Abstract
SUMMARY
OmicCircos is an R software package used to generate high-quality circular plots for visualizing genomic variations, including mutation patterns, copy number variations (CNVs), expression patterns, and methylation patterns. Such variations can be displayed as scatterplot, line, or text-label figures. Relationships among genomic features in different chromosome positions can be represented in the forms of polygons or curves. Utilizing the statistical and graphic functions in an R/Bioconductor environment, OmicCircos performs statistical analyses and displays results using cluster, boxplot, histogram, and heatmap formats. In addition, OmicCircos offers a number of unique capabilities, including independent track drawing for easy modification and integration, zoom functions, link-polygons, and position-independent heatmaps supporting detailed visualization.
AVAILABILITY AND IMPLEMENTATION
OmicCircos is available through Bioconductor at http://www.bioconductor.org/packages/devel/bioc/html/OmicCircos.html. An extensive vignette in the package describes installation, data formatting, and workflow procedures. The software is open source under the Artistic–2.0 license.
Keywords: R package, circular plot, genomic variation
Introduction
Visualization is an essential aspect of the statistical analysis of large-scale genomic data collections. It plays important roles in exploring, analyzing, and presenting patterns and correlations found in multidimensional genomic data.1 The circular plot with multiple “doughnut” tracks is particularly effective in displaying relationships among genes or genomic intervals. The software package Circos, written in Perl, is widely used to generate circular plots.2 It has been cited in more than 300 publications describing the landscape of genomic alterations including mutations, aberrant expression patterns, CNVs, methylation patterns, and other types of structural variation (http://circos.ca). However, while Circos is flexible and quite powerful, it demands that users conquer a steep learning curve. Other less well-known Java-based circular plot tools, such as CGView and DNAplotter,3,4 are also known to be less than completely user-friendly.
In efforts to develop more efficient software programs for generating circular plots, researchers have looked to the R package, a common environment for statistical computing and graphic visualization of large genomic data sets.5 The recently published R package Rcircos incorporates standard R graphics into a circular ideogram, but lacks detailed visualization functions like zoom and independent track drawing.6 Another application, ggbio, has very limited circular plot functionality since it utilizes a third-party graphic tool called ggplot.7 Clearly, a distinct need exists for a software package for generating circular plots with readily customizable features that can help scientists more easily display and communicate complex genomics data.
Here, we present OmicCircos, a simple and comprehensive approach to creating circular plots. The software package offers end users four unique customizable capabilities, all of which are available through a single command:
Independent track plotting and modification that does not affect other tracks in a given plot;
Integrated statistical analysis and the concomitant display of results using boxplot, histogram, or position-independent heatmap representations;
Provision of a zoom function that allows detailed visualization and link-polygon formulations to display for large translocations; and
The storage of genomic annotations and alignments in a way that permits the easy manipulation of genomic intervals.
This last capability results from the integration and use of the GenomicRanges package in Bioconductor within the OmicCircos environment.
Features
The OmicCircos package performs three main functions:
1) segAnglePo() specifies the circle size, number of segments, and segment width. The package includes chromosome size data for human genome hg19/hg18 and mouse genome mm10/mm9. Such data are typically used to plot the outermost circle, which serves as an anchor for the figure. Alternatively, if the figure is intended to represent a set of genes rather than a whole genome, the user can define one segment per gene by simply constructing the frame data with gene names and segment sizes. segAnglePo() can also read data from the GRanges object generated by the GenomicRanges package in Bioconductor, which allows genomic intervals and annotations to be easily manipulated.
2) circos() enables users to sup erimpose graphics on the circle. Data can be visualized in many different shapes and forms. For example, expression and CNV data can be viewed using basic shapes like scatterplots and lines while structural variations such as translocations and fusion proteins can be viewed using curves and polygons to connect different segments. Additionally, multiple sample expression and CNV data sets can be displayed as boxplots, histograms, or heatmaps using standard R functions such as apply. Furthermore, users are able to calculate and illustrate statistical values such as pvalue, ratio, standard deviation, mean, and median on the graph.
3) sim.circos() generates simulation data for drawing circular plots. It allows users to preview the graph quickly with different parameters and design an optimal presentation with desired features. The simulation data input files can be used as templates to format the genomic data for drawing publication-ready figures.
Examples
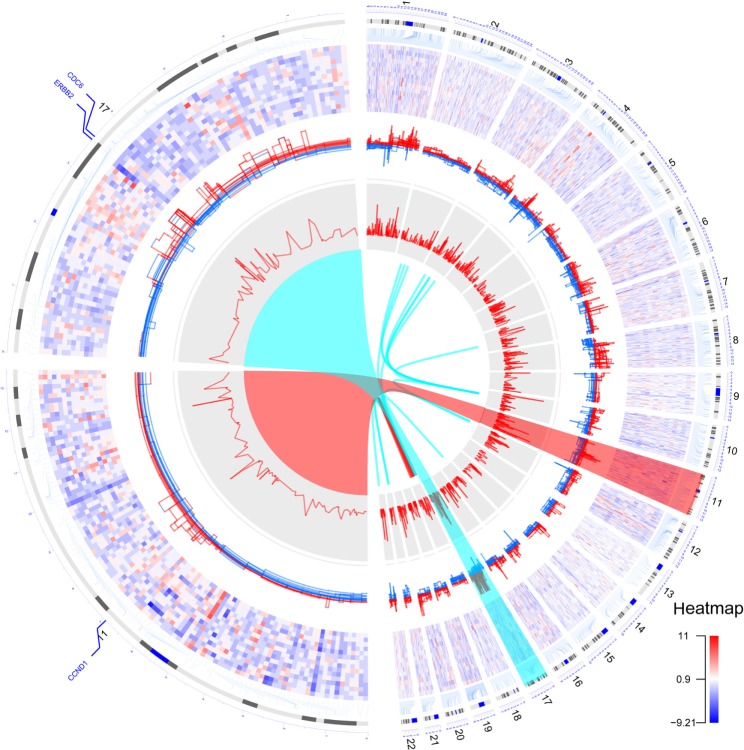
To demonstrate the functionality of the OmicCircos package, we display The Cancer Genome Atlas (TCGA) data describing breast cancer genomic variations such as mutation, expression, and copy number previously reported by the TCGA Network,8 as well as putative fusion proteins that were identified in house from TCGA breast cancer RNAseq data using deFuse.9 Figure 1 illustrates an OmicCircos plot consisting of two halves: the right half displays gene expression, CNV, correlation of CNV and expression, and fusion proteins for 15 Her2 subtype samples; the left half shows the zoom image of chromosomes 11 and 17 to demonstrate the focal copy number amplifications in the region of CDC6, ERBB2, and CCND1. The gene-expression heatmap uses a position-independent display for a better representation of gene-dense areas. The actual gene positions are linked to chromosomes with elbow connectors.
Figure 1.
Circular plots by OmicCircos of expression, CNV, and fusion proteins in 15 Her2 subtype samples from TCGA breast cancer data.8 Circular tracks from outside to inside: genome positions by chromosomes (black lines are cytobands), expression heatmap of 2000 most variable genes whose locations are indicated by elbow connectors, CNVs (red: gain, blue: loss), correlation p-values between expression and CNV, fusion proteins (intra/inter chromosomes: red/blue). Chromosomes 1–22 are shown in the right half of the circle. Zoomed chromosomes 11 and 17 are displayed in the left half. The circle arc was drawn using trigonometric functions. The fusion protein links were plotted with the Bézier curve algorithm.
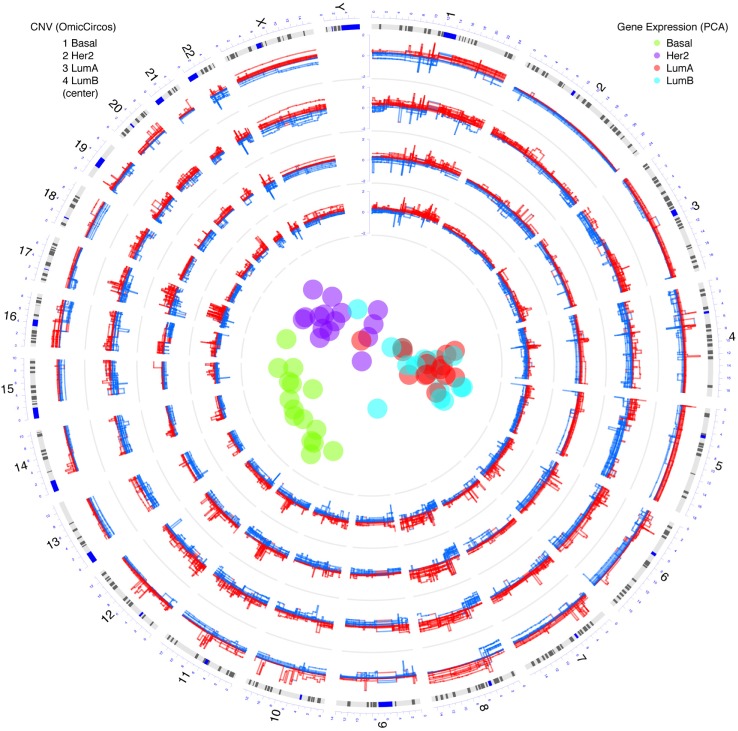
The circular plot from the OmicCircos package can be seamlessly integrated with other types of R statistical graphics. For example, Figure 2 illustrates CNV and gene-expression data for the four breast cancer subtypes: Basal, Her2, Luminal A (LumA) and Luminal B (LumB) with 15 samples in each group. CNVs for each subtype are represented in individual tracks. Principal component analysis (PCA) of gene-expression data for the four subtypes is represented in the center of the figure. Distinct patterns of arm-level CNV gain or loss are visible, eg, 5q loss in Basal, 16q loss in Luminal A/B, and 10p gain in Basal. More examples of circular plots generated using OmicCircos package are available in the package vignette. These show comparative genomic analyses, expression quantitative trait loci (eQTLs), protein-protein interactions (PPIs), and gene interactions within the context of pathways.
Figure 2.
Integration of circular plots by OmicCircos and clusters from R of TCGA breast cancer data. Circular tracks from outside to inside: genome positions by chromosomes (black lines are cytobands), CNVs in Basal, Her2, Luminal A (LumA) and Luminal B (LumB) subtypes with 15 samples in each track (red: gain; blue: loss), in the center, four clusters from principle component analysis (PCA) of the gene expression data with R packages in the bioconductor.
Conclusion
The OmicCircos package offers researchers a highly efficient, user-friendly program and visualization tool that not only illustrates genomic data analyses but also carries out statistical analyses that can be combined to generate publication-ready circular plots for a wide range of data types. Besides these functionalities, OmicCircos also takes advantage of statistical functions and comprehensive genome annotation packages in the R/Biocondutor environment. This makes the package a powerful circular plot tool that provides users with unique functions that include independent track drawing, zooming, easy genome annotation manipulation, and integration of statistical analysis and graphics. OmicCircos has sufficient flexibility to integrate with packages in R/Bioconductor that bring together annotation, analysis, and comparative genomics pipelines.
Supplementary data
This document includes the following:
The Circos-like ideogram drawing
The input file format
Comparison of OmicCircos with other similar R package
The Circos-like ideogram drawing. The trigonometric algorithm and Cubic Bézier curve were used to draw the circular ideogram and polygon links.
The arc of a circular track is defined by the trigonometric algorithm. Given a circular center point (xc, yc), a radius and two angles (start and end points), the sequence points on the arc were calculated by the trigonometric algorithm. The arc was drawn by the line function in R.
The linking curve or polygon between different chromosomal positions are plotted with a cubic Bézier curve. The curve is defined by four points: p0 is the origin/starting point and p3 is the destination/end point; which are corresponding to the positions on the chromosome(s); p1 and p2 are control points defined by the center of the circular.
R code for Figure S1:
## draw circle or arc using trigonometric functions
rm(list = ls ());
## draw.arc function in OmicCircos
draw.arc <- function (xc, yc, r, w1, w2, col = "lightblue", lwd = 1, lend = 1) {
# r = radius
ang.d <- abs(w1–w2);
pix.n <- ang.d * 5;
if (pix.n, 2){
pi x.n <- 2;
}
ang.seq <- rev(seq(w1,w2,length.out = pix.n));
ang.seq <- ang.seq/360*2*pi;
fan.i.x <- xc + cos(ang.seq) * r;
fan.i.y,- yc-sin(ang.seq) * r;
## lend = 0(round); lend = 1(butt); lend = 2(square)
lines(fan.i.x, fan.i.y, col = col, lwd = lwd, type = "l", lend = lend);
}
## colors
cols <- rainbow(20, alpha = 0.5);
pdf("trig_function_curve.pdf", 6, 6);
par(mar = c (1,1,1,1));
plot(c(1,800), c(1,800), type = "n", axes = F, xlab = "", ylab = "", main = "");
## circle center
xc <- 400;
yc <- 400;
## arc number
arc.num <- 20;
for (i in 1:arc.num){
## radius
r <- sample(c(10:3 0 0),1);
## two angles
w1 <- sample(c(0:180),1);
w2 <- sample(c(w 1:360),1);
## arc width
lwd <- sample(c(4:20),1);
## arc color
col <- sample(cols,1);
draw.arc(xc, yc, r, w1, w2, col = col, lwd = lwd);
}
dev.off();
The input data file format. mapping data for expression, copy number, and mutation et al.:
column 1: chromosome number
column 2: chromosome position
column 3 (option): gene symbol
column 4-n: values
For example:
| CHR | Po | NAME | TCGA.A1. | TCGA.A1. | TCGA.A2. | |
|---|---|---|---|---|---|---|
| AOSK.01A | A0S0.01A | A04W.01A | ||||
| 1 | 1 | 939245.5 | ISG15 | −3.618 | −2.286 | −0.998 |
| 2 | 1 | 2 5 3 314 0.5 | MMEL1 | −2.832 | −3.093 | 1.037 |
| 3 | 1 | 6446321.0 | TNFRSF25 | 2.559 | −0.660 | −0.705 |
| 4 | 1 | 7832974.5 | UTS2 | 1.708 | −0.726 | −0.102 |
| 5 | 1 | 7912164.5 | TNFRSF9 | −0.189 | −0.768 | 1.020 |
| 6 | 1 | 9989253.0 | RBP7 | 0.817 | −0.463 | −1.472 |
> head(TCGA.BC.cnv.2k.60[,1:6])
linking data for fusion protein and translocation
column 1: chromosome number
column 2: chromosome position
column 3: gene symbol
column 4: other chromosome number
column 5: other chromosome position
column 6: other gene symbol
For example:
| CHR1 | PO1 | GENE1 | CHR2 | PO2 | GENE2 | |
|---|---|---|---|---|---|---|
| 1 | 2 | 63456333 | WDPCP | 10 | 37493749 | ANKRD30A |
| 2 | 18 | 14563374 | PARD6G | 21 | 14995400 | POTED |
| 3 | 10 | 37521495 | ANKRD30A | 3 | 49282645 | CCDC36 |
| 4 | 10 | 37521495 | ANKRD30A | 7 | 100177212 | LRCH4 |
| 5 | 18 | 18539803 | ROCK1 | 18 | 112551 | PARD6G |
| 6 | 12 | 4618159 | C12orf4 | 18 | 1514414 | PARD6G |
>head(TCGA.BC.fus[,c(1:6)]);
The R codes of figures 1 and 2.
rm(list = ls());
options(stringsAsFactors = FALSE);
library(OmicCircos);
data(“TCGA.PAM50_genefu_hg18”);
data(“TCGA.BC.fus”);
data(“TCGA.BC.cnv.2k.60”);
data(“TCGA.BC.gene.exp.2k.60”);
data(“TCGA.BC.sample60”);
data(“TCGA.BC_Her2_cnv_exp”);
pvalue <- -1 * log10(TCGA.BC_Her2_cnv_exp[,5]);
pvalue <- cbind(TCGA.BC_Her2_cnv_exp[,c(1:3)], pvalue);
Her2.i <- which(TCGA.BC.sample60[,2] = = “Her2”);
Her2.n <- TCGA.BC.sample60[Her2.i,1];
Her2.j <- which(colnames(TCGA.BC.cnv.2k.60)%in%Her2.n);
cnv <- TCGA.BC.cnv.2k.60[,c(1:3,Her2.j)];
cnv.m <- cnv[,c(4:ncol(cnv))];
cnv.m[cnv.m > 2] <- 2;
cnv.m[cnv.m < -2] <- -2;
cnv <- cbind(cnv[,1:3], cnv.m);
Her2.j <- which(colnames(TCGA.BC.gene.exp.2k.60)%in% Her2.n);
gene.exp <- TCGA.BC.gene.exp.2k.60[,c(1:3,Her2.j)];
colors <- rainbow(10, alpha = 0.5);
pdf(“OmicCircos4vignette10_paper1.pdf”, 8,8);
par(mar = c(2, 2, 2, 2));
plot(c(1,800), c(1,800), type = “n”, axes = FALSE, xlab = “”, ylab = “”, main = “”);
zoom <- c(1, 22, 939245.5, 154143883, 0, 180);
circos(R = 400, cir = “hg18”, W = 4, type = “chr”, print.chr.lab = TRUE, scale = TRUE, zoom = zoom);
circos(R = 300, cir = “hg18”, W = 100, mapping = gene.exp, col.v = 4, type = “heatmap2”, cluster = FALSE, col.bar = TRUE, lwd = 0.01, zoom = zoom);
circos(R = 220, cir = “hg18”, W = 80, mapping = cnv, col.v = 4, type = “ml3”, B = FALSE, lwd = 1, cutoff = 0, zoom = zoom);
circos(R = 140, cir = “hg18”, W = 80, mapping = pvalue, col.v = 4, type = “l”, B = TRUE, lwd = 1, col = colors[1], zoom = zoom);
circos(R = 130, cir = “hg18”, W = 10, mapping = TCGA.BC.fus, type = “link”, lwd = 2, zoom = zoom);
## zoom in links by using the hightlight functions
## highlight
the.col1 = rainbow(10, alpha = 0.5)[1];
highlight <- c(140, 400, 11, 282412.5, 11, 133770314.5, the.col1, the.col1);
circos(R = 110, cir = “hg18”, W = 40, mapping = highlight, type = “hl”, lwd = 2, zoom = zoom);
the.col2 = rainbow(10, alpha = 0.5)[6];
highlight <- c(140, 400, 17, 739525, 17, 78385909, the.col2, the.col2);
circos(R = 110, cir = “hg18”, W = 40, mapping = highlight, type = “hl”, lwd = 2, zoom = zoom);
## highlight link
highlight.link1 <- c(400, 400, 140, 376.8544, 384.0021, 450, 540.5);
circos(cir = “hg18”, mapping = highlight.link1, type = “highlight.link”, col = the.col1, lwd = 1);
highlight.link2 <- c(400, 400, 140, 419.1154, 423.3032, 543, 627);
circos(cir = “hg18”, mapping = highlight.link2, type = “highlight.link”, col = the.col2, lwd = 1);
## zoom in chromosome 11
zoom <- c(11, 11, 282412.5, 133770314.5, 180, 270);
circos(R = 400, cir = “hg18”, W = 4, type = “chr”, print.chr.lab = TRUE, scale = TRUE, zoom = zoom);
circos(R = 300, cir = “hg18”, W = 100, mapping = gene.exp, col.v = 4, type = “heatmap2”, cluster = FALSE, lwd = 0.01, zoom = zoom);
circos(R = 220, cir = “hg18”, W = 80, mapping = cnv, col.v = 4, type = “ml3”, B = FALSE, lwd = 1, cutoff = 0, zoom = zoom);
circos(R = 140, cir = “hg18”, W = 80, mapping = pvalue, col.v = 4, type = “l”, B = TRUE, lwd = 1, col = colors[1], zoom = zoom);
gene.names <- c(“ERBB2”,”CDC6”);
PAM50.17 <- which(TCGA.PAM50_genefu_hg18[, 3] = = gene.names);
TCGA.PAM50 <- TCGA.PAM50_genefu_hg18[PAM50.17,];
TCGA.PAM50 <- rbind(TCGA.PAM50, TCGA.PAM50[2,]);
TCGA.PAM50[3,1] <- 11;
TCGA.PAM50[3,2] <- 69165000;
TCGA.PAM50[,3] <- as.character(TCGA.PAM50[,3]);
TCGA.PAM50[3,3] <- c(“CCND1”);
circos(R = 410, cir = “hg18”, W = 40, mapping = TCGA.PAM50, type = “label”, side = “out”, col = “blue”, zoom = zoom);
## zoom in chromosome 17
zoom <- c(17, 17, 739525, 78385909, 274, 356);
circos(R = 400, cir = “hg18”, W = 4, type = “chr”, print.chr.lab = TRUE, scale = TRUE, zoom = zoom);
circos(R = 300, cir = “hg18”, W = 100, mapping = gene.exp, col.v = 4, type = “heatmap2”, cluster = FALSE, lwd = 0.01, zoom = zoom);
circos(R = 220, cir = “hg18”, W = 80, mapping = cnv, col.v = 4, type = “ml3”, B = FALSE, lwd = 1, cutoff = 0, zoom = zoom);
circos(R = 140, cir = “hg18”, W = 80, mapping = pvalue, col.v = 4, type = “l”, B = TRUE, lwd = 1, col = colors[1], zoom = zoom);
## gene label
gene.names <- c(“ERBB2”,”CDC6”);
PAM50.17 <- which(TCGA.PAM50_genefu_hg18[, 3] = = gene.names);
TCGA.PAM50 <- TCGA.PAM50_genefu_hg18[PAM50.17,];
TCGA.PAM50 <- rbind(TCGA.PAM50, TCGA.PAM50[2,]);
TCGA.PAM50[3,1] <- 11;
TCGA.PAM50[3,2] <- 69165000;
TCGA.PAM50[,3] <- as.character(TCGA.PAM50[,3]);
TCGA.PAM50[3,3] <- c(“CCND1”);
circos(R = 410, cir = “hg18”, W = 40, mapping = TCGA.PAM50, type = “label”, side = “out”, col = “blue”, zoom = zoom);
dev.off()
rm(list = ls());
options(stringsAsFactors = FALSE);
library(OmicCircos);
data(“TCGA.BC.fus”);
data(“TCGA.BC.cnv.2k.60”);
data(“TCGA.BC.gene.exp.2k.60”);
data(“TCGA.BC.sample60”);
## gene expression data for PCA
exp.m <- TCGA.BC.gene.exp.2k.60[,c(4:ncol(TCGA.BC.gene.exp.2k.60))];
cnv<- TCGA.BC.cnv.2k.60;
## PCA
type.n <- unique(TCGA.BC.sample60[,2]);
colors <- rainbow(length(type.n), alpha = 0.5);
pca.col <- rep(NA, nrow(TCGA.BC.sample60));
for (i in 1:length(type.n)){
n <- type.n[i];
n.i <- which(TCGA.BC.sample60[,2] = = n);
n.n <- TCGA.BC.sample60[n.i,1];
g.i <- which(colnames(exp.m)%in% n.n);
pca.col[g.i] <- colors[i];
}
exp.m <- na.omit(exp.m);
pca.out <- prcomp(t(exp.m), scale = TRUE);
## subtype cnv
cnv.i <- c();
for (i in 1:length(type.n)){
n <- type.n[i];
n.i <- which(TCGA.BC.sample60[,2] = = n);
n.n <- TCGA.BC.sample60[n.i,1];
cnv.i <- which(colnames(cnv)%in% n.n);
}
## main
pdf(“OmicCircos4vignette7.pdf”, 8,8);
par(mar = c(5, 5, 5, 5));
plot(c(1,800), c(1,800), type = “n”, axes = FALSE, xlab = “”, ylab = “”, main = “”);
legend(680,800, c(“Basal”,”Her2”,”LumA”,”LumB”), pch = 19, col = colors[c(2,4,1,3)], cex = 0.5,
title = “Gene Expression (PCA)”, box.col = “white”);
legend(5,800, c(“1 Basal”, “2 Her2”, “3 LumA”, “4 LumB”, “(center)”), cex = 0.5,
title = “CNV (OmicCircos)”, box.col = “white”);
circos(xc = 400, yc = 400, R = 390, cir = “hg18”, W = 4, type = “chr”, print.chr.lab = TRUE, scale = TRUE);
R.v <- 330;
for (i in 1:length(type.n)){
n <- type.n[i];
n.i <- which(TCGA.BC.sample60[,2] = = n);
n.n <- TCGA.BC.sample60[n.i,1];
cnv.i <- which(colnames(cnv)%in% n.n);
c nv.v <- cnv[,cnv.i];
cnv.v[cnv.v < 2] <- 2;
cnv.v[cnv.v < -2] <- -2;
cnv.m <- cbind(cnv[,c(1:3)], cnv.v);
circos(xc = 400, yc = 400, R = R.v, cir = “hg18”, W = 60, mapping = cnv.m, col.v = 4, type = “ml3”, B = FALSE, lwd = 1, cutoff = 0, scale = TRUE);
R.v <- R.v—60;
}
points(pca.out$x[,1]*3.6+400, pca.out$x[,2]*3.6+400, pch = 19, col = pca.col, cex = 2);
de v.of ()
Comparison of omicCircos with other similar R package. To demonstrate the features of OmicCircos, we compared the circular plot generated by OmicCircos (Fig. 1) with the plot by the recently published R package RCircos (Fig. S2) using the same TCGA breast cancer dataset (Zhang, Meltzer et al. 2013). The main differences between OmicCircos and RCircos were track design, zoom, sample number, heatmap, run time and output file size (Table S1).
Demonstration of drawing circular ideograms by OmicCircos.
Circular plot generated by RCircos using the same data set as Figure 1 generated by OmicCircos.
Table S1.
Comparison between OmicCircos and RCircos.
| SOFTWARE | OMICCIRCOS | RCIRCOS |
|---|---|---|
| FEATURES | ||
| Track Design | Independent-track drawing | Tracks fixed by one set of parameters |
| Zoom | Enabled | Not available |
| Sample size | Multiple samples in a track | One sample/per track |
| Heatmap | Standard heatmap | Heatmap-like with one sample per track and unfilled spaces for non-gene region |
| Run Time | 30.139 s | 4 m28.754 s |
| Output File Size (PDF) | 1.2 MB | 23.7 MB |
REFERENCE
- 1.Zhang H, Meltzer P, et al. RCircos: an R package for Circos 2D track plots. BMC Bioinformatics. 2013;14:244. doi: 10.1186/1471-2105-14-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Acknowledgments
We thank Dr. Laura K. Fleming, our scientific writer and editor in the NCI Center for Biomedical Informatics and Information Technology, for her critical reading of the manuscript.
Footnotes
Academic Editor: JT Efird, Editor in Chief
FUNDING: Author(s) disclose no funding sources.
COMPETING INTERESTS: Author(s) disclose no potential conflicts of interest.
Author Contributions
Conceived and designed the experiments: YH, CY. Analyzed the data: YH, C-HH, KN. Wrote the first draft of the manuscript: YH, CY. Contributed to the writing of the manuscript: YH, CY, C-HH, Q-RC, DM. Agree with manuscript results and conclusions: YH, CY, C-HH, Q-RC, KN, GAK, DM. All authors reviewed and approved of the final manuscript.
DISCLOSURES AND ETHICS
As a requirement of publication the authors have provided signed confirmation of their compliance with ethical and legal obligations including but not limited to compliance with ICMJE authorship and competing interests guidelines, that the article is neither under consideration for publication nor published elsewhere, of their compliance with legal and ethical guidelines concerning human and animal research participants (if applicable), and that permission has been obtained for reproduction of any copyrighted material. This article was subject to blind, independent, expert peer review. The reviewers reported no competing interests.
REFERENCES
- 1.Schroeder MP, Gonzalez-Perez A, Lopez-Bigas N. Visualizing multidimensional cancer genomics data. Genome Med. 2013;5(1):9. doi: 10.1186/gm413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krzywinski M, Schein J, Birol I, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21:537–9. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- 4.Carver T, Thomson N, Bleasby A, et al. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–20. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reimers M, Carey VJ. Bioconductor: an open source framework for bioinformatics and computational biology. Methods Enzymol. 2006;411:119–34. doi: 10.1016/S0076-6879(06)11008-3. [DOI] [PubMed] [Google Scholar]
- 6.Zhang H, Meltzer P, Davis S. RCircos: an R package for Circos 2D track plots. BMC Bioinformatics. 2013;14:244. doi: 10.1186/1471-2105-14-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin T, Cook D, Lawrence M. ggbio: an R package for extending the grammar of graphics for genomic data. Genome Biol. 2012;13:R77. doi: 10.1186/gb-2012-13-8-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The Cancer Genome Atlas Network (TCGA) Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McPherson A, Hormozdiari F, Zayed A, et al. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol. 2011;7:e1001138. doi: 10.1371/journal.pcbi.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
| CHR | Po | NAME | TCGA.A1. | TCGA.A1. | TCGA.A2. | |
|---|---|---|---|---|---|---|
| AOSK.01A | A0S0.01A | A04W.01A | ||||
| 1 | 1 | 939245.5 | ISG15 | −3.618 | −2.286 | −0.998 |
| 2 | 1 | 2 5 3 314 0.5 | MMEL1 | −2.832 | −3.093 | 1.037 |
| 3 | 1 | 6446321.0 | TNFRSF25 | 2.559 | −0.660 | −0.705 |
| 4 | 1 | 7832974.5 | UTS2 | 1.708 | −0.726 | −0.102 |
| 5 | 1 | 7912164.5 | TNFRSF9 | −0.189 | −0.768 | 1.020 |
| 6 | 1 | 9989253.0 | RBP7 | 0.817 | −0.463 | −1.472 |
| CHR1 | PO1 | GENE1 | CHR2 | PO2 | GENE2 | |
|---|---|---|---|---|---|---|
| 1 | 2 | 63456333 | WDPCP | 10 | 37493749 | ANKRD30A |
| 2 | 18 | 14563374 | PARD6G | 21 | 14995400 | POTED |
| 3 | 10 | 37521495 | ANKRD30A | 3 | 49282645 | CCDC36 |
| 4 | 10 | 37521495 | ANKRD30A | 7 | 100177212 | LRCH4 |
| 5 | 18 | 18539803 | ROCK1 | 18 | 112551 | PARD6G |
| 6 | 12 | 4618159 | C12orf4 | 18 | 1514414 | PARD6G |
Demonstration of drawing circular ideograms by OmicCircos.
Circular plot generated by RCircos using the same data set as Figure 1 generated by OmicCircos.
Table S1.
Comparison between OmicCircos and RCircos.
| SOFTWARE | OMICCIRCOS | RCIRCOS |
|---|---|---|
| FEATURES | ||
| Track Design | Independent-track drawing | Tracks fixed by one set of parameters |
| Zoom | Enabled | Not available |
| Sample size | Multiple samples in a track | One sample/per track |
| Heatmap | Standard heatmap | Heatmap-like with one sample per track and unfilled spaces for non-gene region |
| Run Time | 30.139 s | 4 m28.754 s |
| Output File Size (PDF) | 1.2 MB | 23.7 MB |