Abstract
Facioscapulohumeral muscular dystrophy (FSHD), a common hereditary myopathy, is characterized by atrophy and weakness of selective muscle groups. FSHD is considered an autosomal dominant disease with incomplete penetrance and unpredictable variability of clinical expression within families. Mice overexpressing FRG1 (FSHD region gene 1), a candidate gene for this disease, develop a progressive myopathy with features of the human disorder. Here, we show that in FRG1-overexpressing mice, fast muscles, which are the most affected by the dystrophic process, display anomalous fast skeletal troponin T (fTnT) isoform, resulting from the aberrant splicing of the Tnnt3 mRNA that precedes the appearance of dystrophic signs. We determine that muscles of FRG1 mice develop less strength due to impaired contractile properties of fast-twitch fibers associated with an anomalous MyHC-actin ratio and a reduced sensitivity to Ca2+. We demonstrate that the decrease of Ca2+ sensitivity of fast-twitch fibers depends on the anomalous troponin complex and can be rescued by the substitution with the wild-type proteins. Finally, we find that the presence of aberrant splicing isoforms of TNNT3 characterizes dystrophic muscles in FSHD patients. Collectively, our results suggest that anomalous TNNT3 profile correlates with the muscle impairment in both humans and mice. On the basis of these results, we propose that aberrant fTnT represents a biological marker of muscle phenotype severity and disease progression.
Keywords: muscular dystrophy, aberrant splicing, muscle weakness, FRG1, troponin T
facioscapulohumeral muscular dystrophy (FSHD), one of the three most common hereditary myopathies, presents progressive atrophy and weakness of a highly specific set of muscle groups (10, 11, 31). FSHD is characterized by insidious onset and unpredictable progression with high variability of clinical expression, even within the same family (36, 37, 42). At present, no molecular mechanisms have been implicated in the development of these disease phenotypes, and no biological markers are available to define and monitor disease expression.
More than 50 genes have been associated with muscular dystrophies (MD), whose genetic defects have been molecularly defined, and numerous animal models have been developed (30). In many cases, invaluable insights into disease mechanisms, structure and function of gene products, and approaches for therapeutic interventions have benefited from the study of animal models of the different MDs (1). In this context, FSHD is unique because no mutations have been found in any protein-coding gene. Instead, FSHD has been associated with reduction of the number of tandemly repeated 3.3-kb DNA segments (named D4Z4 repeats) located at the subtelomeric region of chromosome 4q (2, 44, 48). The number of D4Z4 repeats varies from 11 to 150 in the general population, whereas less than 11 repeats are present in sporadic and familial FSHD patients (48). The current model to explain FSHD is that the reduction of D4Z4 repeats in FSHD subjects initiates inappropriate overexpression of the diverse 4q35 genes, ultimately leading to disease onset and progression (2, 12, 50).
However, patients' studies have revealed healthy subjects carrying the same D4Z4 reduced allele of their affected relatives, as well as FSHD patients carrying D4Z4 alleles of normal size on both chromosomes 4 (37), suggesting that FSHD is a complex genetic disease, in which more than one gene probably contributes to the molecular mechanism leading to muscle dystrophy (37). Among the 4q35 genes reported to be inappropriately expressed in muscles of FSHD patients, DUX4 and FRG1 seem the best candidates for the disease (13, 23). To better understand the function of these genes and their potential role in FSHD pathogenesis, animal models have been generated (13, 20). Although the misregulation of DUX4 expression in zebrafish, whose genome does not contain a DUX4 homologue, seems to recapitulate some of the phenotypes seen in human FSHD patients (29), overexpression of DUX4 in mice, at extents similar to that of FSHD patients, does not show any obvious muscle phenotype (20). On the other hand, FRG1 is a highly conserved gene whose proper expression has been shown to be crucial for muscle development in vertebrates and invertebrates (17, 24). More importantly, FRG1 overexpression in mice causes a dystrophic phenotype, which recapitulates several features of FSHD (13). Four important similarities between FRG1-overexpressing mice and human FSHD patients have been documented. First, as in FSHD patients, FRG1 overexpression induces a progressive myopathy in which sarcolemma is not damaged and no elevated creatine kinase levels are detected. Second, FRG1-overexpressing mice display a reduced tolerance to exercise, taking a shorter time to reach exhaustion, similarly to the muscle fatigue described by FSHD patients (13, 19). Third, only selective muscles in FRG1-overexpressing mice are dystrophic and display altered accumulation of myosin heavy chain (MyHC) (8). Finally, it has been shown that in muscles of both FSHD patients and FRG1 transgenic mice, specific pre-mRNAs undergo aberrant alternative splicing (13, 34). In particular, our previous studies revealed the aberrant splicing of the fast skeletal troponin T (TNNT3) mRNA in muscles from FRG1 transgenics, as well as in myoblasts of FSHD patients (13). We considered this finding of great importance since the TNNT3 gene encodes one of the three subunits of the troponin complex, which governs striated muscle contraction (14, 16, 32). We, therefore, reasoned that its altered splicing might be relevant to the selective muscle weakness, which affects a subset of muscles in FSHD and in FRG1-overexpressing mice.
The present work was aimed at further investigating the causal relationship between the altered splicing caused by FRG1 overexpression and the onset of the dystrophic phenotype. Here, we demonstrated that aberrant splicing of the Tnnt3 mRNA produces aberrant isoforms of the protein that compromises muscle contractility in muscles of FRG1-overexpressing mice and identifies fast-twitch fibers as the specific target of FSHD.
METHODS
Mice specimens.
Mice were killed by neck dislocation. For all analyses, mice expressing high levels of human FRG1 in a C57BL/6 background and wild-type littermates were used (13). Soleus and vastus lateralis from WT and FRG1 transgenic mice were used to isolate single chemically skinned muscle fibers. Soleus and extensor digitorum longus (EDL) muscles were used for whole muscle experiments. All experiments were performed using wild-type and transgenic female 13-wk-old mice. In addition, wild-type and transgenic female 4-wk-old mice were examined for Tnnt3 splicing profiling, myosin-actin ratio, and muscle strength measurements.
RT-PCR splicing analysis.
For splicing analysis, total RNA was extracted from FRG1 and wild-type mice vastus lateralis and soleus, and from human muscle biopsies, using TRIzol (Invitrogen). The RNA was further purified with the RNeasy Mini Kit (Qiagen) RNA clean-up protocol and on-column DNase treatment. First-strand cDNA synthesis was performed using SuperScript II reverse transcriptase (Invitrogen), random hexamers, and 500 ng of total RNA. For amplification of mouse Tnnt3, forward primer spanning exons 2 and 3, and reverse primer annealing on exon 11 were as follows: 5′-TCTGACGAGGAAACTGAACAAG-3′ and 5′-TGTCAATGAGGGCTTGGAG-3′. For amplification of human TNNT3 forward and reverse primers, annealing on exons 2 and 10, were as follows: 5′-TTCACCATGTCTGACGAGGAAG-3′ and 5′-CTTCTGGGATCTTAGGAGCAGTG-3′. For radioactive PCR, [32P]ATP was added to PCR mix, and 20 amplification cycles were performed (13). PCR products were resolved on a 12% polyacrylamide gel, and radioactivity was quantified using a Typhoon Trio Scanner (GE Healthcare).
Troponin T splicing isoforms identification.
For the identification of the Tnnt3 splicing isoforms, the total RNA was purified, and cDNA was generated as described in RT-splicing analysis section of methods from vastus lateralis and soleus of wild-type and FRG1 transgenic mice. The whole Tnnt3-coding sequence was amplified using the following forward and reverse primers: 5′-TCTGACGAGGAAACTGAACAAG-3′ and 5′-TGATGGTCTCTGCTGCAGTG-3′. PCR products were cloned into pCR2.1 vector (Invitrogen), using the TA cloning kit (Invitrogen). Between 20 and 50 colonies were isolated for each sample, and each insert was fully sequenced. Tnnt3 splicing isoforms were identified by comparison with sequences of the Tnnt3 genomic locus and Tnnt3 RNAs in the National Center for Biotechnology Information database.
Cross-linking immunoprecipitation.
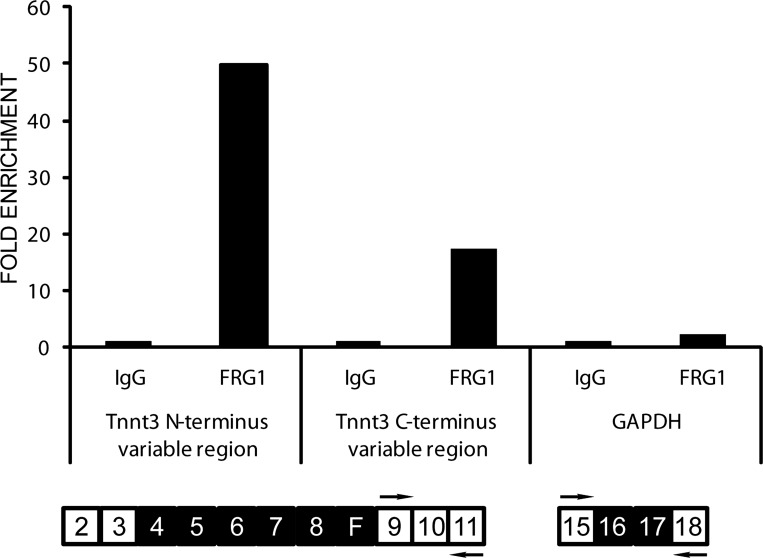
Cross-linking-immunoprecipitation (CLIP) was performed, as previously described, with some modifications (18, 43). In particular, vastus lateralis from 13-wk-old FRG1 transgenic mice was sliced into 1-mm-thick sections in the cold room and irradiated three times at 200 mJ/cm2 in the ultraviolet cross-linker. The tissue was homogenized in lysis buffer (50 mM Tris-HCl pH 7.4, 100 mM NaCl, 1 mM MgCl2, 0.1 mM CaCl2, 1% NP-40, 0.1% SDS, 0.5% Na deoxycholate) and RNA partially digested using RNase I (Ambion) diluted 1:100 to allow small RNA fragments to remain attached to protein. The RNA-protein complexes were purified by immunoprecipitation using A-coated Dynabeads (Invitrogen). The beads were washed two times with lysis buffer and incubated for 1 h at room temperature with the rabbit anti-FRG1 antibody (13) or control rabbit IgG (Jackson Laboratory). The beads were washed two times with lysis buffer and kept in the last-wash solution until the addition of cross-linked lysate. The lysate was added to beads and incubated for 1 h at 4°C. Then the beads were washed two times with lysis buffer, two times with high-salt wash buffer (50 mM Tris-HCl pH 7.4, 1 M NaCl, 1 mM EDTA, 0.1% SDS, 0.5% Na-deoxycholate, 1% NP-40), and 2 times with PNK wash buffer (20 mM Tris-HCl pH 7.4, 10 mM MgCl2, 0.2% Tween-20). Proteinase K was preincubated in PK buffer (100 mM Tris-HCl pH 7.5, 50 mM NaCl, 10 mM EDTA) at 2 mg/ml for 5 min at 37°C. Diluted proteinase K was added to the beads and incubated for 30 min at 55°C. Incubation was repeated after the addition of 130 μl PK/7 M urea buffer (100 mM Tris·HCl pH 7.5, 50 mM NaCl, 10 mM EDTA, and 7 M urea). Samples were cooled to 37°C, mixed with 400 μl RNA phenol (Ambion) and 130 μl chloroform, and incubated for 5 min at 37°C. After centrifugation, the aqueous phase was transferred into a new microcentrifuge tube and again subjected to centrifugation. The supernatant was mixed with 0.75 μl glycogen (Ambion), 50 μl 3 M sodium acetate (pH 5.5), and 1 ml 1:1 ethanol: isopropanol, and incubated overnight at −20°C. RNA precipitate was recovered by centrifugation, washed with 500 μl 80% ethanol, and resuspended in 10 μl water. The RNAs associated with FRG1 were reverse transcribed using Superscript III (Invitrogen), amplified, and analyzed by quantitative PCR (qPCR). We performed two different CLIP reactions, one using rabbit polyclonal antibody against FRG1 and one using IgG as a negative control (NC). Specific fast skeletal troponin T (Tnnt3) primers used were exon 9 forward primer: 5′-AGGAGAAACCAAGACCCAAAC-3′, exon 11 reverse primer: 5′-GGAGCTCCATGAGGTCCTTG-3′, exon 15 forward primer: 5′-ATTCGAGTT TGGGGAGAA GC-3′, and exon 18 reverse primer: 5′-GACTTTGCCCTTGGCTGTGG-3′. Data obtained from qPCR were analyzed using the fold enrichment method; data are represented as the fold increase in signal relative to the background signal.
Immunoblot analysis of fast TnT.
For conventional Western blot analysis, muscle fragment homogenates or single fibers were dissolved in SDS-PAGE buffer supplemented with Complete protease inhibitor cocktail (Roche), resolved by SDS-PAGE (14%, acrylamide/bis-acrylamide ratio 180:1), and transferred to a nitrocellulose filter. Filters were probed with the monoclonal antibody specific for fTnT isoforms (JLT12; Sigma), diluted 1:3,000 in 5% low-fat milk, 0.2% Tween-20, in TBS, and incubated for 2 h. The secondary antibody used was an anti-mouse peroxidase-conjugated antibody (Dako) diluted 1:3,000 in the same buffer as the cognate primary antibody, and incubated for 1 h. The monoclonal antibody specific for α-actinin (EA-53 Sigma) was used as a loading control in Western blots at a dilution of 1:1,000 in 5% low-fat milk, 0.2% Tween-20, in TBS, and incubated for 2 h. Visualization was performed by diaminobenzidine. Signal intensities were evaluated by densitometry (GS-700 Imaging Densitometer, Bio-Rad). Human biceps samples from control C05 and patient FSHD08 (see Fig. 11) were not analyzed by immunoblot due to insufficient amount of starting material.
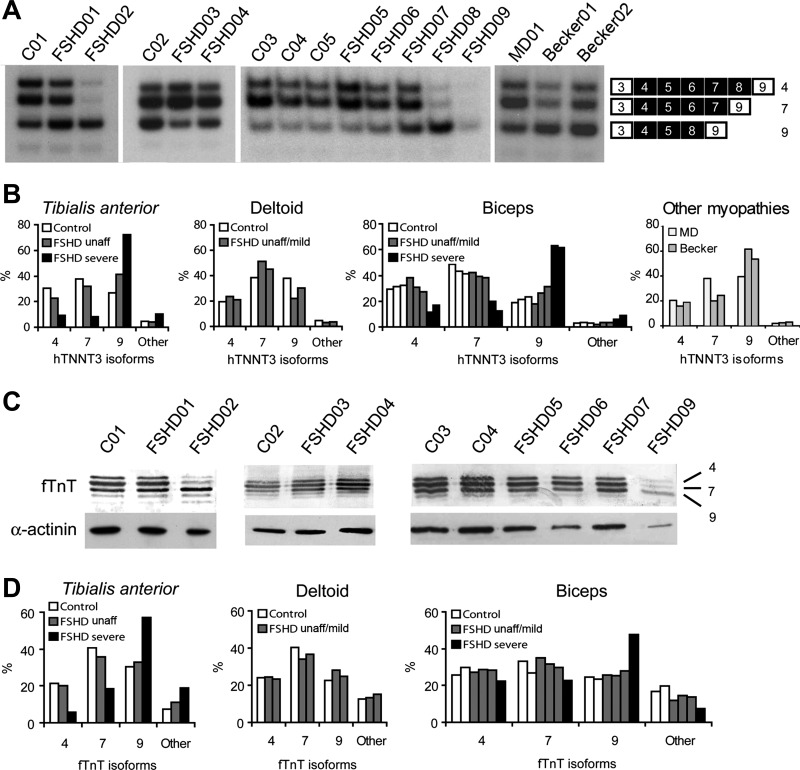
Fig. 11.
Dystrophic muscles from FSHD patients display altered troponin T splicing pattern. A: RT-PCR analysis of TNNT3 splicing pattern in muscle biopsies of FSHD patients, healthy controls, and an unaffected carrier of the molecular defect. One myotonic dystrophy (MD01) and two Becker dystrophy (Becker01 and Becker02) patients biopsies were also analyzed. Controls and FSHD patients codes are as in Fig. 10A. Identities of alternatively spliced products are indicated on the right of the gel. B: quantitative analysis of the bands identified in A. Patients were grouped in FSHD unaffected/mild (FSHD01, FSHD03, FSHD04, FSHD05, FSHD06, and FSHD07) and FSHD severe (FSHD02, FSHD08, and FSHD09), according to histological analysis and clinical features (see Fig. 10). C: immunoblotting analysis of fTnT in muscle biopsies of FSHD patients, healthy controls, and unaffected carrier. The isoform numbers are indicated on the right (upper panels). In the lower panel anti-α-actinin expression is monitored as a loading control. Controls and patients codes are as in Fig. 10A. D: quantitative analysis of the fTnT isoforms detected by immunoblotting. Patients were grouped in FSHD unaffected/mild (FSHD01, FSHD03, FSHD04, FSHD05, FSHD06, and FSHD07) and FSHD severe (FSHD02 and FSHD09), according to histological analysis and clinical features (see Fig. 10). Human biceps samples from control C05 and patient FSHD08 (see Fig. 10A) were not analyzed by immunoblotting due to insufficient amount of starting material.
Recombinant troponin T isoforms production.
Coding sequences of the identified Tnnt3 isoforms, including the stop codon, were amplified using the proper pCR2.1/Tnnt3 plasmids as a template and introducing a NdeI restriction site at the 5′ and a SapI restriction site at the 3′. The PCR products were cloned into pTYB1 plasmid (New England Biolabs) between NdeI and SapI restriction sites. All plasmids were sequence-verified and transformed into Escherichia coli BL21 CP strain for protein expression. For each isoform, a 1-ml culture was induced with 1 mM IPTG for 1 h and lysed in SDS-PAGE buffer supplemented with Complete protease inhibitors cocktail (Roche) for Western blot analysis.
MyHC-actin ratio.
Muscle homogenate and single fibers were examined by SDS-PAGE for MyHC and actin content. The samples were solubilized in SDS-PAGE buffer (62.5 mM Tris pH 6.8, 2.3% SDS, 5% β-mercaptoethanol, 10% glycerol) containing the Complete protease inhibitor cocktail (Roche) and analyzed on 10% polyacrylamide gels. Protein bands were revealed by Coomassie blue staining. The MyHC-actin ratio was determined by using a Bio-Rad imaging densitometer (GS-670). Human biceps samples from control C05 and patient FSHD09 (see Fig. 10A) were not analyzed for the MyHC-actin ratio due to insufficient amount of starting material.
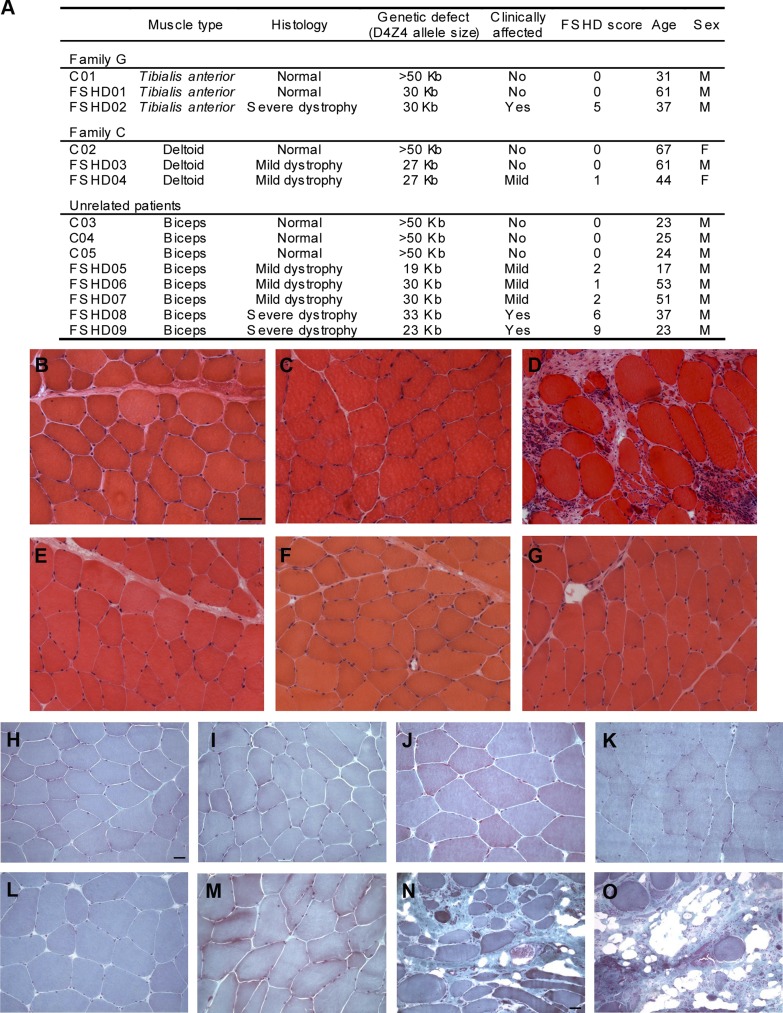
Fig. 10.
Muscles from facioscapulohumeral muscular dystrophy (FSHD) patients are affected differently by the dystrophic process. A: summary of patients' clinical characteristics. Individuals C01, C02, C03, C04, and C05 are normal controls; FSHD01 and FSHD03 carry the D4Z4 reduction but are clinically unaffected; patients FSHD02, FSDH04, FSHD05, FSHD06, FSHD07, FSHD08, and FSHD09 show a variable muscle impairment with FSHD with a clinical score ranging from 1 to 9. B–G: hematoxylin-and-eosin (H&E)-stained cross-cryosections of muscle biopsies from tibialis anterior (B–D) and deltoid (E–G). B and E: normal controls C01 and C02, respectively. C: unaffected carrier of the molecular defect showing no morphological alterations (FSHD01). D: severely affected FSHD02 patient showing increase of connective tissue and variability in myofiber size. F and G: mildly affected patients showing slight fiber-size variation, FSHD03, and FSHD04 respectively. B–G: Scale bar = 50 μm. H–O: Gomori trichrome-stained cryosections of biceps brachii. H–J: Normal controls C03, C04, and C05, respectively. K–M: mildly affected FSHD05, FSHD06, and FSHD07 showing normal morphology. N and O: biopsies of FSHD08 and FSHD09 patients with increase of connective tissue, prominent variability in myofiber size and a few fibers with centrally located nuclei, cell infiltration, and necrotic fibers. H–M: scale bar = 20 μm; N and O: scale bar = 50 μm.
SDS-PAGE analysis of MyHC.
Muscle homogenate and single fibers were examined by SDS-PAGE for MyHC content. The samples were dissolved in SDS-PAGE buffer (62.5 mM Tris pH 6.8, 2.3% SDS, 5% β-mercaptoethanol, 10% glycerol) containing the Complete protease inhibitor cocktail (Roche). A fragment from each chemically skinned single fiber used for physiological analyses was dissolved in 20 μl of the sample buffer, and an aliquot (5 μl) was analyzed on 8% polyacrylamide gels by the method described by Talmadge and Roy (41) to determine MyHC composition. The MyHC protein pattern was revealed by silver staining. Isoform composition was evaluated by densitometry using a Bio-Rad imaging densitometer (GS-670) and expressed as a percentage.
Whole muscle mechanical properties.
The experiments were performed in vitro at 30°C in a vertical muscle apparatus (300B, Aurora Scientific) containing a Ringer solution (120 mM NaCl, 4.7 mM KCl, 2.5 mM CaCl2, 3.15 mM MgCl2, 1.3 mM NaH2PO4, 25 mM NaHCO3, 11 mM glucose, 30 μM d-tubocurarine, pH 7.2–7.4) continuously bubbled with 95% O2-5% CO2. The muscles were stretched to the optimal length, i.e., the length that allowed maximal tension development in response to a single pulse. The muscles were then electrically stimulated, through two parallel electrodes, with supramaximal pulses (0.5-ms duration) delivered by a Grass S44 electronic stimulator through a stimulus isolation unit (Grass SIU5). Muscle response was recorded by isometric force transducer (Grass FT03) connected to AT-MIO 16AD acquisition card (National Instruments). Data were analyzed by the specific module of the National Instruments LabView software (15). Tetanic stimulation was obtained by applying trains of supramaximal stimuli at 80- and 150-Hz frequency for soleus and EDL, respectively. Tetanic tension was normalized to the muscle wet weight (specific tension, N/g). Muscles were weighed at the end of each experiment.
Single fiber preparation.
Muscle fibers were chemically skinned as previously described (7, 28, 38). Briefly, muscles were tied to a wooden stick, stretched to 110–120% of slack length, and quickly immersed into an ice-cold skinning solution containing 170 mM K-propionate, 2.5 mM Mg-propionate, 2.5 mM Na2K2ATP, 5 mM K2EGTA, 10 mM imidazole buffer, pH 7.0. After 24 h, the skinned fibers were stored at −20°C in a skinning solution supplemented with 50% (vol/vol) glycerol. Measurements were made within 4 wk.
pCa-tension relationship.
pCa-tension curves (pCa = −log[Ca2+]) were obtained by exposing the fiber sequentially to solutions with different free Ca2+ concentration (5). Single fiber segments (about 1.5 mm length) were inserted between two clamps, one fiber end fixed and the other connected to a tension transducer (403A force transducer, Aurora Scientific). Fibers were immersed in a relaxing solution (R solution) at room temperature (22–24°C) with the following composition: 170 mM K-propionate, 2.5 mM Mg-propionate, 5 mM Na2K2ATP, 5 mM K2EGTA, 10 mM imidazole buffer, at pH 7.0. Fibers were stretched to a sarcomere length of 2.7 μm, as measured by a video sarcomere length system (Aurora Scientific). Preliminarily, fiber was incubated in 0.2% (wt/vol) BRIJ-58 for 1 min to eliminate any possible contribution of the Ca2+ released from the sarcoplasmic reticulum (38). The tension generated in each pCa solution was continuously recorded, and the baseline tension was established as the steady-state voltage output recorded with the fiber in the relaxing solution. The following parameters were measured: pCa threshold (the minimum tension accepted to identify the pCa tension level was at least 5% of the maximum tension), pCa50 (the pCa value corresponding to 50% of maximum tension), N (the Hill coefficient), and the maximal tension produced at pCa 4.8. Maximum specific tension for each single fiber was calculated by normalizing the tension measured at pCa 4.8 to the fiber cross-sectional area, as calculated by three different diameter determinations along the fiber length, considering the fiber immersed in solution as a cylinder (6).
Purification of troponin complex.
The whole ternary troponin complex was purified from FRG1 and wild-type mice vastus lateralis, as previously described (35), with minor modification. Briefly, the frozen muscles were grinded in liquid nitrogen with mortar and pestle and homogenized at least 10 times in wash solution (1% Triton X-100, 50 mM NaCl, and 5 mM Tris pH 8.0) with the addition of protease inhibitor cocktail (Roche). The myofibrillar proteins were extracted from the final pellet in extraction solution (1 M NaCl, 25 mM Tris pH 8.0, 0.1 mM CaCl2, 0.1 mM DTT) with the addition of protease inhibitors cocktail (Roche). The solubilized proteins were purified by isoelectric point precipitation, the pH was lowered to 4.6, and the contaminant proteins were pelleted. The supernatant containing the troponin complex was adjusted to pH 8.0 and dialyzed against buffer A (100 mM NaCl, 5 mM MgCl2, 0.1 mM CaCl2, 0.1 mM DTT, 10 mM Tris, at pH 8.0). The troponin complex was further purified by anion exchange chromatography using a Resource Q column (GE Healthcare) and a AKTA prime plus system (GE Healthcare), with a NaCl gradient from buffer A to buffer B (500 mM NaCl, 5 mM MgCl2, 0.1 mM CaCl2, 0.1 mM DTT, 10 mM Tris pH 8.0). Fractions containing troponin complex were identified by SDS-PAGE and Coomassie staining, mixed together, concentrated to 1 mg/ml, and dialyzed against the exchange buffer (10 mM imidazole pH 7, 170 mM NaCl, 5 mM MgCl2, 25 mM EGTA, 3 mM CaCl2, and protease inhibitor cocktail).
Troponin exchange experiment.
A fiber fragment either from WT or FRG1 vastus lateralis muscle was mounted in the measure chamber, above described, filled with 100 μl R solution (170 mM K-propionate, 2.5 mM Mg-propionate, 5 mM Na2K2ATP, 5 mM K2EGTA, 10 mM imidazole buffer, pH 7.0). A pCa tension curve was generated by exposing the fiber to progressively higher free calcium solutions. After two washes in rigor solution (170 mM K-propionate, 2.5 mM Mg-propionate, 5 mM K2EGTA, 20 mM imidazole buffer, at pH 7.0) containing 10 mM BDM, the fiber was incubated for 2 h at 21 ± 1°C in rigor exchange solution (10 mM imidazole pH 7, 170 mM NaCl, 5 mM MgCl2, 2.5 mM EGTA, 3 mM CaCl2) containing 1 mg/ml of troponin complex, either from WT or FRG1 vastus lateralis muscle (modified from Ref. 33). After three washings in R solution, a second pCa tension curve was generated. Fibers from WT mice were used as control.
FSHD patients biopsies and clinical evaluation.
Clinical severity of the disease was numerically defined according to the standardized protocol developed by Lamperti et al. (21). All recruited subjects underwent DNA testing. Muscle functionality was evaluated in each subject. In the two families, selection for muscle biopsy was based on the functional impairment observed in the most severely affected member of the family, the tibialis anterior for family G and the deltoid for family C. Healthy controls or unaffected carriers underwent muscle biopsies on the same muscle as their affected relatives. For the group of unrelated patients and controls, biopsies were received from the Biobank of Muscle Tissue, Peripheral Nerve, DNA, cell line at the Foundation IRCCS Ca' Granda-Ospedale Maggiore-Policlinico Milano (Milan, Italy). All biopsies were taken from biceps.
Morphological analysis.
Skeletal muscle tissue specimens were frozen in isopentane cooled in liquid nitrogen, and stored in liquid nitrogen until use. Cross-cryosections (8 μm thick) were prepared and stained with hematoxylin-and-eosin staining or Gomori trichrome, as previously described (9).
Statistical analysis.
Means ± SE were calculated according to standard procedures. Student's t-test was used to test for statistical significance of differences between mean values following ANOVA. Statistical significance was set at P < 0.05. pCa-tension data were fitted by a least squares method using the table curve-fitting program (Jandel Scientific), according to the following equation: y = max xN/(xN + kN), where max is the maximal value of pCa-tension curve, which was normalized to 1, x is [Ca2+], k is the [Ca2+] at 50% of maximum tension, and N is the Hill coefficient. In figures Ca2+ concentration is expressed as −log [Ca2+] = pCa.
Ethics statement.
The experimental protocols on mice were approved by the Ethics Committee of the Medical Faculty of the University of Padova and by the Italian Health Ministry. Human biopsies of family G and family C members were approved by the Ethics Committee of the Medical Faculty of the University of Modena and Reggio Emilia. All other biopsies were obtained from the Telethon Genetic Biobank. All human subjects involved in this study provided informed consent prior to inclusion in the study. All procedures were performed in accordance with the Helsinki Declaration and Italian laws.
RESULTS
Alternative splicing of fast skeletal troponin T gene characterizes FRG1-overexpressing muscles.
The main clinical feature of FSHD is the selective and asymmetric involvement of specific muscle groups. Our previous studies revealed the aberrant splicing of the fast skeletal troponin T (TNNT3) mRNA in muscles from FRG1 transgenics, as well as in myoblasts of FSHD patients (13). Since the TNNT3 gene encodes one of the three subunits of the troponin complex, which governs striated muscle contraction (14, 16, 32), we reasoned that TNNT3 anomalous splicing might be one determinant of the selective muscle weakness that affects a subset of muscles in FSHD and in FRG1-overexpressing mice. If this were true, we would expect to detect an anomalous Tnnt3 splicing pattern in affected muscles. To investigate this possibility, we analyzed the splicing profile of Tnnt3 in vastus lateralis, a fast-twitch muscle, and in soleus, a mainly slow-twitch muscle in mice at 13 wk of age. These muscles were selected because at that age in FRG1 transgenics, vastus lateralis is severely dystrophic, whereas soleus is only slightly affected (8, 13).
The different Tnnt3 splicing isoforms were sequenced and quantified in each muscle type from wild-type and transgenic animals (Fig. 1). To characterize both alternative exons at 5′ end and mutually exclusive exons at 3′ end of the Tnnt3 transcript, we cloned the complete sequence of the different Tnnt3 isoforms from vastus lateralis and soleus of wild-type and FRG1 mice. The relative frequency of each isoform in each muscle is shown in Fig. 2, A and B and is consistent with quantitative PCR results shown in Fig. 1, B and C. We identified 23 Tnnt3 splicing isoforms whose arbitrary classification is reported in Table 1. Different Tnnt3 splicing isoforms characterized the diverse muscles. First, we found that diverse muscle types displayed different splicing profiles: in wild-type vastus lateralis 12A/B isoforms were the most abundant, whereas isoform 6B was prevalent in wild-type soleus (compare Fig. 1B with 1C). Second, as shown in Fig. 1B and Fig. 2A, transgenic vastus lateralis presented radical rearrangement of Tnnt3 isoform composition with the appearance of novel isoforms, named 9B and 13A/B, and disappearance of the isoforms 12A/B characteristic of the wild-type muscle. Third, in transgenic soleus, more limited variations were present, with a decrease of 4B and 6B isoforms and an increase of 9B isoform (Fig. 1C and Fig. 2B). Collectively, this analysis shows that specific Tnnt3 mRNA splicing patterns characterize different muscle types, with vastus lateralis, the most dystrophic muscle, displaying the greatest alterations of Tnnt3 mRNA splicing.
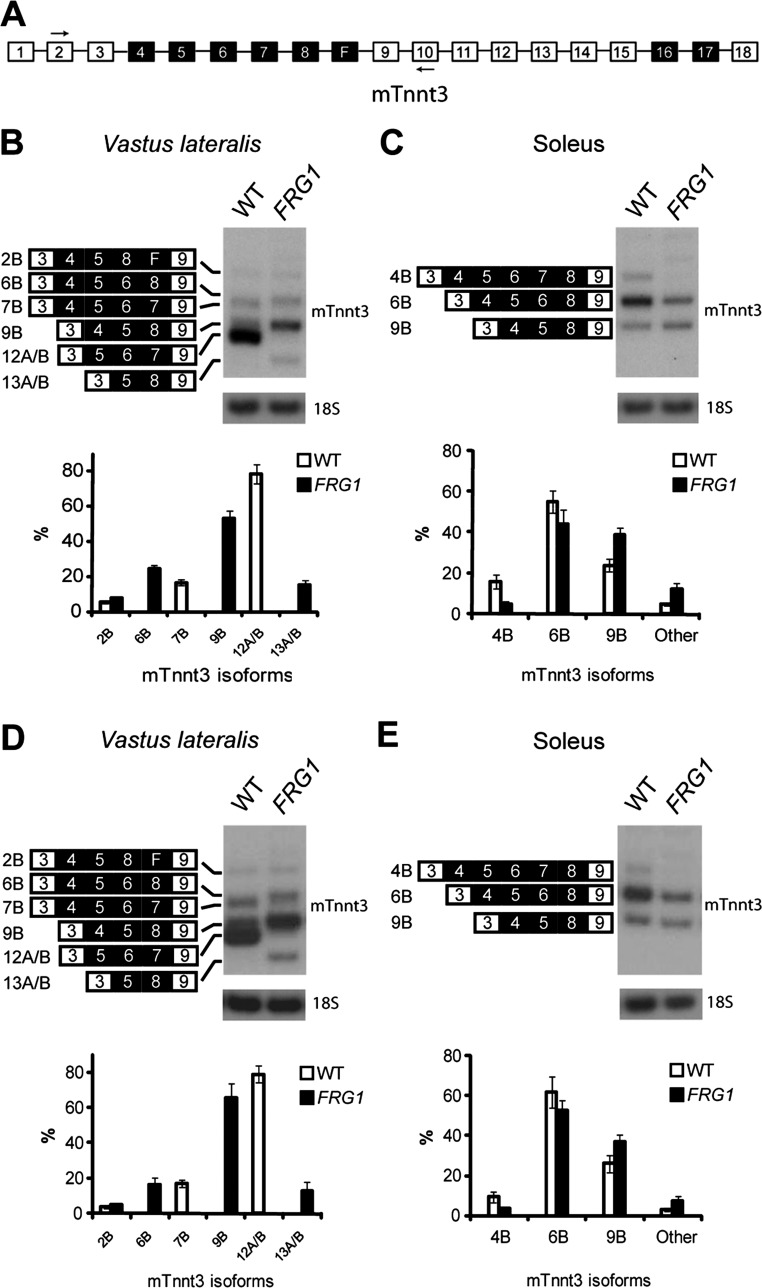
Fig. 1.
Differentially affected muscles of FRG1 mice show altered fast skeletal troponin T-splicing patterns. A: Scheme of Tnnt3 primary transcript. Open boxes illustrate constitutive exons, while black boxes indicate alternatively spliced exons, and the position of primers used for the RT-PCR analysis is indicated by arrows. “F” denotes the fetal exon. B–E: RT-PCR analysis of Tnnt3 splicing pattern in vastus lateralis (B and D) or soleus (C and E) muscles of 13-wk-old (B and C) or 4-wk-old (D and E) wild-type (WT), or FRG1-overexpressing mice. Identities of alternatively spliced products are indicated on the left of the gels. RT-PCR on 18S RNA is shown as a control. Quantitative analysis of the bands identified in the gels is shown in the charts below the gels. The analysis was conducted on three mice for each genotype. Values are expressed as means ± SE. WT (white bars) and FRG1 (solid bars).
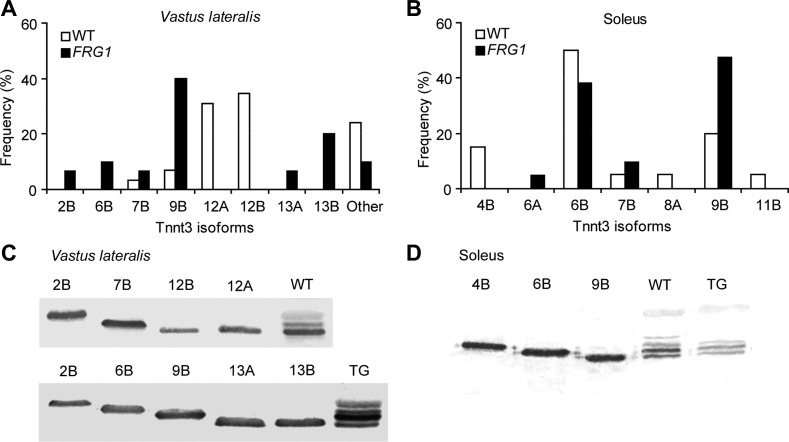
Fig. 2.
Identification of the Tnnt3 splicing isoforms in vastus lateralis and soleus of WT and FRG1 mice. A: frequency of the Tnnt3 isoforms cloned from vastus lateralis cDNA of in 13-wk-old WT (open bars) or FRG1 (solid bars) mice. Twenty-nine and thirty clones from two cDNA libraries representing Tnnt3 transcripts from WT and transgenic muscles, respectively, were full-length sequenced. Isoform identification numbers were assigned on the basis of the predicted protein length, and letters A or B were assigned on the basis of the presence of exon 16 or exon 17. B: frequency of the Tnnt3 isoforms cloned from soleus of 13-wk-old WT or FRG1 mice. Twenty clones from the WT and 21 from the transgenic cDNA libraries were full-length sequenced. A complete list of all the isoforms identified is shown in Table 1. C and D: The most abundant isoforms from WT and FRG1 vastus lateralis or soleus have been expressed as recombinant proteins, and their migration was compared with the fast skeletal troponin T pattern in muscle protein extracts by SDS-PAGE and immunoblotting.
Table 1.
List of Tnnt3 splicing isoforms identified in vastus lateralis and soleus of WT and FRG1 mice
| Isoform | Exon Composition | AA | MW, kDa | pI |
|---|---|---|---|---|
| fTnT-1B | 2 3 .4 5 6 8 F. 9 10 11 12 13 14 15 .17. 18 | 268 | 31.90 | 5.38 |
| fTnT-2B | 2 3 .4 5 8 F. 9 10 11 12 13 14 15 .17. 18 | 263 | 31.29 | 5.60 |
| fTnT-3AB | 2 3 .5 6 7. 9 10 11 12 13 14 15 .16 17. 18 | 262 | 30.58 | 9.04 |
| fTnT-4A | 2 3 .4 5 6 7 8. 9 10 11 12 13 14 15 .16. 18 | 259 | 30.77 | 6.09 |
| fTnT-4B | 2 3 .4 5 6 7 8. 9 10 11 12 13 14 15 .17. 18 | 259 | 30.77 | 6.19 |
| fTnT-5B | 2 3 .5 7 F. 9 10 11 12 13 14 15 .17. 18 | 256 | 30.26 | 6.92 |
| fTnT-6A | 2 3 .4 5 6 8 F. 9 10 11 12 13 14 15 .17. 18 | 255 | 30.50 | 6.38 |
| fTnT-6B | 2 3 .4 5 6 8. 9 10 11 12 13 14 15 .16. 18 | 255 | 30.37 | 6.47 |
| fTnT-7A | 2 3 .4 5 6 7. 9 10 11 12 13 14 15 .16. 18 | 254 | 30.15 | 7.71 |
| fTnT-7B | 2 3 .4 5 6 7. 9 10 11 12 13 14 15 .17. 18 | 254 | 30.15 | 7.73 |
| fTnT-8A | 2 3 .5 6 7 8. 9 10 11 12 13 14 15 .16. 18 | 253 | 29.96 | 8.40 |
| fTnT-8B | 2 3 .5 6 7 8. 9 10 11 12 13 14 15 .17. 18 | 253 | 29.96 | 8.40 |
| fTnT-9A | 2 3 .4 5 8. 9 10 11 12 13 14 15 .16. 18 | 250 | 29.76 | 8.39 |
| fTnT-9B | 2 3 .4 5 8. 9 10 11 12 13 14 15 .17. 18 | 250 | 29.76 | 8.39 |
| fTnT-10B | 2 3 .5 6 8. 9 10 11 12 13 14 15 .17. 18 | 249 | 29.57 | 8.69 |
| fTnT-11B | 2 3 .4 5 7. 9 10 11 12 13 14 15 .17. 18 | 249 | 29.54 | 8.86 |
| fTnT-12A | 2 3 .5 6 7. 9 10 11 12 13 14 15 .16. 18 | 248 | 29.34 | 9.01 |
| fTnT-12B | 2 3 .5 6 7. 9 10 11 12 13 14 15 .17. 18 | 248 | 29.35 | 9.01 |
| fTnT-13A | 2 3 .5 8. 9 10 11 12 13 14 15 .16. 18 | 244 | 28.95 | 9.12 |
| fTnT-13B | 2 3 .5 8. 9 10 11 12 13 14 15 .17. 18 | 244 | 28.95 | 9.12 |
| fTnT-14A | 2 3 .5 7. 9 10 11 12 13 14 15 .16. 18 | 243 | 28.73 | 9.29 |
| fTnT-14B | 2 3 .5 7. 9 10 11 12 13 14 15 .17. 18 | 243 | 28.73 | 9.29 |
| fTnT-15B | 2 3 .5. 9 10 11 12 13 14 15 .17. 18 | 239 | 28.34 | 9.36 |
Complete exon composition of all the identified isoforms. Black highlights indicate alternatively spliced exons. For each isoform aminoacid number (AA), predicted molecular weight (MW) and isoelectric point (pI) are indicated.
To investigate whether Tnnt3 splicing alteration is a direct consequence of FRG1 overexpression or a result of muscle deterioration, we evaluated the Tnnt3 splicing isoforms in fast and slow muscles of 4-wk-old FRG1 mice, an age when muscles do not show histological signs of dystrophy (49). At this age, the Tnnt3 isoforms detected in vastus lateralis and in soleus paralleled the splicing profile observed in 13-wk-old transgenic muscles (compare Fig. 1D with 1B and Fig. 1E with 1C). Thus, Tnnt3 splicing alteration occurs before the appearance of morphological signs of muscle degeneration and apparently results from the aberrant overexpression of FRG1. This idea is supported by the demonstration that FRG1 binds directly the troponin T transcript, as shown by cross-linking immunoprecipitation assay (Fig. 3).
Fig. 3.
FRG1 interacts directly with the troponin T transcripts in mouse muscle. Interaction of FRG1 protein with two different regions of the Tnnt3 mRNA and with GAPDH mRNA as a control was analyzed by cross-linking-immunoprecipitation (CLIP). The amount of RNA immunoprecipitated by the anti-FRG1 antibodies is expressed as a fold enrichment with respect to the RNA immunoprecipitated by a control IgG (NC).
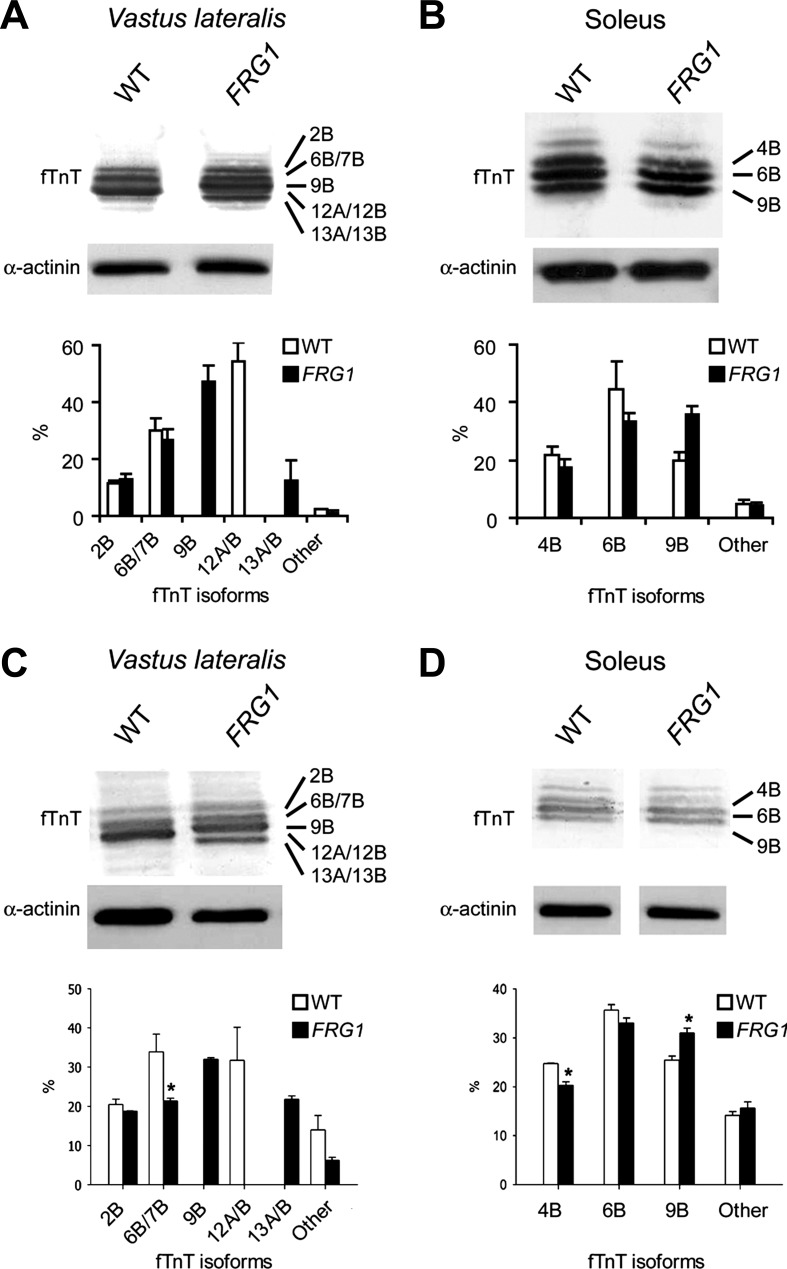
We then determined by immunoblotting the composition of fTnT protein isoforms in vastus lateralis and soleus from wild-type and transgenic animals (Fig. 4, A and B). Identities of the differentially expressed fTnT isoforms were defined by comparing the endogenous isoforms with recombinant fTnT proteins based on the sequence of the splicing isoforms, as shown in Fig. 2, C and D. As expected, the fTnT profile paralleled the splicing pattern with the appearance of the 9B and 13A/B isoforms in transgenic vastus lateralis (compare Fig. 1B with Fig. 4A). Consistent with the results of the splicing analysis, similar expression profile of fTnT isoforms was detected in vastus lateralis and soleus muscles from 4-wk-old transgenic mice (Fig. 4, C and D).
Fig. 4.
Immunoblot analysis of fTnT in vastus lateralis and soleus muscles of WT and FRG1 transgenics. A: vastus lateralis muscle (WT: n = 4; FRG1: n = 4) from 13-wk-old mice. Isoform identities are indicated on the right. Some isoforms comigrate (top). Immunoblotting with anti-α-actinin antibody is shown as a loading control. Quantitative analysis of fTnT isoforms is shown in the lower panel. B: soleus muscle muscle (WT: n = 4; FRG1: n = 4) from 13 wk-old-mice. C: vastus lateralis muscle (WT: n = 4; FRG1: n = 4) from 4-wk-old mice. D: soleus muscle (WT: n = 4; FRG1: n = 4) from 4 wk-old mice. WT (open bars) and FRG1 (solid bars). Values are expressed as means ± SE. *P < 0.05.
Interestingly, the 9B isoform, the most abundant isoform in transgenic vastus lateralis, contains an increased number of glutamic acid residues in the variable region at the NH2 terminus and an increased number of polar amino acids in the COOH terminus region compared with isoform 12A, which is typical of wild-type vastus lateralis.
Molecular reorganization of muscle-specific myofibrillar proteins in FRG1 transgenic mice.
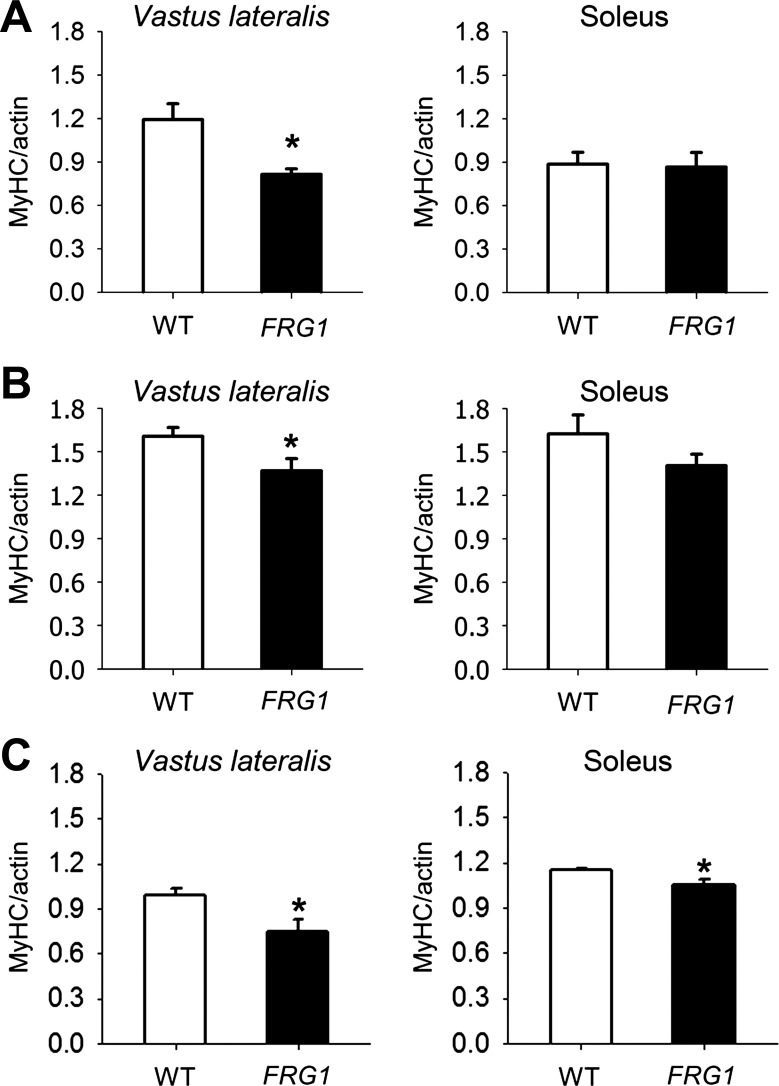
Muscle force is generated by the interaction between the motor protein myosin and actin and is strictly regulated by the troponin complex (14, 16). In addition to troponin T, we analyzed the expression of tropomyosin, troponin I, and troponin C. These analyses failed to detect significant differences of expression in the vastus lateralis and soleus muscles, comparing wild-type and FRG1-overexpressing mice (data not shown). However, we reasoned that, beside troponin T, the altered stoichiometry of myosin and actin also might contribute to the development of muscle weakness in FRG1 transgenics. To verify this possibility, we measured the amount of MyHC and actin in transgenic muscles at 13 wk of age and found that the MyHC-actin ratio was altered in murine FRG1-overexpressing vastus lateralis compared with wild-type muscle. By contrast, transgenic soleus did not display significant differences (Fig. 5A). These results were confirmed by the analysis of single muscle fibers (Fig. 5B), demonstrating that the altered MyHC-actin ratio is exclusively associated with rearrangements in muscle fibers and is not due to changes in nonsarcomeric actin. Importantly, we found that the MyHC-actin ratio was reduced also in FRG1 mice at 4 wk of age, when muscles do not display dystrophic signs (Fig. 5C). This observation supports the idea that FRG1 overexpression might determine the molecular reorganization of muscle-specific myofibrillar proteins that precedes the structural changes associated with the dystrophic process.
Fig. 5.
Dystrophic muscles of FRG1 transgenic mice show anomalous MyHC/actin ratio. A: MyHC-to-actin ratio in vastus lateralis and soleus muscles from 13-wk-old wild-type (n = 4) and transgenic mice overexpressing FRG1 (n = 4). B: MyHC-to-actin ratio in single fibers isolated from vastus lateralis (WT: n = 18; FRG1: n = 17) and soleus (WT: n = 17; FRG1: n = 18) muscles from 13-wk-old mice. C: MyHC/actin ratio in vastus lateralis (WT: n = 4; FRG1: n = 4) and soleus (WT: n = 4; FRG1: n = 4) muscles from 4-wk-old WT and transgenic mice overexpressing FRG1. Values are expressed as means ± SE. *P < 0.05.
Dystrophic FRG1 muscles display reduced force generation.
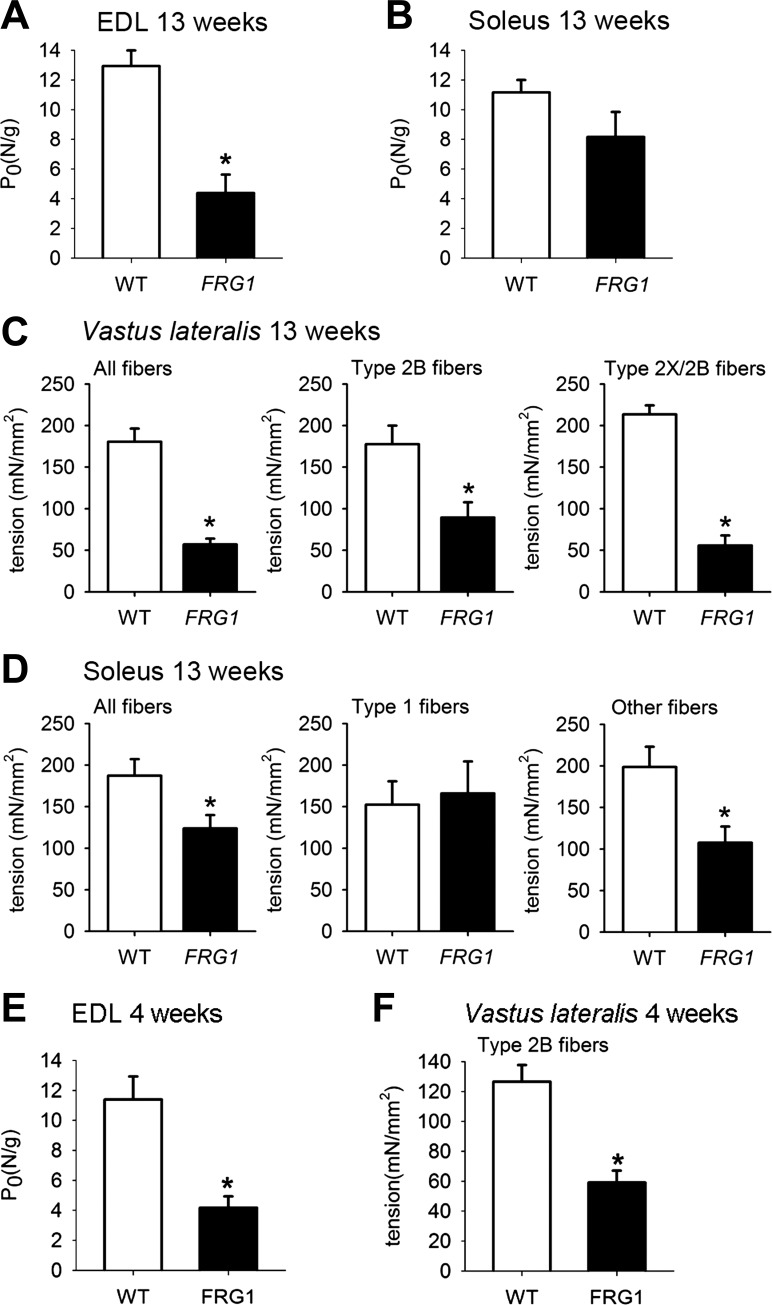
A possible consequence of the molecular reorganization of fTnT isoforms and the anomalous MyHC-actin ratio observed in FRG1 transgenics might be the alteration of muscle strength of the dystrophic muscle. To verify this possibility, the maximal isometric force was measured in soleus and EDL (extensor digitorum longus), a fast muscle with a myosin isoform composition similar to that of vastus lateralis (data not shown). Vastus lateralis was excluded from this analysis because, being part of the quadriceps, it is not possible to accurately separate it from the other muscle heads. Fig. 6A shows that EDL from 13-wk-old FRG1 transgenic mice developed less than half of maximal specific tension (normalized to the wet weight) of age-matched controls. In contrast, maximal specific tension of soleus muscle isolated from FRG1-overexpressing transgenics was not significantly different from the wild-type mice (Fig. 6B). Thus, molecular changes in myofibrillar proteins seem to translate into a defect of muscle contractility.
Fig. 6.
Dystrophic muscles of FRG1 transgenics show reduced force generation. A: maximal tetanic tension of extensor digitorum longus (EDL) (WT: n = 10; FRG1: n = 6) and soleus (WT: n = 8; FRG1: n = 5) (B) muscles from 13-wk-old mice. C: maximal specific tension of all fibers (WT: n = 28; FRG1: n = 18), type 2B fibers (WT: n = 24; FRG1: n = 10) and type 2X/2B fibers (WT: n = 4; FRG1: n = 8) isolated from vastus lateralis muscle of 13-wk-old mice. D: maximal specific tension of all fibers (WT: n = 37; FRG1: n = 25), type 1 fibers (WT: n = 6; FRG1: n = 20) and other fiber types (WT: n = 31; FRG1: n = 5) isolated from soleus muscle of 13-wk-old mice. E: maximal tetanic tension of EDL muscles from 4-wk-old wild-type mice and FRG1 transgenics (left panel) (WT: n = 6; FRG1: n = 11). Maximal specific tension of single fibers isolated from 4-wk-old wild-type mice and FRG1 transgenic vastus lateralis (WT: n = 9; FRG1: n = 9). Values are expressed as means ± SE. *P < 0.05.
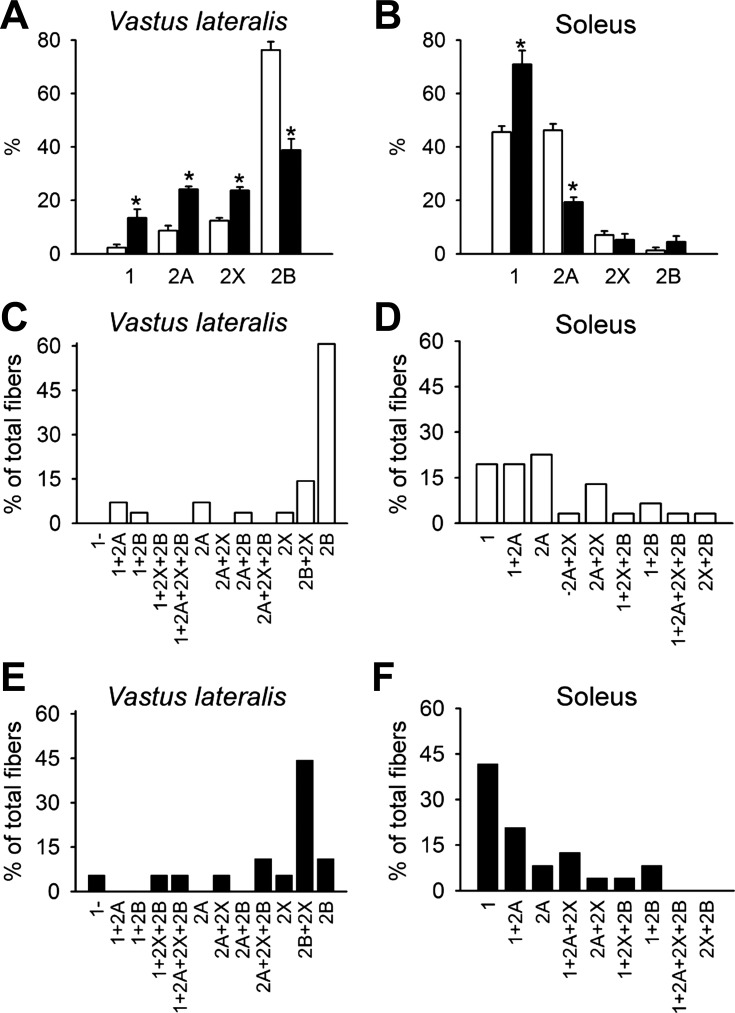
The main physiological difference between vastus lateralis and soleus resides in fiber-type composition. Vastus lateralis is mainly composed by fast-twitch fibers, whereas the soleus contains 50% of slow-twitch fibers. Therefore, we investigated whether the overall MyHC isoforms composition of fast and slow muscles is modified in FRG1 transgenics. In agreement with previous results (8), present results confirm the reduction of type 2B MyHC isoform in the fast transgenic muscle vastus lateralis (Fig. 7A), whereas a significant increase of type 1 isoform was present in transgenic soleus (Fig. 7B). In addition, the analysis of fiber-type composition in the transgenic vastus lateralis revealed a reduction of pure type 2B fibers (exclusively containing the type 2B MyHC) and increased levels of hybrid 2B/2X fibers and of mixed fibers containing type 2X, 2A, and 1 MyHC isoforms (Fig. 7, C and E). In transgenic soleus, we found a significant increase of pure type 1 fibers, associated with a decrease of type 2A fibers (Fig. 7, D and F). Collectively, these results indicated that a composite molecular reorganization occurs in transgenic muscles.
Fig. 7.
FRG1 transgenic muscles show a general fast-to-slow transition of MyHC isoforms. A: MyHC isoform expression in vastus lateralis (WT: n = 6 and FRG1: n = 5) and soleus (WT, n = 6 and FRG1, n = 7) (B) muscle of 13-wk-old mice. MyHC isoform composition in single fibers isolated from vastus lateralis (C and E) and soleus (D and F) of WT (open bars) and FRG1 mice (solid bars), respectively. Values are expressed as means ± SE. *P < 0.05.
To further investigate this possibility, we compared the contractile performance of isolated single fibers identified by their MyHC content. Figure 6, C and D (left panels) show that the mean maximal tension developed by pooled fibers isolated from transgenic vastus lateralis and soleus was significantly lower than the tension developed by control fibers from wild-type animals; the decrease was more marked in the highly affected transgenic vastus lateralis than in transgenic soleus. Remarkably, Fig. 6C (middle and right panel) and Fig. 6D (right panel) show that fast or mixed (fast + slow) fibers were weaker than control fibers in both transgenic muscle types. Most importantly, type 1 fibers from transgenic soleus developed tension comparable to that of control fibers (Fig. 6D, middle panel). These results indicate that FRG1 overexpression exerts its effects in type 2 fast fibers, but not in type 1 fibers, making the fast muscle more susceptible to force loss than the slow muscle.
We also measured the maximal specific isometric force in EDL muscle, and the maximal specific force in single fibers isolated from vastus lateralis of 4-wk-old mice. Consistent with the alteration of sarcomeric proteins (shown in Fig. 1, D and E, Fig. 4, C and D, and in Fig. 5C), we determined that the force reduction was already present in EDL muscle and type 2B skinned fibers from vastus lateralis of FRG1 transgenic mice at 4 wk of age (Fig. 6E). These results support the notion that in FRG1 dystrophic mice, muscle weakness is not due to degeneration of muscle cells caused by the dystrophic process but is associated with molecular rearrangements of muscle proteins occurring at a young age before the appearance of dystrophic changes.
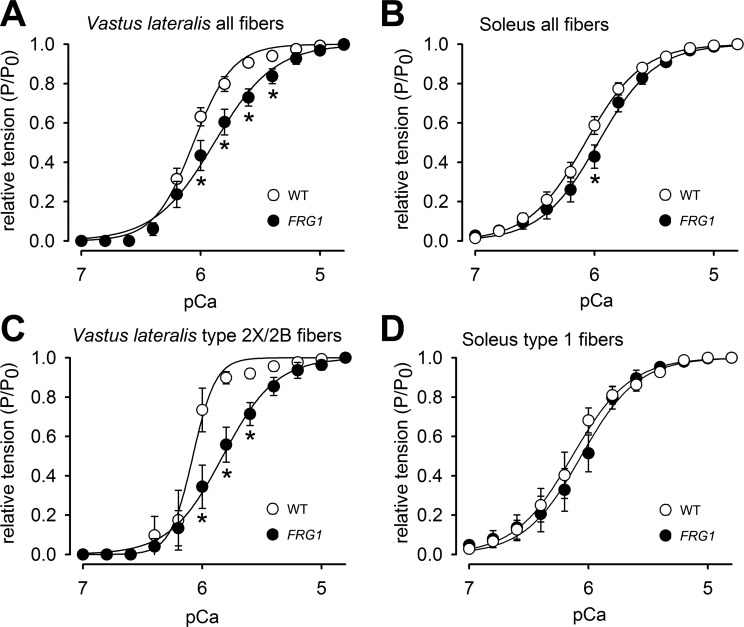
The troponin complex composition influences calcium sensitivity in single muscle fibers.
Since fTnT is a subunit of the trimeric troponin complex, which regulates skeletal muscle contraction in response to calcium, we further investigated whether the altered composition of the fTnT isoforms in myofibers of FRG1 transgenics might translate into alteration of the fibers' calcium sensitivity. To test this possibility, we measured single-fiber contraction after stimulation with increasing calcium concentrations (Table 2). Consistent with our hypothesis, single fibers isolated from FRG1-overexpressing vastus lateralis displayed a reduced ability to develop force in the presence of activating calcium concentrations (Fig. 8A). In contrast, fibers derived from FRG1-overexpressing soleus displayed only minor changes (Fig. 8B). Notably, a net reduced sensitivity to calcium was displayed by transgenic single fibers expressing fast MyHC isoforms from the vastus lateralis, as shown by the lower tension developed at each calcium concentration compared with wild-type (Fig. 8C). By contrast, the calcium sensitivity of type 1 fibers from FRG1 soleus was not changed (Fig. 8D).
Table 2.
Calcium sensitivity of single muscle fibers of WT and FRG1 mice
| Vastus Lateralis |
Soleus |
|||
|---|---|---|---|---|
| WT (20;3) | FRG1 (18;2) | WT (31;4) | FRG1 (24;3) | |
| pCa TH | 6.26 ± 0.03 | 6.12 ± 0.05* | 6.65 ± 0.06 | 6.37 ± 0.08* |
| pCa50 | 6.09 ± 0.04 | 5.89 ± 0.07* | 6.12 ± 0.04 | 5.78 ± 0.22 |
| N | 5.40 ± 0.63 | 2.82 ± 0.26* | 2.96 ± 0.26 | 3.05 ± 0.33 |
| Tension, mN/mm2 | 181 ± 16 | 57.1 ± 6.9* | 187 ± 20 | 132 ± 20* |
| Diameter, μm | 56.8 ± 1.6 | 41.4 ± 3.0* | 35.2 ± 1.1 | 32.5 ± 1.2 |
Values are expressed as means ± SE. pCaTH is the lowest pCa giving a detectable tension; pCa50 is the pCa value corresponding to 50% of maximum tension; N is the Hill coefficient; tension corresponds to the maximal tension produced at pCa 4.8. In parenthesis is the number of fibers and animals utilized.
P < 0.05 indicates statistical significance with respect to WT fibers.
Fig. 8.
Fast-twitch dystrophic fibers show altered ability to contract in response to calcium. Relative pCa/tension relationships of all single fibers (A) and type 2X/2B fibers (C) isolated from vastus lateralis of WT (open circles) and FRG1-overexpressing mice (solid circles). A: values are the mean of 20 fibers for the WT and 18 fibers for the transgenic. C: values are the mean of four fibers for the WT and eight fibers for the transgenic. Relative pCa/tension relationships of all single fibers (A) and type 1 fibers (D) isolated from soleus of WT (open circles) and FRG1-overexpressing mice (solid circles). B: values are expressed as the mean of 31 fibers for the WT and 24 fibers for the transgenic. D: values are the mean of six fibers for the WT and 20 fibers for the transgenic. Values are expressed as means ± SE. *P < 0.05. The mean curve fitting was generated by the data reported in Table 2.
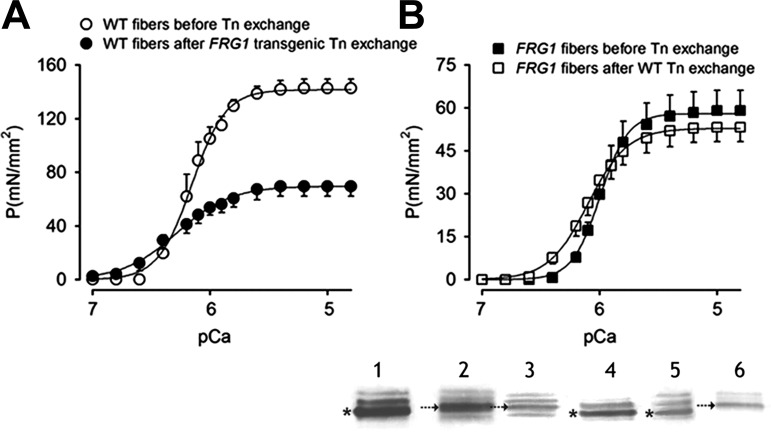
Since the troponin complex regulates muscle contraction in response to calcium, we reasoned that the altered fTnT isoforms might be responsible for the reduction of calcium sensitivity in FRG1 transgenics. We, therefore, considered that if this hypothesis were true, the exchange of the transgenic troponin complex into wild-type fibers would alter their calcium sensitivity. To test this possibility, we isolated the troponin complex from transgenic muscles and replaced it in the isolated wild-type fibers by mass exchange. The success of the troponin replacement was evaluated in each fiber after the exchange by immunoblot (Fig. 9, inset). Force development was then measured in response to increasing calcium concentrations. As expected, Fig. 9A shows that exchange of transgenic troponin complex into wild-type muscle fibers caused more than 50% loss of the maximal tension without significant modification of pCa50 (Table 3). In parallel, we exchanged the wild-type troponin complex into transgenic fibers to verify whether the substitution of the anomalous troponin complex might rescue the calcium sensitivity to normal and improve tension development. Fig. 9B and Table 3 show that replacement of the aberrant troponin complex in transgenic vastus lateralis with the wild-type complex generated a rescue of calcium sensitivity, shown by the significant leftward shift of the pCa/tension curve (Fig. 9B). However, we did not observe a recovery of force. To explain this result, we considered that the complete rescue of the physiological phenotype with an increase of maximal tension in transgenic fibers could be prevented by other alterations occurring in the contractile apparatus of FRG1 transgenic muscles. Consistent with this possibility, the exchange of the wild-type troponin complex into wild-type muscle fibers did not generate significant differences in force development and calcium sensitivity (data not shown). Collectively, these molecular exchanges confirm that aberrant splicing of fast troponin T mRNA in dystrophic muscles produces aberrant fast troponin T isoforms, leading to less efficient muscle contraction. Further, our ability to restore physiological sensitivity to calcium with wild-type troponin T is consistent with physiological relevance for the observed changes in Tnnt3 splicing and suggests a basis for future therapeutic approaches.
Fig. 9.
Substitution of transgenic troponin complex into wild-type fibers and of wild-type troponin complex into dystrophic fibers. A: pCa/tension curves of WT vastus lateralis single fibers (n = 5) before (open circles) and after (solid circles) FRG1 transgenic troponin exchange. B: pCa/tension curves of FRG1 vastus lateralis single fibers (n = 11) before (solid squares) and after (open squares) WT troponin exchange experiments. Values are expressed as means ± SE. The mean curve fitting was generated by the data reported in Table 3. In the panel at the bottom is shown the analysis of fTnT isoforms composition by immunoblotting both in lysates of whole vastus lateralis muscle and of single fibers. Lane 1, fTnT isoforms purified from WT muscle; lane 2, fTnT isoforms purified from FRG1 over-expressing muscle; lane 3, FRG1 single fiber; lane 4, WT single fiber; lane 5, FRG1 single fiber after troponin exchange with a complex isolated from WT muscle; and lane 6, WT single fiber after troponin exchange with a complex isolated from FRG1 muscle. Asterisks and arrows indicate fTnT isoforms 12A/B and 9B, respectively.
Table 3.
Troponin complex exchange in single muscle fibers from vastus lateralis
| WT + FRG1 Tn Complex |
FRG1 + WT Tn Complex |
FRG1+ Exchange Buffer |
||||
|---|---|---|---|---|---|---|
| Before (5;2) | After (5;2) | Before (11;2) | After (11;2) | Before (5;2) | After (5;2) | |
| pCa TH | 6.28 ± 0.05 | 6.64 ± 0.10‡ | 6.22 ± 0.02 | 6.44 ± 0.04* | 6.13 ± 0.02 | 6.20 ± 0.00 |
| pCa50 | 6.18 ± 0.04 | 6.28 ± 0.05 | 5.98 ± 0.02 | 6.08 ± 0.03† | 5.88 ± 0.04 | 5.89 ± 0.04 |
| N | 3.88 ± 0.66 | 2.22 ± 0.32‡ | 3.62 ± 0.40 | 2.98 ± 0.29 | 3.18 ± 0.39 | 2.49 ± 0.23 |
| Tension, mN/mm2 | 143 ± 6 | 70 ± 7* | 59 ± 7 | 53 ± 5‡ | 53 ± 6 | 49 ± 6‡ |
Values are expressed as means ± SE. pCaTH is the lowest pCa giving a detectable tension; pCa50 is the pCa value corresponding to 50% of maximum tension; N is the Hill coefficient; Max tension corresponds to the tension produced at pCa 4.8. In parenthesis is the number of fibers and animals utilized.
P < 0.0001;
P < 0.001;
P < 0.05 indicate statistical significance with respect to the values before the related treatment.
Alternative splicing of TNNT3 gene characterizes healthy and dystrophic muscles in FSHD patients.
Recent studies showed the selective reduction of force in type 2 muscle fibers from FSHD patients (22). This similarity with our findings in FRG1 transgenics, led us to investigate whether TNNT3 splicing profile can be correlated with clinical features such as muscle weakness or histological features found in FSHD patients. Thus, we analyzed the TNNT3 splicing isoforms in human muscle biopsies characterized by different degrees of damage. As shown in Fig. 10, muscle samples were obtained from two FSHD family groups and eight unrelated subjects; the D4Z4 repeat size at 4q35 was molecularly defined in all subjects. Individuals carrying the FSHD molecular defect, as well as normal controls, were clinically examined, using a standardized protocol, which generates the FSHD clinical score (21). The total score can range from 0, when no signs of muscle weakness are present, to 15, when all muscle groups tested are severely impaired (21). On this basis, all subjects were categorized as shown in Fig. 10A. In families G and C two subjects, FSHD01 and FSHD03, carried the molecular defect and displayed no muscle weakness (FSHD score 0). As shown in Fig. 10B–O, biopsies from individuals carrying D4Z4 repeat reduction displayed varying extents of muscle damage (compare D, N, O with C, F, G, K–M and with B, E, H–J, normal controls, in Fig. 10), which reflected the clinical status of each individual (Fig. 10A). Interestingly, analysis of TNNT3 splicing profile (Fig. 11, A and B) and fTnT protein isoform composition (Fig. 11, C and D) highlighted a correlation between the clinical status and the TNNT3 splicing pattern. In particular, major alterations were found in muscles from subjects FSHD02, FSHD08, and FSHD09, whose muscle biopsies display clear dystrophic morphology (Fig. 10, D, N, and O). Notably, these FSHD patients presented a more severe clinical phenotype, as shown by the high severity FSHD score received at clinical examination (Fig. 10A). To further define the clinical significance of the TNNT3 splicing profile, muscle samples obtained from patients affected by Myotonic Dystrophy1 (DM1) and Becker muscular dystrophy (BMD) were analyzed. Fig. 11, A and B shows that muscle from the DM1 patient did not display any alteration compared with healthy controls, while the two samples from BMD patients displayed only a slight shift in isoform composition. These findings support the idea that alteration of fTnT isoforms might reflect muscle impairment and, therefore, correlate with clinical severity in myopathic patients.
DISCUSSION
The molecular complexity of FSHD has still to be unraveled, because no single gene can be considered fully responsible for FSHD pathogenesis (39). However, FRG1-overexpressing mice display several characteristics typical of FSHD patients, including an obvious dystrophic phenotype, involvement of specific muscle groups, absence of regeneration, and lack of dystroglycan complex abnormalities. Here, we demonstrated that the aberrant splicing of Tnnt3 mRNA contributes to the molecular mechanism leading to muscle weakness, the major clinical feature of FSHD.
Using FRG1 transgenics, we analyzed functional impairment in muscles that show different degrees of tissue damage. As expected (8), we observed that force drop is more pronounced in severely dystrophic muscles such as vastus lateralis and EDL, than in soleus, which is only slightly affected by the dystrophic process. Interestingly, this phenomenon is already present in transgenic muscles at 4 wk of age when no histological signs of muscle dystrophy are present. Moreover, we found that transgenic fast fibers, the largest components of vastus lateralis, are significantly weaker than those from wild-type mice. In contrast, tension developed by pure type 1 fibers, which prevail in soleus muscle, is not reduced in transgenic mice, consistent with the minimal force reduction observed in this muscle. This is consistent with the report that muscle fibers isolated from FSHD patients produced a significant force reduction, specifically involving fast-twitch fibers (22). Moreover, our analysis reveals a fast-to-slow shift in fiber-type composition in some FRG1 transgenic muscles. This transition, which has been observed in FSHD patients (3), seems to be more effective in FRG1 slow muscles, such as soleus, that maintain most of its force and contractile properties. In contrast, this transformation is less effective in FRG1 fast muscles, such as the vastus lateralis, where the number of type 1 fibers is very low. Indeed, it seems that transgenic muscles mainly composed of slow-twitch fibers are favored in enduring the pathogenic noxa. This observation raises the possibility that fiber-type composition can be one component of selective muscle weakness characteristic of FSHD and might provide an explanation to the wide variability of the clinical severity of the disease among subjects carrying an identical D4Z4 mutation.
Our study also shows that a reorganization of contractile apparatus, including an altered MyHC-actin ratio and severe rearrangements of fTnT isoforms, occurs in transgenic fast fibers. In particular, we identified one isoform that becomes highly expressed in dystrophic vastus lateralis. Interestingly, this isoform is characterized by an increased number of glutamic acid residues in the variable region at the NH2 terminus and an increased number of polar amino acids in the COOH terminus region. As it is known that fTnT isoforms finely regulate the affinity of the ternary troponin complex for calcium and, ultimately, the contraction capability of the sarcomeric unit following calcium stimulation (31), we speculate that these changes may influence the assembly of the ternary troponin complex and/or finely tune its interactions with other myofibrillar proteins, such as tropomyosin, providing novel characteristics to the contractile unit (4, 46, 47).
The mechanistic relationship between fTnT variations and altered contractile properties of FRG1 transgenic muscles is also provided. By substituting the anomalous troponin complex with the wild-type proteins, we were able to restore the sensitivity to Ca2+ for tension development in dystrophic muscle fibers. Conversely, substitution of the wild-type troponin complex with complexes isolated from FRG1-overexpressing muscle diminishes muscle fiber tension. These results demonstrate for the first time that modifications of troponin T isoforms contribute to reduce muscle contractility in fibers from affected muscles and identify fast-twitch fibers as the specific target of FRG1 overexpression.
In clinical practice, progressive weakness and functional decline define the status and progression of disease and documents the natural course of myopathy (25, 26, 27, 40). Although necrosis of muscle fiber represents the major cause of muscle weakness in muscular dystrophies (45), it is a late consequence of disease, which does not account for the early events of the dystrophic process. In contrast, our study shows that force reduction, as well as alteration of MyHC-actin ratio, Tnnt3 splicing, and fiber-type composition already occur in mice at 4 wk of age before any morphological signs of muscle degeneration are detected (49). These novel phenotypes are consistent with the finding that the aberrant splicing of other alternatively spliced transcripts occurs in muscles of 4-wk-old FRG1 mice before the appearance of dystrophic signs (35). Thus, in FRG1 transgenics, molecular rearrangements of the contractile apparatus, as well as reduced muscle strength, are not a consequence of dystrophic muscle wasting but are early events, possibly involved in the initiation of FSHD pathogenesis.
In this context, the anomalous TnnT3 isoform patterns in predystrophic muscles of 4-wk-old FRG1 transgenic mice opens the possibility that altered TnnT3 isoforms might predict a severe clinical outcome. Consistent with this possibility, the TNNT3 splicing pattern and the fTnT profile correlate with the clinical and histological phenotypes in carriers of the FSHD molecular defect (Figs. 10 and 11). In contrast, we did not find TNNT3 alterations in a subgroup of carriers (older than 40 years) who had no signs of FSHD or very little muscle impairment (Figs. 10 and 11). These observations suggest that subjects without altered TNNT3 or fTnT profiles will not develop severe FSHD; thus, we propose that there is predictive value to these analyses.
Perspectives and Significance
Collectively, our study not only provides new evidence reinforcing the value of FRG1 mice in understanding FSHD pathophysiology, but also confirms that FRG1 overexpression mainly affects fast-twitch fibers and, thereby, causes weakness of fast muscles. These results are of particular interest for myopathic patients since fiber-type composition may be modulated by several factors, including the genetic background and environmental factors, such as hormones, daily activity, exercise, aging, immobilization, and chronic diseases. In addition, the demonstration that modifications of the troponin complex might have a beneficial effect on calcium sensitivity and fiber contraction provides a possible target for therapeutic interventions. Finally, our study opens the possibility that analysis of the profile of TNNT3 isoforms might become a biomarker of muscle wasting and a predictor of phenotype severity.
GRANTS
This work was supported by National Institutes of Health (Grant NIAMS-5RO1AR056129 and NINDS-RO1NS047584), Association Française contre les Myopathies (Grants 12055, 13470, and 13578), Telethon Grant GGP05050, Italian Ministry of Education, University and Research, Associazione Amici del Centro Dino Ferrari-University of Milan, and Telethon Genetic Biobanks Network (project number GTB07001ER) and Eurobiobank (project number QLTR-200102769). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Author contributions: V.S., D.D.-B., and R.G.T. conception and design of research; V.S., E.G., A.E., E.M., and S.P. performed experiments; V.S., E.G., A.E., E.M., S.P., M.M., and G.T. analyzed data; V.S., E.G., M.M., G.T., D.D.-B., and R.G.T. interpreted results of experiments; V.S. and E.G., prepared figures; V.S., E.G., D.D.-B., and R.G.T. edited and revised manuscript; V.S., E.G., A.E., E.M., S.P., M.M., G.T., D.D.-B., and R.G.T. approved final version of manuscript; D.D.-B. and R.G.T. drafted manuscript.
ACKNOWLEDGMENTS
We thank all the FSHD patients and families for their help and support. We thank Pieter deTombe, Brandon Biesiadecki, and Mariapaola Costi for helping in troponin complex isolation; Umberto Muscatello for critical support; and Paul D. Kaufman for reading and critically reviewing the manuscript.
Present address for V. Sancisi: Department of Oncology and Advanced Technology, Arcispedale Santa Maria Nuova- IRCCS, 42123 Reggio Emilia, Italy.
REFERENCES
- 1.Arnett AL, Chamberlain JR, Chamberlain JS. Therapy for neuromuscular disorders. Curr Opin Genet Dev 19: 290–297, 2009 [DOI] [PubMed] [Google Scholar]
- 2.Cabianca DS, Gabellini D. The cell biology of disease: FSHD: copy number variations on the theme of muscular dystrophy. J Cell Biol 191: 1049–1060, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Celegato B, Capitanio D, Pescatori M, Romualdi C, Pacchioni B, Cagnin S, Vigano' A, Colantoni L, Begum S, Ricci E, Wait R, Lanfranchi G, Gelfi C. Parallel protein and transcript profiles of FSHD patient muscles correlate to the D4Z4 arrangement and reveal a common impairment of slow to fast fibre differentiation and a general deregulation of MyoD-dependent genes. Proteomics 6: 5303–5321, 2006 [DOI] [PubMed] [Google Scholar]
- 4.Chaudhuri T, Mukherjea M, Sachdev S, Randall JD, Sarkar S. Role of the fetal and α/β exons in the function of fast skeletal troponin T isoforms: correlation with altered Ca2+ regulation associated with development. J Mol Biol 352: 58–71, 2005 [DOI] [PubMed] [Google Scholar]
- 5.Danieli-Betto D, Betto R, Midrio M. Calcium sensitivity and myofibrillar protein isoforms of rat skinned skeletal muscle fibres. Pflügers Arch 417: 303–308, 1990 [DOI] [PubMed] [Google Scholar]
- 6.Danieli-Betto D, Germinario E, Esposito A, Biral D, Betto R. Effects of fatigue on sarcoplasmic reticulum and myofibrillar properties of rat single muscle fibers. J Appl Physiol 89: 891–898, 2000 [DOI] [PubMed] [Google Scholar]
- 7.Danieli-Betto D, Esposito A, Germinario E, Sandonà D, Martinello T, Jakubiec-Puka A, Biral D, Betto R. Deficiency of α-sarcoglycan differently affects fast-and slow-twitch skeletal muscles. Am J Physiol Regul Integr Comp Physiol 289: R1328–R1337, 2005 [DOI] [PubMed] [Google Scholar]
- 8.D'Antona G, Brocca L, Pansarasa O, Rinaldi C, Tupler R, Bottinelli R. Structural and functional alterations of muscle fibres in the novel mouse model of facioscapulohumeral muscular dystrophy. J Physiol 584: 997–1009, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubowitz V. Muscle Biopsy. A Practical Approach. London: Balliére-Tindall, 1985 [Google Scholar]
- 10.Emery AE. Muscular dystrophy into the new millennium. Neuromusc Disord 12: 343–349, 2002 [DOI] [PubMed] [Google Scholar]
- 11.Flanigan KM. Facioscapulohumeral muscular dystrophy and scapuloperoneal syndromes In: Myology, 3rd ed., edited by Engel A, Franzini-Armstrong C. New York: McGraw Hill Professional Press; 2004, p. 1123–1133 [Google Scholar]
- 12.Gabellini D, Green MR, Tupler R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell 110: 339–348, 2002 [DOI] [PubMed] [Google Scholar]
- 13.Gabellini D, D'Antona G, Moggio M, Prelle A, Zecca C, Adami R, Angeletti B, Ciscato P, Pellegrino MA, Bottinelli R, Green MR, Tupler R. Facioscapulohumeral muscular dystrophy in mice overexpressing FRG1. Nature 439: 973–977, 2006 [DOI] [PubMed] [Google Scholar]
- 14.Galińska-Rakoczy A, Engel P, Xu C, Jung H, Craig R, Tobacman LS, Lehman W. Structural basis for the regulation of muscle contraction by troponin and tropomyosin. J Mol Biol 379: 929–935, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Germinario E, Esposito A, Midrio M, Peron S, Palade PT, Betto R, Danieli-Betto D. High-frequency fatigue of skeletal muscle. Role of extracellular Ca2+. Eur J Appl Physiol 104: 445–453, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev 80: 853–924, 2000 [DOI] [PubMed] [Google Scholar]
- 17.Hanel ML, Wuebbles RD, Jones PL. Muscular dystrophy candidate gene FRG1 is critical for muscle development. Dev Dyn 238: 1502–1512, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen KB, Darnell RB. CLIP: crosslinking and immunoprecipitation of in vivo RNA targets of RNA-binding proteins. Methods Mol Biol 488: 85–98, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalkman JS, Zwarts MJ, Schillings ML, van Engelen BG, Bleijenberg G. Different types of fatigue in patients with facioscapulohumeral dystrophy, myotonic dystrophy and HMSN-I. Experienced fatigue and physiological fatigue. Neurol Sci 29: S238–S240, 2008 [DOI] [PubMed] [Google Scholar]
- 20.Krom YD, Thijssen PE, Young JM, den Hamer B, Balog J, Yao Z, Maves L, Snider L, Knopp P, Zammit PS, Rijkers T, van Engelen BG, Padberg GW, Frants RR, Tawil R, Tapscott SJ, van der Maarel SM. Intrinsic epigenetic regulation of the D4Z4 macrosatellite repeat in a transgenic mouse model for FSHD. PLoS Genet 9:e1003415, doi: 10.137, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamperti C, Fabbri G, Vercelli L, D'Amico R, Frusciante R, Bonifazi E, Fiorillo C, Borsato C, Cao M, Servida M, Greco F, Di Leo R, Volpi L, Manzoli C, Cudia P, Pastorello E, Ricciardi L, Siciliano G, Galluzzi G, Rodolico C, Santoro L, Tomelleri G, Angelini C, Ricci E, Palmucci L, Moggio M, Tupler R. A standardized clinical evaluation of patients affected by facioscapulohumeral muscular dystrophy: the FSHD clinical score. Muscle Nerve 42: 213–217, 2010 [DOI] [PubMed] [Google Scholar]
- 22.Lassche S, Stienen GJ, Irving TC, van der Maarel SM, Voermans NC, Padberg GW, Granzier H, van Engelen BG, Ottenheijm CA. Sarcomeric dysfunction contributes to muscle weakness in facioscapulohumeral muscular dystrophy. Neurology 80: 733–737, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemmers RJ, van der Vliet PJ, Klooster R, Sacconi S, Camaño P, Dauwerse JG, Snider L, Straasheijm KR, van Ommen GJ, Padberg GW, Miller DG, Tapscott SJ, Tawil R, Frants RR, van der Maarel SM. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science 329: 1650–1653, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Q, Jones TI, Tang VW, Brieher WM, Jones PL. Facioscapulohumeral muscular dystrophy region gene-1 (FRG-1) is an actin-bundling protein associated with muscle-attachment sites. J Cell Sci 123: 1116–1123, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lue YJ, Chen SS. The strength and functional performance in patients with facioscapulohumeral muscular dystrophy. Kaohsiung J Med Sci 16: 248–254, 2000 [PubMed] [Google Scholar]
- 26.Lue YJ, Lin RF, Chen SS, Lu YM. Measurement of the functional status of patients with different types of muscular dystrophy. Kaohsiung J Med Sci 25: 325–333, 2009 [DOI] [PubMed] [Google Scholar]
- 27.Mattar FL, Sobreira C. Hand weakness in Duchenne muscular dystrophy and its relation to physical disability. Neuromuscul Disord 18: 193–198, 2008 [DOI] [PubMed] [Google Scholar]
- 28.Midrio M, Danieli-Betto D, Megighian A, Betto R. Early effects of denervation on sarcoplasmic reticulum properties of slow-twitch rat muscle fibres. Pflügers Arch 434: 398–405, 1997 [DOI] [PubMed] [Google Scholar]
- 29.Mitsuhashi H, Mitsuhashi S, Lynn-Jones T, Kawahara G, Kunkel LM. Expression of DUX4 in zebrafish development recapitulates facioscapulohumeral muscular dystrophy. Hum Mol Genet 22: 568–577, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng R, Banks GB, Hall JK, Muir LA, Ramos JN, Wicki J, Odom GL, Konieczny P, Seto J, Chamberlain JR, Chamberlain JS. Animal models of muscular dystrophy. Prog Mol Biol Transl Sci 105: 83–111, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Padberg G. Facioscapulohumeral Disease (PhD thesis) Leiden University, Leiden, The Netherlands, 1982 [Google Scholar]
- 32.Perry SV. Troponin T: genetics, properties and function. J Muscle Res Cell Motil 19: 575–602, 1998 [DOI] [PubMed] [Google Scholar]
- 33.Piroddi N, Tesi C, Pellegrino MA, Tobacman LS, Homsher E, Poggesi C. Contractile effects of the exchange of cardiac troponin for fast skeletal troponin in rabbit psoas single myofibrils. J Physiol 552: 917–931, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pistoni M, Shiue L, Cline MS, Bortolanza S, Neguembor MV, Xynos A, Ares M, Jr, Gabellini D. Rbfox1 downregulation and altered calpain 3 splicing by FRG1 in a mouse model of Facioscapulohumeral muscular dystrophy (FSHD). PLoS Genet 9: e1003186, doi: 10.1371, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Potter JD. Preparation of troponin and its subunits. Methods Enzymol 85: 241–263, 1982 [DOI] [PubMed] [Google Scholar]
- 36.Ricci E, Galluzzi G, Deidda G, Cacurri S, Colantoni L, Merico B, Piazzo N, Servidei S, Vigneti E, Pasceri V, Silvestri G, Mirabella M, Mangiola F, Tonali P, Felicetti L. Progress in the molecular diagnosis of facioscapulohumeral muscular dystrophy and correlation between the number of KpnI repeats at the 4q35 locus and clinical phenotype. Ann Neurol 45: 751–757, 1999 [DOI] [PubMed] [Google Scholar]
- 37.Ricci G, Scionti I, Sera F, Govi M, D'Amico R, Frambolli I, Mele F, Filosto M, Vercelli L, Ruggiero L, Berardinelli A, Angelini C, Antonini G, Bucci E, Cao M, Daolio J, Di Muzio A, Di Leo R, Galluzzi G, Iannaccone E, Maggi L, Maruotti V, Moggio M, Mongini T, Morandi L, Nikolic A, Pastorello E, Ricci E, Rodolico C, Santoro L, Servida M, Siciliano G, Tomelleri G, Tupler R. Large-scale genotype-phenotype analyses indicate that novel prognostic tools are required for facioscapulohumeral muscular dystrophy families. Brain 136: 3408–3417, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salviati G, Sorenson MM, Eastwood AB. Calcium accumulation by the SR in two populations of chemically skinned human muscle fibers. J Gen Physiol 79: 603–632, 1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scionti I, Greco F, Ricci G, Govi M, Arashiro P, Vercelli V, Berardinelli A, Angelini A, Antonini G, Cao M, Di Muzio A, Moggio M, Morandi Ricci E, Rodolico C, Ruggiero L L, Santoro L, Siciliano G, Tomelleri G, Trevisan CP, Galluzzi G, Wright W, Zatz M, Tupler R. Large-scale population analysis challenges the current criteria for the molecular diagnosis of fascioscapulohumeral muscular dystrophy (FSHD). Am J Hum Genet 90: 628–635, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stübgen JP. Limb girdle muscular dystrophy: an interval study of weakness and functional impairment. J Clin Neuromusc Dis 9: 333–340, 2008 [DOI] [PubMed] [Google Scholar]
- 41.Talmadge RJ, Roy RR. Electrophoretic separation of rat skeletal muscle myosin heavy-chain isoforms. J Appl Physiol 75: 2337–2340, 1993 [DOI] [PubMed] [Google Scholar]
- 42.Tonini MM, Passos-Bueno MR, Cerqueira A, Matioli SR, Pavanello R, Zatz M. Asymptomatic carriers and gender differences in facioscapulohumeral muscular dystrophy (FSHD). Neuromuscul Disord 14: 33–38, 2004 [DOI] [PubMed] [Google Scholar]
- 43.Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods 37: 376–386, 2005 [DOI] [PubMed] [Google Scholar]
- 44.van Deutekom JC, Wijmenga C, van Tienhoven EA, Gruter AM, Hewitt JE, Padberg GW, van Ommen GJ, Hofker MH, Frants RR. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Mol Genet 2: 2037–2042, 1993 [DOI] [PubMed] [Google Scholar]
- 45.Wallace GQ, McNally EM. Mechanisms of muscle degeneration, regeneration, and repair in the muscular dystrophies. Annu Rev Physiol 71: 37–57, 2009 [DOI] [PubMed] [Google Scholar]
- 46.Wang J, Jin JP. Primary structure and developmental acidic to basic transition of 13 alternatively spliced mouse fast skeletal muscle troponin T isoforms. Gene 193: 105–114, 1997 [DOI] [PubMed] [Google Scholar]
- 47.Wang J, Jin JP. Conformational modulation of troponin T by configuration of the NH2 terminal variable region and functional effects. Biochemistry 37: 14519–14528, 1998 [DOI] [PubMed] [Google Scholar]
- 48.Wijmenga C, Hewitt JE, Sandkuijl LA, Clark LN, Wright TJ, Dauwerse HG, Gruter AM, Hofker MH, Moerer P, Williamson R, van Ommen G-J B, Padberg G W, Frants R R. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet 2: 26–30, 1992 [DOI] [PubMed] [Google Scholar]
- 49.Xynos A, Neguembor MV, Caccia R, Licastro D, Nonis A, Di Serio C, Stupka E, Gabellini D. Overexpression of facioscapulohumeral muscular dystrophy region gene 1 causes primary defects in myogenic stem cells. J Cell Sci 126: 2236–2245, 2013 [DOI] [PubMed] [Google Scholar]
- 50.Zeng W, de Greef JC, Chen YY, Chien R, Kong X, Gregson HC, Winokur ST, Pyle A, Robertson KD, Schmiesing JA, Kimonis VE, Balog J, Frants RR, Ball AR, Jr, Lock LF, Donovan PJ, van der Maarel SM, Yokomori K. Specific loss of histone H3 lysine 9 trimethylation and HP1γ/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD). PLoS Genet 5: e1000559, doi: 10.1371, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]