45 selected hESC specific miRNAs.
Supplemental Figure S5: “traffic light” H7 cells: (A-B) Bright field, GFP and Tomato fluorescence channel images of undifferentiated traffic light H7 cells. Pictures represented in A were taken 4 days after infection without the CK7-CRE virus, while pictures represented in B show undifferentiated traffic light cells H7 cells with CRE virus infection. (C-D) Bright field and green florescent channel images of traffic light H7 cells “de-differentiated” during 7 or 10 days in hypoxia. (E) Bright field, GFP and Tomato fluorescence channel images of traffic light H7 cells cultured during 15 days in hypoxia after the 6-day differentiation process. Some cells resume a hESC-like colony morphology (“de-differentiated” cells, high magnification of the colony presented in Fig.4G). Bars represent 100μm.
Supplemental Figure S6: H1 Hypoxia-“de-differentiated” cells are able to differentiate: (A) Bright field and fluorescence microscopy images of hypoxia-“de-differentiated” Oct4-GFP cells and 4-6 day serum-induced differentiated Oct4-GFP “de-differentiated” cells. (B) RT-qPCR analysis of retinal stem cell markers (PAX6, LHX2 and SIX3) in hypoxia-“de-differentiated” cells after one week of retinal induction. Results from 2 independent experiments are shown.
Supplemental Table 1: list of top 65 target mRNAs up-regulated in 4day-differentiated cells compared to undifferentiated hESCs: Presented are the fold changes and p-values of the top 65 mRNAs up-regulated after 4 days of serum-induced differentiation in H7 cells (Stadler et al study) and H1 cells (this study and Stadler et al study). Fold change of those mRNAs between 4 day differentiated H1 cells and cells “de-differentiated” for 2 weeks in hypoxia are also shown. In red are indicated the differentiation markers up-regulated in 4day-differentiated cells vs. undifferentiated hESC lines in both this study and Stadler et al. study. Those markers are presented in Fig.1G.
Supplemental Table 2: list of mRNA in H1 “de-differentiation” experiment: List of genes presented in Figure 2D (significantly differentially expressed genes upon differentiation and between cells grown under 2% O2 or 20% O2). Sets with similar gene signature are indicted for genes found in cluster 1 and 3.
Supplemental Table 3: list of microRNA in H1 “de-differentiated” experiment: List of 57 microRNAs presented in Figure 2E (P<0.01 in at least 1 experiment).
Supplemental Table 4: list of HIF target mRNAs up-regulated in 2% O2 “de-differentiated” cells compared to cells cultured for 2 weeks in normoxia: Lists of HIF1 and HIF2 target genes [11] found up-regulated in cells cultured during 2 weeks under 2% O2 compared to cells cultured during 2 weeks under normoxia. GO signature for each of those genes is indicated. Top 24 genes are presented in Figure 6A.
Supplemental Table 5: Sequences of primers used for RT-qPCR


