Abstract
Staphylococcus aureus is a main cause of bovine mastitis and a major pathogen affecting human health. The emergence and spread of methicillin-resistant Staphylococcus aureus (MRSA) has become a significant concern for both animal health and public health. This study investigated the incidence of MRSA in milk samples collected from dairy cows with clinical mastitis and characterized the MRSA isolates using antimicrobial susceptibility tests and genetic typing methods. In total, 103 S. aureus isolates were obtained from dairy farms in 4 different provinces in China, including Gansu, Shanghai, Sichuan, and Guizhou. Antimicrobial susceptibility testing of these isolates revealed that the resistance rates to penicillin and sulfamethoxazole were high, while the resistance rates to ciprofloxacin and vancomycin were low. Among the 103 isolates, 49 (47.6%) were found to be mecA-positive, indicating the high incidence of MRSA. However, 37 of the 49 mecA-positive isolates were susceptible to oxacillin as determined by antimicrobial susceptibility assays and were thus classified as oxacillin-susceptible mecA-positive S. aureus (OS-MRSA). These isolates could be misclassified as methicillin susceptible Staphylococcus aureus (MSSA) if genetic detection of mecA was not performed. Molecular characterization of selected mecA-positive isolates showed that they were all negative with Panton-Valentine leukocidin (PVL), but belonged to different spa types and SCCmec types. These results indicate that OS-MRSA is common in bovine mastitis in China and underscore the need for genetic methods (in addition to phenotypic tests) to accurately identify MRSA.
Introduction
Staphylococcus aureus causes infections in both people and animals. Methicillin-resistant S. aureus (MRSA) is a prominent pathogen in nosocomial and community acquired infections, and is a major threat to human health worldwide due to its antimicrobial resistance, infectivity and possession of virulence factors [1]–[5]. Methicillin resistance in S. aureus is mainly mediated by the expression of the mecA gene, which is located on a mobile genetic element, staphylococcal cassette chromosome mec (SCCmec), and encodes an altered penicillin-binding protein (PBP2a) with an extremely low affinity to β-lactam antibiotics, making it possible for S. aureus to survive the treatment of β-lactam antibiotics [6]. S. aureus that either have the mecA gene or show a minimum inhibitory concentration (MIC) of oxacillin higher than 4 µg/ml are defined as MRSA. However, S. aureus that are positive for mecA and PBP2a, but phenotypically susceptible to oxacillin have been reported [7]–[9]. It is generally accepted that such S. aureus isolates should be defined as oxacillin-susceptible mecA-positive S. aureus (OS-MRSA). Identification of OS-MRSA has clinical implications as precautions have been proposed for the treatment of OS-MRSA infections. Although OS-MRSA is phenotypically susceptible to oxacillin, it may be prone to the development of highly resistant MRSA under antibiotic selection due to the possession of mecA [10] [11].
China has recently experienced significant growth in dairy industry and the total milk production in China reached to the third place worldwide in 2012 [12]. In China, bovine mastitis is a serious problem for dairy industries and the average incidence rate is about 33%, incurring considerable economic losses [13]. For example, it is estimated that 5–10% cows are culled annually in the Shanghai region due to mastitis [14]. Although multiple pathogens are associated with bovine mastitis, S. aureus is a common and important cause of the disease [15]. Antimicrobial treatment is often used to decrease the incidence or shorten the duration of bovine mastitis; however, treatment failure occurs due to development of antibiotic resistance [16].
Despite the fact that S. aureus is commonly associated with bovine mastitis, MRSA isolates have been infrequently reported with the disease. There have been a few reports of MRSA colonization and/or infections in dairy cattle since the very first report of MRSA in mastitis in 1972 [17]–[21]. Recently, a highly divergent mecA gene (now named mecC) in a typeXI SCCmec was found in bovine mastitis S. aureus [22] [23]. Mastitic MRSA strains from different continents may share similar or different molecular characteristics. For example, reports from some European countries indicated that ST398 MRSA with SCCmec type IV or V played an important part in clinical or subclinical bovine mastitis, although it was not the only clonal lineage associated with mastitis [24]. Several genotypes including ST1/t286 MRSA with SCCmec type IVa, ST72/t324 MRSA with SCCmec type IV or IVa, and ST72/untypeable spa-type with SCCmec type IV were reported in Korea [25]. The majority of reported MRSA isolates in Turkey belonged to ST239/spa-type t30 with SCCmec type III, while others belonged to ST8/spa-type t190/SCCmec type IV, or ST329/spa-type t30/SCCmec type III [26]. These data indicated that various MRSA clones or genotypes were associated with bovine mastitis in different countries.
In China, there have been a few reports on MRSA of animal origin. Cui et al. reported presence of MRSA from swine and swine farm workers in four Chinese provinces, all of which belonged to ST9 and spa type t899, contained a type III SCCmec element, and lacked the Panton-Valentine Leukocidin (PVL) gene [27]. There was a report on MRSA from pet animals and veterinary staff in China, in which 22 MRSA isolates were identified by using the API Staph Ident System, MIC tests and mecA-specific PCR assay [28]. Another study reported that MRSA of ST97 with SCCmec typeIV, ST965 with SCCmec typeIV, ST6 with SCCmec typeIV and ST9 with untypeable SCCmec were found in milk samples collected from bovine mastitis cases [29].
Despite the numerous reports of MRSA of human and animal origins in different countries, there have been few published studies on OS-MRSA associated with bovine mastitis. Due to the rising significance of OS-MRSA in clinical treatment, enhanced efforts are needed to identify this type of MRSA by combining phenotypic tests with genetic methods. The objective of this study was to determine the incidence of OS-MRSA in bovine mastitis and to characterize the MRSA isolates using various methods.
Materials and Methods
1. Ethics
Milk samples were obtained on farms from dairy cows with naturally occurring clinical mastitis with consent from farm owners under the ethical approval by Lanzhou Institute of Husbandry and Pharmaceutical Sciences of CAAS. The collections were done by professional veterinarians and permitted by the owners of the dairy farms under investigation. Conventional milking methods were used and no invasive or pain-causing procedures were involved. This study did not involve endangered or protected species and did not use animals for experiments.
2. Sample collection and bacterial isolation
Milk samples were taken from cows with clinical mastitis which was manifested with decreased milk production, color change of the milk, and inflammation of the udder. These samples were collected from farms in 4 different and geographically diverse regions in China including Gansu, Shanghai, Sichuan, and Guizhou in 2008. These farms represent typical dairy production practices in China. For milk collection, udders of the clinical mastitis cow were washed with clean water and dried. Cotton swabs soaked with 70% ethanol were used to disinfect the surfaces of teats. The first few streams of milk were discarded. Then a milk sample was collected into a 10-ml sterile plastic tube. The collected samples were kept in a cooler with ice and transported to the laboratory within 8 hours. Then the samples were stored at −20°C for bacterial isolation.
Isolation of S. aureus was done in the Key Laboratory of Veterinary Pharmaceutical Development, Ministry of Agriculture, Lanzhou, China. Briefly, the milk samples were inoculated on 5% sheep blood agar plates and inoculated at 37°C for 24 h. S. aureus identification was based on Gram staining, morphology, and traditional biochemical tests, including catalase, coagulase and mannitol fermentation tests [30] [31].
In total, 103 S. aureus isolates were obtained, including 17 from Gansu, 52 from Shanghai, 16 from Sichuan, and 18 from Guizhou. The 17 isolates from Gansu were isolated from 150 clinical mastitic milk samples, which were collected from three different commercial dairy farms (50 samples per farm). The 52 S. aureus strains from Shanghai were isolated from 200 milk samples of two different farms (100 samples per farm); the 16 isolates from Sichuan were derived from 50 milk samples; and the 18 isolates from Guizhou were obtained from 50 milk samples. All S. aureus isolates were stored at −80°C. S. aureus ATCC43300 and ATCC25923 were used as quality control organisms.
3. Phenotypic identification of MRSA
S. aureus isolates were tested for methicillin resistance using the cefoxitin and oxacillin disk diffusion methods outlined by the Clinical and Laboratory Standards Institute [32]. Cefoxitin disk (30 µg, Oxoid) and oxacillin disk (1 µg, Oxoid) were used in this study. The zones of inhibition were measured after 18–20 hours incubation at 35°C. Isolates with zone sizes less than 21 mm for cefoxitin and 10 mm for oxacillin were considered methicillin resistant according to the criteria of CLSI [32].
4. Antimicrobial susceptibility tests
The mecA-positive MRSA isolates were tested for susceptibility to various commonly used antimicrobial agents by using two different methods. The disk diffusion test was performed with penicillin (10 IU/disk), gentamicin (10 µg/disk), tetracycline (30 µg/disk), erythromycin (15 µg/disk), ciprofloxacin (5 µg/disk), sulfamethoxazole (300 µg/disk), cefazolin (30 µg/disk) and clindamycin (2 µg/disk). The agar dilution method was used to measure the MICs of vancomycin and oxacillin. Both disk diffusion and agar dilution were performed according to the recommendations of CLSI [32]. The breakpoints of CSLI for the tested antibiotics (for both disk diffusion and agar dilution) were used to determine the susceptibility profiles. All antimicrobial susceptibility testing assays were repeated at least 3 times.
5. Genotypic identification of MRSA
A single colony of S. aureus was inoculated into LB culture medium, and the culture was shaken overnight at 37°C. Then the culture was used for preparation of genomic DNA using the TIANamp Bacteria DNA Kit [TIANGEN BIOTECH (Beijing) CO., LTD] according to the manufacturer's instructions. The genomic DNA was used as template for PCR for typing of SCCmec and spa and for detection of mecA and the PVL toxin gene (lukF-lukS).
The primers used to amplify the mecA gene (310 bp) of MRSA were P1: 5′-TGGCATTCGTGTCACAATCG-3′ and P2: 5′-CTGGAACTTGTTGAGCAGAG-3′ as previously described [33]. Each PCR mixture was composed of 2 µl DNA template, 0.5 µl of each primer (10 µM), 12.5 µl ExTaq buffer mix [TaKaRa Biotechnology (Dalian) Co., Ltd], and 9.5 µl sterile distilled H2O. PCR program began with an initial denaturation step at 94°C for 4 min followed by 34 cycles of 92°C for 1 min, 53°C for 50 seconds, and 72°C for 1 min, and ended with a final extension step at 72°C for 10 min. The mecA-positive strain ATCC43300 and the mecA-negative ATCC25923 were included as positive and negative controls, respectively. The amplified PCR products were electrophoresed in 2% agarose gel at 120 V for 1 hour, stained with ethidium bromide (0.5 µg/ml), and photographed under UV light.
6. Spa-typing
Using the Ridom StaphType standard protocol(www.ridom.com), the MRSA strains from Gansu and Shanghai were PCR amplified for analyzing the polymorphic X-region of Staphylococcus protein A (spa) gene. The amplicons were purified using a TIANgel Midi Puification Kit [TIANGEN BIOTECH (Beijing) CO., LTD] and sequenced using the same PCR primers at Sangon Biotech (Shanghai) Co., Ltd. The spa types were assigned by using an online spa database [34].
7. SCCmec typing
SCCmec types were determined using the primers as described by Zhang et al [35]. A combination of different PCR reactions was performed to type the SCCmec elements. For specific SCCmec types, the SCCmec M-PCR typing assay contained 2 pairs of primers including one pair specific for the mecA gene and another pair specific for SCCmec types and subtypes I, II, III, IVa, IVb, IVc, IVd and V. Then a single PCR reaction was performed to identify the related mec and ccr gene complexes using specific primers. In addition, the PCR amplicon of SCCmec of isolate B5 was sequenced and compared with the standard SCCmec sequences in the NCBI database.
8. Detection of the Panton-Valentine Leukocidin gene lukF-lukS
PCR was performed to determine the presence of the PVL toxin gene lukF-lukS as previously described by Lina et al [36].
Results
1. The overall antimicrobial susceptibility profiles of the bovine S. aureus isolates
The antibiotic susceptibility of the analyzed S. aureus isolates were shown in Table 1. The overall resistance rates were high with penicillin (97.1%) and sulfafurazole (83.5%), while the overall resistance rates were generally low with gentamicin (11.7%), ciprofloxacin (2.9%), cefazolin (6.8%), vancomycin (0%), and oxacillin (12.6%). The resistance rates were at moderate levels to tetracycline (35%), erythromycin (31.1%), and clindamycin (29.1%). Although there were variations in the resistance rates among the 4 regions, it was not obvious that one region showed more incidence of resistance than others (Table 1).
Table 1. The overall drug susceptibility patterns of the S. aureus isolates from bovine mastitis cases*.
| Penicillin | Gentamicin | Tetracycline | Erythromycin | Clindamycin | Ciprofloxacin | Sulfafurazole | Cefazolin | Vancomycin | Oxacillin | ||||||||||||
| Region# | R | S | R | S | R | S | R | S | R | S | R | S | R | S | R | S | R | S | R | S | |
| Shanghai(52) | Number of isolates | 52 | 0 | 11 | 37 | 23 | 29 | 17 | 35 | 24 | 21 | 3 | 48 | 49 | 3 | 6 | 46 | 0 | 52 | 11 | 41 |
| Percentage (%) | 100 | 0 | 21.2 | 71.2 | 44.2 | 55.8 | 32.3 | 67.3 | 46.9 | 39.5 | 5.7 | 92.3 | 94.2 | 5.8 | 11.5 | 88.5 | 0 | 100 | 21.2 | 78.8 | |
| Sichuan(16) | Number of isolates | 16 | 0 | 0 | 15 | 6 | 10 | 3 | 12 | 2 | 14 | 0 | 15 | 12 | 4 | 1 | 15 | 0 | 16 | 2 | 14 |
| Percentage (%) | 100 | 0 | 0 | 93.75 | 37.5 | 62.5 | 18.75 | 75 | 14.3 | 85.7 | 0 | 93.75 | 75 | 25 | 6.67 | 93.33 | 0 | 100 | 12.5 | 87.5 | |
| Guizhou(18) | Number of isolates | 18 | 0 | 0 | 18 | 4 | 14 | 7 | 11 | 2 | 16 | 0 | 18 | 12 | 6 | 0 | 18 | 0 | 18 | 0 | 18 |
| Percentage (%) | 100 | 0 | 0 | 100 | 22.22 | 77.78 | 38.89 | 61.11 | 12.5 | 87.5 | 0 | 100 | 66.67 | 33.33 | 0 | 100 | 0 | 100 | 0 | 100 | |
| Gansu(17) | Number of isolates | 14 | 1 | 1 | 16 | 3 | 13 | 5 | 6 | 2 | 10 | 0 | 17 | 13 | 3 | 0 | 17 | 0 | 17 | 0 | 17 |
| Percentage (%) | 82.4 | 5.88 | 5.88 | 94.1 | 17.7 | 76.5 | 29.4 | 35.3 | 11.8 | 58.8 | 0 | 100 | 76.5 | 17.7 | 0 | 100 | 0 | 100 | 0 | 100 | |
| Total(103) | Number of isolates | 100 | 1 | 12 | 86 | 36 | 66 | 32 | 64 | 30 | 61 | 3 | 98 | 86 | 16 | 7 | 96 | 0 | 103 | 13 | 90 |
| percentage (%) | 97.1 | 0.97 | 11.7 | 83.5 | 34.95 | 64.1 | 31.1 | 62.1 | 29.1 | 59.2 | 2.9 | 95.1 | 83.5 | 15.5 | 6.8 | 93.2 | 0 | 100 | 12.6 | 87.4 | |
R: resistant; S: susceptible.
The number in parenthesis indicate the total number of isolates from a given region.
2. Identification of mecA by PCR
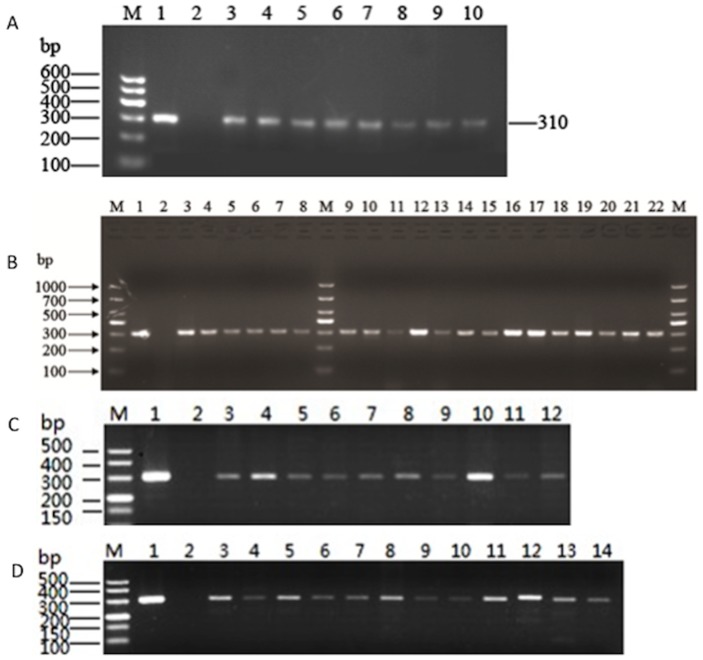
Using primers P1 and P2, we analyzed the presence of mecA in the S. aureus isolates. As shown in Fig. 1, the positive control strain ATCC43300 showed a distinct 310-bp band, while the negative control strain ATCC 25923 did not show a PCR product, indicating the specificity of the PCR assay. Among the S. aureus isolates examined in this study, 8, 20, 11, and 10 mecA-positive strains were identified for Gansu, Shanghai, Sichuan, and Guizhou, respectively (Fig. 1). The overall detection rate of mecA is 47.6% (49 out of 103), indicating the high prevalence of MRSA in S. aureus isolates derived from bovine mastitis in China.
Figure 1. PCR detection of mecA-positive S. aureus isolates.
The results were analyzed by Agarose gel electrophoresis. (A) mecA-positive isolates from Gansu province. Lane M: DNA size Marker. Lane 1: positive control strain ATCC43300. Lane 2: negative control strain ATCC25923. Lanes 3–10: isolate QY4, QY6, QY8, QY10, HG2, HG3, HG4 and HG5, respectively. (B) mecA-positive isolates from Shanghai. M: DNA size Marker. Lane 1: positive control strain ATCC43300. Lane 2: negative control strain ATCC25923. Lane 3–8: isolate SX5, SX6, SX10, SX11, SX13 and SX15, respectively; and lanes 9–22: isolates SH1, SH2, SH3, SH4, SH7, SH8, SH9, SH10, SH13, SH14, SH16, SH17, SH18 and SH20, respectively. (C) mecA-positive isolates from Guizhou. M: DNA size Marker. Lane 1positive control strain ATCC43300. Lane 2: negative control strain ATCC25923. Lanes 3–12: isolates zy1, zy2, zy4, zy5, zy6, zy8, zy11, zy12, zy14, and zy15, respectively. (D). mecA-positive isolates from Sichuan. M: DNA size Marker. Lane 1positive control strain ATCC43300. Lane 2: negative control strain ATCC25923. Lanes 3–14: isolates cx1, cx2, cx5, cx6, cx8, cx9, cx10, cx13, cx14, cx17, cx18, and cx19, respectively.
3. Identification of OS-MRSA
According to the oxacillin disk diffusion tests, only 12.6% of the isolates were resistant to this antibiotic (Table 1), but 47.6% of the isolates were mecA-positive, suggesting the presence of OS-MRSA. In detail, all 17 S. aureus isolates from Gansu were susceptible to the antibiotic, but 8 (47.06%) of them were found carrying the mecA gene by PCR (Fig. 1; Table 2) and were classified as OS-MRSA. Four of the eight mecA-positive isolates were from farm A, and the other 4 from farm B. Among the 52 isolates from Shanghai, 11 were resistant to oxacillin. However, the PCR assay revealed that 20 of the 52 isolates were positive for mecA (Fig. 1). These mecA-positive isolates included 11 oxacillin-resistant strains and 9 oxacillin-susceptible strains (Table 3). Thus the 9 mecA-positive but oxacillin-susceptible isolates were classified as OS-MRSA. Among the 16 S. aureus isolates from Sichuan, 2 were resistant to oxacillin, but 11 were positive for mecA by PCR (Table 4), which included 1 oxacillin-resistant strain and 10 oxacillin-susceptible isolates (classified as OS-MRSA). Thus OS-MRSA accounted for 62.5% of the isolates from Sichuan. For the 18 isolates from Guizhou, none was resistant oxacillin, but 10 were positive for mecA and were considered OS-MRSA (Table 5). Among the 49 mecA-positive isolates, 37 were susceptible to oxacillin, indicating that the majority (75.5%) of the bovine MRSA are OS-MRSA. In total, OS-MRSA accounted for 35.9% (37 out of 103) of the total S. aureus isolates, indicating the high prevalence of OS-MRSA in clinical bovine mastitis cases in China.
Table 2. Genotyping and antibiotic susceptibility patterns of the mecA-positive S. aureus isolates from Gansu Province.
| OXA MIC | ccr gene | Resistance profileb | |||||||||||||
| Isolatea | mecA | (µg/mL) | PVL | spa | complex | SCCmec | PEN | GENTA | TETR | ERYTH | CLIN | CIP | SULF | CEFA | VAN |
| QY4 | + | <0.5 | − | t267 | 5 | V | R | S | R | R | I | S | R | S | S |
| QY6 | + | <0.5 | − | t267 | 5 | V | R | S | S | R | S | S | S | S | S |
| QY8 | + | 1 | − | t267 | 5 | V | R | S | S | I | R | S | R | S | S |
| QY10 | + | 0.5 | − | t267 | 5 | V | R | S | S | I | I | S | I | S | S |
| HG2 | + | <0.5 | − | t267 | 5 | V | S | S | S | I | S | S | R | S | S |
| HG3 | + | <0.5 | − | t267 | 5 | V | R | S | S | I | S | S | R | S | S |
| HG4 | + | <0.5 | − | t267 | 5 | V | R | S | S | S | S | S | R | S | S |
| HG5 | + | <0.5 | − | t267 | 5 | V | R | S | S | S | S | S | R | S | S |
QY and HG represent isolates from two different farms.
R, resistant; S, susceptible.
OXA, Oxacillin ; PEN, penicillin; GENTA, gentamicin; TETR, tetracycline; ERYTH, erythromycin; CLIN, clindamycin ; CIP, ciprofloxacin; SULF, sulfamethoxazole; CEFA, cefazolin; VAN, vancomycin.
Table 3. Genotyping and antibiotic susceptibility profiles of the mecA-positive isolates identified in Shanghai.
| OXA MIC | Resistance profileb | |||||||||||||
| Isolatesa | mecA | (µg/mL) | PVL | spa | SCCmec | PEN | GENTA | TETR | ERYTH | CLIN | CIP | SULF | CEFA | VAN |
| SX5 | + | ≥8 | − | t1234 | II | R | R | R | R | R | R | R | R | S |
| SX6 | + | ≥8 | − | t1234 | II | R | R | R | R | R | S | R | R | S |
| SX10 | + | ≥8 | − | NT | V | R | R | R | S | R | R | R | R | S |
| SX11 | + | ≥8 | − | NT | II | R | R | S | R | R | R | R | R | S |
| SX13 | + | ≥8 | − | t267 | V | R | R | R | R | R | S | R | R | S |
| SX15 | + | ≥8 | − | t267 | V | R | R | R | R | R | S | R | R | S |
| SH1 | + | ≤1 | − | t267 | V | R | S | S | S | R | S | R | S | S |
| SH2 | + | ≥8 | − | t267 | V | R | S | R | R | R | S | R | S | S |
| SH3 | + | ≥8 | − | t267 | V | R | S | R | R | R | S | R | S | S |
| SH4 | + | ≤1 | − | t267 | V | R | S | S | S | I | S | R | S | S |
| SH7 | + | ≤1 | − | t267 | V | R | S | S | S | I | S | R | S | S |
| SH8 | + | ≥8 | − | t267 | V | R | S | S | R | R | S | R | S | S |
| SH9 | + | ≤1 | − | t267 | V | R | S | S | S | R | S | R | S | S |
| SH10 | + | ≤1 | − | t267 | V | R | S | S | S | R | S | R | S | S |
| SH13 | + | ≤1 | − | t1234 | II | R | S | S | S | R | S | R | S | S |
| SH14 | + | ≤1 | − | t1234 | II | R | S | R | S | R | S | R | S | S |
| SH16 | + | ≥8 | − | t1234 | II | R | I | S | R | R | S | R | S | S |
| SH17 | + | ≥8 | − | t1234 | II | R | I | S | R | R | S | R | S | S |
| SH18 | + | ≤1 | − | t1234 | II | R | S | S | S | R | S | R | S | S |
| SH20 | + | ≤1 | − | t1234 | II | R | S | S | S | R | S | R | S | S |
SX and SH represent isolates from two different farms. Bold indicates OS-MRSA.
R, resistant; S, susceptible.
OXA, Oxacillin ; PEN, penicillin; GENTA, gentamicin; TETR, tetracycline; ERYTH, erythromycin; CLIN, clindamycin ; CIP, ciprofloxacin; SULF, sulfamethoxazole; CEFA, cefazolin; VAN, vancomycin.
Table 4. Antibiotic susceptibility profiles of the mecA-positive isolates identified in Sichuan.
| OXA MIC | Resistance profilea | ||||||||||
| Isolates | mecA | (µg/mL) | PEN | GENTA | TETR | ERYTH | CLIN | CIP | SULF | CEFA | VAN |
| CX1 | + | <0.5 | R | S | S | S | I | S | R | S | S |
| CX2 | + | <0.5 | R | S | S | S | S | S | S | S | S |
| CX5 | + | ≤1 | R | S | R | R | R | S | R | S | S |
| CX6 | + | ≤2 | R | S | S | S | I | S | R | S | S |
| CX8 | + | <0.5 | S | S | R | S | S | S | R | S | S |
| CX9 | + | ≥8 | R | I | S | I | S | S | R | R | S |
| CX10 | + | ≤2 | R | S | S | S | S | S | R | S | S |
| CX13 | + | ≤2 | R | S | R | S | S | S | S | S | S |
| CX14 | + | ≤2 | R | S | R | S | S | S | S | S | S |
| CX17 | + | ≤2 | R | S | R | S | S | S | R | S | S |
| CX19 | + | ≤2 | R | S | S | R | R | S | S | S | S |
R, resistant; S, susceptible.
OXA, Oxacillin ; PEN, penicillin; GENTA, gentamicin; TETR, tetracycline; ERYTH, erythromycin; CLIN, clindamycin ; CIP, ciprofloxacin; SULF, sulfamethoxazole; CEFA, cefazolin; VAN, vancomycin.
Table 5. Antibiotic susceptibility profiles of the mecA-positive isolates identified in Guizhou.
| OXA MIC | Resistance profilea | ||||||||||
| Isolates | mecA | (µg/mL) | PEN | GENTA | TETR | ERYTH | CLIN | CIP | SULF | CEFA | VAN |
| ZY1 | + | <0.5 | R | S | R | S | I | S | R | S | S |
| ZY2 | + | <0.5 | R | S | S | S | S | S | R | S | S |
| ZY4 | + | <0.5 | R | S | S | R | R | S | S | S | S |
| ZY5 | + | <0.5 | R | S | S | S | I | S | R | S | S |
| ZY6 | + | ≤2 | S | S | R | S | S | S | R | S | S |
| ZY8 | + | <0.5 | R | R | S | S | S | S | R | S | S |
| ZY11 | + | ≤2 | R | S | S | R | S | S | S | S | S |
| ZY12 | + | <0.5 | R | S | R | S | S | S | R | S | S |
| ZY14 | + | <0.5 | R | S | S | R | S | S | S | S | S |
| ZY15 | + | <0.5 | R | S | S | S | S | S | R | S | S |
R, resistant; S, susceptible.
OXA, Oxacillin ; PEN, penicillin; GENTA, gentamicin; TETR, tetracycline; ERYTH, erythromycin; CLIN, clindamycin ; CIP, ciprofloxacin; SULF, sulfamethoxazole; CEFA, cefazolin; VAN, vancomycin.
4. Antimicrobial susceptibility patterns of the mecA-positive S. aureus isolates
Antibiotic susceptibility patterns of the mecA-positive isolates from different provinces are shown in Tables 2–5. For the mecA-positive isolates from Gansu, their oxacillin MICs were all lower than 2 µg/ml, consistent with the result from the disk diffusion test. They were also all susceptible to gentamicin, ciprofloxacin, cefazolin, and vancomycin, and most of them were non-resistant to tetracycline, erythromycin, and clindamycin (Table 2). However, the isolates were generally resistant to penicillin and sulfafurazole.
For the mecA-positive isolates from Shanghai (Table 3), 11 were resistant and 9 were susceptible to oxacillin, consistent with the result from the disk diffusion test. All of the 20 isolates were resistant to penicillin and sulfafurazole, but were susceptible to vancomycin. Most of them were also resistant to clindamycin. However, there were major differences in the resistance to other antibiotics between the isolates from farm SX and those from farm SH (Table 3). For example, all of the SX isolates were resistant to gentamycin and cefazolin, while all of the SH isolates were susceptible to the two antibiotics. Additionally, the SX isolate were more resistant to tetracycline, erythromycin, and ciprofloxacin than the SH isolates. These results suggest farm-to-farm variations in the antimicrobial susceptibility patterns.
Among the 11 mecA-positive isolates from Sichuan, 10 were OS-MRSA (oxacillin MICs<2 µg/ml) (Table 4). High resistance rates were observed with penicillin and sulfafurazole, but the isolates were generally susceptible to other tested antibiotics. The 10 mecA-carrying S. aureus from Guizhou were all susceptible to oxacillin (Table 5) and are considered OS-MRSA. Similar to the isolates from Sichuan, the Guizhou isolates were generally resistant to penicillin and sulfafurazole, but susceptible to other examined antibiotics (Table 5).
5. Molecular characterization of selected MRSA isolates
Molecular typing analysis was done with the mecA-positive isolates from Gansu and Shanghai. The ones from Gansu were spa-type t267 (allelic profile: 07-23-12-21-17-34-34-34-33-34), SCCmec-type V, ccr complex 5, and PVL negative (Table 2). The sequence of the SCCmec amplicon of isolate HG5 was determined and it was found that it shared 99% identity to SCCmec-type V sequences (GenBank Accession No. AB505629.1), further confirming the PCR result. For the MRSA from Shanghai, all were PVL negative; spa types included t1234 (8 isolates), t267(10 isolates), and two non-typeable; and their SCCmec types were II and V (Table 3). The isolates from Sichuan and Guizhou were not typed. These results suggest the genetic diversity of the mecA-positive isolates from cases of bovine mastitis.
Discussion
In this study, we characterized S. aureus isolates from bovine mastitis milk samples collected from 4 different province/regions in China and identified the high prevalence of OS-MRSA. To our best knowledge, this is the first comprehensive investigation of OS-MRSA of bovine origin, following our initial report [37] on the presence of OS-MRSA on dairy farms in the Inner Mongolia region of China. Although S. aureus is a major cause of bovine mastitis, previously published reports revealed low prevalence of bovine MRSA, implying that MRSA was not commonly associated with mastitis [38]. However, most of previous studies were based on phenotypic tests for identifying MRSA, which may misidentify OS-MRSA as MSSA and underestimate the true prevalence of MRSA. In this study, 47.6% (49 out of 103) of the S. aureus isolates were found carrying mecA, which is unexpectedly higher than the highest reported incidence (17.5%) of MRSA from mastitic milk samples [39]. Presence of mecA is generally recognized as the most reliable method for detection of methicillin resistance, and mecA-positive staphylococcal strains are considered to be resistant to all β-lactam antibiotics [32]. Nevertheless, S. aureus that carry the mecA gene but appear phenotypically susceptible to oxacillin and vice versa have been reported recently [40]–[42] [37]. Thus, combination of genotypic and phenotypic tests is necessary to avoid false positive or false negative results in identifying MRSA.
All of the OS-MRSA isolates had an oxacillin MIC<2 µg/ml (Tables 2–5), indicating that presence of the mecA gene did not confer a high-level resistance to oxacillin. The reason for this phenotype remains to be elucidated. A recent study suggested that amino acid mutations in the FemXAB proteins (involved in cell wall synthesis) might contribute to the OS-MRSA phenotype [43], but the association of the mutations with the phenotype has not been formerly proven. Without testing the mecA gene, these isolates could be misclassified as MSSA based on the result of conventional antimicrobial susceptibility tests. OS-MRSA that carry mecA may result in the emergence of highly resistant MRSA under treatment with β-lactam antibiotics, which underscores the need for precautions when treating OS-MRSA infections [10] [42]. Due to this risk, treatment of OS-MRSA should avoid β-lactam antibiotics. As shown in Tables 2–5, the identified MRSAs were generally susceptible to other classes of antibiotics, such as gentamicin, ciprofloxacin, kanamycin, and vancomycin. Therefore, treatment of mastitis caused by OS-MRSA using non-β-lactam antibiotics may prevent the unwanted consequence of antibiotic resistance development.
Based on the results of molecular characterization, all the OS-MRSA isolates from Gansu belonged to spa-type t267, SCCmec-type V, and ccrC complex 5, and were PVL negative (Table 2), suggesting that these isolates may be a single lineage that was common in the surveyed region. However, the MRSA isolates from Shanghai belonged to different spa and SCCmec types (Table 3), suggesting their genetic diversity. Among the previously reported MRSA isolates associated with bovine mastitis, lineages with spa-type t267 and SCCmec-type V have not been identified [24]. However, a clinical MRSA isolate with the group V SCCmec element and t267 polymorphic X-region of spa was recovered in American University of Beirut Medical Center (AUB-MC) by Tokajian et al [44]. Another mecA-positive oxacillin-resistant S. aureus carrying SCCmec type V and spa type t267 was identified from an inpatient in England [45]. These reports plus findings from this study suggest that this lineage of MRSA may cause infections in both human and bovine and can be either susceptible or resistant to oxacillin. On the other hand, OS-MRSAs belonging to SCCmec types I, II, III IIIv, and IV were reported previously [10] [11], suggesting that MRSA of different genetic backgrounds could be phenotypically susceptible to oxacillin.
None of the OS-MRSA isolates identified in this study harbored the PVL gene. This finding is similar to that reported by Türkyilmaz et al. and Aires-de-Sousa et al. [26] [46]. In contrast, over 50% of bovine S. aureus isolates were determined to carry the PVL gene in another study [47]. PVL is a known virulence factor in S. aureus and is involved in the development of soft tissue infections [48]. The absence of PVL in the identified OS-MRSA isolates and their susceptibility to certain antimicrobial agents suggest that they may be eliminated from infected animals by appropriate antibiotic treatments.
Conclusions
This study demonstrates a high incidence of OS-MRSA on dairy farms in different regions of China. These OS-MRSA isolates carried the mecA gene, were susceptible to oxacillin, and could be mistakenly identified as MSSA if testing of the mecA gene were not conducted. These results suggest that MRSA is more commonly associated with bovine mastitis than previously realized, which is the case at least in China. Findings from this study and the increasing reports of OS-MRSA in clinical settings underscore the need for genetic tests as well as phenotypic assays to accurately identify MRSA.
Funding Statement
This study was funded by grants from the Science and Technology Project Funds of GanSu Province (1304WCG172, http://www.gsstc.gov.cn/Guide/detail.php?n_no=86734&dir=/), and China Agriculture Research System In Dairy Cow (CARS-37;http//119.253.58.231/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, et al. (2007) Invasive methicillin-resistant Staphylococcus aureus infections in the United States. J Am Med Assoc 298: 1763–1771. [DOI] [PubMed] [Google Scholar]
- 2. Lowy FD (2003) Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Invest 111: 1265–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cosgrove SE, Sakoulas G, Perencevich EN, Schwaber MJ, Karchmer AW, et al. (2003) Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin Infect Dis 36: 53–59. [DOI] [PubMed] [Google Scholar]
- 4. Tiemersma EW, Bronzwaer SLAM, Lyytikäinen O, Degener JE, Schrijnemakers P, et al. (2004) Methicillin-resistant Staphylococcus aureus in Europe, 1999–2002. Emerg Infect Dis 10: 1627–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chambers HF (2005) Community-associated MRSA—resistance and virulence converge. New Engl J Med 352: 1485–1487. [DOI] [PubMed] [Google Scholar]
- 6. Hiramatsu K, Cui LZ, Kuroda M, Ito T (2001) The emergency and evolution of methicillin-resistant Staphylococcus aureus. TRENDS Microbiol 9: 486–493. [DOI] [PubMed] [Google Scholar]
- 7. Sakoulas G, Gold HS, Venkataraman L, DeGirolami PC, Eliopoulos GM, et al. (2001) Methicillin-resistant Staphylococcus aureus: comparison of susceptibility testing methods and analysis of mecA-positive susceptible strains. J Clin Microbiol 39: 3946–3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Labrou M, Michail G, Ntokou E, Pittaras TE, Pournaras S, et al. (2012) Activity of oxacillin versus that of vancomycin against oxacillin-susceptible mecA-positive Staphylococcus aureus clinical isolates evaluated by population analyses, time-kill assays, and a murine thigh infection model. Antimicrob Agents Chemother 56: 3388–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Saeed K, Dryden M, Parnaby R (2010) Oxacillin-susceptible MRSA, the emerging MRSA clone in the UK? J Hosp Infect 76: 267–268. [DOI] [PubMed] [Google Scholar]
- 10. Hososaka Y, Hanaki H, Endo H, Suzuki Y, Nagasawa Z, et al. (2007) Characterization of oxacillin-susceptible mecA-positive Staphylococcus auerus: a new type of MRSA. J Infect Chemother 13: 79–86. [DOI] [PubMed] [Google Scholar]
- 11. Ikonomidis A, Michail G, Vasdeki A, Labrou M, Karavasilis V, et al. (2008) In vitro and in vivo evaluations of oxacillin efficiency against mecA-positive oxacillin-susceptible Staphylococcus aureus. Antimicrob Agents Chemother 52: 3905–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feedtrade website (2013) Available: http://www.feedtrade.com.cn/whey/milk_market/2013-01-17/2013257.html.Accessed 2013 Jan 17.
- 13. Bai LX, Hao ML, Qing JH (2010) Research advances in the treatment of dairy cow mastitis. Chin Qing Hai J Anim Vet Sci 5: 45–46. [Google Scholar]
- 14. Jin YZ, Wan SP, Jiang FM, Gong ZL, Cao J, et al. (2011) Isolation and susceptibility testing of major pathogenic bacteria and prevalence of cow mastitis. Anim Husb Vet Med 43: 64–67. [Google Scholar]
- 15. Cao LT, Hu SH (2010) Distribution of major pathogenic bacteria of cow mastitis in milk. J Anhui Agri Sci 38: 19541–19542. [Google Scholar]
- 16. Saini V, McClure JT, Scholl DT, DeVries TJ, Barkema HW (2013) Herd-level relationship between antimicrobial use and presence or absence of antimicrobial resistance in gram-negative bovine mastitis pathogens on Canadian dairy farms. J Dairy Sci 96: 4965–4976. [DOI] [PubMed] [Google Scholar]
- 17. Devriese LA, Van Dammme LR, Fameree L (1972) Methicillin- (cloxacillin)-resistant Staphylococcus aureus strains isolated from bovine mastitis case. Zentralbl Veterinarmed B 19: 598–605. [DOI] [PubMed] [Google Scholar]
- 18. Juhász-Kaszanyitzky É, Jánosi S, Somogyi P, Dán Á, Van Der Graaf-van Bloois L, et al. (2007) MRSA transmission between cows and humans. Emerg Infect Dis 13: 630–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Monecke S, Kuhnert P, Hotzel H, Slickers P, Ehricht R (2007) Microarray based study on virulence-associated genes and resistance determinants of Staphylococcus aureus. Vet Microbiol 125: 128–140. [DOI] [PubMed] [Google Scholar]
- 20. Fessler A, Scott C, Kadlec K, Ehricht R, Monecke S, et al. (2010) Characterization of methicillin-resistant Staphylococcus aureus ST398 from cases of bovine mastitis. J Antimicrob Chemother 65: 619–625. [DOI] [PubMed] [Google Scholar]
- 21. Huber H, Koller S, Glezendanner N, Stephan R, Zweifel C (2010) Prevalence and characteristics of methicillin-resistant Staphylococcus aureus in humans in contact with farm animals, in livestock, and in food of animal origin, Switzerland, 2009. Euro Surveill 15: pii = 19542. [PubMed] [Google Scholar]
- 22. Garcia-Alvarez L, Holden MTG, Lindsay H, Webb CR, Brown DFJ, et al. (2011) Methicillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine population in the UK and Denmark: a descriptive study. Lancet Infect Dis 11: 595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Paterson GK, Morgan FJE, Harrison EM, Peacock SJ, Parkhill J, et al. (2013) Prevalence and properties of mecC methicillin-resistant Staphylococcus aureus (MRSA) in bovine bulk tank milk in Great Britain. J Antimicrob Chemother Oct 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Holmes MA, Zadoks RN (2011) Methicillin resistant S. aureus in human and bovine mastitis. J Mammary Gland Biol Neoplasia 16: 373–382. [DOI] [PubMed] [Google Scholar]
- 25. Nam HM, Lee AL, Jung SC, Kim MN, Jang GC, et al. (2011) Antimicrobial susceptibility of Staphylococcus aureus and characterization of methicillin-resistant Staphylococcus aureus isolated from bovine mastitis in Korea. Foodborne Pathog Dis 8: 231–238. [DOI] [PubMed] [Google Scholar]
- 26. Türkyilmaz S, Tekbiyik S, Oryasin E, Bozdogan B (2010) Molecular epidemiology and antimicrobial resistance mechanisms of methicillin-resistant Staphylococcus aureus isolated from bovine milk. Zoonoses Public Health 57: 197–203. [DOI] [PubMed] [Google Scholar]
- 27. Cui SH, Li JY, Hu CQ, Jin SH, Li FQ, et al. (2009) Isolation and characterization of methicillin-resistant Staphylococcus aureus from swine and works in China. J Antimicrob Chemother 64: 680–683. [DOI] [PubMed] [Google Scholar]
- 28. Zhang WJ, Hao ZH, Wang Y, Cao XY, Logue CM, et al. (2011) Molecular characterization of methicillin-resistant Staphylococcus aureus from pet animals and veterinary staff in China. Vet J 190: e125–e129. [DOI] [PubMed] [Google Scholar]
- 29. Wang DF, Duan XH, Wu JY, Yang XY, Li JJ, et al. (2011) The current status of the drug resistance and evolutionary relationship of MSSA and MRSA isolates from bovine of China. Acta Veterinaria Et Zootechnica Sinica 42: 1416–1425. [Google Scholar]
- 30.Bannerman TL (2003) Staphylococcus, Micrococcus, other catalase-positive cocci that grow aerobically. In Manual of Clinical Microbiology, pp: 384–404. Edited by Murray PR, Baron EJ, Jorgensen JH, Pfaller MA, Yolken RH. Washington, DC: ASM Press. [Google Scholar]
- 31.Turk DC, Porter IA (1978) A Short Textbook of Medical Microbiology, 4th edn London: Hodder and Stoughton. [Google Scholar]
- 32.Clinical and Laboratory Standards Institute (2010) Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals: Informational Supplement. CLSI Document M100-S20. Clinical and Laboratory Standards Institute, Wayne PA. [Google Scholar]
- 33. Galdiero E, Liguori G, Isanto MD, Damiano N, Sommese L (2003) Distribution of mecA among methicillin-resistant clinical staphylococcal strains isolated at hospitals in Naples, Italy. Eur J Epidemiol 18: 139–145. [DOI] [PubMed] [Google Scholar]
- 34.Ridom SpaServer (2003) Available: http://www.spaserver.ridom.de/
- 35. Zhang KY, McClure JN, Elsayed S, Louie T, Conly JM (2005) Novel Multiplex PCR assay for characterization and concomitant subtyping of Staphylococcus cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol 45: 5026–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter MO, et al. (1999) Involvement of Panton-Valentine Leukocidin-Producing Staphylococcus aureus in primary skin infection and pneumonia. Clin Infect Dis 29: 1128–1132. [DOI] [PubMed] [Google Scholar]
- 37. Su Y, Pu WX, Chen ZH, Deng HP (2012) Antimicrobial resistance analysis and detection of methicillin-resistant Staphylococcus aureus (MRSA) among Staphylococcus aureus strains isolated from bovine mastitis. Scientia Agricultura Sinica 45: 3602–3607. [Google Scholar]
- 38. Hendriksen RS, Mevius DJ, Schroeter A, Teale C, Meunier D, et al. (2008) Prevalence of antimicrobial resistance among bacterial pathogens isolated from cattle in different European countries: 2002–2004. Acta Vet Scan 50: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Turutoglu H, Ercelik S, Ozturk D (2006) Antibiotic resistance of Staphylococcus aureus and coagulase-negative Staphylococci isolated from bovine mastitis. Bull Vet Inst Pulawy 50: 41–45. [Google Scholar]
- 40. Turutoglu H, Hasoksuz M, Ozturk D, Yildirim M, Sagnak S (2009) Methicillin and aminoglycoside resistance in Staphylococcus aureus isolates from bovine mastitis and sequence analysis of their mecA genes. Vet Res Commun 33: 945–956. [DOI] [PubMed] [Google Scholar]
- 41. Jannati E, Arzanlou M, Habibzadeh S, Mohammadi S, Ahadi P, et al. (2013) Nasal colonization of mecA-positive, oxacillin-susceptible, methicillin-resistant Staphylococcus aureus isolates among nursing staff in an Iranian teaching hospital. Am J Infect Control 41: 1122–1124. [DOI] [PubMed] [Google Scholar]
- 42. Sharff KA, Monecke S, Slaughter S, Forrest G, Pfeiffer C, et al. (2012) Genotypic resistance testing creates new treatment challenges: two cases of oxacillin-susceptible methicillin-resistant Staphylococcus aureus. J Clin Microbiol 50: 4151–4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Giannouli S, Labrou M, Kyritsis A, Ikonomidis A, Pournaras S, et al. (2010) Detection of mutations in the FemXAB protein family in oxacillin-susceptible mecA-positive Staphylococcus aureus clinical isolates. J Antimicrob Chemother 65: 626–633. [DOI] [PubMed] [Google Scholar]
- 44. Tokajian S, Haddad D, Andraos R, Hashwa F, Araj G (2011) Toxins and antibiotics resistance in Staphylococcus aureus isolated from a major hospital in Lebanon. ISRN Microbiol 812049: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ellington MJ, Yearwood L, Ganner M, East C, Kearns AM (2008) Distribution of the ACME-arcA gene among methicillin-resistant Staphylococcus aureus from England and Wales. J Antimicrob Chemother 61: 73–77. [DOI] [PubMed] [Google Scholar]
- 46. Aires-de-Sousa M, Parente CESR, Vieira-da-Motta O, Bonna ICF, Silva DA, et al. (2007) Characterization of Staphylococcus aureus isolates from Buffalo, Bovine, Ovine, and Caprine milk samples collected in Rio de Janeiro State, Brazil. Appl Environ Microbiol 73: 3845–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zecconi A, Cesaris L, Liandris E, Daprà V, Piccinini R (2006) Role of several Staphylococcus aureus virulence factors on the inflammatory response in bovine mammary gland. Microb Pathogenesis 40: 177–183. [DOI] [PubMed] [Google Scholar]
- 48. Boyle-Vavra S, Daum RS (2007) Community-acquired methicillin-resistant Staphylococcus aureus: the role of Panton-Valentine leukocidin. Lab Invest 87: 3–9. [DOI] [PubMed] [Google Scholar]