Abstract
Plague is a major health concern and Yersinia pestis plays the central causal role in this disease. Yersinia pestis has developed resistance against the commonly available drugs. So, it is now a key concern to find a new drug target. Cysteine protease YopT enzyme is an important factor used by Yersinia pestis for pathogenesis in its host and it has the anti-phagocytic function of removal of C-termini lipid modification. The 3D structure of cysteine protease YopT of Yersinia pestis was determined by means of homology modeling through multiple alignments followed by intensive optimization and validation. The modeling was done by Phyre 2 and refined by ModRefiner. The obtained model was verified with structure validation programs such as PROCHECK, verify 3D and ERRAT for reliability. Interacting partners and active sites were also determined. PROCHECK analysis showed that 93% of the residues are in the most favored region, 5.9% are in the additional allowed region and 1.1% are in the generously allowed region of the Ramachandran plot. The verify 3D value of 0.78 indicates that the environmental profile of the model is good. SOPMA is employed for calculation of the secondary structural features of cysteine protease YopT. Active site determination through CASTp proposes that this protein can be utilized as a potential drug target. However, these findings should further be confirmed by wet lab studies for a targeted therapeutic agent design against Yersinia pestis.
Keywords: plague, Yersinia pestis, homology modeling, active site, potential drug
Introduction
Plague, a disease that can affect humans as well as other mammals, is caused by Yersinia pestis and classified as a Category A agent of bioterrorism.1 In large historic pandemics, this infectious disease caused the death of millions of people, and there have been numerous other deadly but localized outbreaks.2 Usually, plague is transmitted subcutaneously to humans by the bite of an infected flea. But especially during pandemics of the disease, it can also be transmitted by air. The disease has recently been recognized as a re-emerging disease by the World Health Organization.3 Currently there is no available FDA-licensed plague vaccine in the USA. Subunit vaccine processing is a current research focus, followed by the search for different hybrid vaccines. Upon immunization with these vaccines, there is no significant difference obtained for host plague protection.4 In another report it has been found that effective antibiotics have to be given within 24 hours after exposure to the aerosolized form of pathogen.5 There are three types of plague: pneumonic, septicemic and bubonic plagues. All these forms are also responsible for numerous epidemics in human history, as well as three catastrophic pandemics. First, Justinian’s plague (6th–8th centuries) spread from Egypt to areas surrounding the Mediterranean.6 Second, the “Black Death” spread from the Caspian Sea to almost all European countries, causing the demise of one third of the European population over the period of just a few years in the mid-14th century. The third one is a modern plague pandemic, which began in the Yunnan region of China in the mid-19th century, and spread globally via shipping from Hong Kong in 1894. The etiological cause of plague was identified as Yersinia pestis during this last pandemic.7 Clinically, plague is characterized by swollen lymph nodes (bubonic plagues), fever, chills, headaches, body aches (septicemic plague), weakness, vomiting, and nausea (pneumonic plague).8
Yersinia pestis, is a rod-shaped, coccobacillus, Gram negative bacterium and facultative anaerobe that infects humans and animals.9 There are two complete genome sequences for two of three subspecies of Yersinia pestis, strains KIM and CO92. The total size of chromosome of Yersinia pestis is 4,653,728 bp. Gene acquisition has been important in the evolution of Yersinia pestis. In all pathogenic Yersinia, a 70 kb virulence plasmid (pYV/pCD1) is present, but Yersinia pestis has acquired two unique plasmids that encode a variety of virulence determinants. 10 A 9.5 kb plasmid (pPst/pPCP1) encodes the plasminogen activator Pla and 100–110 kb plasmid (pFra/pMT1) encodes murine toxin Ymt and the F1 capsular protein.11
Yersinia pestis harbors a 70 kb extra chromosomal plasmid DNA that encodes Type III secretion system (T3SS), which is one of the most important virulence mechanisms of the lethal plague pathogen.12 This T3SS consists of a sophisticated translocation apparatus that is highly conserved among a number of Gram-negative pathogenic bacteria.13 To contact with the host cell’s receptor, the type III secretion system of Yersinia delivers a set of effector proteins termed Yops (Yersinia outer proteins) into the host cell. To date, six Yop effectors (YopH, YopE, YopJ/P, YpkA/YopO, YopT, and YopM) have been identified. They function to attenuate the host’s immune response during infection, and phagocytes are the main target of T3SS for these Yops effector injections.14 At least four Yops-YopH, YopE, YopO and YopT, are involved in inhibiting phagocytosis, either by targeting the host cell’s actin regulation of Rho GTPase (YopT, YopE, YopO) or by rapidly and specifically targeting host proteins associated with signal transducing from receptor to actin (YopH), which is also responsible for suppressing reactive oxygen intermediates production by macrophages and PMNs.15,16 Our targeted study is on YopT, also known as cysteine protease, which induces cytotoxicity in mammalian cells. This cytotoxicity is characterized by disruption of the cytoskeleton as well as rounding up cells.17 Disruption of the host cell cytoskeleton by cysteine protease YopT contributes to the antiphagocytic effect of Yersinia. Though biochemical function of cysteine protease YopT is poorly understood, it is conserved in all Yersinia species, suggesting that it is important in pathogenesis.18 Cysteine protease YopT also induces an isoelectric point shift of RhoA, a small GTPase known to regulate the actin cytoskeleton.19 YopT causes the release of RhoA from cell membranes or artificial vesicles. Additional insights into the function of cysteine protease YopT were obtained from the observation that Rho family GTPases, including Rac, RhoA, and Cdc42, are all known to undergo post-translational modifications in a sequential manner at their C-terminal CaaX box (C, cysteine; a, aliphatic residue; X, any residue).20 The CaaX box provides the recognition elements for prenylation of the cysteine.21 Cysteine protease YopT carries out a proteolytic cleavage near the C termini of RhoA, Rac1, Cdc42. This cleavage results in the removal of the lipid modification (by removal of prenylated group) from the GTPase at their C-termini, which leads to their subsequent membrane detachment. By this mechanism YopT impairs the ability of host cell to execute phagocytosis and bacterial cell internalization by causing cytoskeletal rearrangement of phagocytic cell. Cleavage of the Rho GTPase by YopT is responsible for the disruption of actin cytoskeleton as well as the proteolytic activity of cysteine protease YopT, which is dependent upon the invariant Cys, His, Asp residues that are conserved in a novel family of cysteine proteases involved in both animal and plant bacterial pathogenesis.19 Our present study aims to predict the 3D structure of cysteine protease YopT, finding out its interacting networks, and also to predict the active sites of cysteine protease YopT for possible drug target by computer simulation.
Materials and Methods
Sequence retrieval
A sequence of cysteine protease YopT in Yersinia pestis was retrieved from the UniProt Knowledge Base (UniProtKB), which is the central hub for the collection of functional information on proteins, with consistent, accurate and rich annotation. The accession ID of cysteine protease is O68703 [UniParc] and it contains 322 amino acids.
Physiochemical properties analysis
The amino acid composition, theoretical isoelectric point (pI), molecular weight, number of positively and negatively charged residues, instability and aliphatic index, extinction coefficient and Grand Average of Hydropathy (GRAVY) was assessed by Protparam [http://web.expasy.org/protparam/].
Functional properties analysis
For functional analysis of this protein, a specialized tool PFP from Kihara Bioinformatics laboratory (http://kiharalab.org/web/pfp.php) was used. The retrieved amino acid sequence in FASTA format was used as input data.
Prediction of disease causing regions and proteolytic cleavage sites
GlobPlot 2.322 was used for the prediction of disease causing regions. Proteolytic cleavage sites were identified by using a web-based tool peptide cutter (http://web.expasy.org/peptide_cutter/), which predicts the proteolytic cleavage sites and sites cleaved by chemicals in a given protein sequence.
Analysis of interacting networks
Alibaba23 and Search Tool for the Retrieval of Interacting Genes/Proteins (STRING)24 were used to identify the networking partners of cysteine protease YopT. AliBaba can be used to fit unstructured text into structured data records. It can be used specifically for increasing accuracy and efficiency in the process of discovering relationships between important biological objects, eg, protein-to-disease associations. STRING quantitatively assimilates interaction data from these sources for a large number of organisms, and transfers information among these organisms where applicable. The database presently covers 5,214,234 proteins from 1133 organisms.
Model building and refinement
To predict the 3D homology model of cysteine protease YopT, the most popular online protein fold recognition server Phyre225 (Protein Homology/Analogy Recognition Engine) was used. The input data was in FASTA format. ModRefiner26 was used to refine the predicted model.
Evaluation and validation of model
The accuracy and stereo chemical quality of the predicted model was evaluated with PROCHECK27 by Ramachandran Plot analysis.28 The best model was selected on the basis of overall G-factor, number of residues in core, allowed, additional allowed, generously allowed and disallowed regions. The selected model was further analyzed with verify 3D,29 ERRAT30 and 3D Match from Softberry (http://linux1.softberry.com/). Finally, the protein was visualized with Swiss-PDB viewer.31
Secondary structure analysis
Secondary structural properties of the protein including alpha helix, 310 helix, Pi helix, beta bridge, extended strand, beta turns, bend region, random coil, ambiguous states and other states were calculated by the use of the self-optimized prediction method with alignment (SOPMA).32
Active site analysis
Active site analysis was done with the help of Computed Atlas of Surface Topography of Protein (CASTp).33 Analysis of active sites is important for the modeled protein as a precursor to further work on its docking studies, and to shape the process of making a grid before docking.
Results
Physicochemical properties
Analysis of physicochemical properties using ProtParam reveals that the protein has 38640 extinction coefficient, 31.1 instability index, 75.71 aliphatic index, −0.390 grand average of hydrophobicity with more positively charged residue than those of negatively charged amino acids.
Function prediction
The functions of protein under study were predicted by PFP from Kihara Bioinformatics Laboratory, which uses references from Gene Ontology. These are shown in Table 1.
Table 1.
Predicted functions of cysteine protease YopT.
| MOLECULAR FUNCTION | BIOLOGICAL PROCESS | CELLULAR COMPONENT |
|---|---|---|
| 1. Cysteine type endopeptidase activity | 1. Cellular protein metabolism | 1. Cell |
| 2. Catalytic activity | 2. Drug metabolism | 2. Inner membrane |
| 3. Binding | 3. Regulation of physiological process | 3. External encapsulating structure |
| 4. Purine nucleotide binding | 4. Immune cell mediated cytotoxicity | 4. Intracellular |
| 5. Antigen binding | 5. Regulation of Type I hypersensitivity | 5. Organelle membrane |
Disease causing region and cleavage site prediction
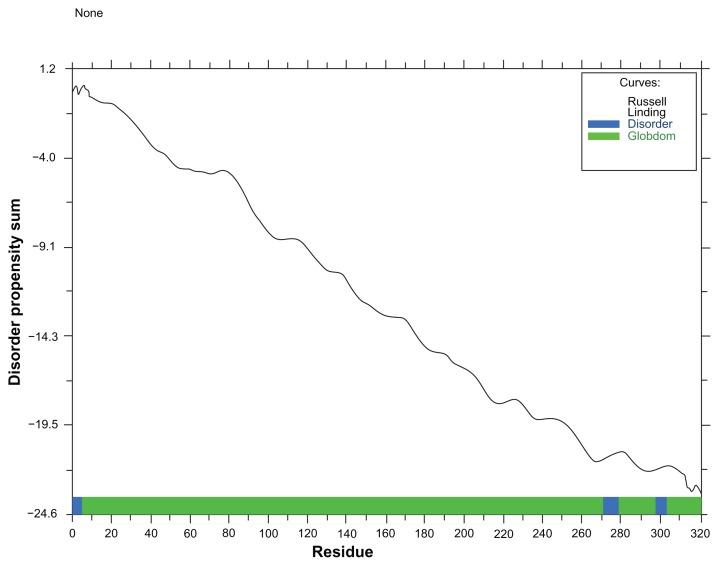
The result from globplot 2.3 was used to identify three disease causing regions (see Fig. 1). Protease digestion is a useful process that is used to identify the presence of any cutting site in protein sequence. A tool from Expasy was used here to determine the proteolytic cleavage site. According to results from peptide cutter there are several cleavage sites for 22 different enzymes (see Table 2).
Figure 1.
Globplot result shows the disease causing regions of this protein.
Table 2.
Result from peptide cutter.
| NAME OF ENZYME | NO. OF CLEAVAGES | POSITIONS OF CLEAVAGE SITE |
|---|---|---|
| Arg-C proteinase | 15 | 35 62 67 78 89 98 103 148 165 207 211 216 289 311 313 |
| Asp-N endopeptidase | 16 | 65 71 132 157 171 182 187 189 191 196 218 229 237 273 283 317 |
| Asp-N endopeptidase + N-terminal Glu | 30 | 18 27 38 65 71 103 132 139 157 171 182 187 189 191 192 196 205 209 218 224 226 229 237 265 273 278 283 285 317 320 |
| BNPS-Skatole | 4 | 146 198 291 297 |
| CNBr | 8 | 1 64 85 87 90 208 255 301 |
| Chymotrypsin-high specificity | 33 | 8 15 81 83 88 93 109 115 117 124 146 157 161 169 175 198 199 244 246 263 272 273 277 280 282 288 291 292 296 297 302 312 317 |
| Chymotrypsin-low specificity | 80 | 1 5 7 8 9 12 15 21 26 34 42 44 47 51 57 64 65 69 81 83 85 88 90 92 93 95 109 115 117 124 125 126 131 142 146 150 156 157 160 161 169 174 175 180 196 198 199 208 212 214 215 229 232 233 237 240 244 246 250 251 255 258 263 272 273 277 280 281 282 288 291 292 296 297 302 303 306 312 315 317 |
| Clostripain | 15 | 35 62 67 78 89 98 103 148 165 207 211 216 289 311 313 |
| Formic acid | 16 | 66 72 133 158 172 183 188 190 192 197 219 230 238 274 284 318 |
| Glutamyl endopeptidase | 14 | 19 28 39 104 140 193 206 210 225 227 266 279 286 321 |
| Hydroxylamine | 1 | 202 |
| Iodosobenzoic acid | 4 | 146 198 291 297 |
| LysC | 23 | 37 49 50 56 68 74 116 121 127 130 166 168 178 186 200 201 247 248 267 285 287 290 320 |
| LysN | 23 | 36 48 49 55 67 73 115 120 126 129 165 167 177 185 199 200 246 247 266 284 286 289 319 |
| NTCB [2-nitro-5-thiocyanobenzoic acid] | 4 | 138 142 184 212 |
| Pepsin [pH 1.3] | 91 | 8 12 14 15 20 21 25 26 41 42 47 50 65 68 81 82 83 87 88 92 93 95 108 109 114 115 116 117 124 124 125 141 142 145 146 155 156 156 157 159 160 160 161 169 173 174 174 175 179 195 196 197 198 198 199 228 229 231 232 232 233 236 237 243 244 245 246 251 262 263 271 272 272 277 279 280 281 282 288 290 295 296 296 297 301 302 304 312 314 316 317 |
| Pepsin [pH > 2] | 64 | 12 20 21 25 26 41 42 47 50 65 68 81 82 83 92 93 95 114 115 116 117 124 124 125 141 142 155 156 156 157 159 160 169 173 174 179 195 196 198 199 228 229 231 232 232 233 236 237 251 271 272 272 277 279 280 281 282 288 295 296 312 314 316 317 |
| Proline-endopeptidase | 1 | 217 |
| Proteinase K | 148 | 4 8 10 12 15 17 19 21 24 25 26 27 28 30 31 33 36 38 39 40 41 42 47 51 53 54 55 65 69 70 75 81 82 83 86 88 91 93 94 95 97 99 101 102 104 106 107 109 113 115 117 118 120 123 124 125 128 129 134 135 138 140 141 142 144 146 147 151 156 157 160 161 162 169 171 173 174 175 177 180 182 187 189 193 194 195 196 198 199 204 206 209 210 214 215 218 220 221 223 224 225 227 229 232 233 235 236 237 239 242 244 246 249 251 257 259 260 261 262 263 264 266 270 271 272 273 277 279 280 282 286 288 291 292 293 296 297 302 304 306 308 312 314 315 316 317 321 322 |
| Staphylococcal peptidase I | 14 | 19 28 39 104 140 193 206 210 225 227 266 279 286 321 |
| Thermolysin | 101 | 3 9 11 16 20 23 25 29 30 32 35 37 40 41 46 50 52 54 63 64 68 74 80 82 84 85 86 89 90 92 93 94 100 101 105 106 112 114 116 117 122 123 124 127 128 134 137 141 143 146 150 155 156 159 161 168 170 173 176 179 181 186 195 198 203 207 208 213 214 217 228 231 232 234 235 236 241 248 250 254 256 258 259 260 261 263 269 271 272 276 281 287 291 295 300 305 307 311 313 314 316 |
| Trypsin | 37 | 35 37 49 50 56 62 67 68 74 78 89 98 103 116 121 127 130 148 165 166 168 178 186 200 201 207 211 247 248 267 285 287 289 290 311 313 320 |
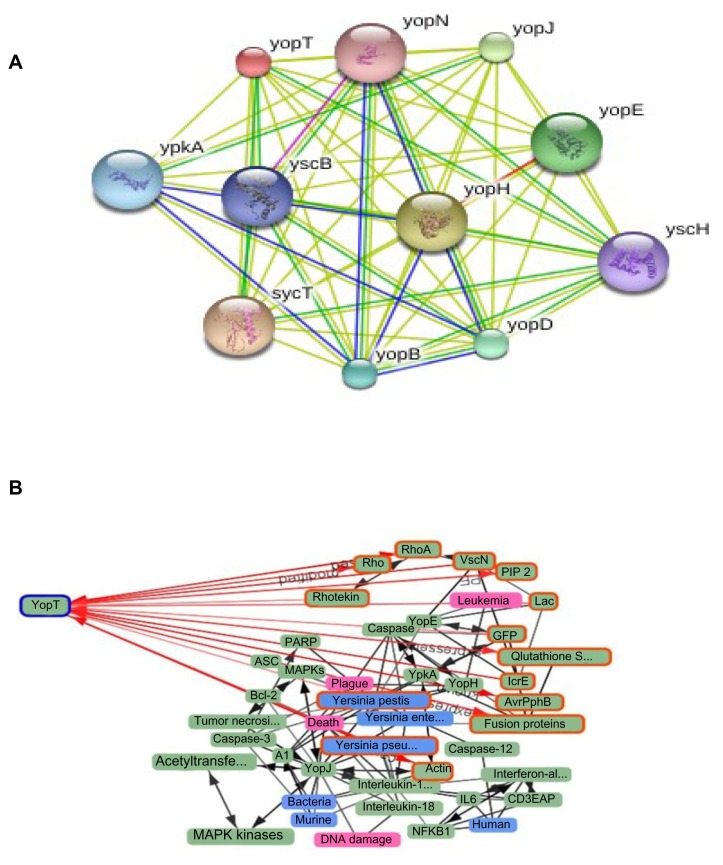
Interacting network
The interacting partners of protein under investigation were determined by STRING (see Fig. 2A). Alibaba was also used to find the interacting proteins, related diseases, species and types of cell where YopT was found (see Fig. 2B).
Figure 2.
STRING (2A) and ALIBABA (2B) shows the interacting networks of cysteine protease YopT.
Model building and refining
Three-dimensional (3D) protein structures provide valuable insights into the molecular basis of protein function. The 3D structure of the protein being investigated was determined by Phyre 2. For further refinement to determine the high resolution structure of the protein, ModRefiner was used.
Model validation
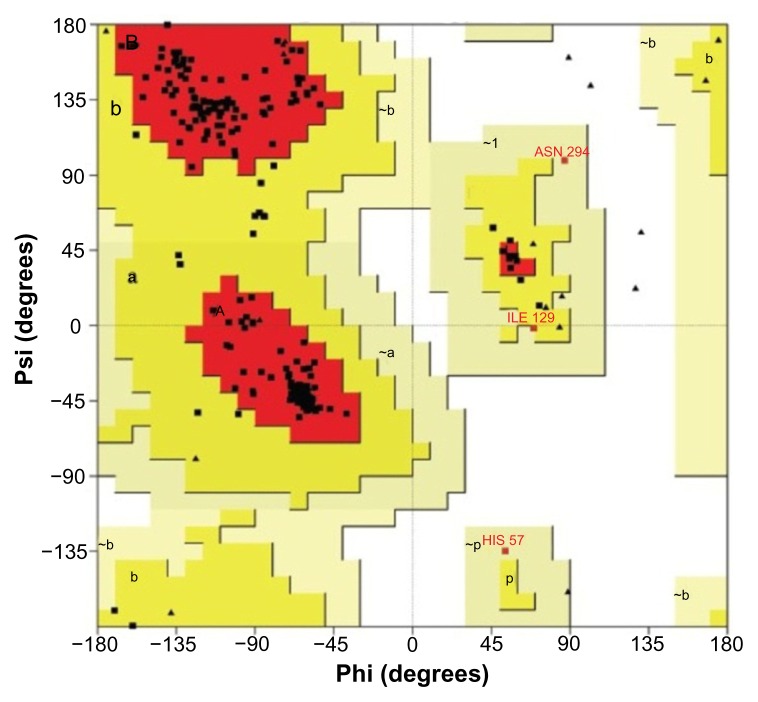
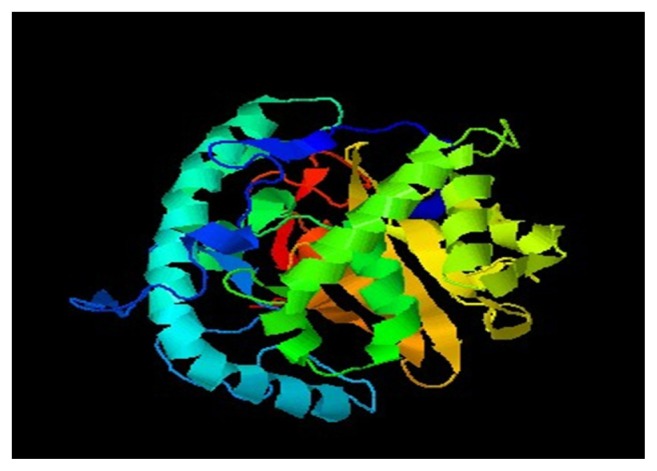
For validation of the predicted structure. Ramachandran plot analysis was done by PROCHECK server. The results of this analysis are depicted in Figure 3 and Table 3. The selected model was then verified by using ERRAT, Verify 3D, and 3D match from Softberry. Validated 3D structure of cysteine protease YopT was determined by SWISS-PDB (see Fig. 4).
Figure 3.
Ramachandran plot of cysteine protease YopT from Yersinia pestis obtained through the modeling tool.
Table 3.
Ramachandran plot statistics of cysteine protease YopT from Yersinia pestis.
| RAMACHANDRAN PLOT STATISTICS | NO. OF RESIDUE | % |
|---|---|---|
| Residues in the most favored regions [A, B, L] | 269 | 93 |
| Residues in the additional allowed regions [a, b, l, p] | 17 | 5.9 |
| Residues in the generously allowed regions [a, b, l, p] | 3 | 1 |
| Residues in the disallowed regions | 0 | 0 |
| Number of non-glycine and non-proline residues | 289 | 100 |
| Number of end-residues [excl. Gly and Pro] | 2 | |
| Number of glycine residues [shown in triangles] | 27 | |
| Number of proline residues | 4 | |
| Total number of residues | 322 |
Figure 4.
Final model of cysteine protease YopT from Yersinia pestis.
Secondary structure analysis
Secondary structure predictions are increasingly becoming the work horse for numerous methods aimed at predicting protein structure and function. SOPMA was used to analyze the secondary structure. The result was found by using standard parameters, given in Table 4.
Table 4.
Computed secondary structure elements of cysteine protease YopT of Yersinia pestis by SOPMA.
| STRUCTURE | NO OF RESIDUE | % |
|---|---|---|
| Alpha helix [Hh]: | 145 | 45.03% |
| 310 helix [Gg]: | 0 | 0.00% |
| Pi helix [Ii]: | 0 | 0.00% |
| Beta bridge [Bb]: | 0 | 0.00% |
| Extended strand [Ee]: | 39 | 12.11% |
| Beta turn [Tt]: | 30 | 9.32% |
| Bend region [Ss]: | 0 | 0.00% |
| Random coil [Cc]: | 108 | 33.54% |
| Ambigous states [?]: | 0 | 0.00% |
| Other states: | 0 | 0.00% |
Active site prediction
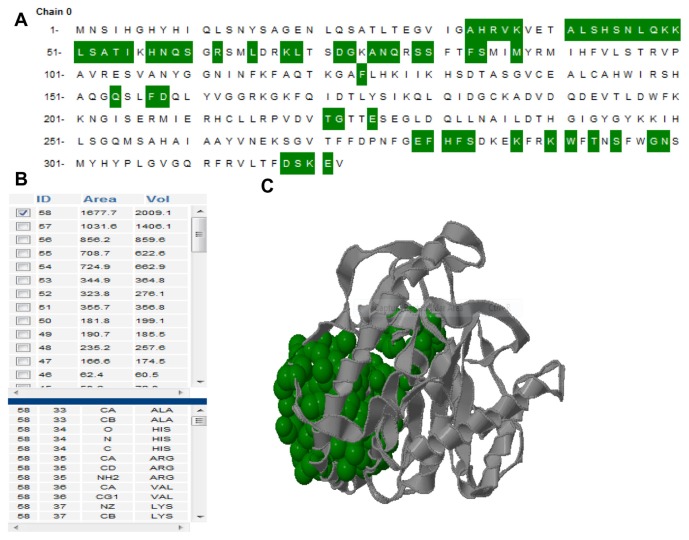
The active site of cysteine protease YopT was predicted using CASTp server. Further, in this study we have also reported the best active site area of the experimental enzyme as well as the number of amino acid involved in it. [Fig. 5] shows the number of pockets, with their area and volume. The best active site is marked with 1677.7 areas and a volume of 2009.1.
Figure 5.
(A) Active site information by CASTp. Green color shows the active site position from 33 to 321 with the beta-sheet in between them. (B) The table shows the area and the volume for different active sites of cysteine protease YopT and the best active site remains in an area of 1677.7 and a volume of 2009.1 amino acid. (C) The 3D structure of best active site.
Discussion
The UniProt Knowledge Base [UniProtKB] delivers a single, centralized, authoritative resource for protein sequences and functional information.34 Sequence of cysteine protease YopT of Yersinia pestis was obtained from UniProtKB. Manual annotation is one of the landmarks of the Swiss-Prot section of UniProtKB. Physicochemical properties of protein under this study were analyzed through ProtParam in respect of different parameters. According to the result, this protein has a high extinction coefficient (at 280 nm and in water), low instability index, high aliphatic index, which is the volume of protein occupied by side chains, is a positive factor for increasing thermo stability, and has negative GRAVY with molecular weight 36308.1 and pI 9.19. The magnitudes of all parameters indicate the stability of the protein. Protein stability is related to its function.35 A major hurdle to annotating function of genomes of different organisms is a lack of coherence in functional annotation.36 A novel algorithm, named PFP from Kihara Bioinformatics Laboratory, which extends a PSI-BLAST search and extracts and scores GO annotations based on the frequency of their occurrence in retrieved sequences, was used to predict the functions of target protein.37 Function prediction was done in terms of molecular function, cellular components and biological process (see Table 1). Aberrant function of protein is due to the disordered regions in it.
Lack of regular secondary structure can be described as protein disorder.38 GlobPlot 2.3 was used to identify disordered region within protein based on a running sum of the propensity for amino acids to be in an ordered or disordered state. This tool can identify such regions by searching domain databases and known disordered proteins.22 Cysteine protease YopT contains three disease causing regions in it from sequence 1 to 5, 271 to 279 and 298 to 303 respectively with decreasing propensity sum [Fig. 1]. Protease digestion can be useful if one wants to carry out experiments on a portion of a protein, separates the domains in a protein, removes a tag protein when expressing a fusion protein, or makes sure that the protein under investigation is not sensitive to endogenous proteases.39 According to results from peptide cutter, there are several cleavage sites for 22 different enzymes in our target and most of the enzymes are with several cutting sites [Table 2]. So, for any type of alteration in its sequences, these cleavage sites are the potential target.
Proper functioning of a protein generally requires interaction with other proteins. Protein–protein interactions have a vital role in functions and the structural organization of a cell. Detailed view of these interactions helps in elucidation of cellular activities, drug target and whole cell engineering.40
Interaction of cysteine protease YopT with its partner determined through STRING is given in [Fig. 2A]. STRING predicts results with a confidence score, protein domains and 3D structures. Predicting functions of interacting protein is also possible from STRING that uses references from UniProt. Network shows that cysteine protease YopT interacts with nine other proteins among which YscB is a hypothetical protein; YopH and YpkA both are virulence determinant. sycT is a membrane bound protein that plays a role in regulating calcium concentration.41 Analysis of the protein with Alibaba [Fig. 2B] entails that it is linked with 62 proteins, present in 22 species in 14 different cell types and causes 7 different diseases. Red path indicates the direct link of protein with objects. The protein is present in three species of Yersinia. According to network generated by Alibaba, YopT is responsible for plague, Black Death, DNA damage and leukemia. YopT is directly linked with actin and Rho family because it disrupts cytoskeleton by cleaving Rho GTPases protein to produce disorders as Rho family involves in organization of actin filament and in different signaling processes.19,20
3D structure determination is the most important part of proteomics. Three-dimensional [3D] protein structures provide valuable insights into the molecular basis of protein function, permitting an effective design of experiments, such as site-directed mutagenesis, studies of disease-associated mutations or the structure based design of specific inhibitors.42 Therefore, the high resolution 3D structure of a protein is the key to understand and manipulate of its biochemical and cellular functions.43 The theoretical structure of cysteine protease YopT from Yersinia pestis is generated using Phyre 2. 100% confidence match was obtained, which is a high confidence match. This implies that, overall fold of the model was almost certainly correct and the central core of the model tends to be accurate.44 According to results from phyre 2, the protein contains 37% disorder, 43% alpha helix and 15% beta strand. In protein structure prediction, refinement for finding structures that are close to the native state is a great challenge. Commonly used structure prediction methods can predict the correct topology but such approximate models are often not with resolution required for many important applications, including virtual ligand screening and studies of reaction mechanisms. 45 Structural validation of target model was performed by PROCHECK, ERRAT and verify 3D. PROCHECK generated Ramachandran plot shows the distribution of φ and ψ angle in the model [Fig. 3]. According to plot statistics, 93.0% of the residues are located in the most favored region, 5.9% in additional region and 1.1% generously allowed region [Table 3] and all non-glycine and non-proline residues are in the allowed region of plot. Verification was also done with verify 3D model, ERRAT, 3D match program from softberry.
Findings of verify 3D model, ERRAT, 3D match program and Ramachandran plot from PROCHECK reveals that all residues are within the limits of the Ramachandran plot and hence it can be considered as a good model. The high score of 0.78 in the verify 3D graph indicates that the environment profile of the model is good and the overall quality of model in ERRAT analysis was 85.357, expressed as the percentage of the protein for which the calculated error value falls below 95% rejection limit. The final model of target protein determined through SWISS PDB viewer is shown in [Fig. 4].
Secondary structure analysis of the modeled protein with SOPMA reveals that it contains 45.03% alpha helix, 12.54% extended strand and 33.54% random coil [Table 4]. The identification and characterization of functional sites on proteins has increasingly become an area of interest. Analysis of the active site residues for the binding of ligands provides insight into towards the design of inhibitors of an enzyme. In this study, we have also reported the best active site area of the experimental enzyme as well as the number of amino acids involved in it. [Fig. 5] shows the area and the volume for different active sites of cysteine protease YopT and the best active site remains in an area of 1677.7 and a volume of 2009.1 amino acids.
In addition, it is desirable to identify which properties of active residues are required for catalysis and/or recognition, in order to understand how one might engineer these properties into a protein scaffold to design an enzymatic species with a specified type of reactivity.46
Conclusion
In this study, the 3D structure of cysteine protease YopT from Yersinia pestis was predicted and validated by various bioinformatics tools and software. It is obvious that this enzyme is the most important factor used by Yersinia pestis for its pathogenesis in host. On the basis of our findings, it could be concluded that advance characterization of cysteine protease YopT from Yersinia pestis will be important for the regulation of plague. Further broad screening inhibitors against this enzyme will help for effective drug designing in future.
Footnotes
COMPETING INTERESTS: Author(s) disclose no potential conflicts of interest.
Author Contributions
Conceived and designed the experiments: MAH. Analyzed the data: MAH, SMA. Wrote the first draft of the manuscript: MAH, MAA, SMN. Contributed to the writing of the manuscript: MAH, MAA, AM. Agree with manuscript results and conclusions: MAH, MAA, SMN, SMA, AM. Jointly developed the structure and arguments for the paper: MAH, SMN, AM. Made critical revisions and approved in all version: MAH. All authors reviewed and approved of the in all manuscript.
DISCLOSURES AND ETHICS
As a requirement of publication the authors have provided signed confirmation of their compliance with ethical and legal obligations including but not limited to compliance with ICMJE authorship and competing interests guidelines, that the article is neither under consideration for publication nor published elsewhere, of their compliance with legal and ethical guidelines concerning human and animal research participants (if applicable), and that permission has been obtained for reproduction of any copyrighted material. This article was subject to blind, independent, expert peer review. The reviewers reported no competing interests.
FUNDING: Author(s) disclose no funding sources.
REFERENCES
- 1.Riedel S. Plague: from natural disease to bioterrorism. Proc (Bayl Univ Med Cent) 2005;18:116–124. doi: 10.1080/08998280.2005.11928049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stenseth NC, Atshabar BB, Begon M, et al. Plague: past, present and future. PloS Med. 2008;5:e3. doi: 10.1371/journal.pmed.0050003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parkhill J, Wren BW, Thomson NR, et al. Genome sequence of Yersinia pestis, the causative agent of plague. Nature. 2001;413:523–27. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- 4.Quenee LE, Ciletti NA, Elli D, Hermanas TM, Schneewind O. Prevention of pneumonic plague in mice, rats, guinea pigs and non-human primates with clinical grade rV10, rV10–2 or F1-Vvaccines. Vaccine. 2011;29:6572–83. doi: 10.1016/j.vaccine.2011.06.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swietnicki W, Carmany D, Retford M, et al. Identification of Small-Molecule Inhibitors of Yersinia pestis Type III Secretion System YscN ATPase. PLoS One. 2011;6:e19716. doi: 10.1371/journal.pone.0019716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryan KJ, Ray CG, editors. Sherris Medical Microbiology. 4th edition. McGraw Hill; 2004. pp. 484–8. [Google Scholar]
- 7.Yersin A. La peste bubonique à Hong Kong. Ann Inst Pasteur Paris. 1894;8:662–7. [Google Scholar]
- 8.Yersin A. La peste bubonique à Hong Kong. Archives de Médecine Navales. 1894;62:256–61. [Google Scholar]
- 9.Perry RD, Fetherston JD. Yersinia pestis etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng W, et al. Genome Sequence of Yersinia pestis KIM. Journal of Bacteriology. 2002;184:4601–11. doi: 10.1128/JB.184.16.4601-4611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cowan C, Jones HA, Kaya YH, Perry RD, Straley SC. Invasion of epithelial cells by Yersinia pestis: evidence for a Y. pestis-specific invasion. Infect. Immun. 2000;68:4523–30. doi: 10.1128/iai.68.8.4523-4530.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornelis GR, Boland A, Boyd AP, et al. The virulence plasmid of Yersinia, an antihost genome. Microbiol Mol Biol Rev. 1998;62:1315–52. doi: 10.1128/mmbr.62.4.1315-1352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hueck CJ. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornelis GR. The Yersinia Ysc-Yop ‘type III’ weaponry. Nat Rev Mol Cell Biol. 2002;3:742–54. doi: 10.1038/nrm932. [DOI] [PubMed] [Google Scholar]
- 15.Aepfelbacher M, Zumbihl R, Heesemann J. Modulation of Rho GTPases and the actin cytoskeleton by YopT of Yersinia. Curr Top Microbiol Immunol. 2005;291:167–75. doi: 10.1007/3-540-27511-8_9. [DOI] [PubMed] [Google Scholar]
- 16.Iriarte M, Cornelis GR. YopT, a new Yersinia Yop effector protein, affects the cytoskeleton of host cells. Mol Microbiol. 1998;9:915–29. doi: 10.1046/j.1365-2958.1998.00992.x. [DOI] [PubMed] [Google Scholar]
- 17.Green SP, Hartland EL, Robins-Browne RM, Phillips WA. Role of YopH in the suppression of tyrosine phosphorylation and respiratory burst activity in murine macrophages infected with Yersinia enterocolitica. J Leukoc Biol. 1995;57:972–7. doi: 10.1002/jlb.57.6.972. [DOI] [PubMed] [Google Scholar]
- 18.Shao F, Dixon JE. YopT is a cysteine protease cleaving Rho family GTPases. Adv Exp Med Biol. 2003;529:79–84. doi: 10.1007/0-306-48416-1_14. [DOI] [PubMed] [Google Scholar]
- 19.Shao F, Merritt PM, Bao Z, Innes RW, Dixon JE. A Yersinia effector and a pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell. 2002;109:575–88. doi: 10.1016/s0092-8674(02)00766-3. [DOI] [PubMed] [Google Scholar]
- 20.Sorg I, Goehring UM, Aktories K, Schmidt G. Recombinant Yersinia YopT leads to uncoupling of RhoA-effector interaction. Infect Immun. 2001;69:7535–43. doi: 10.1128/IAI.69.12.7535-7543.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang FL, Casey PJ. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241–69. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 22.Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31:3701–08. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plake C, Schiemann T, Pankalla M, Hakenberg J, Leser U. Alibaba: Pubmed as a graph. Bioinformatics. 2007;22:2444–45. doi: 10.1093/bioinformatics/btl408. [DOI] [PubMed] [Google Scholar]
- 24.Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:808–15. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–71. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 26.Xu D, Zhang Y. Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys J. 2011;101:2525–34. doi: 10.1016/j.bpj.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–86. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 28.Ramachandran GN, Ramakrishnan C, Sasisekharan V. Stereochemistry of polypeptide chain configurations. J Mol Biol. 1963;7:95–9. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 29.Eisenberg D, Lüthy R, Bowie JU. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;277:396–404. doi: 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- 30.Colovos C, Yeates TO. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sciences. 1993;2:1511–19. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–23. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 32.Geourjon C, Deléage G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci. 1995;11:681–4. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- 33.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:116–8. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider M UniProt Consortium. Poux S. UniProtKB amid the turmoil of plant proteomics research. Front Plant Sci. 2012;3:270–7. doi: 10.3389/fpls.2012.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shoichet BK, Baase WA, Kuroki R, Matthews BW. A relationship between protein stability and protein function. Proc Natl Acad Sci U S A. 1995;92:452–6. doi: 10.1073/pnas.92.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dobson PD, Cai YD, Stapley BJ, Doig AJ. Prediction of protein function in the absence of significant sequence similarity. Curr Med Chem. 2004;11:2135–42. doi: 10.2174/0929867043364702. [DOI] [PubMed] [Google Scholar]
- 37.Hawkins T, Kihara D. Function prediction of uncharacterized proteins. Journal of Bioinformatics and Computational Biology. 2007;5:1–30. doi: 10.1142/s0219720007002503. [DOI] [PubMed] [Google Scholar]
- 38.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–31. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 39.Stein L. Genome annotation: from sequence to biology. Nat Rev Genet. 2001;2:493–503. doi: 10.1038/35080529. [DOI] [PubMed] [Google Scholar]
- 40.Arifuzzaman M, Maeda M, Itoh A, et al. Large-scale identification of protein–protein interaction of Escherichia coli K-12. Genome Res. 2006;16:686–91. doi: 10.1101/gr.4527806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:561–8. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fidanova S, Lirkov I. 3D Protein Structure Prediction. Seria Matematica-Informatica. 2009;XLVII:33–46. [Google Scholar]
- 43.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–71. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 44.Laurie AT, Jackson RM. Methods for the prediction of protein-ligand binding sites for structure-based drug design and virtual ligand screening. Curr Protein Pept Sci. 2006;7:395–406. doi: 10.2174/138920306778559386. [DOI] [PubMed] [Google Scholar]
- 45.Jagielska A, Wroblewska L, Skolnick J. Protein model refinement using an optimized physics-based all-atom force field. Proc Natl Acad Sci U S A. 2008;105:8268–73. doi: 10.1073/pnas.0800054105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ko J, Murga LF, Wei Y, Ondrechen MJ. Prediction of active sites for protein structures from computed chemical properties. Bioinformatics. 2005;21:258–65. doi: 10.1093/bioinformatics/bti1039. [DOI] [PubMed] [Google Scholar]