Abstract
Background
The use of molecular methods to diagnose Clostridium difficile infection (CDI) has improved diagnostic yield compared to conventional methods. However, PCR testing can detect colonization and has introduced several practical challenges pertaining to need for treatment and isolation of cases.
Methods
For all new cases detected by real-time PCR, concurrent cytotoxin assay was performed and genetic characterization with MLVA (multi-locus variable number tandem repeat analysis) was done to determine relatedness. We used PCR cycle threshold (Ct) of detection as surrogate marker for bacterial burden in stool.
Results
Overall, 54 cases of CDI were detected during the study period. 42 were concurrently tested by CYT and characterized by MLVA .MLVA analysis revealed marked genetic diversity with no ongoing outbreaks; four cases were due to NAP1 strain. CYT −/PCR + cases had a higher median Ct value of detection compared to CYT+/PCR + cases (28.2 vs 22.5; p = 0.01). Among 25 strains that were genetically related, 9/11 isolates in this dominant cluster were positive by CYT compared to 4/14 in non-dominant clusters (p = 0.02).
Conclusion
CYT−/PCR+ cases contribute to hospital based transmission. However, the risk of transmission of C. difficile from CYT +/PCR+ cases may be higher than those that are CYT−/PCR+.
Introduction
The introduction of molecular diagnostics into routine hospital care has brought remarkable accuracy and speed into the identification of numerous infections.
However, the increased sensitivity of molecular tests has identified many patients, whose infection would have escaped detection utilizing conventional methods, creating uncertainty about when, and for how long to isolate. Of particular concern is Clostridium difficile, the most common cause of hospital-acquired diarrhea. At many facilities, the overall detection of C. difficile associated diarrhea has increased by 50% or more due to improved sensitivity and favorable operation characteristics of the molecular test [1]–[3]. Yet the contagiousness of patients who are positive on molecular tests but negative by conventional methods is not known.
Recent epidemiologic studies based on CDI cases detected by Enzyme immunoassay (EIA) and culture positive samples have shown that approximately 25% of all CDI cases can be attributed to ward based transmission [4]. Whether additional cases that are detected by molecular methods (PCR) only contribute to hospital based transmission and has thus far been the undetected reservoir of infection, has never been formally studied. To examine this, we compared cases of C. difficile infection detected by PCR only (CYT negative) with cases detected both CYT and PCR in a hyperendemic pediatric population. We based assessment of transmission potential on bacterial carriage and genetic relatedness using the following,
Threshold cycle of detection as a surrogate marker for bacterial load and in turn greater risk of environmental contamination and;
Genetic relatedness using a highly discriminatory MLVA (multi-locus variable number tandem repeat analysis).
Methods
Memorial Sloan Kettering Cancer Center (MSKCC) is a 470-bed tertiary care hospital in New York City with a 39 bed inpatient pediatric unit. Each year, there are approximately 1,500 pediatric admissions and 11,000 pediatric patient days annually. The average length of stay for pediatrics is 7.4 days. The pediatric day hospital (PDH) is a 36 bed facility for outpatient chemotherapy administration and outpatient evaluation and management with about 100 visits per day. Samples were collected from September 2010 until March 2011.
Laboratory methods
All stool samples obtained from pediatric patients that tested positive for C.difficile were stored at −80°C within 24 hours of receipt in the lab. Patients with recurrent CDI or with duplicate specimens obtained within two weeks were excluded from the study.
Xpert C. difficile PCR
The assay was approved by the Food and Drug Administration (FDA) for the detection of C. difficile directly from stool specimens. The assay detects the toxin B gene within 1 hour with minimal hands-on time based on real-time PCR (Cepheid, Sunnyvale, CA). The Xpert C. difficile PCR (Xpert PCR) was performed according to the manufacturer's instructions and as previously described [5]. The cycle threshold (Ct) was defined as the number of PCR cycle required to generate a fluorescent signal above the background fluorescence [6]. It is a relative measure of the concentration of target gene in the PCR reaction.
Cytotoxin neutralization assay (CYT)
The CYT assay was performed as previously described. The assay detects the presence of the toxin B protein as measured through the presence of cytopathic effect in commercially available human lung fibroblast cell line (Diagnostics Hybrids, Athens, OH) [5].
C. difficile culture
C. difficile selective agar (CDSA; BD BBL, Sparks, MD) plates were reduced overnight in an anaerobic chamber prior to use. Stool sample was added to 500 µl of 100% ethanol, vortexed, and incubated at room temperature for 1 to 2 h. The solution was centrifuged at 1,200× g for 5 min, ethanol was removed, and the stool sample was inoculated onto reduced CDSA plates. The plates were incubated for 48 h under anaerobic conditions. Colonies resembling C. difficile (pale yellow to yellow) were sub cultured on sheep blood agar (SBA) plates, and their identity was further confirmed by Remel PRO disk (Thermo Fischer Scientific, Waltham, MA).
MLVA
MLVA and tcdC sequencing were performed as previously described [7]. Resulting tcdC sequences were assigned genotypes by querying the PubMLST database (http://www.pubmlst.org/cdifficile). Minimum spanning trees of the MLVA data were generated using BioNumerics software v6.6 (Applied Maths, Austin TX). The summed tandem repeat difference (STRD) was used as coefficient for determining genetic distance. Based on validation studies performed in an outbreak setting comparing MLVA to REA (restriction enzyme analysis) and whole genome sequencing (WGS) [8] [9], STRD genetic relationships were defined as follows-
Outbreak
Strains with STRD≤2 are considered highly related and representative of an outbreak [9].
Genetically related
Isolates with genotypes having STRD greater than 2 but ≤10 are considered genetically related but not part of an outbreak [8], [9].
Clinical data
C. difficile cases were defined by positive test (PCR or CYT assay). A retrospective chart review was conducted for all patients with positive specimens; clinical, laboratory and demographic data were extracted from the electronic MSKCC clinical information system. Demographic data included age and sex. Clinical data included underlying cancer, transplant type, inpatient stay and duration, presence or absence of diarrhea, previous CDI, and, when applicable, time to recurrence. Cases were defined as healthcare -associated (HA) or community acquired (CA) based on interval between admission and positive test result for CDI. An interval of ≥72 hours after admission was used to define HA cases of CDI. The MSKCC Internal Review Board reviewed the study and granted a HIPAA waiver of authorization.
Statistical analysis
Statistical significance testing was performed using chi-squared tests of independence for categorical variables. The Student's t-test was used for mean age, assuming equal variances. The Wilcoxon Rank Sum test was used for median age.
Results
A total of 54 new C. difficile cases occurred during the study period (September 2010 until March, 2011). Forty-seven samples were available for MLVA typing C. difficile could not be cultured from 5 frozen stool samples, 2 samples were lost during storage. Due to limited sample availability, 42/47 samples that were characterized by MLVA could be concurrently tested with CYT. These 42 samples were included in the final analysis.
For the 42 patients, the median age of the cohort was 11.2 years. 4/42 children were ≤2 years of age, 50% were females. Eighteen patients were allogeneic hematopoietic stem cell transplant (HSCT) recipients. Four patients had underlying ALL and five had neuroblastoma. 19/42 cases were healthcare associated and 38/42 (91%) of patients had diarrhea recorded at the time of testing. In three of the remaining four patients, the presence of diarrhea or change on bowel pattern could not be definitely ascertained due to presence of fecal incontinence, acute lower GI bleeding and chronic lower gastrointestinal GVHD. In one patient, testing was done for fever and abdominal pain that developed during stem cell infusion.
MLVA and tcdC typing
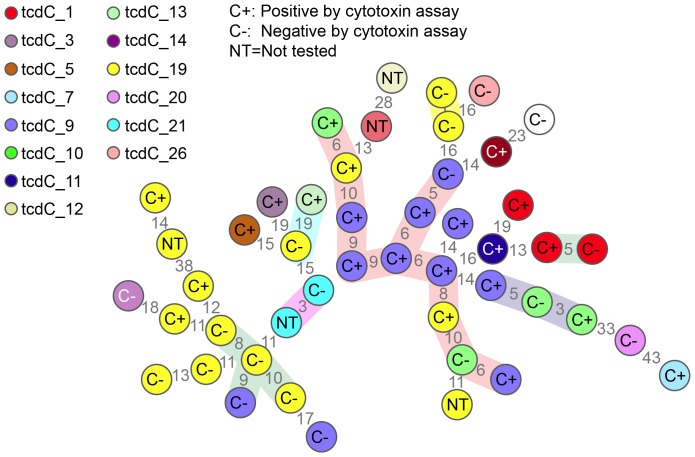
The results of MLVA typing of the study cohort are shown in Figure 1 along with results of CYT testing. For 42 isolates that were tested by CYT and characterized by MLVA, 23 were positive by CYT and 25 were genetically related. Among the 25 strains that were genetically related, 13 were positive by CYT compared to 10/17 strains that were unrelated (p = 0.3).One dominant cluster accounted for almost half of all the related strains (n = 11); isolates within this cluster are related by MLVA but are not part of an outbreak (8). Rather, the strain corresponds to common genetic lineages as defined by tcdC genotyping and represent endemic disease in the hospital setting. 9/11 isolates in this dominant cluster were positive by CYT compared to 4/14 in non-dominant clusters (p = 0.01).
Figure 1. Minimum spanning tree of MLVA data from study isolates (n = 47).
Letter symbol in the center of the circle represents the results for testing by cytotoxin assay for 42 samples included in the analysis (C + and C −). Five samples were not tested by CYT (NT). Each circle represents a distinct MLVA type and numbers between the circles represent the STRD [Summed tandem repeat difference]. Isolates with a STRD<10 are highlighted in colored clouds representing clusters (genetically related clonal complexes). tcdC sequencing is depicted by color coding within circles with tcdC 1 (corresponding to NAP1) strain represented in red (reference in right corner).
The tcdC-1 genotype (corresponding to Ribo type 027) was detected in four samples and was the third most common tcdC genotype isolated among our cohort. Isolates bearing the tcdC-19 genotype were the most common (32%) followed by tcdC-9 genotype which was also highly prevalent (21%).
Testing of PCR positive samples by CYT
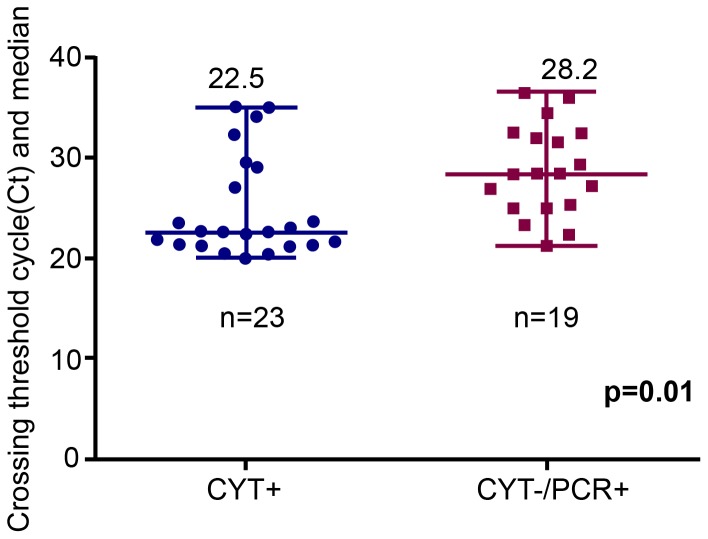
Among 42 samples on which CYT was performed, 23 were found to be positive by CYT. For samples included in the present study on which both CYT and PCR were performed, we found that CYT −/PCR + cases had a higher median Ct value of detection compared to CYT+/PCR + cases (28.2 vs 22.5; p = 0.01; 95%CI 0.85 to 6.8) [Figure 2].
Figure 2. PCR threshold cycle value (Ct) for forty-two samples tested by Cytotoxin assay (CYT) and PCR.
Discussion
The introduction of highly sensitive diagnostic tests has led to identification of additional cases of many diseases with the potential for hospital transmission. This in turn has led to a need to estimate the relative contagiousness of the additional cases detected only by molecular methods. To explore this, we examined the relationship between detection method and genetic relatedness as determined by MLVA in a group of C.difficile isolates collected over a six month period in a closed population of children with cancer.
We found a significant relationship between low Ct value and concurrent positivity on cytotoxin assay. However, we did not detect any difference in genetic relatedness of isolates detected by the different testing methods. The only striking finding is the concomitant positivity on CYT among majority (9/11) of isolates belonging to the dominant endemic strain in the cohort. Our findings establish that cases detected by PCR only contribute towards hospital based transmission. However, dominant endemic clones are likely to be CYT positive, whether this is related to higher bacterial load or strain characteristics, needs to be addressed in future studies.
Children with cancer are a particularly suitable group for examination of the transmission dynamics of CDI. Similar to adults, cancer poses a substantial risk for CDI in children. The rate of CDI is sixteen times higher in children with cancer than other hospitalized children; children at highest risk include those with hematological cancer or those undergoing hematopoietic stem cell transplant [10], [11].Secondly, children with cancer may have diarrhea for multiple reasons and diagnostic tests that detect toxin gene (PCR) rather than toxin itself may detect colonization. For many years, testing and treating infants and children for CDI was discouraged because of recognition that asymptomatic colonization was common [12]–[16]. However, the appearance of the hyper virulent NAP1 strain among older children in the community led to a new appreciation of the potential role of this organism in the young, as did reports of its role as a pathogen among children with cancer and other chronic medical conditions. As a result, over the last decade there has been a renewed interest in examining the epidemiology of C. difficile in healthy and hospitalized children [14], [17]–[19].
At the same time as the apparent shift in epidemiology of CDI [20]–[22], PCR based detection of C. difficile has increasingly been adopted for diagnosis. The implementation of this technology carries uncertainty due to potential over-detection since the test cannot distinguish between colonization and true disease. Many centers find that PCR increases the detection of C. difficile by up to two fold and the incremental cases detected by PCR only, often are clinically mild [2]. Recent study by Curry et al examines transmission pattern from hospitalized patients with asymptomatic C. difficile colonization compared to those with CDI(using found specimens submitted for screening of VRE), their findings attributed 30% of incident cases to CDI patients, whereas 29% cases were associated with carriers. Although PCR was not used for screening, this study highlights the role of asymptomatic colonization with C. difficile in hospital based transmission [23].
Our data in a closed population of children with cancer supports that incremental new cases detected by real-time PCR are genetically related to endemic strains and likely contribute towards previously undetected transmission; however most cases caused by the dominant endemic strain in our cohort were detected using conventional methods. Our study has several limitations; we were unable to retrieve seven samples in the cohort and therefore may have missed an outbreak, although this seems unlikely as the cases were spread out in time and space. We had very few patients less than 24 months old [seven patients] and are unable to draw any meaningful conclusions about strain prevalence and CDI related outcomes in this group of infants. We used a highly discriminatory typing method- although our results were objective, unambiguous and reproducible, comparisons with other studies that have used a wide variety of molecular typing methods, had to made indirectly by correlation with REA, PCR ribotypes and tcdC genotypes. We do not think this limit the findings of our study as establishment of such genetic relationships has been corroborated by other studies [7], [24].Also, Xpert C. difficile assay is only FDA approved as a qualitative assay, its use as a semi-quantitative assay will need further validation. Ct values were derived from stool samples- volume loaded on swabs may have had an impact on the results, although processing of samples is done following standard protocol and our findings support that such bias is unlikely to have influenced the typing results. Finally, the epidemiology of CDI may be different in pediatric patients with cancer as compared to other hospitalized children due to frequent healthcare related exposure, underlying immunosuppression and greater antibiotic use.
In summary, in a closed population of children with cancer, we have found that additional C. difficile cases detected by PCR only are genetically related to endemic strains in the hospital and represent previously undetected transmission, although the dominant endemic clones are likely to be positive when tested using conventional methods.
Acknowledgments
Perminder Khosa and Jeffery Stiles for technical support.
Funding Statement
This study was supported by grant K23 AI083880 [Source NIH/NIAID] awarded to MK. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kaltsas A, Simon M, Unruh LH, Son C, Wroblewski D, et al. (2012) Clinical and laboratory characteristics of Clostridium difficile infection in patients with discordant diagnostic test results. J Clin Microbiol 50: 1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Longtin Y, Trottier S, Brochu G, Paquet-Bolduc B, Garenc C, et al. (2013) Impact of the type of diagnostic assay on Clostridium difficile infection and complication rates in a mandatory reporting program. Clin Infect Dis 56: 67–73. [DOI] [PubMed] [Google Scholar]
- 3. Zahariadis G, Connon JJ, Fong IW (2002) Fulminant Clostridium difficile colitis without diarrhea: lack of emphasis in diagnostic guidelines. Am J Gastroenterol 97: 2929–2930. [DOI] [PubMed] [Google Scholar]
- 4. Walker AS, Eyre DW, Wyllie DH, Dingle KE, Harding RM, et al. (2012) Characterisation of Clostridium difficile hospital ward-based transmission using extensive epidemiological data and molecular typing. PLoS Med 9: e1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Babady NE, Stiles J, Ruggiero P, Khosa P, Huang D, et al. (2010) Evaluation of the Cepheid Xpert Clostridium difficile Epi assay for diagnosis of Clostridium difficile infection and typing of the NAP1 strain at a cancer hospital. J Clin Microbiol 48: 4519–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayden Wa Quantitative Molecular Methods, In Molecular Microbiology.
- 7. Marsh JW, O'Leary MM, Shutt KA, Sambol SP, Johnson S, et al. (2010) Multilocus variable-number tandem-repeat analysis and multilocus sequence typing reveal genetic relationships among Clostridium difficile isolates genotyped by restriction endonuclease analysis. J Clin Microbiol 48: 412–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eyre DW, Fawley WN, Best EL, Griffiths D, Stoesser NE, et al. (2013) Comparison of multilocus variable-number tandem-repeat analysis and whole-genome sequencing for investigation of Clostridium difficile transmission. J Clin Microbiol 51: 4141–4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marsh JW, O'Leary MM, Shutt KA, Pasculle AW, Johnson S, et al. (2006) Multilocus variable-number tandem-repeat analysis for investigation of Clostridium difficile transmission in Hospitals. J Clin Microbiol 44: 2558–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kamboj M, Son C, Cantu S, Chemaly RF, Dickman J, et al. (2012) Hospital-onset Clostridium difficile infection rates in persons with cancer or hematopoietic stem cell transplant: a C3IC network report. Infect Control Hosp Epidemiol 33: 1162–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tai E, Richardson LC, Townsend J, Howard E, McDonald LC (2011) Clostridium difficile infection among children with cancer. Pediatr Infect Dis J 30: 610–612. [DOI] [PubMed] [Google Scholar]
- 12. Brettle RP, Wallace E (1982) Clostridium difficile from stools of normal children. Lancet 1: 1193. [DOI] [PubMed] [Google Scholar]
- 13. Holst E, Helin I, Mardh PA (1981) Recovery of Clostridium difficile from children. Scand J Infect Dis 13: 41–45. [DOI] [PubMed] [Google Scholar]
- 14. Rousseau C, Poilane I, De Pontual L, Maherault AC, Le Monnier A, et al. (2012) Clostridium difficile carriage in healthy infants in the community: a potential reservoir for pathogenic strains. Clin Infect Dis 55: 1209–1215. [DOI] [PubMed] [Google Scholar]
- 15. Stark PL, Lee A, Parsonage BD (1982) Colonization of the large bowel by Clostridium difficile in healthy infants: quantitative study. Infect Immun 35: 895–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Viscidi R, Willey S, Bartlett JG (1981) Isolation rates and toxigenic potential of Clostridium difficile isolates from various patient populations. Gastroenterology 81: 5–9. [PubMed] [Google Scholar]
- 17. Kim J, Shaklee JF, Smathers S, Prasad P, Asti L, et al. (2012) Risk factors and outcomes associated with severe clostridium difficile infection in children. Pediatr Infect Dis J 31: 134–138. [DOI] [PubMed] [Google Scholar]
- 18. Rousseau C, Lemee L, Le Monnier A, Poilane I, Pons JL, et al. (2011) Prevalence and diversity of Clostridium difficile strains in infants. J Med Microbiol 60: 1112–1118. [DOI] [PubMed] [Google Scholar]
- 19. Toltzis P, Kim J, Dul M, Zoltanski J, Smathers S, et al. (2009) Presence of the epidemic North American Pulsed Field type 1 Clostridium difficile strain in hospitalized children. J Pediatr 154: 607–608. [DOI] [PubMed] [Google Scholar]
- 20. Bryant K, McDonald LC (2009) Clostridium difficile infections in children. Pediatr Infect Dis J 28: 145–146. [DOI] [PubMed] [Google Scholar]
- 21. Kim J, Smathers SA, Prasad P, Leckerman KH, Coffin S, et al. (2008) Epidemiological features of Clostridium difficile-associated disease among inpatients at children's hospitals in the United States, 2001–2006. Pediatrics 122: 1266–1270. [DOI] [PubMed] [Google Scholar]
- 22. Pepin J, Valiquette L, Alary ME, Villemure P, Pelletier A, et al. (2004) Clostridium difficile-associated diarrhea in a region of Quebec from 1991 to 2003: a changing pattern of disease severity. CMAJ 171: 466–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Curry SR, Muto CA, Schlackman JL, Pasculle AW, Shutt KA, et al. (2013) Use of multilocus variable number of tandem repeats analysis genotyping to determine the role of asymptomatic carriers in Clostridium difficile transmission. Clin Infect Dis 57: 1094–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Killgore G, Thompson A, Johnson S, Brazier J, Kuijper E, et al. (2008) Comparison of seven techniques for typing international epidemic strains of Clostridium difficile: restriction endonuclease analysis, pulsed-field gel electrophoresis, PCR-ribotyping, multilocus sequence typing, multilocus variable-number tandem-repeat analysis, amplified fragment length polymorphism, and surface layer protein A gene sequence typing. J Clin Microbiol 46: 431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]