Abstract
Heterozygotic loss of SYK, a non-receptor tyrosine kinase, gives rise to mouse mammary tumor formation where Syk protein levels are reduced by about half; loss of SYK mRNA is correlated with invasive cell behavior in in vitro models; and SYK loss has been correlated with distant metastases in patients. Here, allelic loss of the SYK gene was explored in breast ductal carcinoma in situ (DCIS) using fluorescence in situ hybridization and pyrosequencing, respectively, and in infiltrating ductal carcinoma (IDC) using genomic data from The Cancer Genome Atlas (TCGA). Allelic loss was present in a subset of DCIS cases where adjacent IDC was present. SYK copy number loss was found in about 26% of 1002 total breast cancer cases and 30% of IDC cases. Quantitative immunofluorescence revealed Syk protein to be six-fold higher in infiltrating immune cells compared with epithelial cells. This difference distorted tumor cell mRNA and protein levels in extracts. 20% of 1002 IDC cases contained elevated immune cell infiltration as estimated by elevated immune-specific mRNAs. In cases without immune cell infiltration, loss of SYK copy number was associated with a significant reduction of SYK mRNA. Here we define a 55 Gene Set consisting of Syk interacting, motility- and invasion-related genes. We found that overall survival was significantly reduced in IDC and Luminal A+B cases where copy number and mutations of these 55 genes were affected (Kaplan-Meier, Logrank test p-value 0.007141 and Logrank test p-value 0.001198, respectively). We conclude that reduction in Syk expression and contributions of genomic instability to copy number and mutations in the 55 Syk interacting genes significantly contribute to poorer overall patient survival. A closer examination of the role of Syk interacting motility and invasion genes and their prognostic and/or causative association with metastatic disease and patient outcome is warranted.
Introduction
Syk (spleen tyrosine kinase), a 72 kD tyrosine kinase, plays diverse and complicated roles in immunity as well as in epithelial cell biology and is required for activation of immune, integrin, and a growing number of other cell surface receptors [1]. In normal or cancerous mammary epithelial cells, the presence of Syk suppresses malignant growth characteristics including proliferation and invasive cell migration and its loss induces invasion and metastasis [2], [3]. Previously, in patient samples, we demonstrated a loss of SYK mRNA in normal tissue adjacent to breast cancer, and further decreases in ductal carcinoma in situ (DCIS) and intraductal carcinoma (IDC) compared with normal or benign tissues where no cancer was detected [4]. Others have validated the association with Syk loss and breast cancer progression ([5] and references therein).
SYK loss has been attributed to hypermethylation of the promoter in breast cancer tissues and its loss is associated with increased cellular invasiveness [6], [7]. Hypermethylation of the SYK promoter is associated with lower SYK mRNA and poor prognosis and metastasis in various cancers including breast, lung, pancreatic, urinary bladder cancers, mesothelioma, and melanoma; in vitro experiments confirm that re-expression of SYK by transfection or inhibitors of hypermethylation reverses the invasive and metastatic phenotype ([7]–[15] and for review [5]). SYK hypermethylation was noted in 45% of DCIS but only 5% of hyperplasia, thus, hypermethylation in DCIS tissues occurs prior to the development of invasive disease [7]. It was inferred that SYK loss might contribute to the development of invasive breast cancer. Interestingly, SYK mRNA loss in postoperative lung metastases was noted although not directly validated post-surgery in an orthotopic mouse model compared with lung metastases of control mice whose primary tumor was not resected [16]. Recently, several studies have identified SNPs and somatic mutations of SYK associated with breast cancer [17], [18].
Examination of heterozygotic SYK knockdown mice and derivatives of these mice revealed that loss of a single SYK allele was sufficient to reduce Syk protein levels by about half [3]. Syk loss led to accelerated proliferation and ductal outgrowth during puberty and mammary tumor formation in vivo, although the causality of mammary-specific SYK loss was not determined [3]. In vitro, enhanced cell proliferation and invasion were detected in mouse epithelial cells isolated from heterozygotic knockdown mice [3]. In the same study, transient or stable Syk knockdown in a non-transformed human epithelial cell line MCF10A had the same effects; proliferation and invasion were enhanced [3]. These data taken together also significantly strengthen the argument that Syk is a potent breast cancer tumor and metastasis suppressor. Thus, SYK loss results in dramatic effects upon epithelial cell function, promoting proliferative and/or invasive behaviors.
SYK is located on human chromosome 9q22.2 and interestingly, allelic loss on chromosome 9q22 is associated with lymph node metastasis in primary breast cancer [19]. However, in four breast cancer cell lines, Coopman and colleagues found no evidence for alterations in SYK DNA by Southern analysis [2]. To determine whether allelic loss of SYK might be associated with breast cancer invasion and progression in patient samples, we performed fluorescent in situ hybridization (FISH) to detect SYK alleles in a dual color FISH protocol. We focused on DCIS tissues for analysis with comparison to normal or benign tissues. The rationale for this includes the previous observation that single allelic loss of SYK in a mouse model led to enhanced invasion, proliferation, and tumor formation in the mammary gland; Syk knockdown in cells results in an epithelial-to-mesenchymal (EMT)-like transition with enhanced invasiveness; and Syk re-expression in breast cancer cell lines prevents tumor growth and metastasis in mouse models [2], [3], [5]. Thus, SYK allelic loss in human DCIS, particularly in combination with hypermethylation and silencing might result in invasive breast disease ultimately leading to metastasis. If so, SYK status in DCIS tissues might provide a powerful prognostic tool, identifying women most likely to progress to invasive carcinoma. Many women with DCIS will relapse and progress to IDC [20], [21].
Strikingly, the results of our preliminary study of clinical breast cancer tissues revealed that SYK gene loss occurred in 5 out 19 samples of DCIS examined, but in none of 5 normal breast tissues examined. Allelic loss did not occur in DCIS only cases, but rather was exclusively associated with DCIS that was adjacent to IDC. SYK loss in IDC was determined in a large breast cancer data set from The Cancer Genome Atlas (TCGA) using cBioPortal tools. Furthermore, genes relating to motility and invasion that we previously demonstrated to be regulated by SYK were used to query a TCGA breast study. The results indicated that overall survival was significantly influenced by this gene set (51 genes); prediction of improved overall survival could be further enhanced by the addition of TP53, SRC, CTTN, and CDH1, genes that interact with the SYK network or Syk directly.
Materials and Methods
Tissues
Ethics statement: formalin-fixed, paraffin-embedded breast tissue samples were obtained from the Lombardi Comprehensive Cancer Center (LCCC) Histopathology and Tissue Shared Resource (HTSR) using a Georgetown University approved IRB protocol and adjacent sections were used in a previous study [4]. The relevant Georgetown University Institutional Review Board (IRB) protocol was # 1992-048, “Human Tissue Bank”, B. Kallakury, HTSR, and this protocol has been continuously maintained from 1992 to present date. Written consent was obtained from patients for surgery and excess tissue was banked after diagnostic requirements were met. The patients were all pre-HIPAA. Materials and information from patients were de-identified. A serial section from each tissue sample was stained with hematoxylin and eosin and reviewed by pathologists Drs. Metin Ozdemirli and Bhaskar Kallakury to identify the appropriate tissue areas for FISH and protein analysis.
Fluorescence in situ Hybridization (FISH) Probe Development
The Bacterial Artificial Chromosome (BAC) clones, RP11367F29, and RP1183L6 which span the SYK gene region at 9q22.2, were obtained from the BAC/PAC Resource at Children's Hospital Oakland Research Institute (Oakland, CA) for development of a FISH probe. The DNA clones were grown on agarose, colonies were picked for expansion, and DNA isolated from cultures using a Qiagen QIA/Prep mini kit. A 2% agarose gel was run to quantify the DNA for probe mixing.
The SYK DNA clones were labeled via nick translation with Spectrum Orange™ (Vysis, Des Plaines IL.) A chromosome 9, Spectrum Green™ -labeled centromere control probe was obtained from Vysis (Des Plaines, Ill). The labeled BAC clones and control probes were co-hybridized to metaphase preparations of normal lymphocytes and analyzed with fluorescence microscopy to evaluate the FISH signal strength, clarity, and mapping position.
Tissue FISH
A dual label FISH technique was used [22]. The slides with paraffin-embedded breast tissue samples were incubated on a slide warmer at 60°C for at least 1 hour. The breast tumor tissue sections were deparaffinized with a 10 minute xylene wash at room temperature (18°C to 25°C) and dehydrated twice in 100% ethanol for 5 minutes at room temperature (18°C to 25°C). The pretreatment solution was 10% sodium borohydrite in 2× Saline Sodium Citrate (SSC) for 15 minutes. Protein digestion (25 µg/ml) was performed with Proteinase K (Sigma-Aldrich, St Louis, MO, USA) in 2× SSC at 45°C for 10 minutes. The co-denaturation of the test and control probes and target sequence was performed at 85°C for 10 minutes on a hot plate under a coverslip. Following denaturation, the coverslip edges were sealed with rubber cement and hybridization was carried out at 37°C in a humid chamber for 24 hours overnight. Sixteen to 24 hours later the coverslips were removed and slides were then post washed in 2× SSC at 72°C for 5 minutes. The slides were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). The slides were then viewed with fluorescence microscopy with a 100× objective using filters that allow the probe signals to be visualized. 30 cells were counted. The data were expressed as number of SYK signals/number of chromosome 9 signals.
The SYK FISH performed on tumor tissue sections has the advantage over PCR protocols of preserving specimen architecture, allowing the analysis to be focused on neoplastic tissue, without contamination of normal surrounding tissues including immune cells. Also, individual tumor cells can be analyzed for both control and loci of interest. This provides specificity to the analysis and an internal control of the assay. Tissue FISH was performed as a research protocol in our clinical lab and is the approved method for clinical analysis of tissues for diagnosis, prognosis and management by the American College of Medical Genetics (ACMG). The ACMG writes and advised laboratory standards for Clinical Genetics [23].
Methylation
DNA methylation level was assessed by bisulfite pyrosequencing with the Pyromark MD (QIAGEN) instrument, using the Hs_SYK_01_PM PyroMark CpG Assay, a pre-designed assay specific for a CpG rich region within the SYK gene promoter available from QIAGEN. Genomic DNA was extracted from tissue dissected off paraffin section slides as previously described [24] and then subjected to bisulfite pyrosequencing. Pyrosequencing is a sequencing-by-synthesis method that quantitatively monitors the real-time incorporation of nucleotides through the enzymatic conversion of released pyrophosphate into a proportional light signal, thus enabling a quantitative measurement of the methylation extent at each CpG site [25]. Briefly, the DNA was treated with sodium bisulfite using the EpiTect Bisulfite Kit (Qiagen) in order to convert all un-methylated cytosine residues to uracil which will be then converted to thymine during PCR reaction. A target region of 137 bp was then amplified by PCR using primers from the Hs_SYK_01_PM assay (Qiagen) complementary to the bisulfite-treated DNA sequence, amplifying all states irrespective of methylation status. The reverse primer was biotinylated at its 5′-terminus enabling immobilization on streptavidin-coated beads used to purify and render the PCR product single-stranded (as only one strand is biotinylated). The pyrosequencing primer contained in the assay, complementary to the single-stranded template, was then hybridized to the template, and the pyrosequencing reaction was performed by the sequential addition of single nucleotides in a predefined order. The data were then analyzed using the Pyro Q-CpGT Software (Qiagen Inc., Valencia, CA) by calculating the percent methylation for each CpG site in the sequence context.
The assay used herein measured the methylation levels at four CG sites and the mean methylation level was used as a measure of SYK gene promoter methylation. In addition to negative controls, we included in the analysis DNA samples from the Human control DNA set (Qiagen) which contains both bisulfite-converted methylated and unmethylated DNA and unconverted unmethylated normal human DNA). The mean methylation level of the control DNAs were 2.32% for the non-methylated DNA, 70.54% for the methylated DNA, and 4.70% for the normal human DNA.
Immunofluorescence Microscopy
For double immunolabeling of Syk and keratin on paraffin embedded tissue slices, slides were baked overnight at 65°C, and deparaffinized using xylene and a graded series of ethanol. They were then air dried and the tissues were encircled using a PAP pen (Sigma-Aldrich, St. Louis, MO). Slides were rehydrated in phosphate buffered saline, pH 7.4 (PBS) and then submitted to 30 min microwave antigen retrieval using citrate buffer pH 6.0 (Invitrogen cat no. 00-5001) equilibrated in a boiling water bath. The sections were washed for 1 h at room temperature and then endogenous avidin-biotin was blocked (Avidin/Biotin Blocking Kit, Invitrogen cat. No. 00-4303). Non-specific binding of antibodies was blocked for 5 min using PBS containing 10% non-immune goat serum and 0.1% gelatin. Primary antibodies were applied, the sections washed 2× PBS 5 min each, and secondary antibodies were applied in the following steps:
Step 1: 30 min at room temperature: PBS/0.1% gelatin containing 1/100 rabbit monoclonal anti-Syk (clone EP 573Y, Epitomics (1688-1), Burlingame, CA) and 1/100 anti-pan cytokeratin (clone AE1/AE3 mouse monoclonal, Invitrogen cat. No. 18-0132).
Step 2: 15 min at room temperature: PBS/0.1% gelatin containing 1/300 biotin-labeled goat anti-rabbit antibody (Invitrogen cat. No. 50-2352).
Step 3: 15 min at room temperature: PBS/0.1% gelatin containing 1/300 streptavidin AlexaFluor633 (Invitrogen cat. No. S21375, lot 731496), 1/200 rhodamine-labeled goat anti-mouse antibody for 15 min (Sigma), and DAPI (Sigma).
After a final 2× PBS 5 min each wash, the sections were mounted using Fluoro-gel and no. 1.5 coverslips applied (Electron Microscopy Sciences cat. No. 17985-10, Hatfield, PA).
Confocal Imaging and Syk protein quantification
Fluorescently stained sections were imaged using a Zeiss LSM510/META/NLO microscope equipped with a 10×/0.3 N.A. (initial inspection) or a 25×/0.8 N.A. W lens. For quantitative analysis, images were obtained using the 25× objective with identical imaging parameters for all samples. A series of adjacent images was obtained for all available tissue epithelial areas identified by the pathologist (using H/E adjacent sections) for each stained tissue section. Protein analysis was difficult when comparing tissues using many images from the same slide. Uneven staining across sections occurred and different age of tissue blocks affected staining levels between cases. Therefore, image analysis using keratin to normalize Syk epithelial measurements was carried out to increase the accuracy for reporting protein levels.
For image quantification to determine relative protein levels, average background pixel values were first determined and subtracted for each image to produce background subtracted keratin and Syk images. This was accomplished using a custom Metamorph Offline journal (ver. 7.7.1.0) (MetaMorph® Microscopy Automation & Image Analysis Software, Molecular Devices, LLC, Sunnyvale, CA). Another custom automated journal was used to determine values for keratin, epithelial Syk, and immune cell Syk protein as follows. The average intensity and pixel location of keratin was measured, and then Syk was determined using keratin positive pixels for identification of epithelial cells. Next, the average intensity of immune cells only was determined by filtering for the smaller size of immune cells. For each set of images representing one case, the appropriate threshold level was selected for keratin, epithelial Syk, and immune cell Syk. However, objects less than 70 µm2 were excluded from epithelial measurements to minimize the contribution of immune cells to epithelial cell measurements where possible. For selection of immune cells for Syk intensity measurements, areas were excluded that were less than 20 µm2 in size, less than 100 average intensity value or greater than 15,000 µm2 in area to exclude epithelial cells.
Statistical Methods
Single factor ANOVA was performed using Microsoft Excel Tools (Data Analysis). Sigma Plot ver. 7.1 was used to plot the data using box plots (means are indicated by red lines within the box and medians are indicated by black lines). The boundary of the box closest to zero indicates the 25th percentile and the boundary of the box farthest from zero indicates the 75th percentile. Whiskers above and below the box indicate the 95th and 5th percentiles. At least three points are required to compute each set of percentiles.
cBioPortal analysis of TCGA data sets
cBioPortal, a tool developed by the Computations Biology Center at Sloan Kettering, was accessed at http://www.cbioportal.org/public-portal/ [26], [27]. The “Breast Invasive Carcinoma (TCGA, Provisional)” data set was queried (the data set contained 929 total cases at the time of analysis, July, 2013, and 696 cases of IDC were identified with available sequencing and aCGH data). Previously, we determined genes that were up- or down-regulated by SYK siRNA knockdown (> = 1.5 fold) in MCF10A cells cultured on collagen I fibrillar matrices [3]. We subsequently curated 51 genes for their relationship to motility and invasion (ABLIM1, ADAM12, ADAM15, AMOTL2, AP1M2, AXL, CDC42EP4, CHN1, CKAP4, CORO1A, CTNNAL1, CXCL1, CXCL2, ECT2, EGFR, FSCN1, GPER, KIF20A, KIF2C, LAMP1, LAMP2, MARCKSL1, MET, MMP7, MUC1, MYL9, NEBL, PCDH9, RAB11A, RAB11FIP1, RAB20, RAB25, RHOBTB3, RHOD, RND3, RRAS, SMTN, SPRR1A, SPRR1B, SPTBN2, SYK, TIMP1, TIMP2, TNNT1, TPM1, TRAK2, VAMP8, VILL, VIM, WASF1, WASF2). All of these genes were up-regulated or down-regulated > = 1.5- fold following SYK knockdown (Table S1). To this list of SYK-regulated genes, we added SRC (c-Src), CTTN (cortactin), CDH1 (E-cadherin), and TP53 (p53) since the protein products of the first three interact with Syk in a cellular context of motility and invasion; TP53 is part of the interacting network of proteins. The total set of genes is henceforth referred to as the 55 Gene Set. 696 Infiltrating Ductal Carcinoma (IDC) cases were identified using cBioPortal to access de-identified patient information, and cancer subtypes of Her2, basal-like, luminal A, and luminal B were identified as previously determined using PAM50 (known for 675 of the 696 IDC tumor cases) [28]. We obtained 1) Scatter plots for SYK copy number, methylation and protein expression and two-gene comparisons; 2) overall survival curves, and “oncoprints” illustrating copy number and mutation data for each patient and 55 genes; 3) data on the % cases altered for each gene; 4) SYK copy number, methylation, mRNA, and protein, and 5) a gene interaction network map, including 50 nearest interacting genes with the most alterations, for the 55 gene set using cBioPortal.
Results
SYK DNA FISH
BAC probes to detect SYK DNA were tested using a dual color FISH approach on metaphase spreads of normal human lymphocytes with chromosome 9 centromere control labeled in a different fluorophore (Figure 1A) [22]. Next, the protocol was optimized to detect SYK DNA in paraffin embedded sections of breast tissue (Figure 1B). Nineteen cases of breast cancer were selected from a set originally obtained and characterized from the archives at Georgetown University Lombardi Comprehensive Cancer Center [4]. The cases contained DCIS either with or without accompanying evidence of IDC and had the same overall characteristics as the entire set with respect to mRNA Syk expression (Figure 2 in ref. [4]; Figure S1, and Table S2 in this study). De-identified pathology reports and re-examination of sections by an independent pathologist confirmed that 8 cases were DCIS only and 11 cases contained invasive ductal carcinoma (IDC) adjacent to the DCIS (Table S1). For reference, an additional 5 cases were selected that contained benign tissue, but no evidence of DCIS or IDC. Dual FISH analysis for SYK was performed on these 24 cases. For the subset of cases used for FISH (Table S2), on average, SYK mRNA was reduced comparing benign to DCIS and DCIS to IDC (Figure S1A, original study [4]; 1B, Table 1 subset).
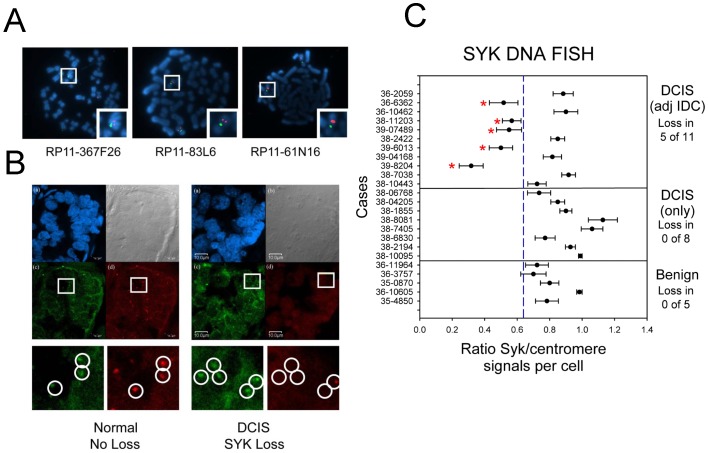
Figure 1. DNA FISH Analysis.
A. Normal control metaphase spreads were used to validate the SYK probe (chromosome 9). The centromeric probe for chromosome 9 is green and the probe for SYK is red. Each BAC was labeled with Spectrum Orange™ (a) RP11-367F26, (b) RP11-83L6, (c) RP11-61N16. B. Confocal microscopy imaging of FISH slides. The panel at left is an example of normal mammary tissue and the panel at right is of DCIS tissue with SYK gene loss. In both, (a) DAPI-stained nuclei, (b) differential interference contrast imaging of section, (c) SYK FISH signal (green), (d) chromosome 9 centromere signal (red). C. The mean value for the ratio of SYK/chromosome 9 centromere signals for each case is illustrated in a bar plot for benign only, DCIS only, and DCIS with adjacent IDC samples. The mean was determined for each case from the ratios obtained individually from 30 cells. Error bars indicate S.E. of the mean. The mean SYK/control ratio of the combined cases was 0.92+/−0.048 S.E. for DCIS only tissues and 0.70+/−0.057 S.E. for DCIS adjacent to IDC cases. The mean for benign only tissues was 0.80+/−0.050 S.E.. The ANOVA P value for the three tissues was 0.035.
All of the benign epithelium (5 cases of benign with no DCIS or IDC detected) and 8 DCIS tissues from DCIS only cases displayed normal Syk/centromere probe ratios indicating no allelic loss, with a range of 0.7 to 1.1 Syk/centromere probe ratios (Figs. 1B–C). In contrast, 5 out of 11 cases of DCIS in which adjacent IDC was detected displayed a clear loss of SYK with a range of 0.3 to 0.6 Syk/centromere probe ratios (Figs. 1B and 1C, asterisks). Thus, for cases in which DCIS was reported to be 100% from the pathology report and subsequent pathologist re-examination, no allelic loss of SYK was detected in DCIS tissue. In contrast, in cases in which DCIS was listed at <100% in the pathology report or in which IDC was identified adjacent to DCIS, allelic loss was detected in 5 of 11 cases (46%) (Table S2, Figure 1C, red asterisks). The mean SYK/centromere probe ratio of the combined cases was 0.92+/−0.048 S.E. for DCIS only tissues and 0.70+/−0.057 S.E. for DCIS adjacent to IDC cases. The mean for benign only tissues was 0.80+/−0.050 S.E.. Benign was significantly different from DCIS only (P = 0.047) and the DCIS only was significantly different from the DCIS adjacent to IDC (P = 0.0079). These data are consistent with the hypothesis that allelic loss of SYK is associated with progression to invasive disease since it occurred more frequently in DCIS with adjacent IDC.
SYK promoter methylation DCIS tissues
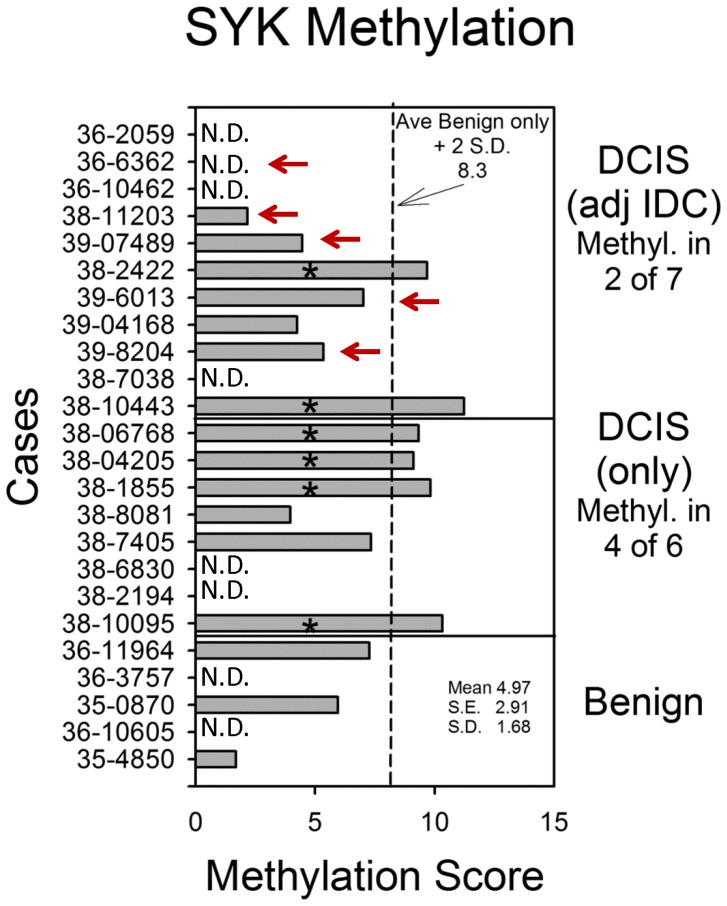
Previously, it was reported that CpG islands in the SYK promoter were methylated resulting in gene silencing and that methylation of SYK was less than 5% in normal or benign tissues, and 47% in node-negative, and 40% in node positive IDC cases [6], [7]. Methylation in DCIS was found to be 45% [7]. We examined the promoter methylation status of SYK in DCIS for the present study using bisulphite pyrosequencing using a subset of cases that were available following the FISH study. In 5 benign cases, only three sections were available or yielded to pyrosequencing, only 6 of 8 DCIS only tissues produced results, and only 7 of 11 DCIS tissues from cases with adjacent IDC produced results (Table S2). If a cutoff of 8.3% methylation was set ( = benign only tissue mean +2 S.D., Figure 2, dashed line), 0 of 3 benign cases, 4 of 6 DCIS (only) cases, and 2 of 7 DCIS (with adjacent IDC) cases were considered positive for CpG island methylation (Table S2). Overall, DCIS tissue was positive in 46% of 13 total cases. When means of methylation were analyzed by tissue, benign, DCIS, or IDC, no overall significant differences were observed between them (Kruskal-Wallis, p = 0.48, data not shown). Taken together with the results revealing SYK allelic loss together with promoter methylation, Syk protein levels would likely be impacted.
Figure 2. SYK promoter methylation.
The mean value for promoter methylation at 4 CpG islands was plotted for each case. DCIS cases are arranged by whether adjacent IDC was absent or present, and benign only cases are shown for comparison (mean 4.97+/−1.68 S.D., 2.91 S.E.). The dashed line indicates the mean of the benign cases plus 2 S.D. (8.3). Red arrows indicate cases where FISH identified allelic loss.
Protein Expression of SYK in DCIS tissues
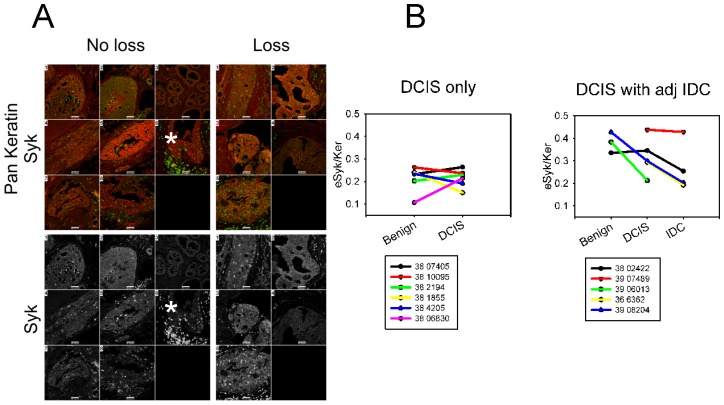
In order to examine whether protein loss occurred in parallel with loss of mRNA as previously reported [4], and whether loss was associated with methylation and/or allelic SYK gene loss, adjacent tissue sections were double labeled with anti-Syk and anti-pan keratin antibodies and then counterstained with DAPI for immunofluorescence confocal microscopy. Representative quantitative images of DCIS tissues are shown in Figure 3A. Images were analyzed using custom image analysis algorithms (see Methods). Determination of the means of average intensities for each case and then pooling all of the means for each tissue, benign, DCIS, and IDC, failed to provide significant data (Figure S2A). Syk-positive immune cells were observed relatively rarely in benign tissue but were particularly abundant infiltrating some DCIS lesions and surrounding IDC (Figure 3A, asterisks). Staining intensity of Syk in immune cells was 6-fold higher than epithelial cells in DCIS, and IDC (Figure S2B).
Figure 3. Immunofluorescence staining of DCIS tissues.
A. Immunofluorescence confocal images of ductal carcinoma in situ (DCIS) tissues from 8 cases with no allelic loss determined by DNA FISH (unless otherwise indicated, DCIS with adjacent IDC): 1) 38 002422 (methylated), 2) 38 06830 (DCIS only, methylation status unknown), 3) 38 10095 (DCIS only, methylated), 4) 38 2194 3 (DCIS only, methylation status unknown), 5) 38 4205 (DCIS only, methylated), 6) 38 7405 A5 (DCIS only, not methylated), 7) 38 8081 8 (DCIS only, not methylated), 8) 39 04168 (not methylated) and 5 cases of DCIS with allelic loss determined by DNA FISH (all DCIS with adjacent IDC: 1) 38 6362, methylation status unknown, 2) 38 11203 (not methylated) 5, 3) 39 06013 (not methylated), 4) 39 07489 (not methylated), 5) 39 08204 (not methylated). Note that anti-Syk staining highlights the presence of a subpopulation of lymphocytes and/or other infiltrating inflammatory cells that is always more intense than tumor epithelium staining (see asterisks). Myoepithelial cells are negative. There was no obvious difference between the intensity of either anti-Syk or anti-pan keratin staining between sections of tissue with or without allelic loss. Scale bars = 50 µm. B. The ratios of eSyk/Ker from individual cases were segregated according to DCIS only versus DCIS with adjacent IDC. DCIS only cases are shown on the left (DCIS) and DCIS with adjacent IDC are shown on the right (DCIS with adj IDC). Data was plotted using eSyk/Ker ratios. All available tissues from each case section were used but were not available for every case. Only cases with two or more tissues present were plotted.
The ratios of eSyk/Ker for each case were plotted separately to see if individual cases demonstrated Syk loss from benign to DCIS to IDC; the cases were also plotted according to whether they were DCIS only or DCIS with adjacent IDC. If normalized against keratin, epithelial Syk loss is evident in cases of DCIS with adjacent IDC (Figure 3B).
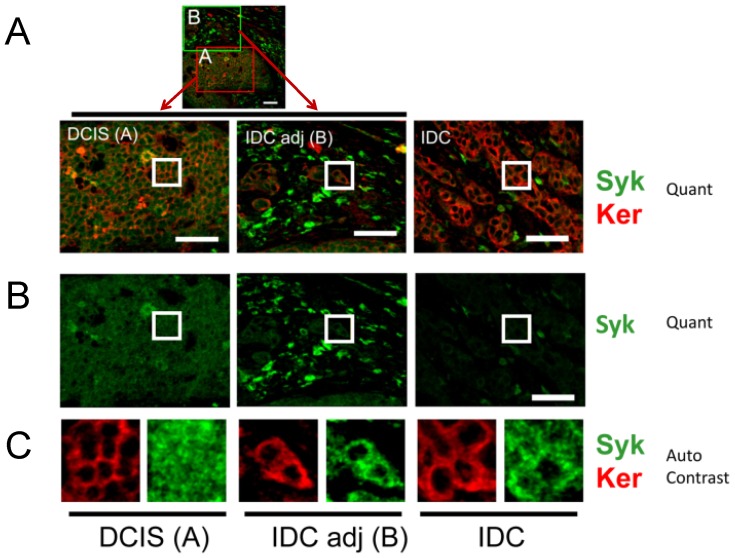
Since overall staining intensities varied between cases and over different areas of the slides, we examined single cases (slides) in more detail and focused on a qualitative observation, namely, that Syk occurred both in the cytoplasm and nucleus. Case number 36 6362 contains DCIS with adjacent IDC, lacks methylation of the promoter (value of 7.7 where the cutoff chosen was 8.3), and displays allelic loss (Table S2). Immunostaining of Syk and keratin in a single location and in adjacent locations is illustrated in Figure 4 where epithelial loss of Syk protein is visually apparent, and intense immune cell staining is present (Figure 4A, Syk, Ker, Quant; Figure 4B, Syk, Quant). Insets from the indicated locations in each image were subjected to autocontrast enhancement and are shown in Figure 4C (Syk, Ker, Auto Contrast), revealing the exclusively cytoplasmic staining of keratin (red) versus the cytoplasmic and nuclear staining of Syk (green). Whereas Syk was present in nucleus and cytoplasm in DCIS tissue, it was generally absent in multiple nuclei in IDC tissue (Figure 4C). Analysis of available images for case 36 6362, revealed a significant loss of Syk comparing DCIS and IDC tissues (Wilcoxon rank sum test p-value 0.008). Taken together, the protein staining studies indicate that Syk is lost comparing DCIS and IDC tissues, including nuclear loss.
Figure 4. Representative confocal immunofluorescence staining of Syk and keratin in a single case, 36 6362.
Two-color images of Syk and keratin staining (Syk,/Ker Quant) are shown for DCIS and IDC tissues within the same confocal image (low magnification view of DCIS with adjacent IDC: “DCIS (A)” and “IDC adj (B)”). IDC from the same slide but a different field of view is shown (“IDC”). The single color, Syk only staining is shown below (Syk, Quant). The background subtracted images were acquired and presented quantitatively for direct comparison (Quant). To qualitatively compare the localization of Syk and keratin, insets are shown on the bottom panels where brightness and contrast were optimized using the auto-contrast function in Metamorph Offline software (Auto Contrast). Keratin is always cytoplasmic, whereas Syk can also be nuclear. The asterisk highlights an area of immune cell infiltration. Scale bars = 50 µm.
Effect of copy number loss on mRNA, methylation, and protein status of SYK invasive breast cancers
To extend the above study on allelic loss of SYK in DCIS to invasive breast cancer cases, we took advantage of the large data set publically available on The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/) to investigate SYK copy number changes. We used cBioPortal tools to access copy number and mutations for SYK in this large breast cancer study [26], [28] and to develop a method to discriminate between immune infiltrated invasive breast cancers and cases depleted of immune infiltration so that mRNA and protein values would more accurately reflect epithelial cells (see Methods).
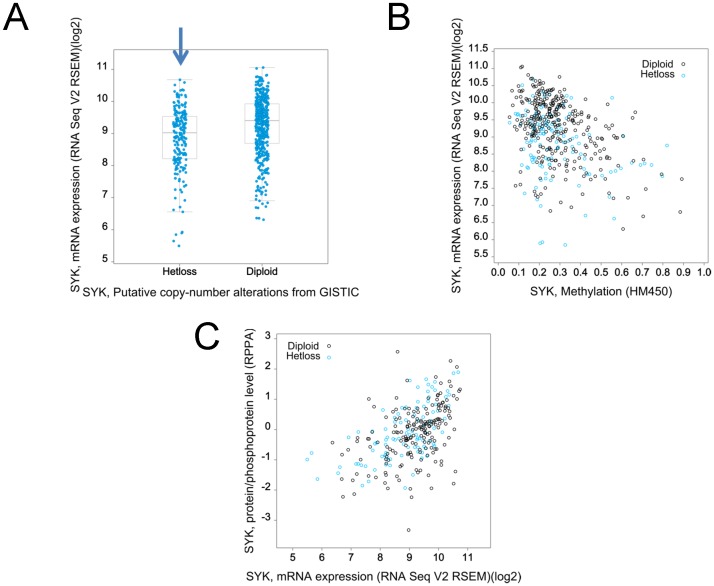
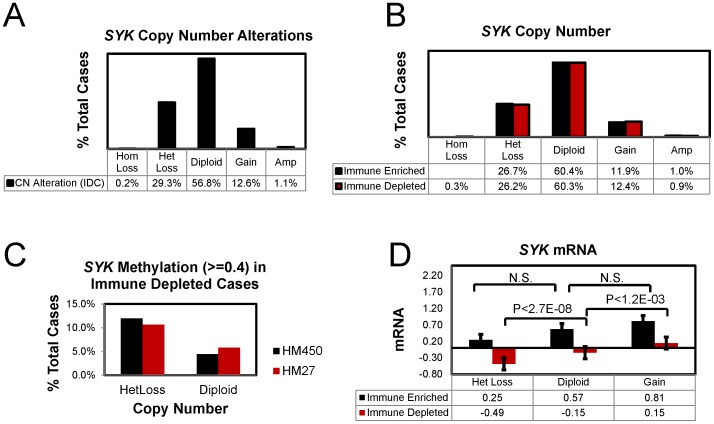
Beginning with a total 1002 invasive breast cancer cases from the TCGA, cases where immune cell infiltration was prominent were identified and removed (see Methods) for a final subset of 800 immune depleted cases. Using the immune depleted subset or using a subset of 696 IDC only cases, we found that only two mutations in SYK were present, one of which would likely have an effect on function, namely A146G [29], [30] (data not shown). The somatic mutation rate for SYK was 0.3% and the overall alteration including copy number (homozygotic deletions or amplifications) was 1.6% of IDC cases (696 cases). Putative copy number loss described as heterozygotic loss (HetLoss) occurs in 26.2% of the immune depleted cases (Figure 5A, 6B) and in 29.3% of IDC cases (Figure 6A). As described previously [6], [7], the SYK promoter is methylated in invasive breast cancers; and, methylation values were correlated with reduction in SYK mRNA levels in immune depleted cases (Figure 5B); similarly protein levels were positively associated with SYK mRNA levels (Figure 5C). In immune depleted cases, higher methylation values (> = 0.4) were found in about 11% of HetLoss cases but only in about 5% of diploid cases using either methylation array data (Figure 6C, HM450 or HM27).
Figure 5. Characterization of the relationship between SYK copy number, mRNA, and protein in immune depleted cases from TCGA Provisional Study.
801 of 1002 breast cancer cases were characterized as immune depleted (see methods) and were used for the following graphs. A. Putative SYK copy number (x-axis) for Diploid and Hetloss is plotted against SYK mRNA in a box plot. The blue arrow indicates the Hetloss group. B. SYK methylation (x-axis) is plotted against SYK mRNA (y-axis) for individual cases of HetLoss or Diploid. Blue circles indicate putative heterozygotic loss and black circles indicate diploid copy number. Dashed line indicates cutoff methylation level. C. SYK mRNA is plotted against Syk protein/phosphoprotein levels for Diploid and Hetloss cases.
Figure 6. Effect of copy number loss of SYK methylation and mRNA expression.
A. Data for IDC cases were extracted using cBioPortal and the percent of cases for each putative copy number category determined (homozygotic deletion (HomLoss, −2), HetLoss (−1), Diploid, Gain (+1), and amplification (Amp, +2). B–D. 1002 invasive breast cancer cases were classified as immune cell depleted (800 cases) or immune cell enriched (202 cases) as described in Methods. B. The percent of cases for each putative copy number category was determined. C. The percent of cases with methylation > = 0.4 using either the HM450 or the HM27 array data were plotted for immune depleted HetLoss and Diploid cases. D. SYK mRNA values for each case were extracted and means + S.E. plotted for each category. Only the differences between HetLoss, Diploid, and Gain for immune depleted cases were significant (ANOVA P value 2E-09). P-values for t-tests assuming unequal variances are shown.
In immune depleted, HetLoss cases, a significantly lower level of SYK mRNA was observed compared with the Diploid cases; similarly a lower level of SYK mRNA was found in Diploid cases compared to Gain cases (Figure 6D). Confirming our identification of a subset of invasive breast cancers where immune infiltration was indicated by alterations in immune specific mRNAs, we found that the mean SYK mRNA level was higher in the immune enriched subset of cases compared with immune depleted cases (Figure 6D).
Poor overall breast cancer survival associated with alterations in genes within the 55 Gene Syk network
Influence of SYK on overall patient survival in IDC
To explore the relationship of SYK copy number and mutational status to overall survival in IDC, we asked whether changes in SYK status might be associated with a change in overall survival of patients. We queried 696 cases (IDC) from the provisional breast cancer study using mutations and copy number and found that overall survival times estimated in a Kaplan-Meier plot with SYK were not significantly different between affected and unaffected cases where affected cases were less than 1% of the total (plot not shown, Logrank test P-value 0.619806).
Syk interacting network of 55 genes
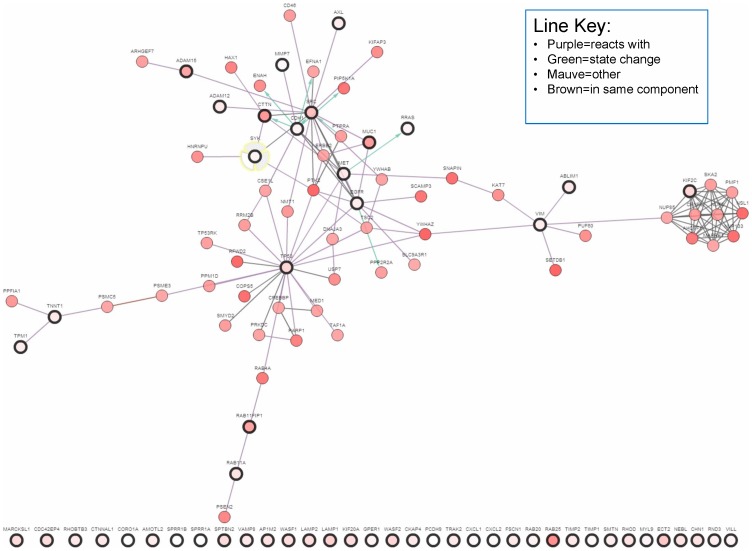
We next asked whether SYK regulation of gene expression, specifically with regard to its role in suppressing motility, invasion, and metastasis in in vitro and in vivo models [2], [3], [5], [31]–[33], might be related to the formation of metastases and ultimately patient survival. In a previous study, we had analyzed significant expression changes (up or down regulated > = 1.5 fold) following SYK siRNA knockdown in a benign human breast cancer cell line, MCF10A, cultured on a matrix of collagen I [3]. From that set of 708 genes, we next performed a literature search and culled a list of 51 genes involved in key activities required for invasion, including cell motility and activities regulating invasion including membrane trafficking (39 up-regulated and 12 down-regulated genes; Table S1). The addition of SRC (c-Src), CTTN (cortactin), TP53 (p53) and CHD1 (E-cadherin) to the list of 51 genes to form the 55 gene set was based on published data relating these gene products with Syk signaling including direct interactions [4], [31], [34], [35] (Table S1). c-Src tyrosine kinase activity is suppressed by Syk in breast cancer cells [4] and both c-Src and Syk phosphorylate cortactin (CTTN) [31], [36]. Syk and cortactin are involved in the development and maintenance of cell-cell contacts and interact with cadherin 1 (CDH1) [31], [37], [38]. The relationship between SYK and the other 54 genes is shown in an interaction network generated by cBioPortal (Figure 7). 36% of the 55 Gene Set members are linked in a network according to cBioPortal (Figure 7, % altered cases signified by pink intensity, dark-rimmed encircled genes are members of the 55 Gene Set).
Figure 7. Network interactions amongst the members of the 55 Gene Set.
Heavy black circles indicate members of the 55 Gene Set; additional nodes were added based on most related with highest copy number alterations (cBioPortal tools).
Functional description of the predictive 55 Gene Set
The 55 genes were annotated using DAVID by investigating Gene Ontology terms (7/19/2013; [39]) to generate a functional annotation table including Biological Function (GOTERM_BP_FAT), Cellular Component (GOTERM_CC_FAT)), and Molecular Function (GOTERM_MF_FAT (Table S3). Prominently, GOTERM terms point to participation in the cytoskeleton, cell adhesion, and association with protein trafficking via intracellular vesicles (Table S3, highlighted terms). Not surprisingly, the actin cytoskeleton and genes that contribute to its regulation are well represented: ABLIM1, CDH1, CORO1A, CTNNAL1, CTTN, CXCL1, EGFR, FSCN1, MET, MYL9, NEB, RND3, RRAS, SMTN, SPTBN2, TNNT1, TPM1, VILL, VIM, WASF1, and WASF2. Many of the GOTERMS are also related to “vesicle-regulating” and “vesicle-associated” proteins (Table S3), for example, EGFR, LAMP1, CORO1A, LAMP2, AP1M2, RAB11A, RAB11FIP1, TIMP1 encode proteins that are vesicle associated proteins and AP1M2, TRAK2, TP53, RAB11A, RAB25, RAB11FIP1, RAB20, KIF20A genes encode products that participate in protein transport within the cell. Others genes code for cell adhesion genes such as EGFR, CTNNAL1, RND3, CORO1A, PCDH9, CDH1, ADAM12, SRC, ADAM15, and SYK. Genes products of EGFR, PCDH9, CDH1, SRC, SYK regulate cell-cell adhesion; while those of EGFR, MET, SRC, MYL9, among other candidate genes, participate in focal adhesion functions.
Characteristics of copy number changes in members of the 55 Gene Set
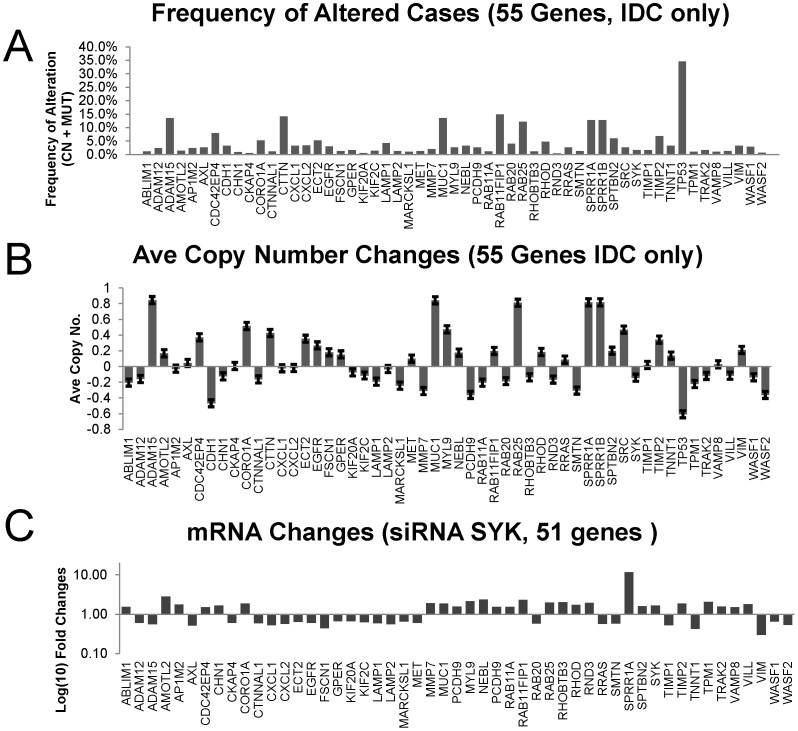
To characterize the frequency of the copy number changes in IDC cases and the average copy number of each of the genes from the 55 Gene Set, means were determined and the results were plotted (Figure 8A–B). Of the 9 genes for which the percent of cases altered for the 55 Gene Set was greater than 4.5% (CN, copy number and MUT, mutations) (Figure 8A), all had increased average copy number (Figure 8B); of those, 2 were from genes whose mRNA was down-regulated by SYK siRNA (ADAM15 and ECT2) and 7 were from genes whose mRNA was up-regulated by SYK siRNA (SPRR1A, RAB11FIP1, MUC1, RHOD, and SPTBN2). The 8 genes whose copy number frequency alterations (% altered cases) were the greatest were TP53, RAB11FlP1, CTTN, ADAM15, MUC1, SPRR1A, SPRR1B, and RAB25 (Figure 8A). Of these genes, all but one had greater than normal average copy number, TP53 being the exception (Figure 8B). The SYK mRNA changes in motility and invasion genes resulting from siRNA treatment of MCF10A cells were plotted for comparison (Figure 8C).
Figure 8. Comparisons of SYK-regulated mRNA changes, average copy numbers, and percent of cases altered.
A. The % altered IDC cases were plotted for each member of the 55 Gene Set. B. The average copy number was plotted for each member of the 55 Gene Set . C. Fold changes in mRNAs (log) of genes regulating motility and invasion following SYK siRNA knockdown in ER-negative, MCF10A breast cells [3] were plotted for 51 mRNA species.
An “oncoprint” illustrates the genomic copy number and mutational changes for each patient and for each of the 55 genes (Figure S3A). This graphic illustration of genes versus patients with amplifications in red, mutations in green, and deletions in blue reveals the pattern of one or more alterations per affected patient (Figure S3A). A curious feature instantly observed upon inspection of the Oncoprint generated by the cBioPortal analysis is that 5 of the amplified genes amongst the 8 with highest frequency of altered cases were co-expressed in the same patient cases (Figure S3A, blue boxed region, and Figure S3B, two gene comparisons). It transpires that these 5 genes are all found on chromosome 1 (SPRR1A: 1q21-q22; SPRR1B: 1q21-q22; ADAM15: 1q21.3; MUC1: 1q21; RAB25: 1q22). Three of these genes, ADAM15, MUC1, and RAB25 are part of an interactive network of genes (Figure S3C).
When cases were stratified according to SYK copy number (−1 and −2, 0, +1 and +2) and then were used to compare differences in the average copy number for each member of the 55 Gene Set, both SYK and CTNNAL1 (alpha-catulin) copy number had parallel changes suggesting that these two genes are linked (Figure S4A). A two gene comparison using cBioPortal for copy number of SYK and CTNNAL1 resulted in a positive correlation in IDC patients for these two genes (Figure S4B). Both are found on chromosome 9 (SYK: 9q22; CTNNAL1 (catenin (cadherin-associated protein), alpha-like 1, alpha-catulin: 9q31.2).
We conclude that co-amplification and co-loss of genes within adjoining stretches of DNA contribute to genetic disruption of the 55 genes.
Identification of breast cancer cases without immune cell infiltration
Syk protein localizes in the luminal ductal epithelium of the breast ([3], [4], therefore we narrowed the cases to include only IDC (infiltrating ductal carcinoma), a subset of 696 cases of the total. Using a set of 9 genes as immune cell markers (IL2RA, CMTM2, FCGR2A, FCER1A, FCER1G, WAS, CD3E, CD22, and CD19) as described in the Methods, we queried the IDC cases for alterations based on mRNA and estimate that tissue from 131 cases were contaminated with immune cell contributions (and the calculated number of altered cases was 90.2% of the total 131). The remaining IDC 565 cases were “immune cell depleted” since the 9 genes were altered in only a calculated 4.3% of those cases. Therefore, about 19% of the IDC cases contained significant immune cell infiltration. When this analysis was also performed on the total collection of invasive breast cancers, we found a similar frequency; 20.2% cases including all breast cancer subtypes were immune enriched leaving 800 cases classified as immune depleted.
Predictive power of the 55 Gene Set
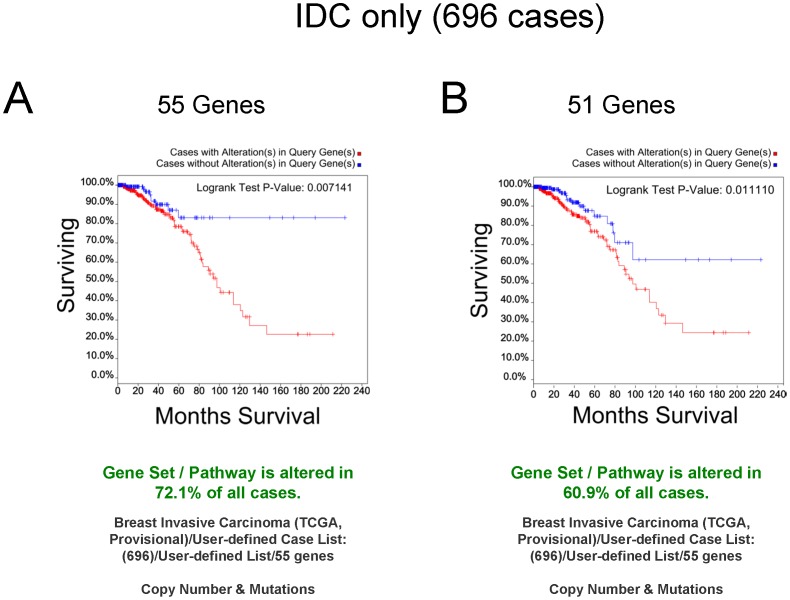
We first analyzed IDC only cases to determine overall survival using copy number and mutation alterations based on the 55 Gene Set. In this analysis, 72.1% of the cases were altered and, impressively, using a Kaplan-Meier analysis, the Logrank test p-value was significant (0.007141) with the curves differentiating between altered cases whose % surviving cases plateaued at ∼20% and unaltered cases at ∼80% (Figure 9A). To ask whether the presence of immune cells might be masking the results in terms of the overall patient survival using the 55 Gene Set, the subset of 565 cases of IDC, depleted of cases with immune contamination, were queried again in the same way. The results were similar curves on the Kaplan-Meier plot with increased significance compared with a query of the entire IDC case set (Immune depleted IDC: Kaplan-Meier plot not shown, Logrank test p-value of 0.003113) and the curve depicting no alterations plateauing at 85% overall survival. Thus, focusing on the copy number and mutational alterations in the 55 Gene Set allows overall survival prediction for the entire set of cases whether or not immune cell infiltration is a prominent component of the extracted tissues. This result indicates a significant decline in survival in affected cases when copy number and mutations (but not mRNA or protein) were considered for these genes. In order to determine the contribution of the four genes whose expression levels were not significantly different according to our previous microarray study, we performed the analysis on just the 51 genes whose mRNAs were significantly regulated by SYK knockdown (Figure 9B). The Logrank test p-value was also significant (0.011110) but the survival plateau for unaffected cases was reduced to 60%. We conclude that the 55 Gene Set has higher predictive value than the set of 51 genes alone.
Figure 9. Kaplan-Meier curves of queries of IDC only cases.
A. Survival curves for 696 cases (IDC cases only) from the TCGA Provisional Breast Cancer Study were generated using cBioPortal querying with the 55 Gene Set. 72.1% of all cases were altered (copy number and mutations). The Logrank test p-value was 0.007141. B. The cBioPortal query was repeated using only the 51 genes from the SYK microarray study [3]. 60.9% of all cases were altered (copy number and mutations). The Logrank test p-value was 0.011110.
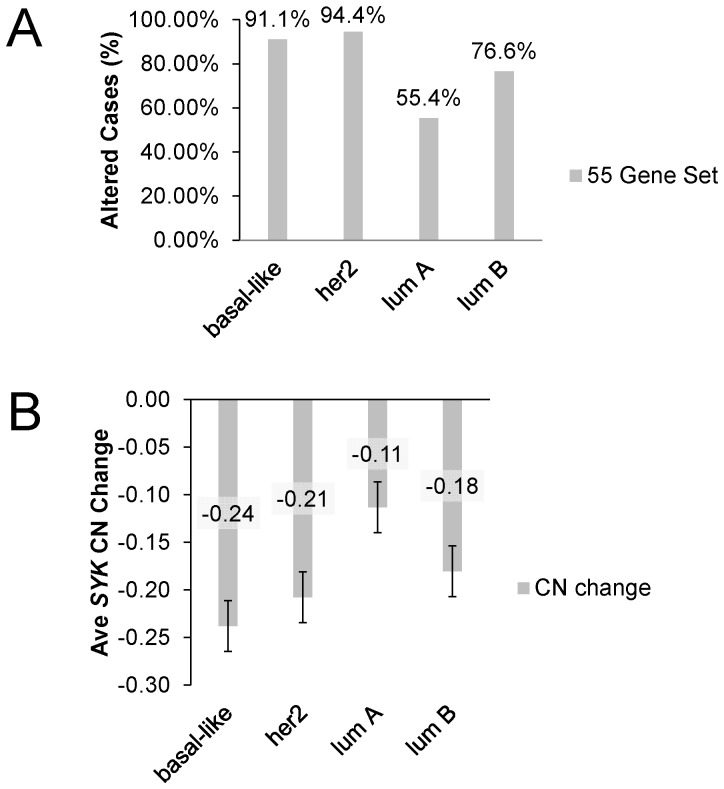
In a previous global characterization of the majority of these breast cancer cases from the TCGA, breast cancer subtypes were characterized using PAM50 ([28], and references therein). We performed queries using the 55 Gene Set to compare basal-like, Her2, Luminal A, and Luminal B tumor types. The Gene Set was altered in 91.1% and 94.4% of basal-like and Her2 cases, respectively, whereas 55.4% and 76.6 percent of luminal A and luminal B cases, respectively, were altered (Figure 10A). Thus, these motility- and invasion-related genes are already altered in nearly all cases of these tumor types with known poorer outcome. A similar trend was observed for the average SYK copy number change in these cancer subtypes (Figure 10B). Average basal-like and Her2 tumor copy number changes were −0.24 and −0.21, respectively, but were −0.11 and −0.18 for luminal A and luminal B tumors, respectively (Figure 10B). In summary, change in the average SYK copy number for each subtype reflects the change in percent of cases with genomic alterations in members of the 55 gene set. We conclude that SYK copy number changes reflect the overall genomic instability in the tumors.
Figure 10. Stratification of cases by breast cancer subtype and by SYK copy number and frequency of alteration.
A. The % of altered IDC cases was determined using the 55 Gene Set for cases identified as basal-like, her2, luminal A or luminal B using the PAM50 gene set [28]. B. The average SYK copy number change (CN) was determined for cases identified as in A.
We next performed queries to determine overall survival in Luminal A + Luminal B subtypes (295) using 55 Gene Set. The Kaplan-Meier Logrank test p value was 0.001198, and % surviving cases plateaued at ∼10% and unaltered cases at ∼90% (Figure 11A). There was no significant predictive power of the 55 Gene Set for the Her2 or basal-like subtypes (not shown). To parse out the effects of the additional 4 genes (CDH1, CTTN, SRC, and TP53), Kaplan-Meier curves were generated revealing significant Logrank p-values of 0.004582 and 0.006403 for the 51 Gene Set and 4 Gene Set, respectively, with associated reductions in the survival plateaus (Figure 11B–C). Thus, the 55 Gene Set significantly predicts overall survival in luminal breast cancer subtypes but not in basal-like or Her2 cases where the majority of cases already have alterations in these genes.
Figure 11. Kaplan-Meier curves of queries of Luminal A + Luminal B cases.
A. A query of 295 cases of luminal A and luminal B (as determined in [28] using the PAM50 gene set) from the Provisional Breast Cancer Study was performed using the 55 Gene Set. 64.1% of all cases were altered (copy number and mutations). The Logrank test p-value was 0.001196. B. The cBioPortal query was repeated using the 51 genes from the SYK microarray study [3]. 53.2% of all cases were altered (copy number and mutations). The Logrank test p-value was 0.004582. C. The cBioPortal query was repeated using 4 genes (CDH1, CTTN, SRC, and TP53). 35.6% of all cases were altered (copy number and mutations). The Logrank test p-value was 0.006403.
Discussion
Significance of results: allelic loss of SYK
For the first time, allelic loss of SYK was detected in DCIS breast tissues using a subset of the tissues in which progressive loss of SYK mRNA from benign to DCIS to IDC tissues was reported [4]. Remarkably, in this small study, 46% of DCIS cases displaying adjacent IDC, but none of the DCIS only cases, were found to have allelic gene loss of SYK. Thus, in this preliminary study of DCIS, there appears to be a strong association of SYK allelic loss in DCIS tissues with adjacent invasive breast cancer. Using a large, publically available data set, we confirm SYK loss at the genomic level in IDC and found that about 30% of IDC cases displayed heterozygotic SYK loss. The frequency of SYK loss in DCIS and IDC tissues suggests that it could play a role in enhancing metastatic progression in a significant number of cases.
SYK mRNA loss in cases of reduced copy number: a method to examine cases depleted of immune cell infiltration
To determine the consequence of SYK copy number loss on Syk expression and thus the potential of Syk to regulate its downstream targets, it was critical to isolate Syk regulated events occurring in epithelial cells from those operative in immune cells. A key source of artifact in studies of Syk arises when tissue samples are taken and used to analyze genomic DNA, mRNA, and protein because of immune cell infiltration. We found that protein levels of Syk were 6-fold higher in immune cells compared with epithelial cells. In this study of TCGA data, we estimated that about 19% of the IDC cases had significant immune cell infiltration that contributed to increased immune cell mRNA and proteins isolated from tissue extracts. Our success at isolating immune enriched versus immune depleted cases was confirmed by observation of elevated levels of SYK mRNA in the immune enriched compared with the immune depleted cases. Analyzing immune depleted cases, we were therefore able to ascertain that a larger proportion of HetLoss SYK cases had SYK promoter methylation compared to diploid SYK cases. Furthermore, and importantly, the mean SYK mRNA level was lower in HetLoss compared with Diploid cases. We therefore conclude that SYK copy number loss and promoter methylation leads to reduction in Syk expression. This evidence from human pathological tissues agrees with the results generated from in vivo mouse studies and in vitro human tissue culture studies that SYK allelic loss results in reduced SYK expression [3]. We propose that SYK loss in human breast tumors in situ may also result in loss of Syk suppression of motility and invasion to support metastatic progression in breast cancer patients via its action upon proteins regulating motility and invasion.
Impact of SYK-regulated motility and invasion gene network on breast cancer survival
Independent of the effect of Syk function on the network of Syk-interacting proteins, increased genomic instability contributing to changes in copy number and mutations of these 55 genes would also have an outcome contributing to metastatic progression. This hypothesis is supported by the observation of a significantly lower overall survival in IDC cases, and specifically in Luminal A plus Luminal B subtypes in cases where members of the 55 Gene Set are altered. Considering the difficulty of performing mRNA and protein studies on breast cancer tissue homogenates, it was fortunate that these genes could be used to assess overall survival based solely on copy number and mutations. Genomic changes would be less sensitive to the presence of immune cell DNA in the tissue lysates compared with the more abundant mRNA or protein species in the cytoplasm. We conclude that Syk-regulated genes represent an epithelial biological network for restraint of motility and invasion that contribute to suppressing metastasis; in turn, loss of this restraint due to genomic instability or loss of Syk expression decreases overall survival of breast cancer patients. Evaluation of the 55 Gene Set for prognostic information or predictive power for treatment outcome should be considered in the future.
Our identification of a 55 Gene Set that potentially represents a new prognostic tool is based on SYK and the identification of the important biological function of Syk to regulate epithelial cellular invasion. This is in contrast to prognostic sets such as the PAM50 and the MammoPrint which were selected based on different strategies. Interestingly, there was no overlap in genes of the 55 Gene Set with 164 of the identified genes in Table S2 reported by Van't Veer et al, and later referred to as MammoPrint [40]. There was some overlap with the PAM50 gene set, namely AP1M2, CDH1, CORO1A, ECT2, LAMP2, MMP7, MUC1, NEBL, RAB11A, SYK, TIMP2, TNNT1, and VIM underscoring the importance of these epithelial invasion/motility genes in patient outcome.
Impact of SYK-regulated motility and invasion gene network on breast cancer survival in cancer subtypes and relationship to estrogen receptor status
The 55 Gene Set appears to be most potent in predicting survival in luminal IDC subtypes (luminal A and luminal B case assignments determined by PAM50 [28]); alterations in the members of the 55 Gene Set resulted in reduced overall survival. However, the 55 Gene Set analyses found that the majority of basal-like and Her2 cases were already altered so that a comparison of altered versus non-altered cases was not useful. A main feature of luminal subtype breast cancers is that they are, for the most part, estrogen receptor (ER) positive. In this study, 478/675 (71%) of IDC cases and 284/295 (96%) of luminal A and luminal B cases were ER+. If IDC cases were stratified by ER+ versus ER− and queried using the 55 Gene Set, only the ER+ cases were significantly different as estimated by Kaplan-Meier curves (Logrank test p-value 0.003212 for ER+, n = 478 cases and 0.700637 for ER−, n = 197 cases). In a study of a series of breast cancer cell lines that were ER+ versus ER−, Syk expression was mostly positive in ER+ and negative in ER− cell lines [2]. Cell lines lacking Syk were invasive in a 3D test of invasion, whereas those with Syk expression were non-invasive [2]. Thus, basal-like cell lines such as MDA-MB-231 are ER− and highly invasive and already are Syk depleted [2]. However, the correlation between ER+ and SYK+ cell lines is not perfect. MCF10A cells used for the SYK knockdown experiments [3] are Syk+ but ER− and so represent one of the exceptions. Even so, the progression series of MCF10A cell lines shows a loss of Syk in the DCIS.COM line compared with MCF10A; and Syk knockdown in either cell line results in increased proliferation and invasion characteristics [3]. Thus, although the functional relationship between Syk and ER is not clear, Syk regulation of motility and invasion activity is independent of ER at least in the case of MCF10A. Interestingly, ESR1 protein and phosphoS118 ESR1 (ERα or estrogen receptor 1) are decreased in cases where alterations in the 55 Gene Set are present (Table S4). That these tumors are more aggressive is also suggested by recent data using anti-phospho S118 ESR1 to identify cases where the ERα pathway is active; mutation of this site leads to more aggressive cell behavior in MCF7 breast cancer cells ([41] and references therein).
Biological functions of the 55 Gene Set Members
Members of the 55 Gene Set had been chosen for their roles in events driving motility and invasion, and thus contributing to metastasis. Many members of these Syk-regulated genes from the 55 Gene Set have already been reported in the context of Cancer and Breast Cancer studies (Figure S5). Src, Syk, EGFR, and MET regulate invasion-promoting activities including invadopodia-mediated matrix degradation via MT1-MMP (MMP14) and cell-cell and cell-extracellular matrix adhesion (including integrin heterodimers and cadherins) to determine the invasive phenotype [42]–[52]. Five of these SYK-regulated genes encode kinases that are proto-oncogenes (EGFR, MET, AXL, ECT2, and SRC) two of which (EGFR and SRC) have previously been linked to Syk in epithelial cells [4], [53]. Specifically, we found earlier that the cell surface expression of invadopodia genes of an integrin heterodimer, alpha6beta1 (CD49f, ITGA6/ITGB1), adhesion receptor CD44, and matrix-degrading protease, MT1-MMP (MMP14), are directly regulated by SYK; SYK knockdown results in their increased expression at the cell surface coupled with the formation of invadopodia and consequent cell invasion [3]. Src and Syk both interact with integrins and interact with each other to regulate tumor cell invasion [4], [54], [55]; Src-substrates are well known contributors to the invasive function of invadopodia [2], [3], [5], [42], [56], [57]. ADAM15, up-regulated following SYK knockdown in MCF10A cells, was identified in this study as frequently altered in copy number. ADAM15 functions in invadopodia by binding TKS5, a Src substrate; variants contribute to mammary cell carcinoma, and increased copy number was found to lead to formation of multiple variants [57]–[59]. RAB25 and RAB11, closely related GTPases, and the RAB-binding protein RAB11FIP1 regulate endosomal traffic, contribute to integrin recycling to the cell surface, to Src positioning in the cell, and to many other endosomal trafficking events related to invasion. Our data agrees and supports other reports of the important role of membrane trafficking for adhesion, motility, and invasion specifically involving RAS family GTPases [60]–[76].
Another major theme is the enrichment of genes whose products are associated with or regulate the actin-cytoskeleton. CTTN (cortactin) is a gene that encodes an-actin binding protein that is also a prominent Src tyrosine kinase substrate. The c-Src-cortactin interaction impacts invasion including its central role in invadopodia formation and function and vesicle trafficking, specifically delivery of matrix-degrading proteases to the cell surface [77]–[80]. CTTN gene amplification in cancer has previously been reported [81], [82].
Another subset of SYK-regulated genes in the 55 Gene Set is important for cell invasion mechanisms including matrix degradation and adhesion. These include MMP7, ADAM12, and ADAM15 metalloproteinases, the latter two of which contribute to the activity of invadopodia and possess disintegrin adhesion domains. Also included are the tissue inhibitors of metalloproteinases TIMP2 and TIMP1; they are metalloproteinase inhibitors that regulate the activity of extracellular matrix degrading MMP-9, MMP-2, and MT1-MMP (MMP14) [83], [84].
Gain of 1q in cancer, specifically in breast cancer has previously been described [28] but is impressively evidenced in this study by frequent co-amplification of the SYK-regulated SPRR1A, SPRR1B, MUC1, RAB25, and ADAM15, genes located within chromosome 1q. We have also made the novel observation of SYK and CTNNAL1 (alpha-catulin, located on chromosome 9q) co-loss in this study. Alpha-catulin is an E-cadherin-regulating cell-cell adhesion protein that has been recently implicated in melanoma invasion and metastasis [85].
Conclusions
In conclusion, we have demonstrated that SYK allelic loss occurs both in DCIS tissues as well as IDC tissues. Immune cell Syk protein is 6-fold greater than epithelial Syk. This complicates analysis of tissue homogenates from the TCGA. However, we demonstrated that heterozygotic SYK is associated with reduction in SYK mRNA compared with diploid cases in breast cancer cases without immune filtration. We accomplished this using our new method to identify cases where immune cell infiltration is present. Finally, we identified a 55 Gene Set, the Syk interacting network of invasion-related genes. This gene set powerfully predicted poorer patient outcome when copy number and mutational alterations of one or more of these genes occurred. In summary, we propose two mechanisms whereby SYK loss together with mutations and copy number changes in the 55 Gene Set results in poorer overall survival of patients.
Supporting Information
Syk mRNA In Situ hybridization. A. In situ data from our original study of SYK mRNA [4] is graphed as mean values for benign only, all DCIS (DCIS), and IDC tissues. ANOVA P-value = 2.91E-11 for the three tissues. Both DCIS and IDC were significantly different from Benign tissues (3.95E-06 and 7.38E-10, respectively) and DCIS was significantly different from IDC (0.013). Scores for benign (mean 2.67+/−0.11 S.E.), DCIS (mean 1.84+/−0.13 S.E.), and IDC (mean 1.29+/−0.17 S.E.) tissues are shown. Box plots were generated using Sigma Plot: red lines indicate the mean, black lines the median, and a 95%/5% range is indicated by whiskers. Boxes represent the 75%/25% range. B. The in situ data for the subset of cases shown in Table S2 were analyzed as in A. and graphed as means of Benign, DCIS and IDC tissues. The results are representative of the data and analysis obtained for the entire set of cases originally published [4]. ANOVA P-value = 0.00027 for the three tissues. Both DCIS and IDC were significantly different from Benign tissues (0.0048 and 0.0072, respectively). DCIS and IDC were not significantly different (0.077184). Scores for benign (mean 2.77+/−0.17 S.E.), DCIS (mean 1.84+/−0.25 S.E.), and IDC (mean 0.75+/−0.48 S.E.) tissues are shown. Box plots were generated using Sigma Plot: red lines indicate the mean, black lines the median, and a 95%/5% range is indicated by whiskers. Boxes represent the 75%/25% range. All t-tests were two-tailed.
(PDF)
Quantitative immunofluorescence determination of protein. A. Mean intensity values were obtained as described in the Methods for each case by tissue type: Benign, DCIS, and IDC. Those means were averaged; the results for Syk in epithelial cells (eSyk), immune cells (eSyk) and keratin (Ker) are shown in the same graph to illustrate the relative staining for Syk in epithelial versus immune cells and as compared with keratin. Box plots were generated using Sigma Plot: red lines indicate the mean, black lines the median, and a 95%/5% range is indicated by whiskers. Boxes represent the 75%/25% range. B. The iSyk/eSyk protein ratios for benign, DCIS and IDC cases were graphed as a box plot. The mean and standard error (S.E.) are shown in the table below.
(PDF)
Co-alterations in 5 genes located on chromosome 1. A. Oncoprint generated using cBioPortal of genomic copy number and mutational changes in the members of the 55 Gene Set. B. Two gene comparisons of ADAM15 (x-axis) against RAB25, SPRR1A, and MUC1, respectively. C. Network of interactions of ADAM15, RAB25, SPRR1A, SPRR1B, and MUC1 including the 50 most mutated genes within the network.
(PDF)
Average copy number of members of the 55 Gene Set in IDC cases stratified by SYK copy number. A. Plot of average copy number (y-axis). B. Two gene plot of copy number for SYK and CTNNAL1 both of which are located on chromosome 9.
(PDF)
PubMed citations for members of the 55 Gene Set AND “Cancer” or members AND “Breast Cancer”. The number of citations for each gene was determined using PubMed for citations including both the gene symbol and the term “Cancer” (black bars) or “Breast Cancer” (red bars) and the results graphed. The results were sorted by Breast Cancer results with the most frequently cited breast cancer gene being EGFR. About half of the genes had been cited for breast cancer at least 5 times. 27 of the Genes were cited less than 5 times. One gene, SPTBN2 was cited 0 times for cancer and breast cancer.
(PDF)
Summary of motility/invasion gene and mRNA data. Microarray data on significantly regulated mRNAs following SYK knockdown in MCF10A cells were obtained in a previous study [3]. Data on number of cases altered, % of cases altered and the average copy number gain or loss were determined from TCGA data as described in the Methods and Results . For number and % cases altered, copy number and mutational changes were evaluated using the cBioPortal software to determine and export this information for the 55 Gene Set. The table is organized by greatest mRNA fold changes to least with losses as numbers >1.5 fold change and gains as <0.67 fold change (VIM the greatest fold increase and SPRR1A the greatest fold decrease in mRNA expression after SYK knockdown).
(PDF)
Summary of FISH, in situ , and methylation data for SYK in tumor epithelium. Cases were separated by whether they contained DCIS only, DCIS with IDC, or benign only tissues. SYK allelic loss, mRNA in situ, methylation, and Syk protein values where available are shown. Scores reflect the tissue types present in each case (slide) and available for analyses. For FISH results, the % DCIS was obtained from available pathology reports from the Histopathology and Tissue Shared Resource (second column). The number of FISH signals from the Syk and chromosome 9 centromere probes was determined microscopically by a licensed cytopathologist and their average ratio calculated which is shown in column 3 (J.B.). A cutoff point for “normal” versus “allelic loss” in DCIS tissues from each case was determined from Figure 1, as indicated. In situ hybridization results for SYK mRNA were obtained previously (Normal, Hyperplasia, DCIS, and Invasive in situ, columns 4–6 [4]. In the next to last set of data (columns 7–9), methylation results are shown. Raw Syk protein values were obtained from image analysis and are shown in the last columns (columns 10–12).
(PDF)
Functional Annotation Table for the 55 Gene Set Members. A functional gene annotation table was generated using DAVID (http://david.abcc.ncifcrf.gov/) to illustrate the groups of genes involved in motility- and invasion-related activities. Some of the important activities are highlighted.
(PDF)
Protein/phosphoprotein results from immune depleted all types or IDC only cases when queried using cBioPortal for the 55 Gene Set. Immune depleted invasive breast cancer cases (800 cases) or immune depleted IDC only cases (565 cases) were queried using the 55 Gene Set for mutation and copy number changes. The significant protein and phosphoprotein changes comparing the altered versus nonaltered cases were extracted using cBioPortal.
(PDF)
Acknowledgments
We thank Deborah Berry for considerable help in immunostaining experiments and Yuriy Gusev for critical reading of the manuscript.
Funding Statement
This work was supported in part by the Lombardi Comprehensive Cancer Center Microscopy & Imaging, Histopathology & Tissue, Tissue Culture & Biorepository for Cell Lines and Biofluids, and the Genomics & Epigenomics Shared Resources funded by U.S. Public Health Service Grant 2P30-CA-51008, 1S10 RR15768-01, and 1 S10 RR019291-01A2; and 9R01 CA112673 to SCM. No additional funding received for this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Mocsai A, Ruland J, Tybulewicz VL (2010) The SYK tyrosine kinase: a crucial player in diverse biological functions. Nat Rev Immunol 10: 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Coopman PJ, Do MT, Barth M, Bowden ET, Hayes AJ, et al. (2000) The Syk tyrosine kinase suppresses malignant growth of human breast cancer cells. Nature 406: 742–747. [DOI] [PubMed] [Google Scholar]
- 3. Sung YM, Xu X, Sun J, Mueller D, Sentissi K, et al. (2009) Tumor Suppressor Function of Syk in Human MCF10A In Vitro and Normal Mouse Mammary Epithelium In Vivo. PLoS ONE 4: e7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Moroni M, Soldatenkov V, Zhang L, Zhang Y, Stoica G, et al. (2004) Progressive loss of Syk and abnormal proliferation in breast cancer cells. Cancer Res 64: 7346–7354. [DOI] [PubMed] [Google Scholar]
- 5. Coopman PJ, Mueller SC (2006) The Syk tyrosine kinase: a new negative regulator in tumor growth and progression. Cancer Letters 241: 159–173. [DOI] [PubMed] [Google Scholar]
- 6. Yuan Y, Mendez R, Sahin A, Dai JL (2001) Hypermethylation leads to silencing of the SYK gene in human breast cancer. Cancer Res 61: 5558–5561. [PubMed] [Google Scholar]
- 7. Yuan Y, Liu H, Sahin A, Dai JL (2005) Reactivation of SYK expression by inhibition of DNA methylation suppresses breast cancer cell invasiveness. Int J Cancer 113: 654–659. [DOI] [PubMed] [Google Scholar]
- 8. Layton T, Stalens C, Gunderson F, Goodison S, Silletti S (2009) Syk tyrosine kinase acts as a pancreatic adenocarcinoma tumor suppressor by regulating cellular growth and invasion. Am J Pathol 175: 2625–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Muthusamy V, Duraisamy S, Bradbury CM, Hobbs C, Curley DP, et al. (2006) Epigenetic silencing of novel tumor suppressors in malignant melanoma. Cancer Res 66: 11187–11193. [DOI] [PubMed] [Google Scholar]
- 10. Dong SW, Ma L, Xu N, Yan HQ, Liu HY, et al. (2011) Research on the reactivation of Syk expression caused by the inhibition of DNA promoter methylation in the lung cancer. Neoplasma 58: 89–95. [DOI] [PubMed] [Google Scholar]
- 11. Ogane S, Onda T, Takano N, Yajima T, Uchiyama T, et al. (2009) Spleen tyrosine kinase as a novel candidate tumor suppressor gene for human oral squamous cell carcinoma. Int J Cancer 124: 2651–2657. [DOI] [PubMed] [Google Scholar]
- 12. Lee HS, Kim BH, Cho NY, Yoo EJ, Choi M, et al. (2009) Prognostic implications of and relationship between CpG island hypermethylation and repetitive DNA hypomethylation in hepatocellular carcinoma. Clin Cancer Res 15: 812–820. [DOI] [PubMed] [Google Scholar]
- 13. Ma L, Dong S, Zhang P, Xu N, Yan H, et al. (2010) The relationship between methylation of the Syk gene in the promoter region and the genesis of lung cancer. Clin Lab 56: 407–416. [PubMed] [Google Scholar]
- 14. Tsou JA, Galler JS, Wali A, Ye W, Siegmund KD, et al. (2007) DNA methylation profile of 28 potential marker loci in malignant mesothelioma. Lung Cancer 58: 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kunze E, Wendt M, Schlott T (2006) Promoter hypermethylation of the 14-3-3 sigma, SYK and CAGE-1 genes is related to the various phenotypes of urinary bladder carcinomas and associated with progression of transitional cell carcinomas. Int J Mol Med 18: 547–557. [PubMed] [Google Scholar]
- 16. Al Sahaf O, Wang JH, Browne TJ, Cotter TG, Redmond HP (2010) Surgical injury enhances the expression of genes that mediate breast cancer metastasis to the lung. Ann Surg 252: 1037–1043. [DOI] [PubMed] [Google Scholar]
- 17. Inayat U, Kayani MA, Faryal R (2012) Sporadic mutations and expression analysis of spleen tyrosine kinase gene in breast cancer: a preliminary report. Pol J Pathol 63: 106–111. [PubMed] [Google Scholar]
- 18. Kanwal S, Kayani MA, Faryal R (2012) Identification of novel SNPs in SYK gene of breast cancer patients: computational analysis of SNPs in the 5′UTR. Mol Biol Rep 39: 8345–8351. [DOI] [PubMed] [Google Scholar]
- 19. Minobe K, Onda M, Iida A, Kasumi F, Sakamoto G, et al. (1998) Allelic loss on chromosome 9q is associated with lymph node metastasis of primary breast cancer. Jpn J Cancer Res 89: 916–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cohen AL, Ward JH (2010) Risk reduction strategies for ductal carcinoma in situ. J Natl Compr Canc Netw 8: 1211–1217. [DOI] [PubMed] [Google Scholar]
- 21. Leonard GD, Swain SM (2004) Ductal carcinoma in situ, complexities and challenges. J Natl Cancer Inst 96: 906–920. [DOI] [PubMed] [Google Scholar]
- 22. Jenkins RB, Qian J, Lieber MM, Bostwick DG (1997) Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer Res 57: 524–531. [PubMed] [Google Scholar]
- 23. Mascarello JT, Hirsch B, Kearney HM, Ketterling RP, Olson SB, et al. (2011) Section E9 of the American College of Medical Genetics technical standards and guidelines: fluorescence in situ hybridization. Genet Med 13: 667–675. [DOI] [PubMed] [Google Scholar]
- 24. Tennis M, Krishnan S, Bonner M, Ambrosone CB, Vena JE, et al. (2006) p53 Mutation analysis in breast tumors by a DNA microarray method. Cancer Epidemiol Biomarkers Prev 15: 80–85. [DOI] [PubMed] [Google Scholar]
- 25. Tost J, Gut IG (2007) DNA methylation analysis by pyrosequencing. Nat Protoc 2: 2265–2275. [DOI] [PubMed] [Google Scholar]
- 26. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, et al. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6: l1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, et al. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cancer Genome Atlas Network (2012) Comprehensive molecular portraits of human breast tumours. Nature 490: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reva B, Antipin Y, Sander C (2007) Determinants of protein function revealed by combinatorial entropy optimization. Genome Biol 8: R232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reva B, Antipin Y, Sander C (2011) Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res 39: e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang X, Shrikhande U, Alicie BM, Zhou Q, Geahlen RL (2009) Role of the protein tyrosine kinase Syk in regulating cell-cell adhesion and motility in breast cancer cells. Mol Cancer Res 7: 634–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Neuhaus B, Buhren S, Bock B, Alves F, Vogel WF, et al. (2011) Migration inhibition of mammary epithelial cells by Syk is blocked in the presence of DDR1 receptors. Cell Mol Life Sci 68: 3757–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mahabeleshwar GH, Kundu GC (2003) Syk, a protein-tyrosine kinase, suppresses the cell motility and nuclear factor kappa B-mediated secretion of urokinase type plasminogen activator by inhibiting the phosphatidylinositol 3′-kinase activity in breast cancer cells. J Biol Chem 278: 6209–6221. [DOI] [PubMed] [Google Scholar]
- 34. Larive RM, Urbach S, Poncet J, Jouin P, Mascre G, et al. (2009) Phosphoproteomic analysis of Syk kinase signaling in human cancer cells reveals its role in cell-cell adhesion. Oncogene 28: 2337–2347. [DOI] [PubMed] [Google Scholar]
- 35. Bailet O, Fenouille N, Abbe P, Robert G, Rocchi S, et al. (2009) Spleen tyrosine kinase functions as a tumor suppressor in melanoma cells by inducing senescence-like growth arrest. Cancer Res 69: 2748–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maruyama S, Kurosaki T, Sada K, Yamanashi Y, Yamamoto T, et al. (1996) Physical and functional association of cortactin with Syk in human leukemic cell line K562. Journal of Biological Chemistry 271: 6631–6635. [DOI] [PubMed] [Google Scholar]
- 37. Helwani FM, Kovacs EM, Paterson AD, Verma S, Ali RG, et al. (2004) Cortactin is necessary for E-cadherin-mediated contact formation and actin reorganization. J Cell Biol 164: 899–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van Rossum AG, Moolenaar WH, Schuuring E (2006) Cortactin affects cell migration by regulating intercellular adhesion and cell spreading. Exp Cell Res 312: 1658–1670. [DOI] [PubMed] [Google Scholar]
- 39. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, et al. (2002) A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med %19 347: 1999–2009. [DOI] [PubMed] [Google Scholar]
- 41. Huderson BP, Duplessis TT, Williams CC, Seger HC, Marsden CG, et al. (2012) Stable inhibition of specific estrogen receptor alpha (ERalpha) phosphorylation confers increased growth, migration/invasion, and disruption of estradiol signaling in MCF-7 breast cancer cells. Endocrinology 153: 4144–4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mueller SC, Artym VV, Kelly T (2008) Invadopodia: Interface for Invasion. In: Dylan E, Hoyer-Hansen G, Blasi F, Sloane BF, editors. The Cancer Degradome – Proteases in Cancer Biology. New York: Springer. pp. 403–432. [Google Scholar]
- 43. Courtneidge SA (2012) Cell migration and invasion in human disease: the Tks adaptor proteins. Biochem Soc Trans 40: 129–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Destaing O, Block MR, Planus E, Albiges-Rizo C (2011) Invadosome regulation by adhesion signaling. Curr Opin Cell Biol 23: 597–606. [DOI] [PubMed] [Google Scholar]
- 45. Linder S (2007) The matrix corroded: podosomes and invadopodia in extracellular matrix degradation. Trends Cell Biol 17: 107–117. [DOI] [PubMed] [Google Scholar]
- 46. Linder S, Wiesner C, Himmel M (2011) Degrading devices: invadosomes in proteolytic cell invasion. Annu Rev Cell Dev Biol 27: 185–211. [DOI] [PubMed] [Google Scholar]
- 47. Machesky LM, Li A (2010) Fascin: Invasive filopodia promoting metastasis. Commun Integr Biol 3: 263–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Poincloux R, Lizarraga F, Chavrier P (2009) Matrix invasion by tumour cells: a focus on MT1-MMP trafficking to invadopodia. J Cell Sci 122: 3015–3024. [DOI] [PubMed] [Google Scholar]
- 49. Sibony-Benyamini H, Gil-Henn H (2012) Invadopodia: the leading force. Eur J Cell Biol 91: 896–901. [DOI] [PubMed] [Google Scholar]
- 50. Struckhoff AP, Rana MK, Worthylake RA (2011) RhoA can lead the way in tumor cell invasion and metastasis. Front Biosci 16:1915–26 1915–1926. [DOI] [PubMed] [Google Scholar]
- 51. Stylli SS, Kaye AH, Lock P (2008) Invadopodia: at the cutting edge of tumour invasion. J Clin Neurosci 15: 725–737. [DOI] [PubMed] [Google Scholar]
- 52. Yamaguchi H (2012) Pathological roles of invadopodia in cancer invasion and metastasis. Eur J Cell Biol 91: 902–907. [DOI] [PubMed] [Google Scholar]
- 53. Ruschel A, Ullrich A (2004) Protein tyrosine kinase Syk modulates EGFR signalling in human mammary epithelial cells. Cell Signal 16: 1249–1261. [DOI] [PubMed] [Google Scholar]
- 54. Destaing O, Block MR, Planus E, Albiges-Rizo C (2011) Invadosome regulation by adhesion signaling. Curr Opin Cell Biol 23: 597–606. [DOI] [PubMed] [Google Scholar]
- 55. Thomas SM, Brugge JS (1997) Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol 13: 513–609. [DOI] [PubMed] [Google Scholar]
- 56. Arias-Salgado EG, Lizano S, Sarkar S, Brugge JS, Ginsberg MH, et al. (2003) Src kinase activation by direct interaction with the integrin beta cytoplasmic domain. Proc Natl Acad Sci U S A 100: 13298–13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Abram CL, Seals DF, Pass I, Salinsky D, Maurer L, et al. (2003) The adaptor protein fish associates with members of the ADAMs family and localizes to podosomes of Src-transformed cells. J Biol Chem 278: 16844–16851. [DOI] [PubMed] [Google Scholar]
- 58. Zhong JL, Poghosyan Z, Pennington CJ, Scott X, Handsley MM, et al. (2008) Distinct functions of natural ADAM-15 cytoplasmic domain variants in human mammary carcinoma. Mol Cancer Res 6: 383–394. [DOI] [PubMed] [Google Scholar]
- 59. Ortiz RM, Karkkainen I, Huovila AP (2004) Aberrant alternative exon use and increased copy number of human metalloprotease-disintegrin ADAM15 gene in breast cancer cells. Genes Chromosomes Cancer 41: 366–378. [DOI] [PubMed] [Google Scholar]
- 60. Aloisi AL, Bucci C (2013) Rab GTPases-cargo direct interactions: fine modulators of intracellular trafficking. Histol Histopathol 28: 839–849. [DOI] [PubMed] [Google Scholar]
- 61. Mitra S, Cheng KW, Mills GB (2012) Rab25 in cancer: a brief update. Biochem Soc Trans 40: 1404–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dozynkiewicz MA, Jamieson NB, Macpherson I, Grindlay J, van den Berghe PV, et al. (2012) Rab25 and CLIC3 collaborate to promote integrin recycling from late endosomes/lysosomes and drive cancer progression. Dev Cell 22: 131–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Subramani D, Alahari SK (2010) Integrin-mediated function of Rab GTPases in cancer progression. Mol Cancer 9: 312 doi: 10.1186/1476-4598-9-312.: 312–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Eva R, Dassie E, Caswell PT, Dick G, Ffrench-Constant C, et al. (2010) Rab11 and its effector Rab coupling protein contribute to the trafficking of beta 1 integrins during axon growth in adult dorsal root ganglion neurons and PC12 cells. J Neurosci 30: 11654–11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Conklin MW, Ada-Nguema A, Parsons M, Riching KM, Keely PJ (2010) R-Ras regulates beta1-integrin trafficking via effects on membrane ruffling and endocytosis. BMC Cell Biol 11: 14 doi: 10.1186/1471-2121-11-14.: 14–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, et al. (2009) Mutant p53 drives invasion by promoting integrin recycling. Cell 139: 1327–1341. [DOI] [PubMed] [Google Scholar]
- 67. Mills GB, Jurisica I, Yarden Y, Norman JC (2009) Genomic amplicons target vesicle recycling in breast cancer. J Clin Invest 119: 2123–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chia WJ, Tang BL (2009) Emerging roles for Rab family GTPases in human cancer. Biochim Biophys Acta 1795: 110–116. [DOI] [PubMed] [Google Scholar]
- 69. Tang BL, Ng EL (2009) Rabs and cancer cell motility. Cell Motil Cytoskeleton 66: 365–370. [DOI] [PubMed] [Google Scholar]
- 70. Mosesson Y, Mills GB, Yarden Y (2008) Derailed endocytosis: an emerging feature of cancer. Nat Rev Cancer 8: 835–850. [DOI] [PubMed] [Google Scholar]
- 71. Caswell PT, Chan M, Lindsay AJ, McCaffrey MW, Boettiger D, et al. (2008) Rab-coupling protein coordinates recycling of alpha5beta1 integrin and EGFR1 to promote cell migration in 3D microenvironments. J Cell Biol 183: 143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sandilands E, Frame MC (2008) Endosomal trafficking of Src tyrosine kinase. Trends Cell Biol 18: 322–329. [DOI] [PubMed] [Google Scholar]
- 73. Nishimura N, Sasaki T (2008) Regulation of epithelial cell adhesion and repulsion: role of endocytic recycling. J Med Invest 55: 9–16. [DOI] [PubMed] [Google Scholar]
- 74. Caswell PT, Spence HJ, Parsons M, White DP, Clark K, et al. (2007) Rab25 associates with alpha5beta1 integrin to promote invasive migration in 3D microenvironments. Dev Cell 13: 496–510. [DOI] [PubMed] [Google Scholar]
- 75. Weaver V, Werb Z (2007) G'rab'bing the microenvironment for invasion. Dev Cell 13: 462–463. [DOI] [PubMed] [Google Scholar]
- 76. Zhang J, Liu X, Datta A, Govindarajan K, Tam WL, et al. (2009) RCP is a human breast cancer-promoting gene with Ras-activating function. J Clin Invest 119: 2171–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Artym VV, Zhang Y, Seillier-Moiseiwitsch F, Yamada KM, Mueller SC (2006) Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res 66: 3034–3043. [DOI] [PubMed] [Google Scholar]
- 78. Artym VV, Matsumoto K, Mueller SC, Yamada KM (2011) Dynamic membrane remodeling at invadopodia differentiates invadopodia from podosomes. Eur J Cell Biol 90: 172–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Weaver AM (2008) Cortactin in tumor invasiveness. Cancer Lett 265: 157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Clark ES, Whigham AS, Yarbrough WG, Weaver AM (2007) Cortactin is an essential regulator of matrix metalloproteinase secretion and extracellular matrix degradation in invadopodia. Cancer Res 67: 4227–4235. [DOI] [PubMed] [Google Scholar]
- 81. Schuuring E, Verhoeven E, Litvinov S, Michalides RJ (1993) The product of the EMS1 gene, amplified and overexpressed in human carcinomas, is homologous to a v-Src substrate and is located in cell-substratum contact sites. Mol Cell Biol 13: 2891–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Schuuring E, Verhoeven E, van Tinteren H, Peterse JL, Nunnink B, et al. (1991) Amplification of genes within the chromosome 11q13 region is indicative of poor prognosis in patients with operable breast cancer Isolation of two distinct epithelial cell lines from a single feline mammary carcinoma with different tumorigenic potential in nude mice and expressing different levels of epidermal growth factor receptors. Cancer Res 51: 4028–4037. [PubMed] [Google Scholar]
- 83. Zitka O, Kukacka J, Krizkova S, Huska D, Adam V, et al. (2010) Matrix metalloproteinases. Curr Med Chem 17: 3751–3768. [DOI] [PubMed] [Google Scholar]
- 84. Murphy G, Nagase H (2008) Progress in matrix metalloproteinase research. Mol Aspects Med 29: 290–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kreiseder B, Orel L, Bujnow C, Buschek S, Pflueger M, et al. (2013) alpha-Catulin downregulates E-cadherin and promotes melanoma progression and invasion. Int J Cancer 132: 521–530. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Syk mRNA In Situ hybridization. A. In situ data from our original study of SYK mRNA [4] is graphed as mean values for benign only, all DCIS (DCIS), and IDC tissues. ANOVA P-value = 2.91E-11 for the three tissues. Both DCIS and IDC were significantly different from Benign tissues (3.95E-06 and 7.38E-10, respectively) and DCIS was significantly different from IDC (0.013). Scores for benign (mean 2.67+/−0.11 S.E.), DCIS (mean 1.84+/−0.13 S.E.), and IDC (mean 1.29+/−0.17 S.E.) tissues are shown. Box plots were generated using Sigma Plot: red lines indicate the mean, black lines the median, and a 95%/5% range is indicated by whiskers. Boxes represent the 75%/25% range. B. The in situ data for the subset of cases shown in Table S2 were analyzed as in A. and graphed as means of Benign, DCIS and IDC tissues. The results are representative of the data and analysis obtained for the entire set of cases originally published [4]. ANOVA P-value = 0.00027 for the three tissues. Both DCIS and IDC were significantly different from Benign tissues (0.0048 and 0.0072, respectively). DCIS and IDC were not significantly different (0.077184). Scores for benign (mean 2.77+/−0.17 S.E.), DCIS (mean 1.84+/−0.25 S.E.), and IDC (mean 0.75+/−0.48 S.E.) tissues are shown. Box plots were generated using Sigma Plot: red lines indicate the mean, black lines the median, and a 95%/5% range is indicated by whiskers. Boxes represent the 75%/25% range. All t-tests were two-tailed.
(PDF)
Quantitative immunofluorescence determination of protein. A. Mean intensity values were obtained as described in the Methods for each case by tissue type: Benign, DCIS, and IDC. Those means were averaged; the results for Syk in epithelial cells (eSyk), immune cells (eSyk) and keratin (Ker) are shown in the same graph to illustrate the relative staining for Syk in epithelial versus immune cells and as compared with keratin. Box plots were generated using Sigma Plot: red lines indicate the mean, black lines the median, and a 95%/5% range is indicated by whiskers. Boxes represent the 75%/25% range. B. The iSyk/eSyk protein ratios for benign, DCIS and IDC cases were graphed as a box plot. The mean and standard error (S.E.) are shown in the table below.
(PDF)
Co-alterations in 5 genes located on chromosome 1. A. Oncoprint generated using cBioPortal of genomic copy number and mutational changes in the members of the 55 Gene Set. B. Two gene comparisons of ADAM15 (x-axis) against RAB25, SPRR1A, and MUC1, respectively. C. Network of interactions of ADAM15, RAB25, SPRR1A, SPRR1B, and MUC1 including the 50 most mutated genes within the network.
(PDF)
Average copy number of members of the 55 Gene Set in IDC cases stratified by SYK copy number. A. Plot of average copy number (y-axis). B. Two gene plot of copy number for SYK and CTNNAL1 both of which are located on chromosome 9.
(PDF)
PubMed citations for members of the 55 Gene Set AND “Cancer” or members AND “Breast Cancer”. The number of citations for each gene was determined using PubMed for citations including both the gene symbol and the term “Cancer” (black bars) or “Breast Cancer” (red bars) and the results graphed. The results were sorted by Breast Cancer results with the most frequently cited breast cancer gene being EGFR. About half of the genes had been cited for breast cancer at least 5 times. 27 of the Genes were cited less than 5 times. One gene, SPTBN2 was cited 0 times for cancer and breast cancer.
(PDF)
Summary of motility/invasion gene and mRNA data. Microarray data on significantly regulated mRNAs following SYK knockdown in MCF10A cells were obtained in a previous study [3]. Data on number of cases altered, % of cases altered and the average copy number gain or loss were determined from TCGA data as described in the Methods and Results . For number and % cases altered, copy number and mutational changes were evaluated using the cBioPortal software to determine and export this information for the 55 Gene Set. The table is organized by greatest mRNA fold changes to least with losses as numbers >1.5 fold change and gains as <0.67 fold change (VIM the greatest fold increase and SPRR1A the greatest fold decrease in mRNA expression after SYK knockdown).
(PDF)
Summary of FISH, in situ , and methylation data for SYK in tumor epithelium. Cases were separated by whether they contained DCIS only, DCIS with IDC, or benign only tissues. SYK allelic loss, mRNA in situ, methylation, and Syk protein values where available are shown. Scores reflect the tissue types present in each case (slide) and available for analyses. For FISH results, the % DCIS was obtained from available pathology reports from the Histopathology and Tissue Shared Resource (second column). The number of FISH signals from the Syk and chromosome 9 centromere probes was determined microscopically by a licensed cytopathologist and their average ratio calculated which is shown in column 3 (J.B.). A cutoff point for “normal” versus “allelic loss” in DCIS tissues from each case was determined from Figure 1, as indicated. In situ hybridization results for SYK mRNA were obtained previously (Normal, Hyperplasia, DCIS, and Invasive in situ, columns 4–6 [4]. In the next to last set of data (columns 7–9), methylation results are shown. Raw Syk protein values were obtained from image analysis and are shown in the last columns (columns 10–12).
(PDF)
Functional Annotation Table for the 55 Gene Set Members. A functional gene annotation table was generated using DAVID (http://david.abcc.ncifcrf.gov/) to illustrate the groups of genes involved in motility- and invasion-related activities. Some of the important activities are highlighted.
(PDF)
Protein/phosphoprotein results from immune depleted all types or IDC only cases when queried using cBioPortal for the 55 Gene Set. Immune depleted invasive breast cancer cases (800 cases) or immune depleted IDC only cases (565 cases) were queried using the 55 Gene Set for mutation and copy number changes. The significant protein and phosphoprotein changes comparing the altered versus nonaltered cases were extracted using cBioPortal.
(PDF)