Abstract
Background
A number of observational studies have been conducted to investigate the association of IL-10 gene polymorphisms with tuberculosis (TB) susceptibility. However, the results of different studies were inconsistent. The aim of this study was to investigate the relationship between IL-10 -1082G/A, -819T/C, and -592A/C polymorphisms and TB risk by meta-analysis.
Methods
A literature search was conducted among six English databases (PubMed, Embase, Web of Science, Science Direct, SpringerLink and EBSCO) and two Chinese databases (Wanfang and Chinese National Knowledge Infrastructure databases) to identify studies involving association between IL-10 −1082G/A, −819T/C, and −592A/C polymorphisms and TB susceptibility before May. 2013. Statistical analysis was performed using Revman 5.0 and Stata 12.0.
Results
A total of 31 studies with 6,559 cases and 7,768 controls were included in this meta-analysis. The results showed that three polymorphisms (-1082G/A, -819T/C, and -592A/C) in the IL-10 gene were not associated with the risk of TB in general population. In the subgroup analysis by ethnicity, IL-10 -1082G/A polymorphism was associated with TB risk in Europeans (AA+AG vs. GG: OR = 0.57, 95% CI = 0. 0.37–0.89, P = 0.01) and Americans (AA+AG vs. GG: OR = 0.39, 95% CI = 0.27–0.57, P<0.01), and IL-10 -819T/C (C allele vs. T allele: OR = 0.83, 95% CI = 0.72–0.96, P = 0.01) and -592A/C (CC+AC vs. AA: OR = 0.65, 95% CI = 0.49–0.85, P = 0.002) polymorphisms were significantly associated with TB risk in Asians.
Conclusion
This meta-analysis provides strong evidence that IL-10-1082G/A polymorphism was associated with TB risk in Europeans and Americans, and IL-10 -819T/C and -592A/C polymorphisms could be risk factors for TB in Asians. Additional well designed large studies were required for the validation of our results.
Introduction
Tuberculosis (TB) is a chronic infectious disease that occurs worldwide, leading to 1.6 million deaths annually worldwide [1]. However, from the estimated 2 billion individuals that have been initially infected with Mycobacterium tuberculosis (M. tuberculosis), only 5% to 10% develop symptomatic TB [2]. The exact reasons as to why only some of the individuals exposed to M. tuberculosis develop uncontrolled disease and others have an effective immune response to limit the spread of the pathogen remains unknown. The genetic influence on TB infection was established by several studies of monozygotic and dizygotic twins, linkage and candidate gene analysis, indicating that genetics may play a role in the susceptibility to TB infection [3]–[5].
A major determinant for the clinical expression of the different forms of TB, and their final outcome, is the interaction between the pathogen and the host immune system. Cytokines play an important role in anti-TB immune response, and cytokines interleukin -10 (IL-10) have been implicated in the pathogenesis of TB [6]. IL-10 is an important immunoregulatory cytokine mainly produced by macrophages, monocytes, T cells, B cells, dendritic cells, mast cells and eosinophils [7]. Turner J, et al. demonstrated that increased susceptibility to reactivation tuberculosis in the mouse model is strongly influenced by the expression of IL-10 during the chronic or latent phase of the infection [8]. IL-10 potentially helps M. tuberculosis persistence in humans by blocking phagosome maturation in macrophages [9]. The ability of IL-10 to down-regulate immune responses and the fact that IL-10 can be detected in tuberculosis patients have led researchers to investigate whether IL-10 plays a role in susceptibility to tuberculosis [10], [11].
Certain single nucleotide polymorphisms within the promoter region of the IL-10 gene have been associated with altered levels of circulating IL-10, such as −1082G/A, −819T/C, and −592A/C [12], [13]. These polymorphisms have been investigated as potential susceptibility factors for TB. Given the functional significance of this genetic variant, a number of case-control studies have been done in different populations to investigate its susceptibility towards TB, but the findings remain conflicting rather than conclusive. Therefore, we conducted a systematic review and meta-analysis to get a more precise estimate of the association between IL-10 polymorphisms and TB risk.
Materials and Methods
Literature Search Strategy
A literature search was conducted among six English databases (PubMed, Embase, Web of Science, Science Direct, SpringerLink and EBSCO) and two Chinese databases (Wanfang and Chinese National Knowledge Infrastructure databases) to identify studies involving association between IL-10 −1082G/A, −819T/C, and −592A/C polymorphisms and TB susceptibility before May. 2013. Key words used in the research included “interleukin”, “interleukin-10”, “cytokine”, “tuberculosis”, “Mycobacterium tuberculosis”, “single nucleotide polymorphism”, “variant”, “genotype”, “mutation”. To minimize potential publication bias, no restrictions were placed on language, sample size, and time period.
Inclusion and Exclusion Criteria
All included studies have to fulfill the following characteristics and inclusion criteria: (a) case-control studies focused on associations between IL-10 -1082G/A, -819T/C, and −592A/C polymorphisms and the risk of TB; (b) the diagnosis of TB should meet the internationally accepted criteria; (c) genotype distribution in both cases and controls were available for estimating an odds ratio (OR) with 95% confidence interval (CI). The exclusion criteria of the meta-analysis were: (a) animal studies; (b) meta-analyses, letters, reviews, meeting abstracts, or editorial comments; (c) studies with duplicate data, incomplete data, and unavailable data. When an individual author published several articles obtained from the same patient population, only the newest or most complete article was included in the analysis.
Data Extraction
Data were independently abstracted by two reviewers (Liang and Li) using a standard protocol and data-collection according to the inclusion criteria. The following data were collected from each study: first author’s name, year of publication, country, ethnicity, source of controls, sample size, genotyping method, and number of cases and controls for IL-10 −1082G/A, −819T/C, and −592A/C polymorphisms. An attempt was made to contact authors if data presentation was incomplete or if it was necessary to resolve an apparent conflict or inconsistency in the article. Any disagreements were resolved by consensus.
Statistical Analysis
Review manager 5.0 program provided by the Cochrane Library and Stata (Version12.0, Stata Corporation) were used to perform all the statistical analysis. The combined odds ratio (OR) with its 95% confident interval (CI) was used to assess the strength of the association between the IL-10 polymorphisms and TB risk. The significance of the combined OR was determined by the Z-test, in which P<0.05 was considered significant. The pooled ORs were calculated for allele model (mutation [M] allele versus wild [W] allele), dominant model (WM+MM versus WW), recessive model (MM versus WM+WW), homozygote comparison (MM versus WW), and heterozygote comparison (WM versus WW), respectively. Two models of pooling data for dichotomous outcomes were conducted: the random-effects model and the fixed-effects model. Heterogeneity assumption was assessed by the Chisquare based Q test and was regarded to be statistically significant if P<0.10. When the P≥0.10, the pooled statistical analysis was calculated by the fixed-effects model, otherwise, a random-effect model was used. To evaluate the ethnicity-specific effects, subgroup analyses were performed by ethnic group. The potential publication bias was assessed by Begg’s funnel plot and Egger’s test [14], [15].
Results
Characteristics of Studies
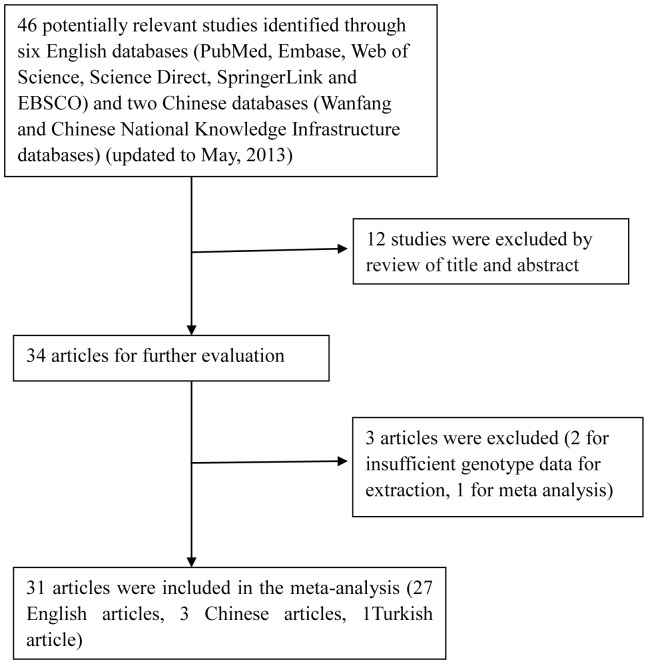
The flow chart that displays the study selection process was shown in Figure 1. A total of 31 case-control studies, including 6,559 cases and 7,768 controls, were finally identified according to inclusion and exclusion criteria. There are 29 case-control studies concerning -1082G/A polymorphism [16]–[44], 14 case-control studies concerning -819T/C polymorphism [16], [18], [20]–[23], [25], [31], [33], [37]–[39], [42], [45], and 16 case-control studies concerning -592A/C polymorphism [16], [20]–[25], [28], [31], [33], [37]–[40], [45], [46]. Among the 31 eligible studies, 17 of them were of Asians [16], [18], [19], [26], [28], [30]–[32], [34], [36], [39], [41]–[44], [46], 6 studies were of Europeans [17], [21], [25], [27], [35], [37], 5 studies were of Africans [20], [22], [29], [33], [38], and 3 studies were of Americans [23], [24], [40]. Controls were selected from healthy population in all studies and most studies used frequency-matched controls to the cases by age, sex, or ethnicity. The genotype distributions among the controls of all studies were in agreement with HWE except for ten studies for the -1082G/A, two studies for the -819T/C, and three studies for the -592A/C. The detailed characteristics of the eligible studies included in this meta-analysis were shown in Table 1.
Figure 1. Flow diagram of the literature search and trial selection process.
Table 1. Baseline characteristics of the 31 eligible studies for the analysis of IL-10 polymorphism.
| Studies | Year | Country | Ethnicity | Source of controls | Sample size | SNP studied | Genotyping method | HWE |
| Wu F | 2008 | China | Asian | HB | 61/122 | -1082G/A, -819T/C, -592A/C | PCR-RFLP | 0.379, 0.125, 0.125 |
| Scola A | 2003 | Italy | European | PB | 45/114 | -1082G/A | ARMS-PCR | <0.001 |
| Selvaraj P | 2008 | India | Asian | PB | 155/183 | -1082G/A, -819T/C | PCR-RFLP | 0.204, 0.174 |
| Sharada RS | 2012 | India | Asian | HB | 104/102 | -1082G/A | ARMS-PCR | 0.057 |
| Bellamy R | 1998 | Gambia | African | HB | 401/408 | -1082G/A, -819T/C, -592A/C | PCR-SSP | 0.824, 0.779, 0.779 |
| Oral HB | 2006 | Turkey | European | HB | 81/50 | -1082G/A, -819T/C, -592A/C | PCR-SSP | 0.060, 0.320, 0.320 |
| Ben-Selma W | 2011 | Tunisian | African | HB | 131/95 | -1082G/A, -819T/C, -592A/C | PCR-RFLP | 0.020, 0.957, 0.957 |
| Heno MI | 2006 | Colombia | American | HB | 190/135 | -1082G/A, -819T/C, -592A/C | PCR-SSP | 0.674, 0.410, 0.518 |
| Garcia-Elorriaga G | 2006 | Mexico | American | HB | 77/60 | -1082G/A, -592A/C | Taqman | 0.728, <0.001 |
| Ates O | 2008 | Turkey | Asian | HB | 128/80 | -1082G/A, -819T/C, -592A/C | ARMS-PCR | 0.978, 0.819, 0.819 |
| Delgado JC | 2002 | Cambodian | Asian | HB | 356/106 | -1082G/A, | PCR-RFLP | <0.001 |
| Ulger M | 2013 | Turkey | European | HB | 84/110 | -1082G/A | PCR-RFLP | <0.001 |
| Shin HD | 2005 | Korea | Asian | HB | 449/851 | -1082G/A, -592A/C | MAPA | 0.168, 0.631, |
| Mosaad YM | 2010 | Egypt | African | HB | 110/118 | -1082G/A | ARMS-PCR | <0.001 |
| Oh JH | 2007 | Korea | Asian | HB | 145/117 | -1082G/A | ARMS-PCR | 0.612 |
| Liang L | 2011 | China | Asian | HB | 235/78 | -1082G/A, -819T/C, -592A/C | SNaPshot assay | 0.589, 0.253, 0.253 |
| Ansari A | 2009 | Pakistan | Asian | PB | 178/376 | -1082G/A | ARMS-PCR | <0.001 |
| Thye T | 2009 | Ghana | African | PB | 2010/2346 | -1082G/A, -819T/C, -592A/C | FRET | 0.542, 0.380, 0.551 |
| Ansari A | 2011 | Pakistan | Asian | PB | 102/166 | -1082G/A | ARMS-PCR | <0.001 |
| Lopez-Maderueyo D | 2003 | Spain | European | HB | 113/100 | -1082G/A | ARMS-PCR | 0.949 |
| Meenakshi P | 2013 | India | Asian | HB | 100/100 | -1082G/A | PCR-RFLP | 0.058 |
| Mhmoud N | 2013 | Sudan | Asian | HB | 191/206 | -819T/C, -592A/C | PCR-RFLP | <0.001, <0.001 |
| Mei | 2012 | China | Asian | HB | 169/156 | -592A/C | PCR-RFLP | 0.622 |
| Ma Z | 2007 | China | Asian | HB | 40/40 | -1082G/A | PCR-SSP | 0.292 |
| Yang H | 2010 | China | Asian | HB | 198/200 | -1082G/A | PCR-SSP | 0.253 |
| Liu XX | 2009 | China | Asian | HB | 141/135 | -1082G/A, -819T/C | PCR-RFLP | <0.001, <0.001 |
| Trajkov D | 2009 | Macedonia | European | PB | 75/299 | -1082G/A, -819T/C, -592A/C | PCR-SSP | <0.001, 0.879, 0.403 |
| Fitness J | 2004 | Malawi | African | HB | 210/705 | -1082G/A, -819T/C, -592A/C | ARMS-PCR | 0.524, 0.062, 0.035 |
| Amirzargar AA | 2006 | Iran | Asian | HB | 41/123 | -1082G/A, -819T/C, -592A/C | PCR-SSP | <0.001, 0.671, 0.671 |
| Taype CA | 2010 | Peru | American | HB | 626/513 | -1082G/A, -592A/C | Taqman PCR | 0.142, 0.055 |
| Prabhu Anand S | 2007 | India | Asian | HB | 132/143 | -1082G/A | PCR-RFLP | 0.123 |
PB, population-based controls, HB, hospital-based controls. HWE, Hardy–Weinberg equilibrium. PCR, polymerase chain reaction; SSP, sequence-specific primers; ARMS, amplification refractory mutation system; RFLP, restriction fragment length polymorphism; FRET, fluorescence resonance energy transfer. MAPA, multiplex automated primer extension analysis.
Quantitative Data Synthesis
The evaluation of association between IL-10 polymorphisms and TB risk was presented in Table 2.
Table 2. Determination of the genetic effects of IL-10 polymorphisms on TB and subgroup analysis.
| Allele model | Dominant model | Recessive model | Homozygous model | Heterozygous model | |||||||
| OR [95% CI] | P | OR [95% CI] | P | OR [95% CI] | P | OR [95% CI] | P | OR [95% CI] | P | ||
| -1082G/A | A allele vs. G allele | AA+GA vs. GG | AA vs. GG+GA | AA vs. GG | GA vs. GG | ||||||
| overall | 0.97 [0.79–1.20] | 0.81 | 0.95 [0.68–1.34] | 0.79 | 0.92 [0.75–1.14] | 0.46 | 0.90 [0.61–1.33] | 0.59 | 0.99 [0.72–1.36] | 0.96 | |
| Asian | 1.15 [0.89–1.47] | 0.27 | 1.48 [0.69–3.17] | 0.32 | 1.24 [0.91–1.69] | 0.17 | 1.65 [0.74–3.72] | 0.22 | 1.39 [0.67–2.90] | 0.38 | |
| European | 0.75 [0.56–1.01] | 0.05 | 0.57 [0.37–0.89] | 0.01 | 0.67 [0.36–1.26] | 0.21 | 0.53 [0.26–1.08] | 0.08 | 0.60 [0.39–0.93] | 0.02 | |
| African | 1.13 [0.62–2.07] | 0.69 | 1.03 [0.85–1.25] | 0.78 | 0.76 [0.53–1.09] | 0.13 | 0.84 [0.56–1.24] | 0.37 | 1.09 [0.90–1.32] | 0.40 | |
| American | 0.58 [0.32–1.06] | 0.07 | 0.39 [0.27–0.57] | <0.01 | 0.53 [0.20–1.41] | 0.20 | 0.31 [0.13–0.77] | 0.01 | 0.46 [0.32–0.68] | <0.01 | |
| -819T/C | C allele vs. T allele | CC+TC vs. TT | CC vs. TC+TT | CC vs. TT | TC vs. TT | ||||||
| overall | 0.98 [0.92–1.05] | 0.59 | 1.03 [0.85–1.25] | 0.77 | 0.87 [0.73–1.05] | 0.15 | 0.98 [0.85–1.12] | 0.73 | 1.09 [0.85–1.41] | 0.49 | |
| Asian | 0.83 [0.72–0.96] | 0.01 | 0.94 [0.57–1.54] | 0.80 | 0.60 [0.40–0.90] | 0.01 | 0.63 [0.45–0.88] | 0.006 | 1.10 [0.57–2.10] | 0.78 | |
| European | 1.09 [0.84–1.41] | 0.51 | 1.27 [0.70–2.31] | 0.43 | 1.08 [0.75–1.56] | 0.67 | 1.33 [0.71–2.49] | 0.38 | 1.19 [0.63–2.24] | 0.59 | |
| African | 1.02 [0.94–1.10] | 0.62 | 1.08 [0.94–1.25] | 0.27 | 0.99 [0.88–1.11] | 0.88 | 1.06 [0.90–1.25] | 0.46 | 1.09 [0.93–1.27] | 0.29 | |
| American | 1.00 [0.72–1.38] | 1.00 | 0.89 [0.48–1.64] | 0.71 | 1.06 [0.68–1.66] | 0.79 | 0.94 [0.49–1.81] | 0.85 | 0.85 [0.44–1.63] | 0.62 | |
| -592A/C | C allele vs. A allele | CC+AC vs. AA | CC vs. AC+AA | CC vs. AA | AC vs. AA | ||||||
| overall | 0.99 [0.83–1.18] | 0.90 | 0.89 [0.74–1.08] | 0.25 | 0.92 [0.78–1.09] | 0.32 | 0.87 [0.68–1.11] | 0.27 | 0.90 [0.76–1.07] | 0.24 | |
| Asian | 0.69 [0.57–0.85] | <0.01 | 0.65 [0.49–0.85] | 0.002 | 0.62 [0.49–0.79] | <0.01 | 0.49 [0.34–0.71] | <0.01 | 0.70 [0.55–0.89] | <0.01 | |
| European | 1.19 [0.92–1.54] | 0.18 | 1.47 [0.81–2.66] | 0.20 | 1.18 [0.85–1.65] | 0.32 | 1.57 [0.85–2.90] | 0.15 | 1.34 [0.72–2.52] | 0.36 | |
| African | 1.01 [0.92–1.10] | 0.90 | 1.10 [0.94–1.30] | 0.24 | 0.95 [0.83–1.08] | 0.45 | 1.06 [0.88–1.27] | 0.54 | 1.14 [0.96–1.35] | 0.14 | |
| American | 1.56 [1.08–2.27] | 0.02 | 1.04 [0.79–1.36] | 0.79 | 1.24 [0.92–1.69] | 0.16 | 1.26 [0.94–1.69] | 0.13 | 0.86 [0.64–1.15] | 0.30 | |
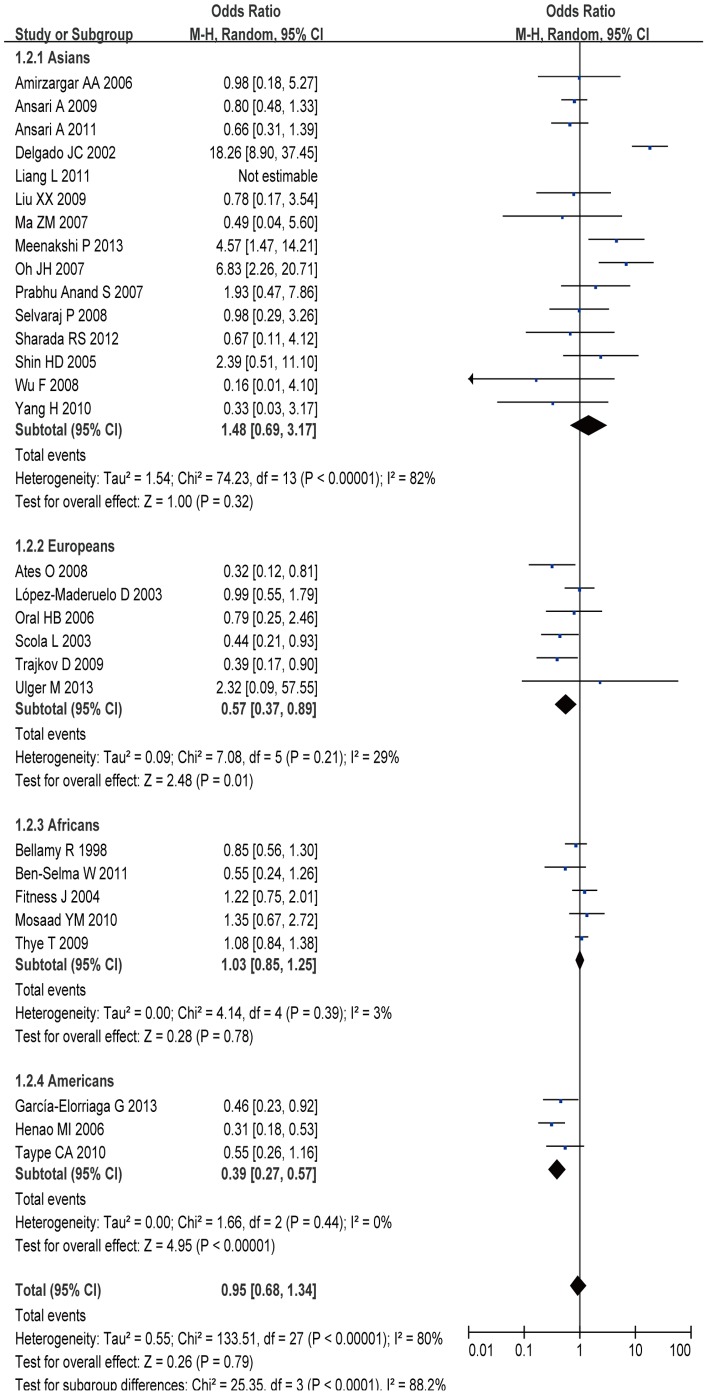
Of the 31 studies investigating the association between IL-10 -1082G/A polymorphism and TB susceptibility, 29 provided enough data to calculate ORs, including 6,199 cases and 7,406 controls. The results of pooling all studies showed that the IL10 -1082 G/A polymorphism was not associated with TB susceptibility in general population under all genetic models (A allele vs. G allele: OR = 0.97, 95% CI = 0.79–1.20, P = 0.81; AA+GA vs. GG: OR = 0.95, 95% CI = 0.68–1.34, P = 0.79; AA vs. GA+ GG: OR = 0.92, 95% CI = 0.75–1.14, P = 0.46; AA vs. GG: OR = 0.90, 95% CI = 0.61–1.33, P = 0.59; GA vs. GG: OR = 0.99, 95% CI = 0.72–1.36, P = 0.96) (Figure 2). In the stratified analysis by ethnicity, we found that TB risk was significant decreased in European group under dominant model (Figure 2) (AA+GA vs. GG: OR = 0.57, 95% CI = 0.37–0.89, P = 0.01) and heterozygous model (GA vs. GG: OR = 0.60, 95% CI = 0.39–0.93, P = 0.02). However, no significant association between this polymorphism and TB risk was observed in other comparison models in European group. Moreover, significant increased TB risk was observed in dominant model (Figure 2) (AA+GA vs. GG: OR = 0.39, 95% CI = 0.27–0.57, P<0.01), homozygous model (AA vs. GG: OR = 0.31, 95% CI = 0.13–0.77, P = 0.01), and heterozygous model (GA vs. GG: OR = 0.46, 95% CI = 0.32–0.68, P<0.01) in American group.
Figure 2. Meta-analysis with a random-effect model for the ORs of tuberculosis risk associated with IL-10 -1082G/A polymorphism in dominant genetic model comparison.
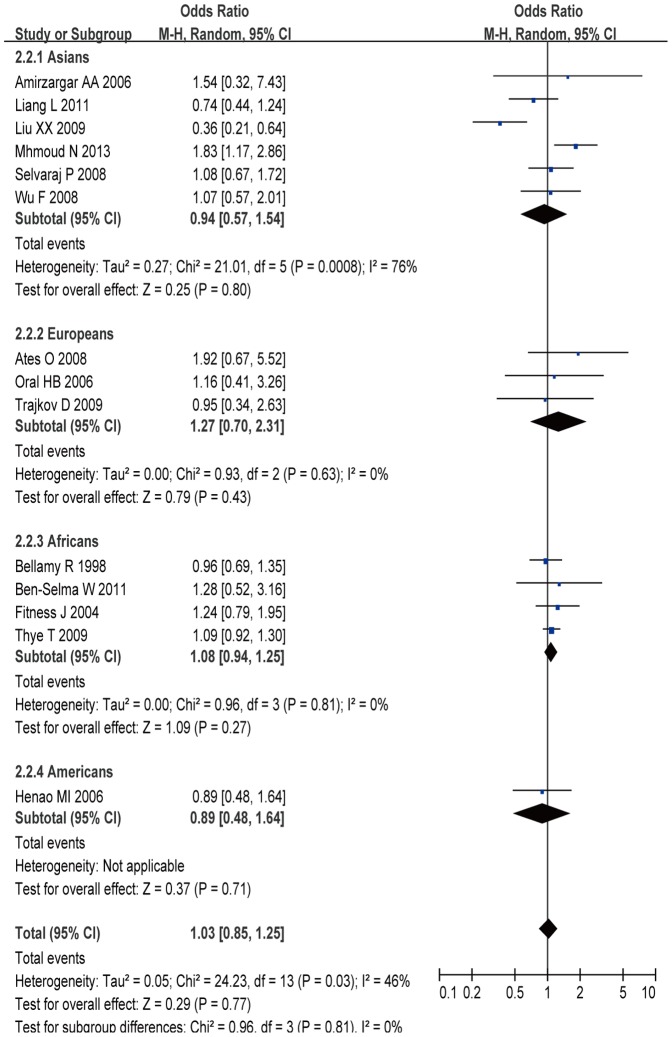
A total of 3,584 cases and 4,584 controls from 14 case-control studies were included for data synthesis. The results showed that there was no statistically significant association between IL-10 -819C/T polymorphism and TB risk in general population (C allele vs. T allele: OR = 0.98, 95% CI = 0.92–1.05, P = 0.59; CC+TC vs. TT: OR = 1.03, 95% CI = 0.85–1.25, P = 0.77; CC vs. TC+TT: OR = 0.87, 95% CI = 0.73–1.05, P = 0.15; CC vs. TT: OR = 0.98, 95% CI = 0.85–1.12, P = 0.73; TC vs. TT: OR = 1.09, 95% CI = 0.85–1.41, P = 0.49) (Figure 3). In the stratified analyses for the -819C/T polymorphism, there was a significantly decreased risk was observed among Asians in allele model (C allele vs. T allele: OR = 0.83, 95% CI = 0.72–0.96, P = 0.01), homozygous model (CC vs. TT: OR = 0.60, 95% CI = 0.40–0.90, P = 0.01), and heterozygous model (TC vs. TT: OR = 0.63, 95% CI = 0.45–0.88, P = 0.006).
Figure 3. Meta-analysis with a random-effect model for the ORs of tuberculosis risk associated with IL-10 -819T/C polymorphism in dominant genetic model comparison.
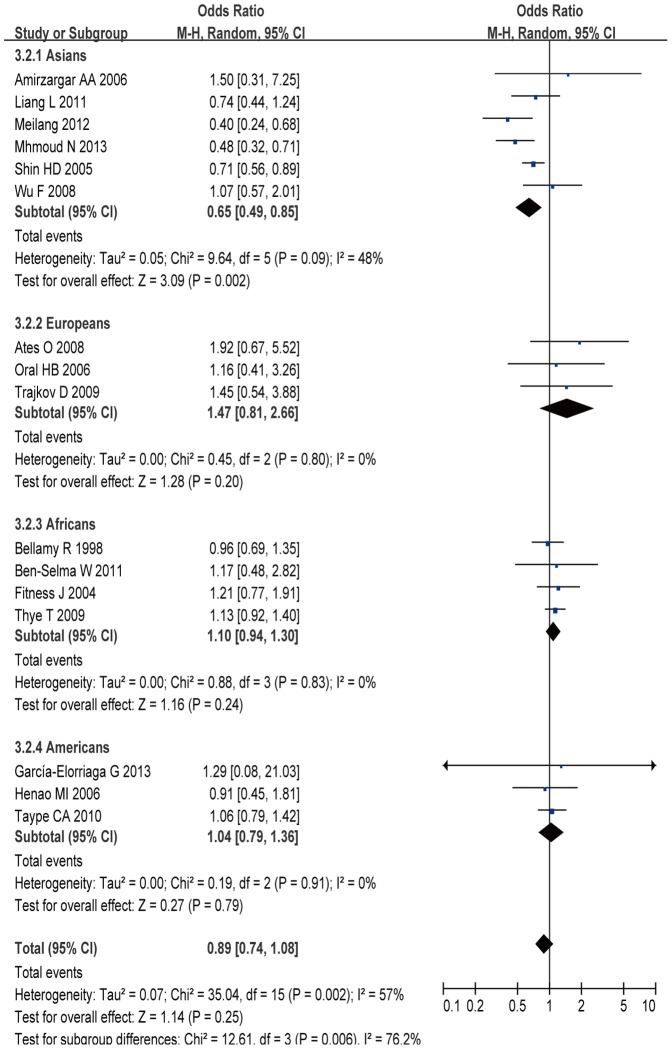
A total of 4,063 cases and 5,326 controls from 16 case-control studies were included for data synthesis. In the current meta-analysis, we did not find a significant relationship between IL-10 -592A/C polymorphism and TB risk (C allele vs. A allele: OR = 0.99, 95% CI = 0.83–1.18, P = 0.90; CC+AC vs. AA: OR = 0.89, 95% CI = 0.74–1.08, P = 0.25; CC vs. AC+AA: OR = 0.92, 95% CI = 0.78–1.09, P = 0.32; CC vs. AA: OR = 0.87, 95% CI = 0.68–1.11, P = 0.27; AC vs. AA: OR = 0.90, 95% CI = 0.76–1.07, P = 0.24) (Figure 4). In the subgroup analysis by ethnicity, the results indicated that there was significant association between IL-10 -592A/C polymorphism and TB risk in Asians under all gene models (C allele vs. A allele: OR = 0.69, 95% CI = 0.57–0.85, P<0.01; CC+AC vs. AA: OR = 0.65, 95% CI = 0.49–0.85, P = 0.002; CC vs. AC+AA: OR = 0.62, 95% CI = 0.49–0.79, P<0.01; CC vs. AA: OR = 0.49, 95% CI = 0.34–0.71, P<0.01; AC vs. AA: OR = 0.70, 95% CI = 0.55–0.89, P<0.01), but not in Europeans, Africans, and Americans, suggesting genetic diversity among ethnicities.
Figure 4. Meta-analysis with a random-effect model for the ORs of tuberculosis risk associated with IL-10 -592A/C polymorphism in dominant genetic model comparison.
Publication Bias
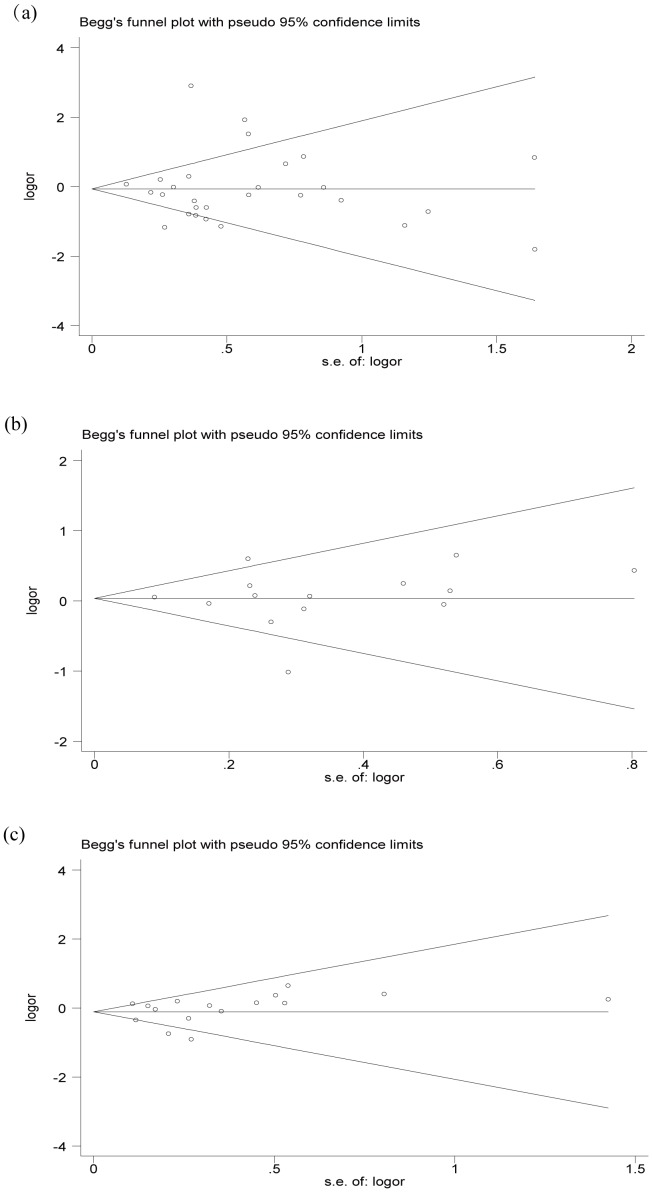
Begg’s funnel plot and Egger’s test were performed to assess the publication bias of included studies. The shapes of the funnel plots did not reveal any evidence of obvious asymmetry under the dominant model (-1082G/A, P = 0.953; -819T/C, P = 0.661; -592A/C, P = 0.685) (Figure. 5). Egger’s test also did not show any significantly statistical evidence of publication bias under the dominant model (-1082G/A, P = 0.992; -819T/C, P = 0.981; -592A/C, P = 0.712), which indicated low risk of publication bias in this meta-analysis.
Figure 5. Funnel plot for publication bias of the meta-analysis of tuberculosis risk and IL-10 polymorphisms in dominant genetic model comparison.
(a) IL-10 -1082G/A polymorphism, (b) IL-10 -819T/C polymorphism, (c) IL-10 -592A/C polymorphism.
Discussion
To date, convincing evidence indicate that the outcome of TB is modulated by the environment as well as bacterial and host genetic components. Many investigations have confirmed that cytokines appear to play the critical roles in the development of TB. Polymorphisms in several cytokine genes have been described and demonstrated to influence gene transcription, leading to interindividual variations in cytokine [47]. IL-10 is a powerful T helper 2 regulatory cytokine and plays an essential role during the latent TB stage, where increased production of this cytokine promotes reactivation of disease in mice [8] and suppression of cell-mediated immunity against the intracellular infection [4]. IL-10 gene is located on the long arm of chromosome 1, where several polymorphisms have been identified within the promoter region, such as −1082G/A, −819T/C, and −592A/C. Genetic studies showed that IL-10 polymorphisms in the promoter region were associated with TB risk, and a number of studies have been performed to investigate that association. However, inconclusive results were obtained. To provide further investigation into these controversial points, a meta-analysis is needed to achieve a more reliable and comprehensive conclusion.
Based on a meta-analysis from 18 studies that contained 4740 cases and 5919 controls, Zhang J, et al. found that -819C/T and -592A/C polymorphisms do not affect susceptibility to TB, while the -1082G/A polymorphism was significantly associated with decreased risk of TB only in Europeans [48]. Our meta-analysis, which involved 31 studies including 6559 cases and 7768 controls, also found that the presence of the -1082G/A, -819T/C, and -592A/C genotypes was not associated with the risk of TB in the general population. In our analysis, there was evidence of heterogeneity between studies. It may be due to some factors, including the ethnicity, the selection of methods, definition of cases, and sample sizes.
As recent reports showed that genotype frequencies at IL-10 vary greatly in different populations, particularly in individuals of different ethnicities [49], a subgroup analysis was conducted in our study. Our subgroup analyses showed that the IL-10 -1082G/A genotype significantly decreased TB risk in Europeans and Americans, but not in Asians or Africans. The individuals who carry variant A allele (AA+GA) had a nearly 43% and 63% decreased risk of TB in Europeans and Americans, respectively, suggesting a possible role of ethnic differences in genetic backgrounds and environmental exposures. In stratified analyses for IL-10 -819 T/C, we observed a significant association between IL-10 -819 T/C polymorphism and TB risk in Asians under allele model, recessive model, and homozygous model. We also observed an association between IL-10 -592A/C polymorphism and TB risk in Asians under all gene models. In the case of European population, there were only 4 studies containing 284 cases and 429 controls for IL-10 -819T/C and IL-10 -592A/C polymorphisms analysis. As for American population, there were only one study containing 190 cases and 235 controls for IL-10 -819T/C polymorphism analysis. Therefore, our results should be interpreted with caution, and more case-control studies based on larger sample size of the different ethnicity population should be carried out in the future.
Some limitations of this meta-analysis should be considered when explaining our results. First, given that only published studies were included in the meta-analysis, publication bias may be present, although our results of publication bias showed no significance. Second, some studies were not in agreement with the HWE, making the sample a poor representation. Third, significant between-study heterogeneity was observed in some comparisons, and as such, results may be distorted. Different ethnic populations and different sources of controls may contribute to the heterogeneity. Forth, the interaction of different susceptibility genes and environment factors leaded to the disease, but our study could not assess gene-gene and gene-environment interactions due to the limited information of included studies. Last, but not the least, meta-analysis remains a retrospective research that is subject to the methodological deficiencies of the included studies. In view of these limitations, further studies should focus on the associations of gene polymorphisms and clinical or laboratory characteristic in a large cohort of TB patients.
Conclusion
In conclusion, despite the several considerations mentioned above, this meta-analysis indicated that three polymorphisms (-1082G/A, -819T/C, and -592A/C) in the IL-10 gene were not associated with the risk of TB in general population. In the subgroup analysis, IL-10 -1082G/A polymorphism was associated with TB risk in Europeans and Americans, and IL-10 -819T/C and -592A/C polymorphisms were significantly associated with TB risk in Asians. In the future, additional large studies are warranted to validate our findings. Future studies should include multi-ethnic groups and use standardized unbiased genotyping methods, and well-matched controls.
Supporting Information
PRISMA 2009 Checklist.
(WIZ)
Funding Statement
This study was supported by Liaoning Natural Science Fund, Liaoning Province, China (Topic No: 201102257). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Zembrzuski VM, Basta PC, Callegari-Jacques SM, Santos RV, Coimbra CE, et al. (2010) Cytokine genes are associated with tuberculin skin test response in a native Brazilian population. Tuberculosis (Edinb) 90: 44–49. [DOI] [PubMed] [Google Scholar]
- 2. Kleinnijenhuis J, Oosting M, Joosten LA, Netea MG, Van Crevel R (2011) Innate immune recognition of Mycobacterium tuberculosis. Clin Dev Immunol 2011: 405310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hill AV (2001) The genomics and genetics of human infectious disease susceptibility. Annu Rev Genomics Hum Genet 2: 373–400. [DOI] [PubMed] [Google Scholar]
- 4. Moller M, Hoal EG (2010) Current findings, challenges and novel approaches in human genetic susceptibility to tuberculosis. Tuberculosis (Edinb) 90: 71–83. [DOI] [PubMed] [Google Scholar]
- 5. Vannberg FO, Chapman SJ, Hill AV (2011) Human genetic susceptibility to intracellular pathogens. Immunol Rev 240: 105–116. [DOI] [PubMed] [Google Scholar]
- 6. Knight J (2001) Polymorphisms in Tumor Necrosis Factor and Other Cytokines As Risks for Infectious Diseases and the Septic Syndrome. Curr Infect Dis Rep 3: 427–439. [PubMed] [Google Scholar]
- 7. Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG (2011) Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu Rev Immunol 29: 71–109. [DOI] [PubMed] [Google Scholar]
- 8. Turner J, Gonzalez-Juarrero M, Ellis DL, Basaraba RJ, Kipnis A, et al. (2002) In vivo IL-10 production reactivates chronic pulmonary tuberculosis in C57BL/6 mice. J Immunol 169: 6343–6351. [DOI] [PubMed] [Google Scholar]
- 9. O'Leary S, O'Sullivan MP, Keane J (2011) IL-10 blocks phagosome maturation in mycobacterium tuberculosis-infected human macrophages. Am J Respir Cell Mol Biol 45: 172–180. [DOI] [PubMed] [Google Scholar]
- 10. Barnes PF, Lu S, Abrams JS, Wang E, Yamamura M, et al. (1993) Cytokine production at the site of disease in human tuberculosis. Infect Immun 61: 3482–3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Verbon A, Juffermans N, Van Deventer SJ, Speelman P, Van Deutekom H, et al. (1999) Serum concentrations of cytokines in patients with active tuberculosis (TB) and after treatment. Clin Exp Immunol 115: 110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Crawley E, Kay R, Sillibourne J, Patel P, Hutchinson I, et al. (1999) Polymorphic haplotypes of the interleukin-10 5' flanking region determine variable interleukin-10 transcription and are associated with particular phenotypes of juvenile rheumatoid arthritis. Arthritis Rheum 42: 1101–1108. [DOI] [PubMed] [Google Scholar]
- 13. Maurer M, Kruse N, Giess R, Toyka KV, Rieckmann P (2000) Genetic variation at position -1082 of the interleukin 10 (IL10) promotor and the outcome of multiple sclerosis. J Neuroimmunol 104: 98–100. [DOI] [PubMed] [Google Scholar]
- 14. Begg CB, Mazumdar M (1994) Operating characteristics of a rank correlation test for publication bias. Biometrics 50: 1088–1101. [PubMed] [Google Scholar]
- 15. Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu F, Qu Y, Tang Y, Cao D, Sun P, et al. (2008) Lack of association between cytokine gene polymorphisms and silicosis and pulmonary tuberculosis in Chinese iron miners. J Occup Health 50: 445–454. [DOI] [PubMed] [Google Scholar]
- 17. Scola L, Crivello A, Marino V, Gioia V, Serauto A, et al. (2003) IL-10 and TNF-alpha polymorphisms in a sample of Sicilian patients affected by tuberculosis: implication for ageing and life span expectancy. Mech Ageing Dev 124: 569–572. [DOI] [PubMed] [Google Scholar]
- 18. Selvaraj P, Alagarasu K, Harishankar M, Vidyarani M, Nisha Rajeswari D, et al. (2008) Cytokine gene polymorphisms and cytokine levels in pulmonary tuberculosis. Cytokine 43: 26–33. [DOI] [PubMed] [Google Scholar]
- 19. Ramaseri Sunder S, Hanumanth SR, Nagaraju RT, Venkata SK, Suryadevara NC, et al. (2012) IL-10 high producing genotype predisposes HIV infected individuals to TB infection. Hum Immunol 73: 605–611. [DOI] [PubMed] [Google Scholar]
- 20. Bellamy R, Ruwende C, Corrah T, McAdam KP, Whittle HC, et al. (1998) Assessment of the interleukin 1 gene cluster and other candidate gene polymorphisms in host susceptibility to tuberculosis. Tuber Lung Dis 79: 83–89. [DOI] [PubMed] [Google Scholar]
- 21. Oral HB, Budak F, Uzaslan EK, Basturk B, Bekar A, et al. (2006) Interleukin-10 (IL-10) gene polymorphism as a potential host susceptibility factor in tuberculosis. Cytokine 35: 143–147. [DOI] [PubMed] [Google Scholar]
- 22. Ben-Selma W, Harizi H, Boukadida J (2011) Association of TNF-alpha and IL-10 polymorphisms with tuberculosis in Tunisian populations. Microbes Infect 13: 837–843. [DOI] [PubMed] [Google Scholar]
- 23. Henao MI, Montes C, Paris SC, Garcia LF (2006) Cytokine gene polymorphisms in Colombian patients with different clinical presentations of tuberculosis. Tuberculosis (Edinb) 86: 11–19. [DOI] [PubMed] [Google Scholar]
- 24. Garcia-Elorriaga G, Vera-Ramirez L, del Rey-Pineda G, Gonzalez-Bonilla C (2013) -592 and -1082 interleukin-10 polymorphisms in pulmonary tuberculosis with type 2 diabetes. Asian Pac J Trop Med 6: 505–509. [DOI] [PubMed] [Google Scholar]
- 25. Ates O, Musellim B, Ongen G, Topal-Sarikaya A (2008) Interleukin-10 and tumor necrosis factor-alpha gene polymorphisms in tuberculosis. J Clin Immunol 28: 232–236. [DOI] [PubMed] [Google Scholar]
- 26. Delgado JC, Baena A, Thim S, Goldfeld AE (2002) Ethnic-specific genetic associations with pulmonary tuberculosis. J Infect Dis 186: 1463–1468. [DOI] [PubMed] [Google Scholar]
- 27. Ulger M, Emekdas G, Aslan G, Tas D, Ilvan A, et al. (2013) [Determination of the cytokine gene polymorphism and genetic susceptibility in tuberculosis patients]. Mikrobiyol Bul 47: 250–264. [DOI] [PubMed] [Google Scholar]
- 28. Shin HD, Park BL, Kim YH, Cheong HS, Lee IH, et al. (2005) Common interleukin 10 polymorphism associated with decreased risk of tuberculosis. Exp Mol Med 37: 128–132. [DOI] [PubMed] [Google Scholar]
- 29. Mosaad YM, Soliman OE, Tawhid ZE, Sherif DM (2010) Interferon-gamma +874 T/A and interleukin-10 -1082 A/G single nucleotide polymorphism in Egyptian children with tuberculosis. Scand J Immunol 72: 358–364. [DOI] [PubMed] [Google Scholar]
- 30. Oh JH, Yang CS, Noh YK, Kweon YM, Jung SS, et al. (2007) Polymorphisms of interleukin-10 and tumour necrosis factor-alpha genes are associated with newly diagnosed and recurrent pulmonary tuberculosis. Respirology 12: 594–598. [DOI] [PubMed] [Google Scholar]
- 31. Liang L, Zhao YL, Yue J, Liu JF, Han M, et al. (2011) Interleukin-10 gene promoter polymorphisms and their protein production in pleural fluid in patients with tuberculosis. FEMS Immunol Med Microbiol 62: 84–90. [DOI] [PubMed] [Google Scholar]
- 32. Ansari A, Talat N, Jamil B, Hasan Z, Razzaki T, et al. (2009) Cytokine gene polymorphisms across tuberculosis clinical spectrum in Pakistani patients. PLoS One 4: e4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thye T, Browne EN, Chinbuah MA, Gyapong J, Osei I, et al. (2009) IL10 haplotype associated with tuberculin skin test response but not with pulmonary TB. PLoS One 4: e5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ansari A, Hasan Z, Dawood G, Hussain R (2011) Differential combination of cytokine and interferon- gamma +874 T/A polymorphisms determines disease severity in pulmonary tuberculosis. PLoS One 6: e27848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lopez-Maderuelo D, Arnalich F, Serantes R, Gonzalez A, Codoceo R, et al. (2003) Interferon-gamma and interleukin-10 gene polymorphisms in pulmonary tuberculosis. Am J Respir Crit Care Med 167: 970–975. [DOI] [PubMed] [Google Scholar]
- 36. Meenakshi P, Ramya S, Shruthi T, Lavanya J, Mohammed HH, et al. (2013) Association of IL-1beta +3954 C/T and IL-10-1082 G/A Cytokine Gene Polymorphisms with Susceptibility to Tuberculosis. Scand J Immunol 78: 92–97. [DOI] [PubMed] [Google Scholar]
- 37. Trajkov D, Trajchevska M, Arsov T, Petlichkovski A, Strezova A, et al. (2009) Association of 22 cytokine gene polymorphisms with tuberculosis in Macedonians. Indian J Tuberc 56: 117–131. [PubMed] [Google Scholar]
- 38. Fitness J, Floyd S, Warndorff DK, Sichali L, Malema S, et al. (2004) Large-scale candidate gene study of tuberculosis susceptibility in the Karonga district of northern Malawi. Am J Trop Med Hyg 71: 341–349. [PubMed] [Google Scholar]
- 39. Amirzargar AA, Rezaei N, Jabbari H, Danesh AA, Khosravi F, et al. (2006) Cytokine single nucleotide polymorphisms in Iranian patients with pulmonary tuberculosis. Eur Cytokine Netw 17: 84–89. [PubMed] [Google Scholar]
- 40. Taype CA, Shamsuzzaman S, Accinelli RA, Espinoza JR, Shaw MA (2010) Genetic susceptibility to different clinical forms of tuberculosis in the Peruvian population. Infect Genet Evol 10: 495–504. [DOI] [PubMed] [Google Scholar]
- 41. Prabhu Anand S, Selvaraj P, Jawahar MS, Adhilakshmi AR, Narayanan PR (2007) Interleukin-12B & interleukin-10 gene polymorphisms in pulmonary tuberculosis. Indian J Med Res 126: 135–138. [PubMed] [Google Scholar]
- 42. Liu XX, Sun YH, Guo M, Feng FM (2009) Study on the relationship of interleukin-10 genetic polymorphisms with the susceptibility of pulmonary tuberculosis. Modern Prev Med 36: 1827–1830. [Google Scholar]
- 43. Yang H, Liang ZH, Liu XL, Wang F (2010) Association between polymorphisms of interleukin-10, interferon-gamma gene and the susceptibility to pulmonary tuberculosis. Chin J Epidem 31: 155–158. [PubMed] [Google Scholar]
- 44. Ma ZM, Xiao F, Tang LG, Liu JX, et al. (2007) A study on the correlation between the polymorphism of interleukin-10 gene and susceptibility to pulmonary tuberculosis. Guangdong Med 28: 1243–1245. [Google Scholar]
- 45. Mhmoud N, Fahal A, Wendy van de Sande WJ (2013) Association of IL-10 and CCL5 single nucleotide polymorphisms with tuberculosis in the Sudanese population. Trop Med Int Health 18: 1119–1127. [DOI] [PubMed] [Google Scholar]
- 46. Meilangqucuo Zaduo, Huang LP, Duolina, et al (2012) Study on association between interleukin-10 gene polymorphism with pulmonary tuberculosis in Tibetans. Modern Prev Med 39: 3607–3610. [Google Scholar]
- 47. Pan F, Tian J, Pan YY, Zhang Y (2012) Association of IL-10-1082 promoter polymorphism with susceptibility to gastric cancer: evidence from 22 case-control studies. Mol Biol Rep 39: 7143–7154. [DOI] [PubMed] [Google Scholar]
- 48. Zhang J, Chen Y, Nie XB, Wu WH, Zhang H, et al. (2011) Interleukin-10 polymorphisms and tuberculosis susceptibility: a meta-analysis. Int J Tuberc Lung Dis 15: 594–601. [DOI] [PubMed] [Google Scholar]
- 49. Meenagh A, Williams F, Ross OA, Patterson C, Gorodezky C, et al. (2002) Frequency of cytokine polymorphisms in populations from western Europe, Africa, Asia, the Middle East and South America. Hum Immunol 63: 1055–1061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA 2009 Checklist.
(WIZ)