Abstract
The aberrantly expressed signal transducer and activator of transcription 3 (STAT3) predicts poor prognosis, primarily in estrogen receptor positive (ER(+)) breast cancers. Activated STAT3 is overexpressed in luminal A subtype cells. The mechanisms contributing to the prognosis and/or subtype relevant features of STAT3 in ER(+) breast cancers are through multiple interacting regulatory pathways, including STAT3-MYC, STAT3-ERα, and STAT3-MYC-ERα interactions, as well as the direct action of activated STAT3. These data predict malignant events, treatment responses and a novel enhancer of tamoxifen resistance. The inferred crosstalk between ERα and STAT3 in regulating their shared target gene-METAP2 is partially validated in the luminal B breast cancer cell line-MCF7. Taken together, we identify a poor prognosis relevant gene set within the STAT3 network and a robust one in a subset of patients. VEGFA, ABL1, LYN, IGF2R and STAT3 are suggested therapeutic targets for further study based upon the degree of differential expression in our model.
Keywords: STAT3 transcriptional regulatory network, prognosis, TAM resistance, tumorigenesis, breast cancer
Introduction
Breast cancer (BC) is a global health problem and, in Taiwan, BC has replaced cervical cancer as the most common female cancer.1 The inherent genetic complexity and heterogeneity of breast cancer limits the prognostication value of many current model systems, as well as the ability to predict specific cancer identities from generalized information. An increasing number of transcription factor regulatory networks play unique roles in mammary epithelial development and tumorigenesis.2 STAT3 is a transcriptional regulator that is involved in mammary gland development, and elevated STAT3 has been widely observed in breast cancers.3–7 Alterations in STAT3 transcription programs may be a major switch in determining roles and clinical outcomes among breast cancer subtypes. To date, the prognostic value of STAT3 in human breast cancer remains unclear.8 Dissection of the global transcriptome in a clinical breast cancer cohort study suggests a role for STAT3 in coupling with MYC. And this, in turn, conditions a broad spectrum of pathophysiological effects in early development of estrogen receptor α negative (ER(−)) breast cancers, typically in triple negatives (TN).9 In silico studies demonstrate that some STAT3 target genes are potentially unfavorable prognostic markers in 77 breast cancer patients with ER(−) IDCs. However, the prognostic features of STAT3 in an ER(−) breast cancer setting have not been identified. In order to assess the predictive value of STAT3 pathway components, we evaluated clinical responses relative to STAT3 activities in 2 breast cancer patient populations with 77 ER(−) IDCs9 and 90 ER(+) IDCs, respectively.
STATs are known downstream targets of non-genomic ER actions in breast cancer cell models.10 Miller et al11 summarized routes for reciprocal crosstalk between estrogen receptor (ERα) and growth factor receptor signaling pathways. They indicated that membrane ER might activate oncogenic kinases to promote endocrine resistance; however, these mechanisms remain to be proven clinically. Importantly, the status of STAT3 as a target to treat ER(+) breast cancer with TAM resistance remains unclear. Using a network approach, it is possible to evaluate interactions between 2 transcription factors (ERα, STAT3) in regulating genes that may be causally associated with de novo or acquired resistance to endocrine therapy. Using this approach, we evaluated the role of STAT3 as a survival predictor gene based upon altered STAT3 transcriptional regulatory activity in an ER(+) breast cancer model system, consisting of 4 subtypes (groups IE, IIE, luminal A and B).
STAT3 can be activated by classical and non-classical mechanisms.12,13 The cooperation of both tyrosine (Tyr705) and serine (Ser727) phosphorylation is necessary for full classical activation of STAT3. For instance, STAT3 can be activated by the 17-β-estradiol-induced pathway via phosphorylation at Tyr705 and Ser727.12 Unphosphorylated STAT3 transcription factor can also bind DNA according to the non-classical model.13 Greten FR et al14 reported that a STAT3-dependent transcriptional program, in part, is triggered by an excess concentration of activating cytokines secreted in an autocrine or paracrine manner by tumor and stromal cells. Cytokine activation of STAT3 is constitutively activated by JAKs and JAK/STAT3 promotes breast cancer progression.15 STAT3 is also down-regulated by the chemodrugs 5-fluorouracil and gemcitabine in a MCF-7 cell model16 and by dehydrocostuslactone (DHE) in MCF-7 and MDA-MB231 cell models.17 At least 3 signal transduction pathways including MAPK, PI3-kinase, and Src-kinase pathways are required for 17-β-estradiol induced activation of a STAT-regulated promoter.10,18 Hart et al19 further showed that STAT3 activation is essential for transformation in PI3K-transformed cells. Inhibition of PI3K prevents STAT3 phosphorylation.
Herein, we find that high STAT3 mRNA levels in tumor tissue is a marker for poor prognosis and dissect the STAT3 network interactions as a basis for developing a predictive model. Finally, we identify major components of the STAT3 transcriptional regulatory network in silico that may be prognostic markers and therapeutic targets in ER(+) breast cancers in a subtype relevant manner.
Materials and Methods
Features of surgical specimens for generating the dataset
We used the immunohistochemical (IHC) status of 3 biomarkers (i.e. estrogen receptor α (ER), progesterone receptor A (PR) and HER-2/neu (HER)) as classifiers to identify 8 intrinsic subtypes. However, for ERBB2 (IHC score: 2+), determination of Her-2/neu gene copy number was done by chromogenic in situ hybridization (CISH) and IHC/CISH status was used for determining HER status.
90 specimens from primary infiltrating ductal breast carcinomas (IDCs) including group IE (i.e. ER(+)PR(+)) (61/90) and group IIE (i.e. ER(+) PR(−)) (29/90) were subgrouped into luminal A (i.e. ER(+)PR(+)HER(−) and ER(+)PR(−)HER(−)) (42/90), luminal B (ER(+)PR(+)HER(+) and ER(+) PR(−)HER(+)) (30/90), ER(+)PR(+)HER(?) (13/90) and ER(+)PR(−)HER(?)(5/90) categories. 13, 38, 34 and 1 samples were from a cancer stage I, II, III and IV, respectively. 3 additional samples did not have a cancer stage classification. Samples were obtained from patients who underwent surgery at the National Taiwan University Hospital (NTUH) between 1995 and 2007. 18 non-tumor samples were also surgically removed from breast tissue adjacent to some of the 90 ER(+) IDC breast tumors as controls. We obtained written consent from the patients or their relatives for the microarray study. Both clinicopathological data and angiograms provided in the article are part of their medical records. These medical records were originally included in the microarray study. Such study has been approved by the institutional review board (IRB) at NTUH (IRB number: 200706039R, Research Ethics Committee at National Taiwan University Hospital, Taipei, Taiwan). The gene expression dataset for this study can be retrieved from the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE24124. The abbreviation for each gene expression array data was “A”. Classification of datasets was based upon group IE (61 A) and group IIE (29 A) for the 90 A dataset and luminal A (42 A) and luminal B (30 A) for 72 A dataset. The cohorts contain 90 A and 72 A datasets that consist of 90 and 72 gene expression profiles, respectively. They were designated as 90 A cohort and 72 A cohort, respectively.
Microarray data analyses
A genome-wide gene expression profile per breast tumor specimen was analyzed using an Agilent Human 1 A (version 2) oligonucleotide microarray (half a genome size: 22 k) (Agilent technologies, USA). Quality control data (QC data) was established for 3 breast cancer biomarker genes—ESR1 (N = 151), PGR (N = 151) and ERBB2 (N = 151) using the same total ribonucleic acid (RNA) samples used for generating the gene expression profiles in 181 infiltrating ductal carcinomas (N = 181). The missing data for 30 samples (N = 30) was due to insufficient RNA for quantitative polymerase chain analysis (qPCR).
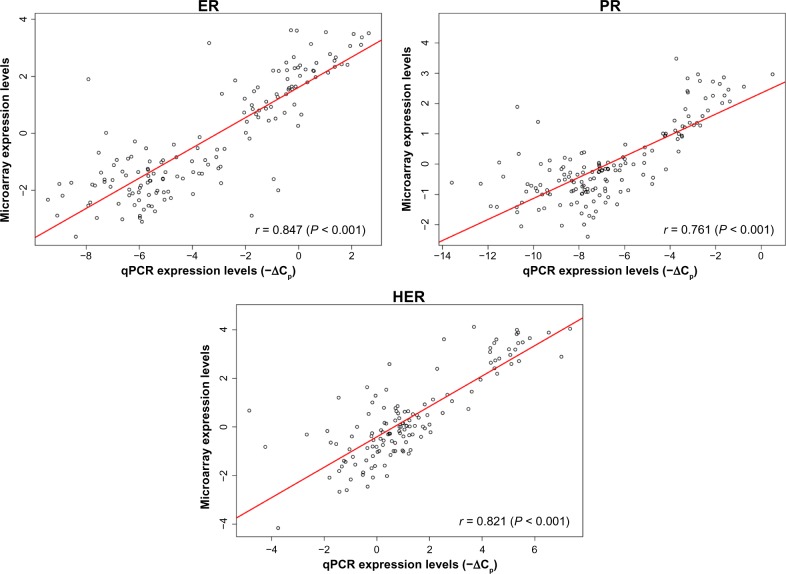
The qPCR procedure was done according to Kuo et al.20 4 primer IDs (Applied Biosystems, Foster City, CA, USA) designated as HT-A003, HT-A004, HT-A006 and a control primer ID as HH-T001 (TIB MOL BIOL, Germany) were used for amplification of the complementary deoxyribonucleic acid (cDNA) for PR, HER-2/neu, ER and the TATA box binding protein (TBP), respectively. Quality control data are shown in Figure 8.
Figure 8.
Quality control evaluation on 151 gene expression dataset.
The evaluation was made by the scatter plot analysis using data from hybridization (log2 ratios) and qPCR (−ΔCps). The Pearson’s correlation coefficient was used to find the linear relationships between mRNA expression levels derived from both log2 ratios and −ΔCps for the gene of interest. A good correlation is indicated between array gene expression (log2 ratio) and their corresponding qPCR data (−ΔCp) for ESR1(5561), PGR(11809) and ERBB2(764), respectively. The Agilent feature numbers are listed within the parenthesis next to the corresponding gene symbols. The 60 mer for PGR on array is for hybridizing with transcripts of PGR. The primer used for qPCR analysis only amplifies transcript variant 1 of PGR.
The heatmaps were displayed after unsupervised hierarchical clustering using R package (version 2.15.1). The “hcluster” function in the “stats” package was utilized to perform the unsupervised clustering. The heatmap was produced by the “rect” function to generate a customized view of the subcohorts. Gene Spring GX7.3.1 was used for generating Venn diagrams. ANOVA tests and the statistical methods for establishing the STAT3 transcriptional regulatory network were performed.9 Kaplan-Meier survival analyses21 were done using the “survival” package in R (version 2.15.1) for the gene profiles of 90 A cohort, 91 A cohort, 181 A cohort or the extracted gene pools of interest in the assigned cohorts. To quantify the weight of hazard ratios associated with the prognostic gene signature and the traditional prognostic factors in a given cohort of interest, both univariate and multivariate COX proportional hazard (COXPH) regression model in R package were performed.
Experimental design
Previously, we demonstrated that a new method, combining the coefficient of intrinsic dependence (CID) and Galton-Pierson’s Correlation Coefficient (GPCC), potentially has significant advantages in predicting network responses at the transcriptome level when using a systems biology approach.22 We found that the nuclear receptor ERα, which is a ligand dependent transcription factor, is activated by the environmental trigger (i.e. estrogen) in the breast cancer specimens. As a result, the relationship between the environmental trigger and a phenotype (i.e. the ERα transcriptional regulatory network) could be functionally dissected from the gene expression profiles in the breast tumor population. To specifically classify regulatory mechanisms impacting ERα functional transcription activities, we established the multivariate space of the ERα transcriptional regulatory network using multivariate CID.23
Although this network analysis has proven to be statistically significant relative to specificity and sensitivity, the sensitivity of interactions among gene products22 was not established. This was due to the complexity and size of TFs that are potentially functioning in the gene expression profile consisting of half a genome size, given that the predicted number of human putative transcription factors genome-wide is between 1,850 and 4,105.24 Moreover, each transcription factor has its unique regulatory mechanism and most have not been studied in breast cancers. Therefore, we designed a series supervised approach to reduce the confounder effects due to sampling, cohort composition and gene expression data after processing.
This network analysis is based on the prior data related to the functional status of a given TF in a given sample population. The method has been modified to increase the specificity and sensitivity of network analysis. Firstly, we have modified the procedure for CID via replacing the quantile clustering by hierarchical clustering before subgrouping the data for CID analysis. Hierarchical clustering mimics the biological event in which a functional TF with relatively similar gene expression levels may regulate its target genes in a similar manner under similar environments. Secondly, we have designed a dataset consisting of 2 subcohorts with different features of interest for network analysis. Thirdly, we established the predicted networks of a TF, which has transcript variants acting as the same target gene regulator, as a function of the whole network of the TF. Finally, we optimized the subgrouping strategy to be 1/10th for CID analysis. This effectively localizes the most relevant transcriptional regulatory mechanism of interest to a small subgroup of tumor specimens (i.e. the highest subCID value) as compared to that in other subgroups.
Herein, we designed specific subcohort combinations for univariate CIDUGPCC analysis based on data from ANOVA tests, hypothesis testing and consideration of the reduction of the confounder effects. For instance, we predict a clinically significant transcriptional regulatory network for a TFx by combining sub-cohorts, which have opposite status of a given clinical parameter, to run CIDUGPCC analysis on the TFx. CIDUGPCC analysis is an established statistical measure for building a network based on significance in non-linear or in linear associations. Biologically, such combined measurements allow the gene expression relationship between a transcription factor and its predicted target gene to be identified in a given population. When a gene pool is identified as the potential target of a transcription factor, the relationship among them can be linked to form a transcriptional regulatory network.9,22 We investigated networks of MYC, STAT3, ESR1, ARNT and FOXC1 in this study.
The mRNA expression levels of ESR1 are not linearly correlated with ERα protein levels within ER(+) IDCs.23 Additionally, STAT3 mRNA levels are elevated preferentially in the HER(−) IDCs group. To predict subtype-enriched transcriptional regulatory programs of ERα and STAT3, we analyzed group IE (61 A), group IIE (29 A), luminal A (42 A) and luminal B (30 A). The counter cohort (29 A) to these subcohorts was ERBB2+ (i.e. ER(−)PR(−) HER(+)). Dataset (119 A), comprised of ERBB2+ (29 A), groups IE (61 A) and IIE (29 A) and dataset (101 A) comprised of ERBB2+ (29 A), luminal A (42 A) and luminal B (30 A) were used for univariate and bivariate network studies.22,23 To evaluate crosstalk between ERα and STAT3 at the transcriptional level, we used the bivariate CID method to extract shared regulatory network.
Venn diagram analysis was performed to identify overlapping and/or non-overlapping gene pools related to the feature of interest. The combinatorial interactions between transcription factors were further investigated to dissect the regulatory mechanisms on their shared target genes (see examples in Suppl. 2 of Additional file 1). Gene annotation was done according to the Gene References Into Function (Gene RIFs of NCBI) and Gene Spring GX7.3.1.
In vitro validation of estrogen actions for predicted ERα target gene expression in a cell model and in vivo validation of sustained angiogenesis by sonograms
Reagents, cell culture and treatments
Estrogen (E2 or 17-β-estradiol) and antiestrogen ICI 183,780 were obtained from Sigma Chemical Co. (St. Louis, MO, USA). Human breast cancer cell line MCF-7 was obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured in DMEM supplemented with 5% heat-inactivated fetal bovine serum from Atlanta Biologicals, Inc. (Norcross, GA, USA). Cells were grown in a humidified atmosphere of 95% O2 and 5% CO2 at 37 °C. 350,000 cells per well were plated in 6-well plates for 2 days in DMEM with L-glutamine and phenol red free supplemented with 5% charcoal-stripped serum before appropriate drugs were added.
Western blot analysis
MCF-7 cells were treated with appropriate amounts of estrogen, anti-estrogen or both for 48 hours before cells were harvested for total protein extraction. Western blot analysis was performed as described25 with a few modifications.
To each well dish in 6-well plates, 0.2 ml of the ice-cold lysis buffer (20 mM Tris, pH7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM sodium orthovanadate, 2.5 mM sodium pyrophosphate, 1% TritonX100, 1 mM β-glycerophosphate, 1 mg/ml leupeptin and aprotinin and 1 mM phenylmethylsulphonyl fluoride) was added. The lysed cells were prepared by pulse-sonication and centrifugation at 14,000 rpm for 10 minutes. Cell lysate samples (60 μg total protein/lane) were loaded in a 7.5% polyacrylamide gel containing 1% SDS (SDS-PAGE) for gel electrophoresis. Fractionated proteins were transferred to a nitrocellulose membrane and probed with primary antibodies in 5% bovine serum albumin dissolved in Tris-buffered saline with 0.05% Tween 20 and 10% non-fat milk. Secondary antibodies conjugated to horseradish peroxidase were then applied. Chemiluminescent substrate of horseradish peroxidase was added at the final step. The specific protein bands labeled with a final chemiluminescent dye were visualized by exposing the membrane to X-ray film. Western blot analysis to detect p67 was done using polyclonal rat p67 antibody.26 β-actin in each lane was probed with antiβ-actin antibody (Sigma Diagnostics, St. Louis, MO, USA) as the loading control.
In vivo validation and supporting documents
Angiosonograms were gathered at NTUH. Sonograms of the cohort study and literature documentations were used to partially validate the network prediction.
Results and Discussion
Network analysis was utilized to biochemical profiling of 12 signal transduction pathway activities, the clinical relevance of ten clinical parameters, prognosis relevant events, and other malignant phenotypes including proliferation, the Warburg effect, sustained angiogenesis, and ES like epithelial mesenchymal transition. STAT3 is projected to differentially regulate these pathophysiological activities and some additional transcription factors are suggested as co-regulators of STAT3 to regulate these cellular activities. However, further time course studies in model systems are required to validate transcriptional roles of STAT3 in regulating tumorigenesis because clinical tumor samples used in our studies were collected at a single time point.
-
STAT3 may be a transcriptional regulator of tumorigenesis and responses to cancer therapies.
-
There are major clinical implications of STAT3 in ER(+) breast cancers.ANOVA tests (Fig. 1) indicate a significant decrease in STAT3 mRNA expression levels during mitotic count progression. STAT3 is significantly elevated in ER(+) HER(−) IDCs.
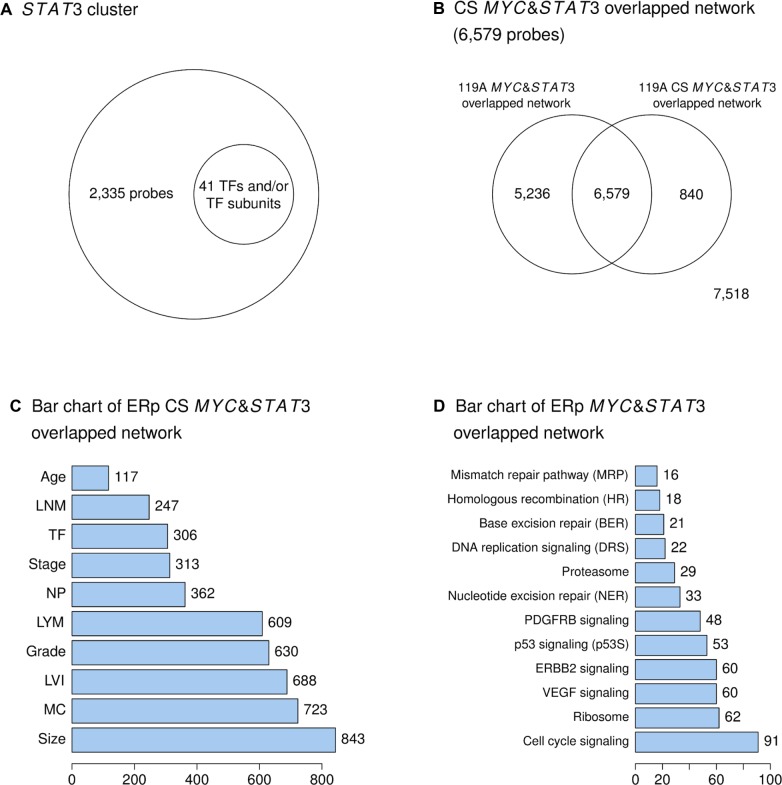
- Clinically relevant activities of STAT3 and its common partner MYC, condition mitotic count, a pathological subindex in ER(+) breast cancers.There are 2,335 probes in the STAT3 cluster selected by the ANOVA test that are potential determining factors of mitotic count (Fig. 2A). Only 41 TFs and/or TF subunits are in this cluster, which includes STAT3 and ARNT (Fig. 2A). However, based upon mitotic count relevant STAT3 regulatory network, MYC but not ARNT is the target gene of STAT3 (Table S1.1 in Suppl. 1 of Additional file 1). Moreover, mitotic count relevant MYC regulatory network predicts that MYC regulates STAT3 but not ARNT (Table S1.2 in Suppl. 1 of Additional file 1). Data in Tables S1.5 and S1.8 in Suppl. 1 indentify STAT3 as the predicted target gene of MYC in both 90 A and 72 A cohorts.
-
Clinically relevant, subtype enriched MYC and STAT3 may co-contribute 10 clinical parameters based on their target gene pools. The regulatory interactions between MYC and STAT3 were further investigated by an overlapping network of MYC and STAT3 (6,579 probes in Fig. 2B and Table S1.7 in Suppl. 1 of Additional file 1), which was determined to be significantly relevant in both ER(+) IDCs and in mitotic count.
Relevant to ER positive breast cancers, the clinically significant MYC and STAT3 overlapping network (identified based upon approximately 122 TFs and/or their subunits in this network as shown in Table S1.7 in Suppl. 1 of Additional file 1) shows increased regulation of genes associated with tumor size (size), mitotic count (MC), lymphovascular invasion (LVI) than regulation of those associated with histological grade (grade) and lymph node metastasis status (LYM). These results indicate relatively less regulation of genes associated with nuclear pleomorphism (NP), cancer stage (stage), tubule formation (TF), number of lymph node metastasis (LNM) and age (Fig. 2C and Suppl. 3). Many TFs (about 144 TFs and/or their subunits in Table S1.3; about 122 TFs and/or their subunits in Table S1.7) are partners of STAT3 and possibly participate in co-regulating those gene pools. We summarize the clinically significant gene pools in Figure 2C that may be relevant to ER(+) tumor development due to MYC and STAT3 regulation of their shared target genes.
STAT3 has a large number of transcription factors other than MYC as potential regulatory partners (Fig. 2B, Table S1.7 in Suppl. 1 of Additional file 1) and these may control tumor fate in multiple pathophysiological events (Fig. 2C) and in deregulated biochemical events (Fig. 2D). Notably, Figure 2C indicates a broad spectrum of early clinically relevant and luminal A subtype enriched pathological features affected by transcriptional regulation of STAT3. Unexpectedly, among 10 clinical parameters we found that tumor size shares the largest gene pool with the overlapping network of STAT3 and MYC (Fig. 2C). This suggests that a pre-programmed transcriptional event of STAT3 in coupling with MYC may slightly shift the preferential influence of STAT3 for a series of pathophysiological features.
In addition, STAT3 with different TF partner pools among different subtypes may offer another mechanism for predicting prognostic features of STAT3 in different subtypes (Fig. 3F, Tables 2 and 3), based on results in Figures 1 and 2, STAT3 may play a central role in ER(+) IDCs similar to that in ER(−) IDCs.9
-
Subtype enriched transcription factors— MYC and STAT3 may co-contribute 12 signal transduction pathways based on their target gene pools.
The MYC and STAT3 overlapping network for an ER(+) breast cancer population (90 A) predicts shared target genes to be involved in regulating the cell cycle signal transduction pathway as compared to those involved in regulating signal transduction pathways of VEGF, ribosome and ERBB2. Breast cancer cell proliferation may be a consequence of STAT3 activation by autocrine/paracrine signals27 and this indirectly supports the network prediction.
-
STAT3 is critical to ER(+) breast tumor development.
A moderate number of genes within this network are predicted gene components in signal transduction pathways of p53, PDGFRB, nucleotide excision repair (NER) and proteasome. Only small numbers of genes within this overlapping network are predicted to regulate DNA replication, base excision repair homologous recombination, and mismatch repair pathways (Suppls. 4, 5 in Additional file 1 and Fig. 2D). Thus, using the heatmap approach, we have identified shared gene pools between the MYC and STAT3 overlapping network and twelve signal transduction pathways among non-tumor component, groups IE and IIE (Suppl. 5 in Additional file 1). STAT3 is critical in ER(+) breast tumor development.
-
Four functional subnetworks are predicted to be involved in malignant phenotypes enriched in luminal A subtype.
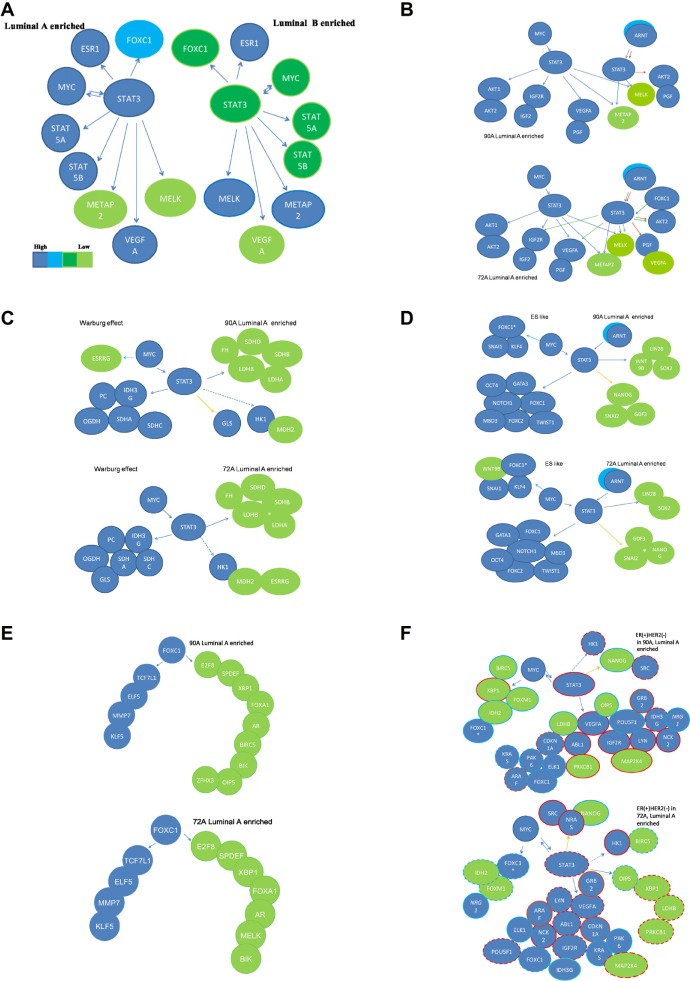
- A metabolic transcriptome involving the STAT3 network.OGDH, PC, IDH3G, SDHA, SDHC, and GLS are predicted to be up-regulated by STAT3 coupled with MYC in the 72 A cohort. The 90 A cohort has the same regulatory subnetwork except GLS is predicted to be regulated by STAT3 alone (Fig. 3C). LDHA and LDHB appear to be down-regulated by STAT3 and MYC in both 90 A and 72 A cohorts and low LDHB mRNA levels in 90 A cohort are a predictor of favorable prognosis (Table 2). High levels of IDH3G are a favorable prognosis predictor in 72 A cohort (Table 3). ESRRG, PC, a transcript variant of MYC, SDHD, and LDHB are highly expressed in the non-tumor component (Fig. 4B).The STAT3 network is predicted to regulate only a subset of genes in the Warburg effect. Relatively low expression levels of LDHB and LDHA indicate that part of the Warburg effect may be suppressed in ER(+)BCs (Figs. 3C and 4B). In addition, MYC and STAT3 differentially regulate the expression of subunits for succinate dehydrogenase (SDH) that may alter the enzyme activities of SDH. However, the expression pattern of PC and GLS are conserved between ER(+) and ER(−) breast cancers (Figs. 5D9 and 3C). They do not follow the same regulatory route within the STAT3 transcriptional regulatory network. Based upon these results, the physiological role for high levels of LDHB, PC, SDHD and a transcript variant of MYC in non-tumor components could be of interest for future study.
- Phenotype-like mesenchymal stem cells in tumor pathogenesis are predicted to be regulated by the STAT3 network.GATA3, OCT4, FOXC1, FOXC2, NOTCH1, TWIST1 and MBD3 are predicted to be up-regulated by MYC and STAT3 or ARNT and STAT3 in both 90 A and 72 A cohorts. STAT3- and/or MYC-mediated regulation of WNT9B is different in 90 A and 72 A cohorts (Fig. 3D). Both invasiveness and proliferation promoting genes are regulated by STAT3 but less in ER(+) BCs than in ER(−) BCs. For instance, MMP7 and MELK are not in the STAT3 network (Figs. 3D and 4C). Both GATA3 and OCT4 (POU5F1) are up-regulated by MYC and STAT3 in 90 A and 72 A cohorts. Siegel PM et al2 demonstrated that high GATA3 mRNA levels corresponded to a strong propensity for luminal breast cancer subtype and Sano H et al28 found that increased OCT4 (POU5F1) levels suppressed invasion and metastasis in MCF−7. A small subset of ER(+)BCs shows 3 core regulatory factors (OCT4, SOX2 and NANOG) to be co-expressed at relatively high levels and this may maintain pluripotency and a self-renewal phenotype (ES-like; Fig. 4C).Based on the network prediction for ES-like phenotypes, STAT3 interactions with HIF and MYC has more impact on ER(+) BCs than on ER(−) BCs relative to development of a less invasive malignant phenotype. Further in vitro studies and a network approach in silico are required to identify the cooperative regulatory relationships among STAT3, OCT4 and GATA3 in order to fully distinguish tumor fate in luminal subtypes and basal subtypes.
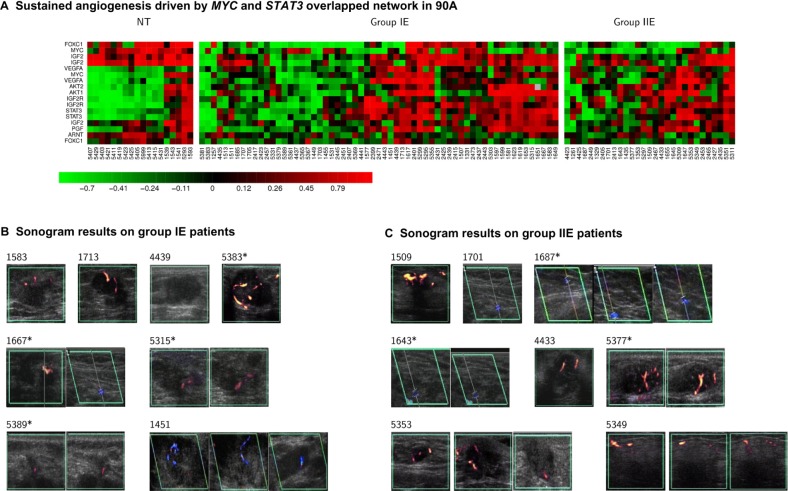
- An increased tumor survival mechanism via sustained angiogenesis involving STAT3.We find functionally sustained angiogenesis to be mainly regulated by STAT3 in conjunction with MYC. Both 90 A and 72 A cohorts share major STAT3 network architecture for sustained angiogenesis, except that FOXC1 is predicted to be an additional partner of STAT3 in the 72 A cohort that includes both the IGF2-IGF2R-PLC_2 axis and VEGF signaling (KEGG database; Fig. 3B). Increased expression of VEGFA, ABL1 and IGF2R, and decreased expression of PRKCB1 (or PRKCB) are poor prognostic factors in the 90 A cohort (Table 2 and Fig. S5.2 in Suppl.5 of Additional file 1).It is of interest that the sustained angiogenesis network in ER(+) BCs shares the same architecture with ER(−) BCs (Figs. 5C9 and 3B) except that FOXC1 is an additional partner of STAT3 (Fig. 3B, Tables S1.10 and S1.11). Importantly, we found that the mechanism for sustained angiogenesis driven by the STAT3 network in ER(−) BCs (Fig. 7B9) is partially relevant in ER(+) BCs (Fig. 3B). This may suggest that different transcriptional regulators interact with STAT3 to control tumor angiogenesis dependent upon different BC subtypes. Other in vitro studies29,30 support the concept that FOXC1, ARNT and FOXC2 are transcriptional regulators in angiogenesis. Further investigations of co-regulatory subnetworks involving these transcription factors and their relationship with patient angiosonograms will be explored in the future. Figure 6 shows that 43.75% of sonograms do not match with the gene expression signature of sustained angiogenesis based upon heatmaps. This could be due to limitations in sampling of gene expression profiles that reduce the accuracy of prediction9 or an alternative mechanism(s) controlling sustained angiogenesis. Therefore, more regulatory components of sustained angiogenesis are expected to be discovered in vitro, in vivo and in silico.
- Tumor proliferative activities are predicted to be regulated by the STAT3 and/or other TF(s) network.Both METAP2 and MELK are positive determinants of mitotic count in 90 A and 72 A cohorts (Figs. S7.1 and S7.2 in Suppl. 7 of Additional file 1). Lower tumor proliferative activities in ER(+) IDCs (Figs. 3A and 4) are indicated by METAP2 and MELK, which are down-regulated by MYC and STAT3 in both 90 A and 72 A cohorts. METAP2 and MELK show high mRNA expression levels when the expression of STAT3 decreases (Figs. 3A and 4A).FOXC1 is a shared target gene of MYC and STAT3 (Fig. 3D). We first reported the overexpression of OIP5, E2F8 and BIRC5 that are possibly regulated by FOXC1 in triple negatives.9 These genes and FOXC1 are positive determinants of histological grade and mitotic count in ER(−) BCs. The proliferation activities in tumors due to FOXC1 transcriptional regulation appear to be less in ER(+) IDCs as compared to ER(−) IDCs. A transcript variant of FOXC1 (FOXC1*) is potentially regulated by MYC (Fig. 3F). Interestingly, high expression levels of one FOXC1 transcript variant (FOXC1*) is a predictor of good prognosis in ER(+) IDCs (Fig. S6.3 in Suppl. 6 of Additional file 1). The STAT3 network in 77 A and 90 A cohorts is similar except that MELK is down-regulated by MYC and STAT3 in the 90 A cohort, and by MYC, STAT3 and FOXC1 in the 72 A cohort. The STAT3 mediated tumor proliferative activity response is different in ER(−) IDCs “(Figs. 5C, F, 6A and D in article,9 Figs, 3B, E, 4A and D).Figure 3E shows partial activities of the FOXC1 subnetwork to be conserved between 90 A and 72 A cohorts. Only E2F8 is down-regulated by FOXC1 in both 90 A and 72 A cohorts as compared to those in the 77 A cohort of ER(−) IDCs (Figs. 5F9 and 3E). Decreased expression of OIP5 is a good prognostic factor (Table 2 and Fig. 3F) in the 90 A cohort. Heatmaps for the FOXC1 subnetwork indicate that FOXC1 is differentially regulating a set of genes between non-tumor and tumor components. However, no obvious differences in the gene expression patterns of FOXC1 subnetwork were observed among subtypes (Fig. 4D).We further compared the functional sub-network of FOXC1, which is an indicator of high mitotic count in triple negatives (Fig. 6D9), between 90 A and 77 A cohorts. FOXC1 is preferentially increased in ER(+)HER(−) breast cancers. However, it down-regulates E2F8, BIRC5 and OIP5 in the 90 A cohort, while, in turn, it up-regulates them in the 77 A cohort. In addition, both SP5 and MTA1 are not in the STAT3 network of ER(+) BCs. Such dramatic changes may reduce its influence on enhancing tumor proliferation and metastasis. Multiple possibilities may explain how a malignant phenotype (e.g. proliferation activities) is altered due to the same regulator(s) in different BC subtypes and this remains to be evaluated by additional research.
-
-
STAT3 may have a critical role in ER(+) breast tumors in response to cancer therapies.
-
Hypothesis testing for cancer-related activities of the STAT3 and/or MYC and STAT3 overlapping network.
-
A hypothesis and the contradictory findings.
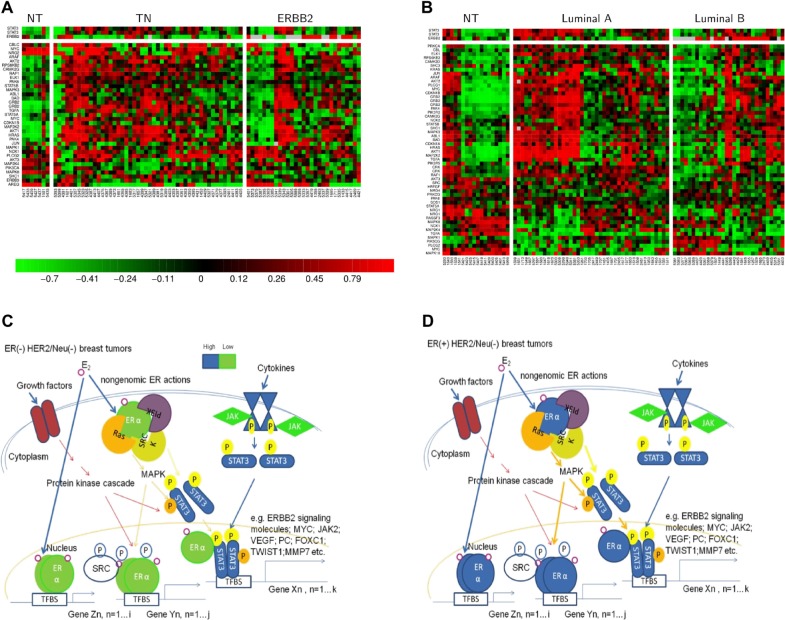
The ANOVA test suggests that STAT3 is preferentially increased in HER(−) BCs. Thus, we hypothesize that TN or luminal A patients have a lower propensity for ERBB2 interaction. Surprisingly, STAT3 regulates 59% (60/101) of ERBB2 signaling molecules in the 90 A cohort. The expression patterns of these signaling molecules shown in heatmaps (Fig. 7B) indicate that STAT3 mediated regulation of these 6 signaling molecules (PRKCB1, MAP2K4, NRG1, NCK2, ABL1 and GRB2) provides a good prognostic indicator in the 90 A cohort (Table 2). This regulatory event may be involved in the crosstalk between ERα and STAT3 that enhances a TAM resistance mechanism. This mechanism may be due to aberrant STAT3 activities on up-regulating key components in the PI3K, MAPK and c-SRC signaling pathways in ER(+) BCs. Interestingly, the gene expression for a few target genes (e.g. ABL1, GRB2, MAP2K4 and ERBB2) in the ERBB2 signal transduction pathway mediated by STAT3 are conserved between ER(+) and ER(−) BCs (Figs. 7A and B). With different TF partners of STAT3 among different subtypes, it may condition different mechanisms for predicting prognostic features of STAT3 in different subtypes. For instance, we found the expression levels of GRB2, NCK2, STAT3, PRKCB1, MAP2K4, ABL1, IGF2R, LYN, and VEGFA in the STAT3 network to be predictors for poor clinical outcome in the 90 A cohort. Alternately, the expression levels of NANOG, OIP5, LDHB, NRG1 and POU5F1 in the STAT3 network are predicted to be good prognostic factors in the 90 A cohort (Fig. 3F and Table 2). Moreover, there are 6 poor and 5 good prognostic factors in the STAT3 network of 72 A cohort (Fig. 3F and Table 3).
- A new hypothesis based on network prediction.Gene expression of ERBB2 signal transduction pathway components may, in part, involve regulation by STAT3. Although STAT3 is predominantly elevated in breast tumors with HER(−), we found that the ERBB2 signal transduction pathway is activated by STAT3 in HER(−) tumors (Fig. 7). For those tumors expressing a low amount of ERBB2 (HER-2/neu), STAT3 enhances the growth factor stimulated basal activity of ERBB2 signaling via regulating expression of its essential components (i.e. 60 probes in Fig. 7B). We reason that the ERBB2 signal transduction pathway shares many enzymes and adaptors with other oncogenic signal transduction pathways, including the ERα-mediated signal transduction pathway that promotes a malignant phenotype.18 As a result, it is proposed that crosstalk between ERα and STAT3 enhances a mechanism of tamoxifen (TAM) resistance. This possible regulatory mechanism may include two parts (Fig. 7D). Part 1 is defined by aberrant gene expression regulation by STAT3 via a loop connecting non-genomic ERα activities, genomic ERα activities, and genomic STAT3 activities after exposure to estrogen: (A) pre-existing protein cascade along with the extranuclear ERα to activate STAT3 via phosphorylation; (B) aberrant transcriptional activities of STAT3 (either following the non-classical ERα pathway or STAT3 alone) up-regulate a subset of key components in the PI3K, MAPK and c-SRC signaling pathways; (C) aberrant protein cascade enhances (A) and (B). Part 2 is defined by the effect of aberrant regulation of ERα and/or cofactors via phosphorylation after exposure to estrogen. Taken together, STAT3 is predicted to regulate the gene expression of key components in the pre-existing protein cascade. It may indirectly induce conformational changes of cofactors and/or ERα via phosphorylation that, in turn, cause tumor resistance to TAM treatment.
-
- Roles of STAT3 in ER(+) breast tumors in response to cancer therapies.Figure 7 shows the hypothesized mechanism of the pathological roles of ERα extranuclear signaling, growth factor receptor signaling, cytokine receptor signaling and non-receptor tyrosine kinases in activation of STAT3 transcriptional activities in HER(−) breast cancer subtypes.A possible effect of elevated STAT3 on responses to chemotherapy that may condition TAM resistance are provided in Figure 7. Elevated STAT3 expression may induce anti-estrogen resistance through up-regulating key components (e.g. adaptors and enzymes) in the MAPK/ERK, PI3K/Akt, PDGFR and ERBB2 signaling pathways.Figure 7 includes the recent research findings by others that predict the constitutive JAK/STAT pathway in all breast cancer subtypes being sensitive to chemo drugs—5FU, Gentamine16 and DHE.17 STAT3 suppresses the expression of NANOG in the cohorts (90 A and 72 A; Fig. 3D). Only a subset of ER(+) BCs show relatively high NANOG and STAT3 levels (Fig. 4C) that may determine drug resistance due to up-regulation on MDR (multiple drug resistance) gene expression.31 Another chemotherapeutic drug resistance marker up-regulated by the STAT3 network is RAF1 (Fig. 7B and Table S1.4 in Suppl. 1 of Additional file 1). In breast cancer cells, activated Raf confers resistance to the chemotherapeutic drugs doxorubicin and paclitaxel. Raf induces the expression of the multidrug resistance protein 1(Mdr-1) and the Bcl-2 anti-apoptotic protein.32More network studies will help in mapping the relevant effects of STAT3 as a poor prognostic factor in ER(+) BCs. For instance, cyclins including CCND1 are STAT3 target genes that have been identified in vitro18 and are indentified herein (Fig. S5.1 in Suppl. 5 of Additional file 1). Ishii Y et al33 suggested a mechanism that STAT3 can stimulate tumor growth in CCND1 overexpressing ER(+) breast cancer cells in response to TAM treatment. This supports our data that TAM resistance occurs in a luminal A subtype relevant manner whereby a subset expresses high levels of CCND1 and STAT3.
-
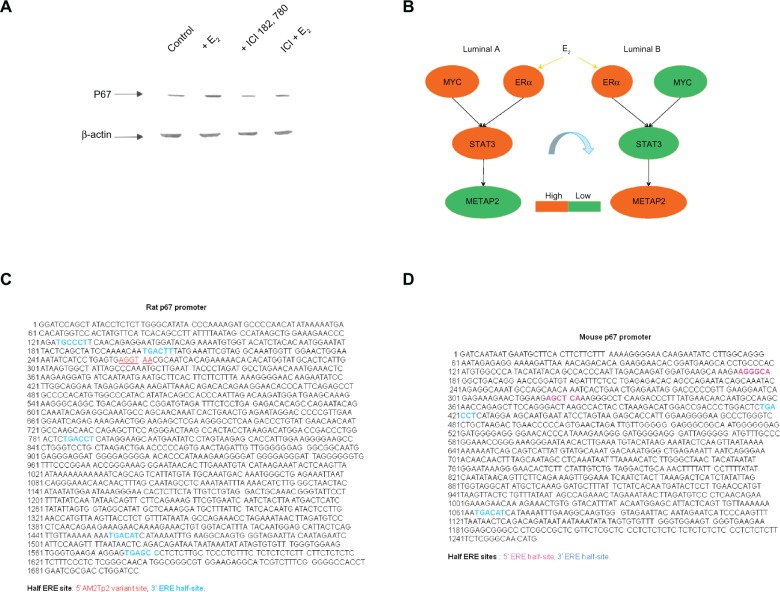
- STAT3 transcriptional regulatory network predicts combinatorial interactions between STAT3 and other transcription partner(s) in a subtype enriched manner based upon Western blot analysis and previous literature.The STAT3 transcriptional regulatory network offers the opportunity for dissecting the potential mechanism(s) of STAT3 as a poor prognostic predictor. The entire array of STAT3 transcriptional activity includes both univariate (STAT3 alone) and multivariate (STAT3 and other TF(s)) regulatory networks. We proposed that the specific prognostic features of STAT3 in a subtype relevant manner may be, in part, due to multiple routes that STAT3 interacts with other TF(s) to differentially regulate target gene expressions among subtypes.In this study, we identify the potential interactions (ERα, STAT3), (MYC, STAT3) and (ERα, MYC, STAT3) in relation to poor prognostic value of STAT3 in an ER(+) breast cancer model system A relatively small gene pool in the STAT3 network is regulated by ERα in ER(+) BCs (Tables S2.3 and S2.4 in Suppl. 2 of Additional file 1) comparing the total probe numbers in the STAT3 network that interact with MYC (13,712 probes) and ESR1 (12,146 probes; Tables S2.5 and S2.6 in Suppl. 2 of Additional file 1). This suggests that STAT3 is a master TF in addition to ESR1 and MYC in ER(+) breast cancers. The multivariate space of the STAT3 network predicts that both ERαnSTAT3 and MYCnSTAT3 promoter pathways may cooperatively suppress the METAP2 mediated proliferative activities in luminal A subtype via down-regulating the METAP2 expression. For example, STAT3 is predicted as the major transcription regulator of METAP2 via active involvement in these promoter use pathways that are functionally significant in the majority of luminal A subtypes. Conversely, ERα is predicted to principally promote proliferative activities by up-regulating METAP2 expression in the luminal B subtype where STAT3 and MYC are expressed at low levels.Western blot analysis of METAP2 (p67) in a luminal B breast cancer cell model-MCF-7 suggests it is an estrogen responsive gene. This is supported by the demonstration of differential METAP2 (p67) expression in estrogen and/or antiestrogen treated MCF-7 (Fig. 5A). Estrogen induces and antiestrogen suppresses METAP2 (p67) expression. Although no published promoter sequence for human METAP2 is available, we analyzed the rat34 and mouse35 DNA sequences and determined that they contain multiple half ERE sites indicating they are ERα target genes (Figs. 5C and D). Another line of indirect evidence for human METAP2 via CART model 2 prediction, suggests it to be an ERα target gene following an indirect tethering mechanism.36 These data support our network prediction that METAP2 is a component of the multivariate portion of the STAT3 transcriptional regulatory network (ESR1n-STAT3) in ER(+) breast cancers. The regulatory mode of both ESR1 and STAT3 on METAP2 appears to be, in part, due to the expression ratio of ESR1 and STAT3. Further evaluation of the detailed mechanisms involved between ERα and STAT3 in regulating METAP2 will be needed to identify the cause(s) determining tumor development in a luminal A subtype-enriched manner.
-
-
Validation of the major functional STAT3 subnet-works in groups IE, IIE, luminal A and luminal B.
-
Identification of functional components of STAT3 network as potential prognostic markers and therapeutic targets in ER(+) breast cancer. We further evaluated 5 functional STAT3 subnetworks and a FOXC1 subnetwork for their prognostic values. Table 2 shows the results from survival analyses of 4 functional STAT3 subnetworks (Figs. 3B–D), the FOXC1 subnetwork (Fig. 3E) and the genes in the ERBB2 signal transduction pathway (Fig. 7B). These pathways are potentially regulated by the STAT3 network in ER(+) IDCs. We found 9 poor prognostic factors and 5 good prognostic factors of the STAT3 network in the 90 A cohort (Fig. 3F and Table 2). Results for the 72 A cohort are listed in Table 3 and Figure 3F.
The prognostic feature for STAT3 in ER(+) breast cancer was demonstrated by the components of functional STAT3 subnetworks that are also predictor(s) for clinical outcomes (Fig. 3F and Suppl. 6 in Additional file 1). Some were already validated by the literature, including BIRC5,37 IGF2R,38 VEGF,39 LYN,40 OCT4 (POU5F1),41 NANOG,41 GATA342 and NRG1.43
Several drug targets, including VEGFA, IGF2R, ABL1, LYN and STAT3, are in the STAT3 network. Blocking their gene activities may improve the clinical outcome for a subset of ER(+)HER(−) BCs containing elevated STAT3. METAP2 and MELK may be drug targets in a subset of ER(+) HER(−) BCs with low STAT3 expression levels. Importantly, ERBB2 is a STAT3 target gene.44 It is down-regulated by STAT3 in HER(−) BCs based upon data using the network approach. However, STAT3 mRNA is moderately expressed in some of HER(+) BCs, which have moderate ERBB2 mRNA levels regardless of ERBB2 amplification based upon chromogenic in situ hybridization (CISH). Therefore, STAT3 and the oncogenic target genes of the STAT3 network may be candidate drug targets for treating a subset of HER(+) patients.
-
-
Dynamic changes of network activities mediated by STAT3 or by STAT3 and MYC in 90 A cohort predominantly show in luminal A subtype.
Figures 3, 4, 7 and 9 identify the STAT3 subnetworks involved in the ERBB2 signal transduction pathway, proliferation, sustained angiogenesis, Warburg effect, ES-like phenotype, and prognosis in ER(+) breast cancers. Gene expression patterns of 90 A and 72 A cohorts were compared in Figures 3, 4 and 9 and comparisons between ER(−) BCs (77 A cohort) and ER(+) BCs (72 A cohort) are in Figure 7.
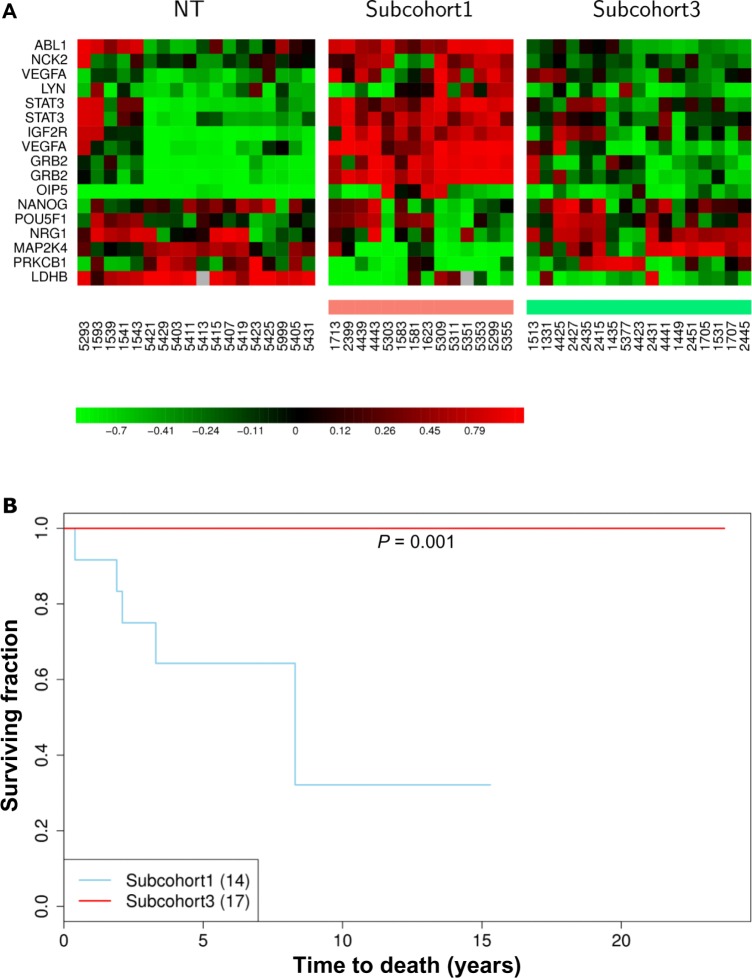
Heatmaps (Fig. 6A) show the dynamic transcriptional activities of MYC and STAT3 conditioning sustained angiogenesis in a luminal A relevant manner. Luminal A subtype locates in a large portion of group IE and a small portion of group IIE. 16 sonographic imaging results for groups IE and IIE are presented. The sonograms in Figures 6B and C with the corresponding heatmaps show partial validation in vivo for a subnetwork of sustained angiogenesis regulated by STAT3. We found that 56.25% (9/16) of sonograms match with the heatmap display. Results in heatmaps that do not match (IDs 5383, 1667, 5315, 5389, 1687, 1643 and 5377) indicate the presence of false negatives (see potential causes described in I_B_a_iii of the “Results and Discussion” section).
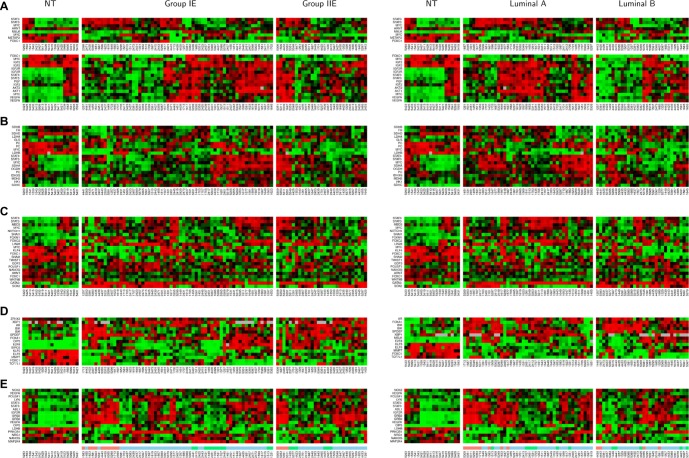
Heatmaps in Figure 4 of network activities following gene expression show small variations in cell proliferation (Fig. 4A), and the Warburg effect (Fig. 4B) among subtypes but are enriched in luminal A. The FOXC1 subnetwork (Fig. 4D) shows increased transcriptional dynamics between non-tumor and tumor components as opposed to among tumor subtypes. However, the heatmaps for the ES-like phenotype (Fig. 4C) in the ER(+) breast cancer population scattered with little dynamic change in gene expression patterns among subtypes. Heatmaps in Figure 4E show that approximately one third of the 90 A population (designated as subcohorts 1 and 3) have a distinct pattern of dynamic changes in a prognostic signature. To elucidate activities of this STAT3 subnetwork in relation to prognostic features of STAT3, we divided ER(+)IDCs (90 A cohort) into three subgroups based on the differential expression patterns of genes within the prognostic signature predicted to be controlled by STAT3 (Fig. 4E).
Here, we suggest this 15 gene signature to be different from other published signatures. First, each functional transcription factor (e.g. STAT3) has its own transcriptional mechanisms predicted by network analysis. The target gene activities of STAT3 may also be regulated by other regulators. For instance, both MYC and STAT3 share target genes within the MYC and STAT3 transcriptional regulatory network. Network analysis allows dissection of STAT3 mediated transcriptional activities, although we only analyzed half a genome due to the microarray limitations. The most relevant STAT3 transcriptional regulatory network is predicted in a breast cancer model system that has a relatively small N number for 8 molecular subtypes. Second, the network analysis is a qualitative method. We observed a variable expression pattern of STAT3 target genes. As a result, the heatmaps (Fig. 9) show the transcriptional dynamics for the prognostic signature ruled by STAT3 only between subcohorts 1 and 3. Kaplan-Meier survival analysis predicts a poor prognostic feature in subcohort 1 (P = 0.001) as compared to subcohort 3. Table S6.1 shows only univariate COXPH analysis of subcohort 1/non 1 to be significant but not those of subcohort 1/3, subcohort 2/3, subcohort 1/2, subcohort 2/non 2, subcohort 3/non 3 and nine major traditional prognostic factors. Additionally, no multivariate COXPH analysis of tested prognostic factors shows significance.There are 15 prognosis predictors identified within the STAT3 subnetwork of the 90 A cohort. Dynamic changing in transcriptional activities of STAT3 for the 15 probes between two tumor sample populations (subcohorts 1 and 3) suggest that STAT3 may differentially regulate the consensus gene cluster that promotes tumor activities, drug resistance, and conditions poor prognosis in subcohort 1 (Fig. 9). Importantly, subcohorts 1 and 3 do not show a dramatic change in the expression pattern of the most relevant STAT3 prognostic subnetwork but are a robust poor prognostic signature. This signature has altered expression patterns of POU5F1, OIP5 and NRG1 compared to the established prognostic subnetwork (Fig. 3F). Such deviation can be due to the difference in each gene expression data distribution within a given cohort. The results from the COXPH model indicate that the 15 gene signature is not prognostic relevant when comparison is made between 2 subsets of ER(+) IDCs that show differential gene expression pattern of this gene signature (i.e. subcohort 1/3). However, there is a significant difference relative to early tumor development with the 15 gene signature versus other expression patterns (i.e. subcohort 1/non 1) in the 90 A cohort. These results suggest that this signature alone may have importance to identify the subset of ER(+) breast tumors that have a poor prognosis. However, based upon the multivariate COXPH model in the 90 A cohort, neither the tested prognostic factors nor the 15 gene signature in the 90 A cohort are independent prognostic factors.The heterogeneity of breast cancers and the unique regulatory mechanisms of STAT3 can be dissected, in part, by network analysis. Kaplan-Meier and COXPH analyses of the 15 gene signature suggest that it may be an unfavorable prognostic factor. However, further investigations of a large population is necessary to establish prognostic value of this gene signature.
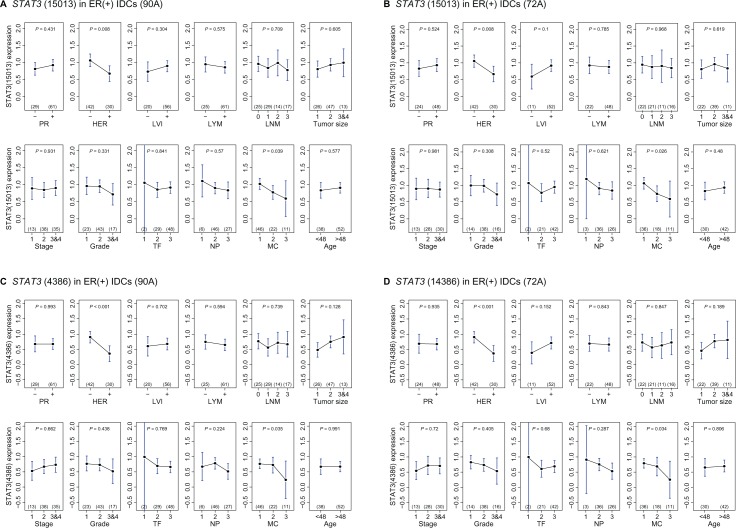
Figure 1.
Clinical impact of STAT3 in two cohorts of ER(+) IDCs analyzed by ANOVA tests.
Upper panel, ANOVA test results of STAT3 (15013) mRNA levels in 8 clinical indices—progesterone receptor (PR), HER-2/neu (HER), lymphovascular invasion (LVI), lymph nodal category (lymph node metastasis status-LYM, No. of lymph node metastasis-LNM), age, tumor size, grade (nuclear pleomorphism (NP), mitotic count (MC), tubule formation (TF)) and cancer stage in ER(+) IDCs (90 A; 72 A) (A and B).
Lower panel, ANOVA test results of STAT3 (4836) mRNA levels in 8 clinical indices in ER(+) IDCs (90 A; 72 A) (C and D). 15013 and 4836 are the Agilent feature number for STAT3, a STAT3 variant respectively. NP, MC and TF are 3 clinical subindices of the histological grade (grade).
Figure 2.
ANOVA tests and Venn diagram analyses for finding the main components of the STAT3 network.
(A) shows a gene pool called STAT3 cluster, which is significantly associated with mitotic count (41TFs including STAT3 and ARNT). (B) demonstrates the gene pools, which include (1) the MYC and STAT3 overlapping network of the ER(+) IDCs with clinicopathologically significant (CS) and luminal A enriched and (2) the non-overlapping gene pools. (C) is a bar chart, which contains 10 bars for the number of probes identified in 10 clinical parameters and in the CS and luminal A enriched MYC and STAT3 overlapping network (6,579 probes), with bars displayed in an ascending order. (D) is a bar chart, which contains 12 bars for the number of probes identified in 12 signal transduction pathways that are also in the MYC and STAT3 overlapping network of the ER(+) IDCs, with bars displayed in an ascending order.
Figure 3.
Functional prediction on roles of STAT3 gene partners in STAT3 subnetworks of ER(+) IDCs.
6 functional subnetworks in 90 A and 72 A cohorts are generated via (1) Predicted networks derived from overlapping genes in Venn diagrams of the STAT3 subnetworks in ER(+) breast cancer gene expression profiles from a 90 A cohort (A); (2) 4 feature functionalities (cell proliferation, sustained angiogenesis, the Warburg effect and ES-like phenotype) of the major STAT3 target genes in 2 STAT3 subnetworks are either commonly co-regulated by MYC (C) and/or differentially co-regulated by FOXC1 and/or ARNT (B and D) in the ER(+) IDCs. A subset of genes, which are predicted to be prognostic factors, is potentially regulated by multiple combined routes of MYC and STAT3, ARNT/HIF1α and STAT3, ARNT/HIF2α and STAT3 or STAT3 for ES-like phenotype (D). E stands for the FOXC1 subnetwork to be a part of activities in cell proliferation. The summary for prognostic features of the STAT3 subnetworks in 90 A cohort and 72 A cohort are shown in F. Solid/dashed lines stand for the specific pathway identified as significant/insignificant in gene expression relationship between a TF and a target gene. Each arrow points toward its downstream target. The combined routes toward the same target gene are labeled with the same color. Relative mRNA expression levels are shown in a color scale (A). Poor prognostic factors are marked by the red rings. Good prognostic factors are marked by the light blue rings. If a probe is not significant shown by Kaplan-Meier survival analysis, it is marked with dotted ring.
Table 2.
The prognostic values for inferred target genes of STAT3 and MYC in the 90 A cohort.
| Gene symbol (feature no.) | Increased expression level | P value | Pathways |
|---|---|---|---|
| ABL1 (8019) | Poor prognosis | 0.046 | ERBB2, PDGFRB, cell cycle and angiogenesis |
| IGF2R (1723) | Poor prognosis | 0.048 | Angiogenesis |
| PRKCB1 (6676) | Good prognosis | 0.033 | ERBB2 and VEGF |
| MAP2K4 (18954) | Good prognosis | 0.034 | ERBB2 |
| NRG1 (11559) | Good prognosis | 0.011 | ERBB2 |
| LYN (19236) | Poor prognosis | 0.001 | PDGFRB and angiogenesis |
| STAT3 (4386) | Poor prognosis | 0.028 | PDGFRB and angiogenesis |
| STAT3 (15013) | Poor prognosis | 0.002 | PDGFRB and angiogenesis |
| VEGFA (1135) | Poor prognosis | 0.02 | VEGF and angiogenesis |
| VEGFA (15367) | Poor prognosis | 0.008 | VEGF and angiogenesis |
| OIP5 (16433) | Poor prognosis | 0.013 | FOXC1 network |
| NCK2 (3851) | Poor prognosis | 0.029 | ERBB2 and PDGFRB |
| LDHB (20259) | Poor prognosis | 0.038 | Warburg effect |
| GRB2 (16731) | Poor prognosis | 0.019 | ERBB2 and PDGFRB |
| NANOG (C12928.2) | Poor prognosis | 0.024 | EMT |
| POU5F1 (6057) | Good prognosis | 0.02 | EMT |
| GRB2 (1952) | Poor prognosis | 0.03 | ERBB2 and PDGFRB |
Table 3.
The prognostic values for inferred target genes of STAT3 and MYC in the 72 A cohort.
| Gene symbol | Regulation status | Biochemical pathway | Prognosis | Regulators |
|---|---|---|---|---|
| GRB2 | Up | ERBB2 and PDGFRB | Poor | MYC and STAT3 |
| GRB2 | Up | ERBB2 and PDGFRB | Poor | MYC and STAT3 |
| CDKN1A | Up | p53, cell cycle and ERBB2 | Poor | MYC and STAT3 |
| ARAF | Up | ERBB2 | Poor | MYC and STAT3 |
| NCK2 | Up | ERBB2 and PDGFRB | Poor | MYC and STAT3 |
| PAK6 | Up | ERBB2 | Good | MYC and STAT3 |
| KRAS | Up | ERBB2 and VEGF | Good | MYC and STAT3 |
| IDH3G | Up | Warburg effect | Good | MYC and STAT3 |
| ELK1 | Up | FOXC1 network and ERBB2 | Good | MYC and STAT3 |
| NANOG | Down | ES like phenotype (EMT) | Good | MYC and STAT3 |
| NRAS | Up | ERBB2 | Poor | MYC and STAT3 |
| SRC | Up | ERBB2, VEGF and PDGFRB | Poor | MYC and STAT3 |
Figure 4.
Heatmaps for the subnetworks of MYC and STAT3 in different subtypes of the ER(+) IDCs. Non-tumor components (NT) serve as the controls. Left panel shows the heatmaps for 90 A cohort, which were generated from 61 group IE, and 29 group IIE breast cancer subtypes. Right panel shows the heatmaps for 72 A cohort which were generated from 42 luminal A and 32 luminal B breast cancer subtypes. The hierarchically clustered gene expression patterns were based on the similar expression levels among genes in the subnetworks of four altered biological events—cell proliferation (upper panel of A), sustained angiogenesis (lower panel of A), Warburg effect (B) and ES-like phenotype (C). A FOXC1 subnetwork (D) contains a gene list to be a part of the FOXC1 subnetwork in the ER(−) IDCs.9 4E stands for heatmaps of 2 cohorts (90 A, 72 A) for the prognostic factors (17 probes) identified in the STAT3 subnetworks (4A–D and Fig. 7B) of 90 A cohort. We located 3 subcohorts based on their similarity in gene expression patterns for a prognosis signature (17 probes) indicated by feature color bars underneath of the heatmap for Figure 4E. Light red color bar stands for subcohort 1 (N = 14). Light green color bar stands for subcohort 3 (N = 17). Light blue color bar stands for subcohort 2 (N = 59).
Figure 5.
In vitro validation of an ERα target gene—METAP2 (p67).
The upper left panel shows the results of western blot analysis on protein expression levels of METAP2 (p67) in MCF-7 cell model (A).
Western blot analysis for METAP2 encoded protein indicates it to be regulated by ERα. We found increased p67 protein in MCF-7 with E2 treatment as compared to that with fulvestrant (ICI 182, 780 or ICI) treatment, (ICI + E2) treatment and control. MCF-7 cells were deprived of estrogen for 2 days and treated with 10−9 M E2 (labeled as + E2), 10−7 M ICI183,780 (labeled as + ICI183,780), or a combination of both (ICI + E2) for 48 hours. Total lysate (60 μg/lane) from MCF-7 cells was resolved in 7.5% SDS-PAGE and immunoblotted with anti-rat p67. β-actin was as the loading control. The lower blot was probed with anti-β-actin. The upper right panel shows that a diagram of the network prediction for interaction between ERα and STAT3 results in a switch in expression mode of their potential target gene-METAP2, which is predicted to be subtype relevant in ER(+) IDCs (B).
Moreover, METAP2 is predicted to be shared target genes due to the combinatorial interaction of 2 given transcription factors (see the overlapping network of MYCnSTAT3 and ESR1nSTAT3 in Table S2.4 of Suppl. 2) but it is neither in the overlapping network of ESR1 and STAT3 nor in that of MYC and STAT3 (Table S2.6 in Suppl. 2). Based on the network analysis results, the proposed interplay between promoter use pathways of ESR1 nSTAT3 and MYCnSTAT3 in luminal A and B in regulating METAP2 is proposed (B).
The lower left panel demonstrates that DNA sequence of promoter region for rat METAP2 (p67) (GenBank: U37710) includes 5 3′ ERE half-sites45 and a 5′Am2Tp2 variant site46 (C). This indicates rat METAP2 to be a target gene of ERα due to the half-ERE sites to be the candidate binding sites of ERα.
The lower right panel demonstrates that DNA sequence of promoter region for mouse METAP2 (p67) includes two 5′ ERE half-sites and two 3′ ERE half-sites (D). This indicates mouse METAP2 to be a target gene of ERα due to the half-ERE sites to be the candidate binding sites of ERα.
Figure 7.
Heatmaps for the subnetworks of MYC and STAT3 and the proposed TAM resistance mechanism.
We described the levels of gene expression for both ERα and STAT3 by coloring with green for low and blue for high. The thickness of line indicates the degree of activities that are predicted to depend on the expression level of ERα when STAT3 is elevated in triple negatives and ER(+) infiltrating ductal breast carcinomas.
ERα is weakly expressed in triple negatives. Thus, it is possible that crosstalk between ERα and STAT3 in TN is relatively weak. To this subset of patients, TAM treatment is not applied as one of the cancer therapies. On the other hand, the high possibility of TAM resistance in a subset of luminal A is proposed. The proposed mechanism of TAM resistance due to crosstalk between ERα and STAT3 has been described in the main text. Figures 7A and B show the gene profilings of ERBB2 signaling molecules predicted to be regulated by STAT3 in coupling with MYC in triple negatives/ERBB2+, groups IE/IIE, respectively. Two corresponding non-tumor (NT) components are as the control gene profilings in this case (A and B).
Both MYC and STAT3 differentially up-regulate the mRNA expression for a subset of ERBB2 signaling molecules among and/or within subtypes (A and B). The estrogen action on crosstalk between ERα and STAT3 is mainly seen in luminal A (see arrows with thick lines colored with orange or dark blue in Fig. 7D) but is weak in TN (see arrows with thin lines colored with orange or dark blue in Fig. 7C). We describe phosphorylation at serine/threonine residues (The p is high-lighted by orange color), tyrosine residues (The p is high-lighted by yellow color) and the mixed types (The p is not high-lighted). (C and D) are summarized from current review articles10,18 with a few modifications derived from our findings. In Figures 7C and D, the example of the most relevant target genes of STAT3 and/or ERα are from our findings in Tables S1.4, S2.1, S2.3, S2.4 and S2.6.
Figure 6.
Further evaluation on a gene set predicted to be involved in tumor angiogenesis.
Upper panel, 2 heatmaps are displayed for the overlapped subnetworks of MYC and STAT3 differentially coupling with ARNT/HIF1α and/or ARNT/HIF2α in two subtypes of breast cancer (group IE and IIE) (A). Non-tumor components (NT) are the controls.
Lower panel, their related clinicopathological phenotype—vascularity (B and C) are demonstrated.
Figure 9.
STAT3 regulated unfavorable prognosis signature in a subset of ER(+) IDCs (subcohort 1).
Upper panel shows the heatmaps of a prognosis relevant gene set in 3 sample groups—NT, subcohorts 1 and 3 (A).
Subcohort 1 has the STAT3 subnetwork (total 17 probes) that is predicted to be activated due to high levels of STAT3. Subcohort 3 has the STAT3 subnetwork (total 17 probes) that is predicted to be suppressed due to low levels of STAT3. NT stands for non-tumor components. Light red color bar stands for subcohort 1 (see Fig. 7E). Light green color bar stands for subcohort 3 (see Fig. 7E). The functional annotation of 17 probes (Y axis) and clinical array IDs (X axis) are displayed along with the heatmaps generated by unsupervised clustering (A).
Lower panel shows the significant difference in the clinical outcome of subcohort 1 as compared to that of subcohort 3.
Kaplan-Meier curves estimate the association between 2 subcohorts and their overall survival probabilities (see method). To correlate clinical outcomes, we calculated the probability of “cancer specific overall survival” in 2 subcohorts (1 and 3) in the ER(+) infiltrating ductal carcinomas. Overall survival was defined as the time interval between the first date of breast tumor surgery and the last follow-up date or date of death. The numerical number within the parentheses next to each subcohort means the total patient number in each subcohort.
Conclusions
The prognostic value of STAT3 in an ER(+) breast cancer cohort model (90 A cohort) is unfavorable. This is indicated by a prognosis relevant gene signature within the STAT3 network, which is also relevant to the development of malignant phenotypes and biochemical responses in an ER(+) breast cancer population enriched in luminal A subtype.
In this study, we have successfully dissected the functional transcriptome, which includes statistically identified STAT3 target genes, to establish poor prognostic features of STAT3 in a subtype enriched breast cancer population. We conclude the most relevant mechanisms contributing to the prognosis and/or subtype relevant features of STAT3 in ER(+) breast cancers are associated with multiple activities mainly crosstalk between and among (STAT3, ERα), (STAT3, MYC), (STAT3, MYC, ERα) and the action of STAT3.
2 novel findings from network analysis establish the mechanisms that support an interpretation for a STAT3 role in treatment response and in ER(+) breast cancer development. First, tamoxifen resistance is enhanced by crosstalk between ERα and STAT3. ERα plays a major role in activating STAT3 via the non-genomic pathway. Activated STAT3 is predicted to preferentially up-regulate genes coding for enzymes and adaptors shared by many key oncogenic signal transduction pathways including MAPK/ERK signaling. As a result, this may enhance ER(+) tumor resistance to 4-hydroxyl tamoxifen treatment predominantly in the luminal A subtype. Second, the competitive regulatory mode between ERα and STAT3 that differentially regulates shared target genes is subtype enriched. For instance, METAP2 is up-regulated in luminal B and down-regulated in luminal A via the subtype relevant promoter pathways (Fig. 5C). Importantly, we have validated this transcriptional regulatory mechanism via the combinatorial interaction between ERα and STAT3 in regulating METAP2 (p67) protein expression in a luminal B breast cancer cell model.
Another major control mechanism is triggered by crosstalk between STAT3 and MYC. We demonstrate that the overlapping network of MYC and STAT3, identified in ER(+) breast cancers, involves 4 malignant phenotypes: proliferation, sustained angiogenesis, ES-like phenotype and the Warburg effect. In addition, we identify network genes that overlap with genes contributing to the development of 10 clinical parameters and 12 cancer related signal transduction pathways. However, these gene expression patterns are less aggressive than those in ER(−) BCs. This may, in part, be due to different pre-programmed TF partners of STAT3 that differentially determine tumor cell fate between ER(+) and ER(−) subtypes.
Additionally, 3 important findings identify STAT3 as a central regulator in ER(+) BCs, including its action contributing to the prognosis and/or subtype relevant features of STAT3 in ER(+) breast cancers. They are (1) the major clinically relevant STAT3 partner is MYC and more than 100 TF partners are the components of STAT3 network; (2) high expression of NANOG or RAF1 in the STAT3 network may cause chemodrug resistance and elevated levels of CCND1 by STAT3 regulation may cause TAM resistance, which have been supported by numerous in vitro studies;34–36 (3) the prognostic relevant gene set, which is also the target gene set of STAT3, is poor clinical outcome predictor in 2 ER(+) breast cancer subcohorts. It contains relatively high levels of GRB2, LYN, IGF2R, VEGFA, STAT3, NCK2, OIP5 and ABL1 and low levels of MAP2K4, PRKCB, POU5F1 and NRG1, indicating poor prognosis. Low levels of LDHB and NANOG predict good prognosis. Several potential therapeutic targets have been identified (e.g. VEGFA, STAT3, ABL1, IGF2R and LYN) within this prognostic relevant gene set.
Supplementary Data
A PDF file contains table of contents and suppls. 1–7. Suppls. 1–7 contain results of Venn diagram analyses, annotated gene lists from Gene Spring GX7.3.1 (March, 2011), heatmaps, results of survival analyses and results of ANOVA tests for genes in the STAT3 subnetworks in this study.
Table of contents—a PDF file contains a brief description for supplementary files.
Supplementary file 1—a PDF file contains Tables S1.1–S1.11.
Supplementary file 2—a PDF file contains Tables S 2.1–S2.7.
Supplementary file 3—a PDF file contains Tables S3.1–S3.10.
Supplementary file 4—a PDF file contains Tables S4.1–S4.15.
Supplementary file 5—a PDF file contains Figures S5.1–S5.11.
Supplementary file 6—a PDF file contains Figures S6.1–S6.3 and Table S6.1.
Supplementary file 7—a PDF file contains Figures S7.1–S7.5 and Table S7.1.
Suppls.1–4 are summarized gene pools in four combined Tables starting with sets of their corresponding Venn diagrams. Some of gene symbols (~100) are left as “blank” within Tables based on annotation provided by Gene Spring GX7.3.1(March, 2011). The gene symbols high-lighted with light blue are either transcription factors or transcription factor subunits. Some of gene symbols high-lighted with light yellow are molecules of interest. We only mark “Y” in each table that means the gene symbol to be one of components listed in the analysis of interest.
Suppl. 5 contains heatmaps for the common gene pools shared by both 11 signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A), respectively.
Suppl. 6 has the prognostic value(s) of component(s) in the STAT3 subnetworks (Figure S6.2) and not in the STAT3 subnetworks (Figure S6.3) in 90A (Groups IE and IIE) and 72A (Luminal A and Luminal B), respectively. Two controls in survival analyses are made for pair-wise comparisons between subtypes used in this study (see Figure S6.1). Additional Survival analyses are provided (Table S6.1).
Suppl. 7 contains ANOVA test results on genes of interest in 90A and 72A, respectively. They are presented by ten mean plots and a table.
Table S 1.1. Clinically significant STAT3 transcriptional regulatory network for mitotic count parameter in 90 ER(+) infiltrating ductal carcinomas (IDCs).
Table S1.2. The gene pool for the MYC transcriptional regulatory network relevant to clinical mitotic count parameter in ER(+)IDCs (i.e. 90A).
Table S1.3. The gene pool for the overlapped network of MYC&STAT3 in ER(+)IDCs (i.e. 90A) relevant to clinical mitotic count parameter.
Table S1.4. The gene pool for the STAT3 transcriptional regulatory network in ER(+) IDCs (i.e. 90A).
Table S1.5. The gene pool for the MYC transcriptional regulatory network in ER(+) IDCs (i.e. 90A).
Table S1.6. The common gene pool for the overlapped network of MYC&STAT3 in ER(+)IDCs (i.e. 90A).
Table S1.7. The shared gene pool between the overlapped network of MYC&STAT3 relevant to ER(+) IDCs and that significant to clinical mitotic count parameter.
Table S1.8. The gene pool for the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 72A).
Table S1.9. The shared gene pool between overlapped networks of MYC&STAT3 in 90A and 72A and their non-overlapping gene pools.
Table S1.10. The FOXC1 transcriptional regulatory network in 90 ER(+) infiltrating ductal carcinomas.
Table S1.11. The FOXC1 transcriptional regulatory network in 72 ER(+) infiltrating ductal carcinomas.
Table S2.1. The shared gene pool between the overlapped networks of MYC&STAT3 in 90A (for ER(+) IDCs) and 77A (for ER(−) IDCs) and their non-overlapping gene pools. The gene pool shared by two univariate overlapped networks of MYC and STAT3 in ER(+) and ER(−) IDCs indicating different percentages of shared network activities for MYC and STAT3 in two subtypes (81.93% (77A) versus 45.81% (90A)), respectively. A cohort (77A) consisting of 48 triple negatives (TN) and 29 ERBB2+was used (see reference 9 in the main text).
Table S2.2. The shared gene pool between networks of MYCnSTAT3 in 90A (for ER(+) IDCs) and 77A (for ER(−) IDCs) and their non-overlapping gene pools. The gene pool shared by two networks of MYCnSTAT3 in ER(+) and ER(−) IDCs indicating different percentages of shared network activities for MYC and STAT3 in two subtypes (73.14% (77A) versus 45.75% (90A)), respectively.
Table S2.3. The common gene pool overlapped by networks of ESR1, MYC and STAT3 in ER(+) IDCs (i.e. 90A).
Table S2.4. The common gene pool overlapped by networks of ESR1n STAT3 and MYCnSTAT3 in ER(+) IDCs (i.e. 90A).
Table S2.5. The common gene pool overlapped by networks of MYC & STAT3 amd MYCnSTAT3 in ER(+) IDCs (i.e. 90A).
Table S2.6. The common gene pool overlapped by networks of ESR1 & STAT3 and ESR1nSTAT3 in ER(+) IDCs (i.e. 90A).
Table S2.7. Venn diagrams and their overlapping and non-overlapping gene pools for both networks of ESR1 and MYC in ER(+) IDCs (i.e. 90A). The common gene pool overlapped by networks of ESR1 & MYC(S2.7a), the potential ESR1 regulated gene pool but not in MYC network (S2.7b) and the potential MYC regulated gene pool but not in ESR1 network (S2.7c) are listed.
Table S3.1. The common gene pool between age-related gene pool in ER(+) IDCs (i.e.90A) and the overlapped network of MYC&STAT3 not only relevant to ER(+) IDCs but to clinical mitotic count parameter.
Table S3.2. The common gene pool between grade-related gene pool in ER(+) IDCs (i.e. 90A) and the overlapped network of MYC&STAT3 not only relevant to ER(+) IDCs but to clinical mitotic count parameter.
Table S3.3. The common gene pool between LNM-related gene pool in ER(+) IDCs (i.e. 90A) and the overlapped network of MYC&STAT3 not only relevant to ER(+) IDCs but to clinical mitotic count parameter.
Table S3.4. The common gene pool between LVI-related gene pool in ER(+) IDCs (i.e. 90A) and the overlapped network of MYC&STAT3 not only relevant to ER(+) IDCs but to clinical mitotic count parameter.
Table S3.5. The common gene pool between LYM-related gene pool in ER(+) IDCs (i.e. 90A) and the overlapped network of MYC&STAT3 not only relevant to ER(+) IDCs but to clinical mitotic count parameter.
Table S3.6. The common gene pool between MC-related gene pool in ER(+) IDCs (i.e. 90A) and the overlapped network of MYC&STAT3 not only relevant to ER(+) IDCs but to clinical mitotic count parameter.
Table S3.7. The common gene pool between NP-related gene pool in ER(+) IDCs (i.e. 90A) and the overlapped network of MYC&STAT3 not only relevant to ER(+) IDCs but to clinical mitotic count parameter.
Table S3.8. The common gene pool between size-related gene pool in ER(+) IDCs (i.e. 90A) and the overlapped network of MYC&STAT3 not only relevant to ER(+) IDCs but to clinical mitotic count parameter.
Table S3.9. The common gene pool between stage-related gene pool in ER(+) IDCs (i.e. 90A) and the overlapped network of MYC&STAT3 not only relevant to ER(+) IDCs but to clinical mitotic count parameter.
Table S3.10. The common gene pool between TF-related gene pool in ER(+) IDCs (i.e. 90A) and the overlapped network of MYC&STAT3 not only relevant to ER(+) IDCs but to clinical mitotic count parameter.
Table S4.1. The common gene pool in both cell cycle signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Table S4.2. The common gene pool in both VEGF signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Table S4.3. The common gene pool in both BER signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Table S4.4. The common gene pool in both DRS signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Table S4.5. The common gene pool in both HR signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Table S4.6. The common gene pool in both MRP signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Table S4.7. The common gene pool in both NER signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Table S4.8. The common gene pool in p53 signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Table S4.9. The common gene pool in both proteasome signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Table S4.10. The common gene pool in both ribosome signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Table S4.11. The common gene pool in both PDGFRB signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Table S4.12. The common gene pool in both ERBB2 signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Table S4.13. The common gene pool shared between the multivariate space of STAT3 network (MYCnSTAT3) and ERBB2 signal transduction pathway in ER(+) IDCs (i.e. 90A).
Table S4.14. The common gene pool in both ERBB2 signal transduction pathway and the MYC & STAT3 overlapped network in ER(−) IDCs (i.e. 77A). ER(−) IDCs contain 48 triple negatives (TN) and 29 ERBB2+.
Table S4.15. The common gene pool shared between the MYCnSTAT3 network and ERBB2 signal transduction pathway in ER(−) IDCs (i.e. 77A).
Figure S5.1. Heatmaps for the common gene pool in both cell cycle signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Figure S5.2. Heatmaps for the common gene pool in both VEGF signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Figure S5.3. Heatmaps for the common gene pool in both BER signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Figure S5.4. Heatmaps for the common gene pool in both DRS signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A)
Figure S5.5. Heatmaps for the common gene pool in both HR signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Figure S5.6. Heatmaps for the common gene pool in both MRP signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Figure S5.7. Heatmaps for the common gene pool in both NER signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Figure S5.8. Heatmaps for the common gene pool in both p53 signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Figure S5.9. Heatmaps for the common gene pool in both proteasome signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Figure S5.10. Heatmaps for the common gene pool in both ribosome signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A). Grey color within the heatmap stands for data not available after data processing.
In this case, we do not use this piece of information for further discussion due to most data not available.
Figure S5.11. Heatmaps for the common gene pool in both PDGFRB signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Figure S6.1. The survival analyses on subtypes of ER(+) infiltrating ductal carcinoma in this study. Figure S6.1a. stands for survival analysis on Group IE vs. Group IIE; Figure S6.1b. stands for survival analysis on Luminal A vs. Luminal B. Both results are not significant.
Figure S6.2. The survival analyses on probes in the STAT3 driven subnetworks in coupling with its major transcription factor partner - MYC for (a) proliferation; (b) sustained angiogenesis; (c) Warburg effect; (d) ES-like phenotype; (e) FOXC1 transcriptional regulatory network and (f) ERBB2 signaling in ER(+) IDCs. We compared the probes in the subnetworks in both 90A and 72A for predicting clinical outcome that was evaluated by overall disease free survival. Their results are summarized in Figure 3F (please see main text).
Figure S6.3. The survival analyses on a probe –FOXC1(12715) not in the STAT3 subnetwork. We compared this probe in the subnetworks in both 90A and 72A for predicting clinical outcome that was evaluated by overall disease free survival. It is a significant favorable prognostic factor in 90A cohort but not in 72A cohort.
Table S6.1. Univariate and multivariate analyses for survival on prognostic factors in 90A and 181A cohorts, respectively. The p values of tests in the Cox proportional hazard model to be less or equal to 0.05 are high-lighted with light blue. Both univariate and multivariate analyses are negative except univariate COXPH analysis on subcohort 1 versus non subcohort1.
Figure S7.1 Mean plot analyses of MELK mRNA levels in eight clinical categories of two cohorts of infiltrating ductal carcinomas (IDCs).
Lymphovascular invasion (LVI), nodal category (lymph node metastasis (LYM), number of nodal metastasis (LNM)), histological grade (Grade) category (nuclear pleomorphism (NP) and tubule formation (TF)) and stage were analyzed. Cohort 1 (90IDCs) has Groups IE and IIE. Cohort 2 (72A) has Luminal A and Luminal B (see the main text for definitions).
Figure S7.2. Mean plot analyses of METAP2 mRNA levels in eight clinical categories of two cohorts of infiltrating ductal carcinomas (IDCs).
Figure S7.3. Mean plot analyses of SRC mRNA levels in eight clinical categories of two cohorts of infiltrating ductal carcinomas (IDCs).
Figure S7.4. Mean plot analyses of a transcript variant SRC mRNA levels in eight clinical categories of two cohorts of infiltrating ductal carcinomas (IDCs).
Figure S7.5. Mean plot analyses of OIP5 mRNA levels in eight clinical categories of two cohorts of infiltrating ductal carcinomas (IDCs).
Table S7.1. ANOVA tests on ARNT mRNA levels in eight clinical categories of two cohorts of infiltrating ductal carcinomas (IDCs).
Table 1.
The pathological information (clinicopathological parameters) for group IE, group IIE, luminal A and luminal B.
| Clinical index | Status | Number of patients
|
|||
|---|---|---|---|---|---|
| Grp IE | Grp IIE | Luminal A | Luminal B | ||
| ER | 0 | 0 | 0 | 0 | 0 |
| 1 | 61 | 29 | 42 | 30 | |
| NA | 0 | 0 | 0 | 0 | |
| PR | 0 | 0 | 29 | 17 | 7 |
| 1 | 61 | 0 | 25 | 23 | |
| NA | 0 | 0 | 0 | 0 | |
| HER | 0 | 25 | 17 | 42 | 0 |
| 1 | 23 | 7 | 0 | 30 | |
| NA | 13 | 5 | 0 | 0 | |
| Stage | 1 | 11 | 2 | 5 | 8 |
| 2 | 23 | 15 | 15 | 13 | |
| 3 | 25 | 10 | 21 | 9 | |
| NA | 2 | 2 | 1 | 0 | |
| LYM | 0 | 17 | 8 | 11 | 11 |
| 1 | 41 | 20 | 29 | 19 | |
| NA | 3 | 1 | 2 | 0 | |
| LVI | 0 | 16 | 4 | 4 | 7 |
| 1 | 36 | 20 | 31 | 21 | |
| NA | 9 | 5 | 7 | 2 | |
| Age | 0 | 30 | 8 | 15 | 15 |
| 1 | 31 | 21 | 27 | 15 | |
| NA | 0 | 0 | 0 | 0 | |
| Grade | 1 | 19 | 4 | 11 | 3 |
| 2 | 31 | 12 | 23 | 15 | |
| 3 | 8 | 9 | 6 | 10 | |
| NA | 3 | 4 | 2 | 2 | |
| TF | 1 | 2 | 0 | 2 | 0 |
| 2 | 21 | 8 | 9 | 12 | |
| 3 | 32 | 16 | 26 | 16 | |
| NA | 6 | 5 | 5 | 2 | |
| NP | 1 | 6 | 0 | 3 | 0 |
| 2 | 33 | 13 | 21 | 15 | |
| 3 | 16 | 11 | 13 | 13 | |
| NA | 6 | 5 | 5 | 2 | |
| MC | 1 | 35 | 11 | 26 | 10 |
| 2 | 13 | 9 | 9 | 9 | |
| 3 | 7 | 4 | 2 | 9 | |
| NA | 6 | 5 | 5 | 2 | |
| Size | 1 | 19 | 7 | 10 | 12 |
| 2 | 33 | 14 | 25 | 14 | |
| 3 | 7 | 6 | 7 | 4 | |
| NA | 2 | 2 | 0 | 0 | |
| LNM | 0 | 17 | 8 | 11 | 11 |
| 1 | 18 | 11 | 11 | 10 | |
| 2 | 10 | 4 | 9 | 2 | |
| 3 | 14 | 3 | 9 | 7 | |
| NA | 2 | 3 | 2 | 0 | |
Acknowledgments
Both Dr. Shih-Ming Jung and Dr. Huang-Chun Lien did the scoring for both ERα and PR proteins in tumor sections of breast cancer specimens. Dr. Shiu-Feng Huang did CISH for some tumor samples with ERBB2 (IHC score: 2+). We owe many thanks to the great assistance from the office of medical record (Cancer Registry, Medical Information Management Office, NTUH) for accessing medical records of those patients who agreed on providing their specimens for microarray study. We appreciate Miss. Fu-Chin Chen at NTUH providing the great assistance in gathering breast sonographic images for this study. In addition, we feel thankful to receive the antibody from Dr. Gupta’s group and thanks to Miss Maria Turner for her technical assistance in p67 project.
Glossary
List of Abbreviations
- ABL1
c-abl oncogene 1; non-receptor tyrosine kinase
- ANOVA
analysis of variance
- ARAF
v-raf murine sarcoma 3611 viral oncogene homolog
- ARNT
aryl hydrocarbon receptor nuclear translocator (also known as HIF1B, the beta subunit of a heterodimeric transcription factors; HIF1 and HIF2)
- CDKN1A
cyclin-dependent kinase inhibitor 1 A (p21, Cip1)
- CID
coefficient of intrinsic dependence
- c-Src, v-src
sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian)
- ELK1
member of ETS oncogene family
- EMT
epithelial mesenchymal transition
- ER(+)
estrogen receptor positive
- ERBB2+
ER(−), PR(−) and HER(+)
- ESR1
estrogen receptor 1
- ESRRG
estrogen-related receptor gamma
- FOXC1
forkhead box C1
- FOXC2
forkhead box C2 (MFH-1, mesenchyme forkhead 1)
- GLS
glutaminase
- GPCC
Galton Pearson’s Correlation Coefficient
- GRB2
growth factor receptor-bound protein 2
- HER(−)
v-erb-b2 erythroblastic leukemia viral oncogene homolog 2 negative
- HIF1
hypoxia inducible factor 1
- HIF1α
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor)
- HIF2
hypoxia inducible factor 2
- HIF2α (EPAS1)
hypoxia inducible factor 2, alpha subunit (other designation was endothelial PAS domain protein 1)
- HK1
hexokinase1
- IDH3G
isocitrate dehydrogenase 3 (NAD+) gamma
- IGF2
insulin-like growth factor 2 (somatomedin A)
- IGF2R
insulin-like growth factor 2 receptor
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KLF4
Kruppel-like factor 4 (gut)
- KRAS
v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog
- LDHA
lactate dehydrogenase A
- LDHB
lactate dehydrogenase B
- LYN
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
- MAP2K4
mitogen-activated protein kinase kinase 4
- MELK
maternal embryonic leucine zipper kinase
- METAP2
methionyl aminopeptidase 2
- MMP7
matrix metallopeptidase 7 (matrilysin, uterine)
- MYC
v-myc myelocytomatosis viral oncogene homolog (avian)
- NANOG
homeobox transcription factor Nanog
- NCK2
NCK adaptor protein 2
- NOTCH1
Notch homolog 1, translocation associated (Drosophila)
- NRAS
neuroblastoma RAS viral (v-ras) oncogene homolog
- NRG1
neuregulin 1
- OGDH
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
- OIP5
Opa interacting protein 5
- PAK6
p21 protein (Cdc42/Rac)-activated kinase 6
- PC
pyruvate carboxylase
- PDGFRB
platelet-derived growth factor receptor, beta polypeptide
- POU5F1
POU class 5 homeobox 1
- PR(−)
progesterone receptor negative
- PRKCB(PRKCB1)
protein kinase C, beta (protein kinase C, beta 1 polypeptide)
- SNAI1
snail homolog 1 (Drosophila)
- SNAI2
snail homolog 2 (Drosophila)
- STAT5a
signal transducer and activator of transcription 5 A
- STAT5b
signal transducer and activator of transcription 5B
- TAM
Tamoxifen
- TWIST1
twist homolog 1 (Drosophila)
- VEGFA
vascular endothelial growth factor A
- ZFHX3(ATBF1)
the official name is zinc finger homeobox 3 (other designation was AT motif binding factor 1)
Footnotes
This article is available from http://www.la-press.com.
Author Contributions
Conceived and designed the experiments: LYC. Analysed the data: LYDL, LYC, YSL, MHJ, DR, WHK, HLH. Wrote the first draft of the manuscript: LYC. Contributed to the writing of the manuscript: LYC, DR, MHJ. Agree with manuscript results and conclusions: LYC, DR, FJH, LYDL, MHJ, HLH, WHK, KJC, YSL. Jointly developed the structure and arguments for the paper: LYC, LYDL. Made critical revisions and approved final version: LYC. All authors reviewed and approved of the final manuscript.
Funding
The financial support of this work was mainly from grants (NSC95-2314-B-002-255-MY3 and NSC98-2314-B-002-093-MY2) (to Dr. Fon-Jou Hsieh).
Competing Interests
Author(s) disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication the authors have provided signed confirmation of their compliance with ethical and legal obligations including but not limited to compliance with ICMJE authorship and competing interests guidelines, that the article is neither under consideration for publication nor published elsewhere, of their compliance with legal and ethical guidelines concerning human and animal research participants (if applicable), and that permission has been obtained for reproduction of any copyrighted material. This article was subject to blind, independent, expert peer review. The reviewers reported no competing interests.
References
- 1.Chang LY, Yang YL, Shyu MK, Hwa HL, Hsieh FJ. Strategy for breast cancer screening in Taiwan: Obstetrician-Gynecologists should actively participate in breast cancer screening. J Med Ultrasound. 2012;20:1–7. [Google Scholar]
- 2.Siegel PM, Muller WJ. Transcription factor regulatory networks in mammary epithelial development and tumorigenesis. Oncogene. 2010;29(19):2753–9. doi: 10.1038/onc.2010.43. [DOI] [PubMed] [Google Scholar]
- 3.Watson CJ, Neoh K. The Stat family of transcription factors have diverse roles in mammary gland development. Semin Cell Dev Biol. 2008;19(4):401–6. doi: 10.1016/j.semcdb.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 4.Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–88. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 5.Frank DA. STAT3 as a central mediator of neoplastic cellular transformation. Cancer Lett. 2007;251(2):199–210. doi: 10.1016/j.canlet.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 6.Turkson J, Bowman T, Garcia R, Caldenhoven E, De Groot RP, Jove R. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol Cell Biol. 1998;18(5):2545–52. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsieh FC, Cheng G, Lin J. Evaluation of potential Stat3-regulated genes in human breast cancer. Biochem Biophys Res Commun. 2005;335(2):292–9. doi: 10.1016/j.bbrc.2005.07.075. [DOI] [PubMed] [Google Scholar]
- 8.Sato T, Neilson LM, Peck AR, et al. Signal transducer and activator of transcription-3 and breast cancer prognosis. Am J Cancer Res. 2011;1(3):347–55. [PMC free article] [PubMed] [Google Scholar]
- 9.Liu LY, Chang LY, Kuo WH, et al. Major functional transcriptome of an inferred center regulator of an ER(−) breast cancer model system. Cancer Inform. 2012;11:87–111. doi: 10.4137/CIN.S8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Björnström L, Sjöberg M. Signal transducers and activators of transcription as downstream targets of nongenomic estrogen receptor actions. Mol Endocrinol. 2002;16(10):2202–14. doi: 10.1210/me.2002-0072. [DOI] [PubMed] [Google Scholar]
- 11.Miller TW, Balko JM, Arteaga CL. Phosphatidylinositol 3-kinase and antiestrogen resistance in breast cancer. J Clin Oncol. 2011;29(33):4452–61. doi: 10.1200/JCO.2010.34.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li L, Shaw PE. Elevated activity of STAT3C due to higher DNA binding affinity of phosphotyrosine dimer rather than covalent dimer formation. J Biol Chem. 2006;281(44):33172–81. doi: 10.1074/jbc.M606940200. [DOI] [PubMed] [Google Scholar]
- 13.Timofeeva OA, Chasovskikh S, Lonskaya I, et al. Mechanisms of unphosphorylated STAT3 transcription factor binding to DNA. J Biol Chem. 2012;287(17):14192–200. doi: 10.1074/jbc.M111.323899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greten FR, Karin M. Peering into the aftermath: JAKi rips STAT3 in cancer. Nat Med. 2010;16(10):1085–7. doi: 10.1038/nm1010-1085. [DOI] [PubMed] [Google Scholar]
- 15.Garcia R, Bowman TL, Niu G, et al. Constitutive activation of Stat3 by the Src and JAK tyrosine kinases participates in growth regulation of human breast carcinoma cells. Oncogene. 2001;20(20):2499–513. doi: 10.1038/sj.onc.1204349. [DOI] [PubMed] [Google Scholar]
- 16.Uluer ET, Aydemir I, Inan S, Ozbilgin K, Vatansever HS. Effects of 5-fluorouracil and gemcitabine on a breast cancer cell line (MCF-7) via the JAK/STAT pathway. Acta Histochem. 2012;114(7):641–6. doi: 10.1016/j.acthis.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 17.Kuo PL, Ni WC, Tsai EM, Hsu YL. Dehydrocostuslactone disrupts signal transducers and activators of transcription 3 through up-regulation of suppressor of cytokine signaling in breast cancer cells. Mol Cancer Ther. 2009;8(5):1328–39. doi: 10.1158/1535-7163.MCT-08-0914. [DOI] [PubMed] [Google Scholar]
- 18.Björnström L, Sjöberg M. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol. 2005;19(4):833–42. doi: 10.1210/me.2004-0486. [DOI] [PubMed] [Google Scholar]
- 19.Hart JR, Liao L, Yates JR, Vogt PK. Essential role of Stat3 in PI3K-induced oncogenic transformation. Proc Natl Acad Sci U S A. 2011;108(32):13247–52. doi: 10.1073/pnas.1110486108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuo WH, Chang LY, Liu DL, et al. The interactions between GPR30 and the major biomarkers in infiltrating ductal carcinoma of the breast in an Asian population. Taiwan J Obstet Gynecol. 2007;46(2):135–45. doi: 10.1016/S1028-4559(07)60007-2. [DOI] [PubMed] [Google Scholar]
- 21.Ray PS, Wang J, Qu Y, et al. FOXC1 is a potential prognostic biomarker with functional significance in basal-like breast cancer. Cancer Res. 2010;70(10):3870–6. doi: 10.1158/0008-5472.CAN-09-4120. [DOI] [PubMed] [Google Scholar]
- 22.Liu LY, Chen CY, Chen MJ, et al. Statistical identification of gene association by CID in application of constructing ER regulatory network. BMC Bioinformatics. 2009;10:85. doi: 10.1186/1471-2105-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu LYD, Chang LY, Kuo WH, et al. In silico prediction for regulation of transcription factors on their shared target genes indicates relevant clinical implications in a breast cancer population. Cancer Inform. 2012;11:113–37. doi: 10.4137/CIN.S8470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng G, Tu K, Yang Q, et al. ITFP: an integrated platform of mammalian transcription factors. Bioinformatics. 2008;24(20):2416–7. doi: 10.1093/bioinformatics/btn439. [DOI] [PubMed] [Google Scholar]
- 25.Jeng MH, Shupnik MA, Bender TP, et al. Estrogen receptor expression and function in long-term estrogen-deprived human breast cancer cells. Endocrinology. 1998;139(10):4164–74. doi: 10.1210/endo.139.10.6229. [DOI] [PubMed] [Google Scholar]
- 26.Datta B, Chakrabarti D, Roy AL, Gupta NK. Roles of a 67-kDa polypeptide in reversal of protein synthesis inhibition in heme-deficient reticulocyte lysate. Proc Natl Acad Sci U S A. 1988;85(10):3324–8. doi: 10.1073/pnas.85.10.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L, Shaw PE. Autocrine-mediated activation of STAT3 correlates with cell proliferation in breast carcinoma lines. J Biol Chem. 2002;277(20):17397–405. doi: 10.1074/jbc.M109962200. [DOI] [PubMed] [Google Scholar]
- 28.Sano H, Leboeuf JP, Novitskiy SV, et al. The Foxc2 transcription factor regulates tumor angiogenesis. Biochem Biophys Res Commun. 2010;392(2):201–6. doi: 10.1016/j.bbrc.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Xiao W, Wang L, Tian Z, Zhang J. Deactivation of signal transducer and activator of transcription 3 reverses chemotherapeutics resistance of leukemia cells via down-regulating P-gp. PLoS ONE. 2011;6(6):e20965. doi: 10.1371/journal.pone.0020965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis JM, Navolanic PM, Weinstein-Oppenheimer CR, et al. Raf-1 and Bcl-2 induce distinct and common pathways that contribute to breast cancer drug resistance. Clin Cancer Res. 2003;9(3):1161–70. [PubMed] [Google Scholar]
- 31.Ishii Y, Waxman S, Germain D. Tamoxifen stimulates the growth of cyclin D1-overexpressing breast cancer cells by promoting the activation of signal transducer and activator of transcription 3. Cancer Res. 2008;68(3):852–60. doi: 10.1158/0008-5472.CAN-07-2879. [DOI] [PubMed] [Google Scholar]
- 32.Chatterjee N, Zou C, Osterman JC, Gupta NK. Cloning and characterization of the promoter region of a gene encoding a 67-kDa glycoprotein. J Biol Chem. 1997;272(19):12692–8. doi: 10.1074/jbc.272.19.12692. [DOI] [PubMed] [Google Scholar]
- 33.Jin VX, Leu YW, Liyanarachchi S, et al. Identifying estrogen receptor alpha target genes using integrated computational genomics and chromatin immunoprecipitation microarray. Nucleic Acids Res. 2004;32(22):6627–35. doi: 10.1093/nar/gkh1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu J, Qin K, Zhang Y, et al. Downregulation of transcription factor Oct4 induces an epithelial-to-mesenchymal transition via enhancement of Ca2+ influx in breast cancer cells. Biochem Biophys Res Commun. 2011;411(4):786–91. doi: 10.1016/j.bbrc.2011.07.025. [DOI] [PubMed] [Google Scholar]
- 35.Nakagawa H, Koyanagi S, Takiguchi T, et al. 24-hour oscillation of mouse methionine aminopeptidase2, a regulator of tumor progression, is regulated by clock gene proteins. Cancer Res. 2004;64(22):8328–33. doi: 10.1158/0008-5472.CAN-04-2122. [DOI] [PubMed] [Google Scholar]
- 36.Seo S, Singh HP, Lacal PM, et al. Forkhead box transcription factor FoxC1 preserves corneal transparency by regulating vascular growth. Proc Natl Acad Sci U S A. 2012;109(6):2015–20. doi: 10.1073/pnas.1109540109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Athanassiadou AM, Patsouris E, Tsipis A, Gonidi M, Athanassiadou P. The significance of Survivin and Nectin-4 expression in the prognosis of breast carcinoma. Folia Histochem Cytobiol. 2011;49(1):26–33. doi: 10.5603/fhc.2011.0005. [DOI] [PubMed] [Google Scholar]
- 38.Esseghir S, Reis-Filho JS, Kennedy A, et al. Identification of transmembrane proteins as potential prognostic markers and therapeutic targets in breast cancer by a screen for signal sequence encoding transcripts. J Pathol. 2006;210(4):420–30. doi: 10.1002/path.2071. [DOI] [PubMed] [Google Scholar]
- 39.Gasparini G. Prognostic value of vascular endothelial growth factor in breast cancer. Oncologist. 2000;5(Suppl 1):37–44. doi: 10.1634/theoncologist.5-suppl_1-37. [DOI] [PubMed] [Google Scholar]
- 40.Choi YL, Bocanegra M, Kwon MJ, et al. LYN is a mediator of epithelial-mesenchymal transition and a target of dasatinib in breast cancer. Cancer Res. 2010;70(6):2296–306. doi: 10.1158/0008-5472.CAN-09-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simões BM, Piva M, Iriondo O, et al. Effects of estrogen on the proportion of stem cells in the breast. Breast Cancer Res Treat. 2011;129(1):23–35. doi: 10.1007/s10549-010-1169-4. [DOI] [PubMed] [Google Scholar]
- 42.Yoon NK, Maresh EL, Shen D, et al. Higher levels of GATA3 predict better survival in women with breast cancer. Hum Pathol. 2010;41(12):1794–801. doi: 10.1016/j.humpath.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raj EH, Skinner A, Mahji U, et al. Neuregulin 1-alpha expression in locally advanced breast cancer. Breast. 2001;10(1):41–5. doi: 10.1054/brst.2000.0182. [DOI] [PubMed] [Google Scholar]
- 44.Qian L, Chen L, Shi M, et al. A novel cis-acting element in Her2 promoter regulated by Stat3 in mammary cancer cells. Biochem Biophys Res Commun. 2006;345(2):660–8. doi: 10.1016/j.bbrc.2006.04.153. [DOI] [PubMed] [Google Scholar]
- 45.Shu FJ, Sidell N, Yang D, Kallen CB. The tri-nucleotide spacer sequence between estrogen response element half-sites is conserved and modulates ERalpha-mediated transcriptional responses. J Steroid Biochem Mol Biol. 2010;120(4–5):172–9. doi: 10.1016/j.jsbmb.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deegan BJ, Bhat V, Seldeen KL, McDonald CB, Farooq A. Genetic variations within the ERE motif modulate plasticity and energetics of binding of DNA to the ERα nuclear receptor. Arch Biochem Biophys. 2011;507(2):262–70. doi: 10.1016/j.abb.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file 1—a PDF file contains Tables S1.1–S1.11.
Supplementary file 2—a PDF file contains Tables S 2.1–S2.7.
Supplementary file 3—a PDF file contains Tables S3.1–S3.10.
Supplementary file 4—a PDF file contains Tables S4.1–S4.15.
Supplementary file 5—a PDF file contains Figures S5.1–S5.11.
Supplementary file 6—a PDF file contains Figures S6.1–S6.3 and Table S6.1.
Supplementary file 7—a PDF file contains Figures S7.1–S7.5 and Table S7.1.
Suppls.1–4 are summarized gene pools in four combined Tables starting with sets of their corresponding Venn diagrams. Some of gene symbols (~100) are left as “blank” within Tables based on annotation provided by Gene Spring GX7.3.1(March, 2011). The gene symbols high-lighted with light blue are either transcription factors or transcription factor subunits. Some of gene symbols high-lighted with light yellow are molecules of interest. We only mark “Y” in each table that means the gene symbol to be one of components listed in the analysis of interest.
Suppl. 5 contains heatmaps for the common gene pools shared by both 11 signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A), respectively.
Suppl. 6 has the prognostic value(s) of component(s) in the STAT3 subnetworks (Figure S6.2) and not in the STAT3 subnetworks (Figure S6.3) in 90A (Groups IE and IIE) and 72A (Luminal A and Luminal B), respectively. Two controls in survival analyses are made for pair-wise comparisons between subtypes used in this study (see Figure S6.1). Additional Survival analyses are provided (Table S6.1).
Suppl. 7 contains ANOVA test results on genes of interest in 90A and 72A, respectively. They are presented by ten mean plots and a table.
Table S 1.1. Clinically significant STAT3 transcriptional regulatory network for mitotic count parameter in 90 ER(+) infiltrating ductal carcinomas (IDCs).
Table S1.2. The gene pool for the MYC transcriptional regulatory network relevant to clinical mitotic count parameter in ER(+)IDCs (i.e. 90A).
Table S1.3. The gene pool for the overlapped network of MYC&STAT3 in ER(+)IDCs (i.e. 90A) relevant to clinical mitotic count parameter.
Table S1.4. The gene pool for the STAT3 transcriptional regulatory network in ER(+) IDCs (i.e. 90A).
Table S1.5. The gene pool for the MYC transcriptional regulatory network in ER(+) IDCs (i.e. 90A).
Table S1.6. The common gene pool for the overlapped network of MYC&STAT3 in ER(+)IDCs (i.e. 90A).
Table S1.7. The shared gene pool between the overlapped network of MYC&STAT3 relevant to ER(+) IDCs and that significant to clinical mitotic count parameter.
Table S1.8. The gene pool for the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 72A).
Table S1.9. The shared gene pool between overlapped networks of MYC&STAT3 in 90A and 72A and their non-overlapping gene pools.
Table S1.10. The FOXC1 transcriptional regulatory network in 90 ER(+) infiltrating ductal carcinomas.
Table S1.11. The FOXC1 transcriptional regulatory network in 72 ER(+) infiltrating ductal carcinomas.
Table S2.1. The shared gene pool between the overlapped networks of MYC&STAT3 in 90A (for ER(+) IDCs) and 77A (for ER(−) IDCs) and their non-overlapping gene pools. The gene pool shared by two univariate overlapped networks of MYC and STAT3 in ER(+) and ER(−) IDCs indicating different percentages of shared network activities for MYC and STAT3 in two subtypes (81.93% (77A) versus 45.81% (90A)), respectively. A cohort (77A) consisting of 48 triple negatives (TN) and 29 ERBB2+was used (see reference 9 in the main text).
Table S2.2. The shared gene pool between networks of MYCnSTAT3 in 90A (for ER(+) IDCs) and 77A (for ER(−) IDCs) and their non-overlapping gene pools. The gene pool shared by two networks of MYCnSTAT3 in ER(+) and ER(−) IDCs indicating different percentages of shared network activities for MYC and STAT3 in two subtypes (73.14% (77A) versus 45.75% (90A)), respectively.
Table S2.3. The common gene pool overlapped by networks of ESR1, MYC and STAT3 in ER(+) IDCs (i.e. 90A).
Table S2.4. The common gene pool overlapped by networks of ESR1n STAT3 and MYCnSTAT3 in ER(+) IDCs (i.e. 90A).
Table S2.5. The common gene pool overlapped by networks of MYC & STAT3 amd MYCnSTAT3 in ER(+) IDCs (i.e. 90A).
Table S2.6. The common gene pool overlapped by networks of ESR1 & STAT3 and ESR1nSTAT3 in ER(+) IDCs (i.e. 90A).
Table S2.7. Venn diagrams and their overlapping and non-overlapping gene pools for both networks of ESR1 and MYC in ER(+) IDCs (i.e. 90A). The common gene pool overlapped by networks of ESR1 & MYC(S2.7a), the potential ESR1 regulated gene pool but not in MYC network (S2.7b) and the potential MYC regulated gene pool but not in ESR1 network (S2.7c) are listed.
Table S3.1. The common gene pool between age-related gene pool in ER(+) IDCs (i.e.90A) and the overlapped network of MYC&STAT3 not only relevant to ER(+) IDCs but to clinical mitotic count parameter.
Table S3.2. The common gene pool between grade-related gene pool in ER(+) IDCs (i.e. 90A) and the overlapped network of MYC&STAT3 not only relevant to ER(+) IDCs but to clinical mitotic count parameter.
Table S3.3. The common gene pool between LNM-related gene pool in ER(+) IDCs (i.e. 90A) and the overlapped network of MYC&STAT3 not only relevant to ER(+) IDCs but to clinical mitotic count parameter.
Table S3.4. The common gene pool between LVI-related gene pool in ER(+) IDCs (i.e. 90A) and the overlapped network of MYC&STAT3 not only relevant to ER(+) IDCs but to clinical mitotic count parameter.
Table S3.5. The common gene pool between LYM-related gene pool in ER(+) IDCs (i.e. 90A) and the overlapped network of MYC&STAT3 not only relevant to ER(+) IDCs but to clinical mitotic count parameter.
Table S3.6. The common gene pool between MC-related gene pool in ER(+) IDCs (i.e. 90A) and the overlapped network of MYC&STAT3 not only relevant to ER(+) IDCs but to clinical mitotic count parameter.
Table S3.7. The common gene pool between NP-related gene pool in ER(+) IDCs (i.e. 90A) and the overlapped network of MYC&STAT3 not only relevant to ER(+) IDCs but to clinical mitotic count parameter.
Table S3.8. The common gene pool between size-related gene pool in ER(+) IDCs (i.e. 90A) and the overlapped network of MYC&STAT3 not only relevant to ER(+) IDCs but to clinical mitotic count parameter.
Table S3.9. The common gene pool between stage-related gene pool in ER(+) IDCs (i.e. 90A) and the overlapped network of MYC&STAT3 not only relevant to ER(+) IDCs but to clinical mitotic count parameter.
Table S3.10. The common gene pool between TF-related gene pool in ER(+) IDCs (i.e. 90A) and the overlapped network of MYC&STAT3 not only relevant to ER(+) IDCs but to clinical mitotic count parameter.
Table S4.1. The common gene pool in both cell cycle signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Table S4.2. The common gene pool in both VEGF signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Table S4.3. The common gene pool in both BER signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Table S4.4. The common gene pool in both DRS signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Table S4.5. The common gene pool in both HR signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Table S4.6. The common gene pool in both MRP signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Table S4.7. The common gene pool in both NER signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Table S4.8. The common gene pool in p53 signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Table S4.9. The common gene pool in both proteasome signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Table S4.10. The common gene pool in both ribosome signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Table S4.11. The common gene pool in both PDGFRB signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Table S4.12. The common gene pool in both ERBB2 signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Table S4.13. The common gene pool shared between the multivariate space of STAT3 network (MYCnSTAT3) and ERBB2 signal transduction pathway in ER(+) IDCs (i.e. 90A).
Table S4.14. The common gene pool in both ERBB2 signal transduction pathway and the MYC & STAT3 overlapped network in ER(−) IDCs (i.e. 77A). ER(−) IDCs contain 48 triple negatives (TN) and 29 ERBB2+.
Table S4.15. The common gene pool shared between the MYCnSTAT3 network and ERBB2 signal transduction pathway in ER(−) IDCs (i.e. 77A).
Figure S5.1. Heatmaps for the common gene pool in both cell cycle signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Figure S5.2. Heatmaps for the common gene pool in both VEGF signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Figure S5.3. Heatmaps for the common gene pool in both BER signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Figure S5.4. Heatmaps for the common gene pool in both DRS signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A)
Figure S5.5. Heatmaps for the common gene pool in both HR signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Figure S5.6. Heatmaps for the common gene pool in both MRP signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Figure S5.7. Heatmaps for the common gene pool in both NER signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Figure S5.8. Heatmaps for the common gene pool in both p53 signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Figure S5.9. Heatmaps for the common gene pool in both proteasome signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Figure S5.10. Heatmaps for the common gene pool in both ribosome signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A). Grey color within the heatmap stands for data not available after data processing.
In this case, we do not use this piece of information for further discussion due to most data not available.
Figure S5.11. Heatmaps for the common gene pool in both PDGFRB signal transduction pathway and the overlapped network of MYC&STAT3 in ER(+) IDCs (i.e. 90A).
Figure S6.1. The survival analyses on subtypes of ER(+) infiltrating ductal carcinoma in this study. Figure S6.1a. stands for survival analysis on Group IE vs. Group IIE; Figure S6.1b. stands for survival analysis on Luminal A vs. Luminal B. Both results are not significant.
Figure S6.2. The survival analyses on probes in the STAT3 driven subnetworks in coupling with its major transcription factor partner - MYC for (a) proliferation; (b) sustained angiogenesis; (c) Warburg effect; (d) ES-like phenotype; (e) FOXC1 transcriptional regulatory network and (f) ERBB2 signaling in ER(+) IDCs. We compared the probes in the subnetworks in both 90A and 72A for predicting clinical outcome that was evaluated by overall disease free survival. Their results are summarized in Figure 3F (please see main text).
Figure S6.3. The survival analyses on a probe –FOXC1(12715) not in the STAT3 subnetwork. We compared this probe in the subnetworks in both 90A and 72A for predicting clinical outcome that was evaluated by overall disease free survival. It is a significant favorable prognostic factor in 90A cohort but not in 72A cohort.
Table S6.1. Univariate and multivariate analyses for survival on prognostic factors in 90A and 181A cohorts, respectively. The p values of tests in the Cox proportional hazard model to be less or equal to 0.05 are high-lighted with light blue. Both univariate and multivariate analyses are negative except univariate COXPH analysis on subcohort 1 versus non subcohort1.
Figure S7.1 Mean plot analyses of MELK mRNA levels in eight clinical categories of two cohorts of infiltrating ductal carcinomas (IDCs).
Lymphovascular invasion (LVI), nodal category (lymph node metastasis (LYM), number of nodal metastasis (LNM)), histological grade (Grade) category (nuclear pleomorphism (NP) and tubule formation (TF)) and stage were analyzed. Cohort 1 (90IDCs) has Groups IE and IIE. Cohort 2 (72A) has Luminal A and Luminal B (see the main text for definitions).
Figure S7.2. Mean plot analyses of METAP2 mRNA levels in eight clinical categories of two cohorts of infiltrating ductal carcinomas (IDCs).
Figure S7.3. Mean plot analyses of SRC mRNA levels in eight clinical categories of two cohorts of infiltrating ductal carcinomas (IDCs).
Figure S7.4. Mean plot analyses of a transcript variant SRC mRNA levels in eight clinical categories of two cohorts of infiltrating ductal carcinomas (IDCs).
Figure S7.5. Mean plot analyses of OIP5 mRNA levels in eight clinical categories of two cohorts of infiltrating ductal carcinomas (IDCs).
Table S7.1. ANOVA tests on ARNT mRNA levels in eight clinical categories of two cohorts of infiltrating ductal carcinomas (IDCs).