Abstract
A fundamental task in neuroscience is to understand how neural ensembles represent information. Population decoding is a useful tool to extract information from neuronal populations based on the ensemble spiking activity. We propose a novel Bayesian decoding paradigm to decode unsorted spikes in the rat hippocampus. Our approach uses a direct mapping between spike waveform features and covariates of interest and avoids accumulation of spike sorting errors. Our decoding paradigm is nonparametric, encoding model-free for representing stimuli, and extracts information from all available spikes and their waveform features. We apply the proposed Bayesian decoding algorithm to a position reconstruction task for freely behaving rats based on tetrode recordings of rat hippocampal neuronal activity. Our detailed decoding analyses demonstrate that our approach is efficient and better utilizes the available information in the nonsortable hash than the standard sorting-based decoding algorithm. Our approach can be adapted to an online encoding/decoding framework for applications that require real-time decoding, such as brain-machine interfaces.
Keywords: neural decoding, spatial-temporal Poisson process, population codes, kernel density estimation, spike sorting
features of sensory stimuli and intended motor actions are reflected in the activity of neuronal ensembles that are distributed across the brain (Boloori et al. 2010; Huys et al. 2007; Sanger 2003). A fundamental goal in neuroscience is to understand how the information about external stimuli is transformed into neural activity patterns and how this information is represented in the brain. The relationship between stimuli and neural activity can be described by statistical encoding models (Brown et al. 1998; Paninski et al. 2007; Sanger 2003; Truccolo et al. 2005). Inversion of these encoding models, i.e., extraction of information about the stimulus from observed neural activity (“neural decoding”), aids in revealing the principles of the encoding process (Quian Quiroga and Panzeri 2009). Neural decoding can also be applied to uncover internal neural representations in the absence of an overt stimulus, for example, the reexpression of spatial sequences in the hippocampus (Davidson et al. 2009; Gupta et al. 2010) or movement intentions in motor cortices (Georgopoulos et al. 1986; Zhuang et al. 2010). Decoding motor plans in particular is an important part of the development of neural prosthetics and brain-machine interfaces that may restore motor function in patients with neurological damage (Chapin 2004; Hochberg et al. 2012; Schwartz et al. 2006).
The principal goal of neural decoding is to extract as much information about a stimulus as possible from a neural signal. As with all signal processing, any additional operation on the raw signal in neural decoding adds complexity and leads to possible loss of information. Most approaches for decoding neural spiking activity rely on the intermediate step of sorting spike waveforms into groups of single units. The spike sorting process is subject to at least two problems that could affect decoding performance. The first issue is that the goal of neural decoding to minimize decoding error is substantially different from the goal to label each spike uniquely and confidently with the identity of the cell that emitted it. In particular, spike sorting is generally conservative, and many spikes are left unclassified in an attempt to minimize classification errors. However, spikes thrown out during the sorting process could still convey information about the stimulus and hence contribute to decoding performance. Second, inherent to spike sorting are misclassification errors, that is, incorrect assignment of spikes to a unit. Theoretical analysis has shown that different spike sorting errors have various impacts on information capacity, with false positive errors having the most serious effect (Goodman and Johnson 2008). Another potential source of misclassification is the use of hard decision boundaries—it has been suggested that a soft decision boundary is more appropriate for evaluating neural ensemble codes (Wood and Black 2008).
To maximize the stimulus information extracted from neural spiking activity, we propose a novel Bayesian decoding paradigm that does not require the intermediate step of spike sorting. Key in our approach is a direct mapping between the raw data (i.e., spike waveform features) and the stimulus in a joint probability distribution. In contrast to previous work, our approach does not assume a parametric or biophysical model to describe the relation between stimulus and neural activity.
The performance of the new decoding approach is analyzed by applying it to hippocampal population recordings in order to estimate the location of a rat on a track.
METHODS
Spike Feature Decoding Framework
Encoding model.
The ultimate goal of our method is to reconstruct a sensory stimulus, motor action, or other covariate (from here on referred to as “stimulus”) from neuronal spiking activity recorded from an array of sensors (e.g., single-wire electrodes, stereotrodes, or tetrodes). First, we build an encoding model that relates the neural activity on a single sensor to the stimulus of interest. Let us assume that in the time period (0,T] we recorded the time-varying stimulus vector x(t) as well as N discrete spike events and their waveforms at times tn, with 0 < t1 < t2 < . . . <tN ≤ T. For simplicity we assume that all spikes occur conditionally independent of previous spikes and treat the detected spikes as a spatio-temporal Poisson process, or equivalently as a marked temporal Poisson process, in which the spatial component (the “mark”) is a vector space S of spike waveform features a ∈ S. Examples of typical waveform features are peak amplitude, spike width, extracted principal components (PCs), or other derived features. The same waveform features are generally used by spike sorting processes to extract single units from multiunit activity (MUA). In our approach the spike sorting step is bypassed by creating a direct mapping between spike waveform features and stimulus (Fig. 1). The spatio-temporal Poisson process is fully characterized by its generalized rate function λ(a,t). In cases in which the rate is determined by the stimulus of interest, the rate function can be reexpressed as λ(a,t) = λ(a,x(t)). Here, λ(a,x) can be viewed as a tuning curve that relates the average rate of spike events with waveform features a to the stimulus x.
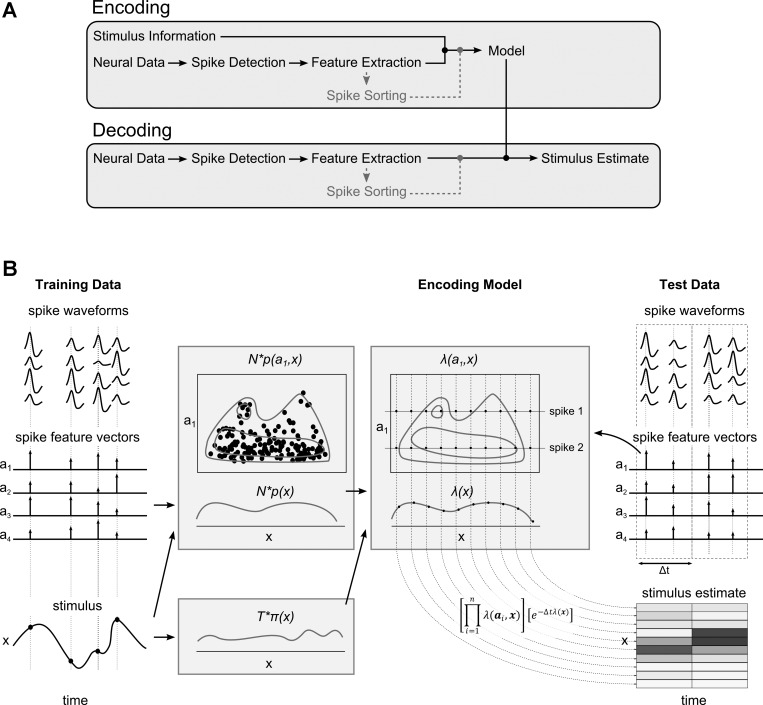
Fig. 1.
A: diagram of spike waveform feature decoding approach. In the encoding stage (top), a model describing the relation between spike waveform features and the stimulus of interest (for example, an animal's position) is constructed. Note that no prior sorting of the recorded spikes is required. In the decoding stage (bottom), the encoding model is used to obtain an estimate of the stimulus from the measured spike waveform features. B: pictographic overview of major steps in the algorithm of the spike feature decoding approach. From spike waveform features computed for the recorded raw spike waveforms and the known stimulus, the probability distributions p(a,x), p(x), and π(x) are derived. From these distributions, the joined rate function λ(a,x) and marginal rate function λ(x) can be derived. During the decoding process, observed spike waveform features in single time bins are used to evaluate the rate functions [indirectly through evaluation of the probability distributions p(a,x), p(x), and π(x)] at a user-defined stimulus grid. The evaluated rates are used to compute the posterior distribution.
To compute the probability that we observe n spikes with associated features a1:n in a small time window [t,t + Δt) in the presence of a known stimulus, the waveform feature space S is first divided into J nonoverlapping regions: S ≡ (S1 ∪ S2 ∪ . . . ∪ SJ). Each region Sj contains nj spikes, which is a subset of the observed n spikes (i.e., ). The expected number of spikes in each region Sj follows a Poisson distribution with rate function λSj(x) = ∫Sjλ(a,x)da. The likelihood of finding exactly n1:J spikes in regions S1:J can be computed from the product of Poisson likelihoods of all regions:
| 1 |
In the limiting case when regions S1:J become sufficiently small such that n1:J are equal to 0 or 1 within the time interval Δt, the likelihood can be rewritten as
| (2) |
This likelihood function, when calculated for all possible waveform feature vectors a, completely characterizes the encoding process for a single sensor. For multiple sensors, assuming conditional spiking independence between sensors (i.e., each sensor records from an independent population of neurons), the joint data likelihood can be computed as a product of individual likelihoods contributed by each sensor. For K sensors and nk spike events on the kth electrode, the joint likelihood is given by
| (3) |
Relation to encoding with spike-sorted units or multiunit activity.
It is possible to choose the spike feature a such that it is a discrete scalar variable that represents cell identity (for example, obtained through a spike sorting procedure). In that case, each region Sj can be constructed such that it corresponds to a single cell c. This means that λc(x) = λSj(x) is the tuning curve of cell c and nc = nj is the number of spikes emitted by cell c. By rewriting Eq. 1 we recover the likelihood for a population of spike-sorted cells as a special case (Zhang et al. 1998):
| (4) |
where C is the total number of cells and is the total number of spike events.
When all spikes are considered part of a single multiunit cluster and spike features a are ignored, then the likelihood can be further simplified to
| (5) |
Evaluation of the likelihood.
To compute the likelihood in Eq. 2, representations of the generalized rate function λ(a,x) and its marginal rate function λ(x) need to be constructed. A parametric model of the rate functions would allow for straightforward evaluation during decoding; however, it is not clear what model, if any, would be appropriate. In contrast, nonparametric models provide a more flexible estimate of the rate functions. Here we used kernel density-based estimators of the rate functions.
To construct the kernel density estimators, the generalized rate function λ(a,x) and the marginal rate function λ(x) are decomposed into spike event probability distributions [p(a,x) and p(x)] and a stimulus probability distribution [π(x)]:
| (6) |
| (7) |
Here, spikecount represents the number of spikes with features a that occurred at stimulus x, occupancy represents the total presentation time of stimulus x, N is the total number of spikes recorded in the time interval (0,T], and μ is the average spiking rate.
The probability distributions p(a,x), p(x), and π(x) can be estimated with the following multivariate kernel density estimators:
| (8) |
| (9) |
| (10) |
Here, represents the set of N spikes with associated feature vectors and stimuli that are collected during the encoding phase. represents the set of R observed (or chosen) stimuli, which are generally sampled at regular time intervals during the encoding stage. KH(·) is a kernel function with bandwidth matrix H. Hx represents the bandwidth matrix for the stimulus only, and Hax represents the combined bandwidth matrix for spike features and stimulus. Examples of kernels that may be used are Gaussian, Epanechnikov, uniform, von Mises (for circular variables), or Kronecker delta (for discrete variables).
The bandwidth of a kernel determines the amount of smoothing that is applied to the underlying data and therefore has a strong influence on the shape of the final density estimate. Determining the optimal full multivariate bandwidth matrix H is computationally and mathematically challenging. In practice, a simplified parameterization of the kernel is generally used in which the orientation of the kernel is ignored and only the individual scaling factors for each of the axes are taken into account (i.e., the diagonal matrix of H). If all variables are similarly distributed (except for a scaling factor), then a further simplification can be made by assuming the same bandwidth for each axis after prescaling the variables such that they have the same variance. Most approaches for bandwidth determination are heuristic or approximate (Scott 1992; Wand and Jones 1995). For low (such as 2)-dimensional space, there are few principled methods to estimate nonisotropic bandwidth parameters (Botev et al. 2010). Adaptive bandwidth selection methods have also been proposed using Markov chain Monte Carlo (MCMC) techniques (Zhang et al. 2006).
Bayesian decoding.
To infer the uncertainty or probability of a hidden stimulus xt at time t given the observed m spike events with associated features a1:m, we resort to Bayes' rule:
| (11) |
Bayes' rule provides a way to combine prior information about the stimulus P(xt) with information obtained from observations through the likelihood function P(a1:m|xt) (taken from Eqs. 2 and 3). The denominator P(a1:m) is a normalizing constant such that the posterior P(xt|a1:m) is a proper probability distribution. The posterior distribution contains all information about the stimulus at time t that can be extracted from the observed spike events. If a noninformative temporal prior is assumed, then the aim of Bayesian decoding is to maximize the product of the likelihood and time-independent prior: P(x|a1:m) ∝ P(a1:m|x)P(x). If a completely noninformative prior is assumed, then Bayesian decoding is analogous to maximum likelihood estimation: P(x|a1:m) ∝ P(a1:m|x).
Implementation.
The spike feature-based decoder was implemented in MATLAB (The MathWorks, Natick, MA) with custom C extensions. In Eqs. 6–10 we developed a representation for the rate functions λ(a,x) and λ(x), which are needed to compute the likelihood in Eq. 2. The rate functions either can be precomputed at a user-defined grid to construct look-up tables or can be evaluated online during decoding. Precomputing the rate functions has the advantage that the computational load for decoding is low, making it suitable for real-time applications. When densities need to be evaluated at a fine grid for high-dimensional feature vectors and/or stimulus vectors, then the precomputing approach becomes impractical. In that case, the rate functions may be evaluated at a coarser grid, at the expense of decoding accuracy. Instead, here we chose to evaluate the rate function λ(a,x) on the fly during decoding at a user-defined grid in stimulus space. λ(x) was precomputed at the same stimulus grid. A small offset (0.1 Hz) was applied to both rate functions to avoid cases in which the rate is zero at all nodes in the stimulus grid (Davidson et al. 2009; Zhang et al. 1998). Decoding is performed separately for predefined time bins. For each detected spike and its associated features within a time bin, the rate function λ(a,x) (Eq. 6) is evaluated, after which the likelihood (Eqs. 2 and 3) and the posterior distributions (Eq. 11) are computed. Computations are performed in log-space, such that products in Eq. 2 are replaced by summations.
Application to Hippocampal Recordings
Electrophysiology and behavior.
Eight male Long-Evans rats, weighing between 400 and 600 g, were implanted with custom-made micro-drive arrays (Kloosterman et al. 2009; Nguyen et al. 2009). Individual arrays carried between 9 and 24 independently movable tetrodes targeted either to the right dorsal hippocampus or bilaterally to both hippocampi (coordinates: 2.5 mm lateral and 4.0 mm posterior to bregma). The tetrodes were slowly lowered into the brain until they reached the cell layer of CA1 2–4 wk after array implantation. A reference electrode was positioned in the white matter overlying the dorsal hippocampus. In the case of bilateral recordings, separate ipsilateral references were used for each hippocampus. Signals were filtered between 300 Hz and 6 kHz, and all extracellular spike waveforms that crossed a preset amplitude threshold (73 μV) on any of the four tetrode channels were sampled at 31,250 Hz (32 samples per spike waveform) and saved to disk.
During 30- to 60-min-long recording sessions rats were allowed to freely explore an ∼3-m-long linear or circular track. While no behavioral restrictions were placed on the rats, they did receive a small food reward (chocolate sprinkle, fruit loop, etc.) to encourage exploration. The position of the rats was tracked at 30 Hz with an overhead video camera and infrared light-emitting diodes mounted on the implanted micro-drive array. All experiments were conducted under protocols reviewed and approved by the Massachusetts Institute of Technology Committee on Animal Care and followed the guidelines of the National Institutes of Health.
Decoding of animal's position.
Data collected in a recording session were divided into a training set for construction of the encoding model and a testing set for evaluation of the decoding. In both data sets only RUN epochs, when the animals were actively moving along the track at a speed higher than 10 cm/s, were selected for further analyses. The stimulus of interest x corresponds to the one-dimensional position of the animals along the track. Only spikes of putative pyramidal cells (spike peak-trough latency > 0.35 ms) with a minimum peak amplitude of 75 μV were selected for decoding analysis. A four-dimensional spike feature vector a was constructed from the spike waveform peak amplitudes on each tetrode. To establish a kernel density-based estimate of the rate functions we used a truncated Gaussian kernel (cutoff at 2 standard deviations) for both the spike amplitude dimensions and the position dimension. For spike amplitude an isotropic kernel was used with the same kernel bandwidth in all four dimensions. The rate functions were evaluated at a regular grid with 2-cm intervals that spanned the whole track. For decoding, the testing data set was divided into nonoverlapping 250-ms-long time bins and spikes within each bin were used to compute the posterior distribution of position according to Eq. 11.
In the Bayesian decoding framework, the decoding performance is determined by both the choice of encoding model and prior knowledge of the stimulus. Here we focus on the contribution of the novel formulation of the encoding model that includes spike waveform features, and hence we chose to use a noninformative prior, making the decoding similar to maximum likelihood estimation.
The decoding error in each time bin was computed as the shortest distance along the track between the true (observed) position at the center of the time bin and the maximum a posteriori (MAP) estimate of position. To assess decoding performance, we analyzed the cumulative distribution of decoding error, the median error, and the confusion matrix.
Spike sorting-based decoding.
Spikes of putative pyramidal neurons (spike peak-trough latency > 0.35 ms, minimum peak amplitude > 75 μV) in RUN epochs were selected prior to sorting. Spikes were automatically sorted with KlustaKwik (version 2.0.1, http://klustakwik.sourceforge.net/; see Harris et al. 2000) using three features per electrode: spike waveform energy and the first two PCs of the energy-normalized spike waveform. For each cluster a set of four quality metrics was computed: Lratio and isolation distance (IsoD; using waveform energy and 1st PC for each channel; see Schmitzer-Torbert et al. 2005), fraction of interspike intervals smaller than 2 ms (ISI), and the Mahalanobis distance of the cluster to the spike amplitude threshold (Dthreshold; using spike amplitudes on the electrode with largest mean spike amplitude). Only clusters that met the following criteria were retained: Lratio < 0.1, IsoD > 20, ISI < 2%, and Dthreshold > 1. The remaining clusters were collapsed into a single noise or “hash” cluster. The spatial tuning curve of each cluster was constructed by kernel-density estimation with a truncated Gaussian kernel with an optimized spatial bandwidth, and the likelihood was computed according to Eq. 4.
Bandwidth selection.
We used a diagonal bandwidth matrix to compute the kernel density estimates in Eqs. 8–10. Since the spike features that were used for decoding all represent the same physical quantity (i.e., spike peak amplitude) with a similar distribution, the same scalar bandwidth (ha) was selected for all the feature dimensions and for all tetrodes. A separate bandwidth (hx) was chosen for the stimulus dimensions (i.e., position on track). For each separate data set, optimal values for ha and hx were determined by a twofold cross-validation procedure on the training set.
RESULTS
A total of 12 data sets from 8 rats were used to test the spike feature-based decoding approach. Table 1 summarizes the experimental data, and Table 2 tabulates the decoding results, as discussed in more detail below.
Table 1.
Summary of experimental data
| Data Set | No. of Tetrodes | No. of Total Spikes | No. of Sorted Spikes | % Sorted Spikes | No. of Clusters |
|---|---|---|---|---|---|
| R1D2 | 14 | 81 K | 5 K | 6 | 9 |
| R2D2 | 18 | 106 K | 7 K | 6 | 12 |
| FK11 | 9 | 195 K | 15 K | 8 | 18 |
| R1D1 | 14 | 170 K | 36 K | 21 | 29 |
| R2D1 | 18 | 206 K | 36 K | 17 | 32 |
| Esm02 | 17 | 307 K | 44 K | 14 | 32 |
| Sat2 | 18 | 355 K | 44 K | 12 | 35 |
| SL13 | 17 | 126 K | 22 K | 17 | 40 |
| SL16 | 16 | 160 K | 22 K | 14 | 40 |
| Esm01 | 18 | 222 K | 30 K | 14 | 48 |
| SL14 | 14 | 215 K | 43 K | 20 | 52 |
| SL15 | 17 | 330 K | 43 K | 13 | 56 |
Data sets are ranked by number of clusters.
Table 2.
Summary of decoding performances across all data sets
| Spike Feature Decoder |
Identity Decoder |
|||||||
|---|---|---|---|---|---|---|---|---|
| Data Set | All spikes | Spikes in isolated clusters | Spikes in hash | MUA Decoder, All Spikes | Isolated clusters + hash | Isolated clusters | Hash only | All clusters |
| R1D2 | 6.0 | 28.0 | 18.5 | 23.0 | 11.0 | 36.3 | 31.3 | 7.5 |
| R2D2 | 6.0 | 18.0 | 13.5 | 12.0 | 7.0 | 21.5 | 17.5 | 7.0 |
| FK11 | 6.2 | 9.9 | 7.1 | 25.3 | 7.2 | 9.1 | 23.5 | 5.9 |
| R1D1 | 4.5 | 7.5 | 7.0 | 13.5 | 5.5 | 8.5 | 24.0 | 4.5 |
| R2D1 | 5.5 | 7.5 | 7.0 | 12.0 | 6.0 | 8.5 | 23.0 | 5.0 |
| Esm02 | 3.6 | 5.0 | 4.1 | 12.2 | 4.5 | 5.8 | 12.6 | 3.0 |
| Sat2 | 6.0 | 6.8 | 6.2 | 12.7 | 6.9 | 7.1 | 16.1 | 5.9 |
| SL13 | 4.7 | 4.9 | 6.2 | 13.9 | 4.6 | 5.5 | 19.8 | 4.9 |
| SL16 | 3.7 | 4.8 | 4.5 | 18.2 | 4.3 | 4.8 | 34.8 | 3.6 |
| Esm01 | 6.0 | 6.2 | 6.3 | 9.0 | 5.3 | 5.8 | 9.9 | 5.0 |
| SL14 | 3.6 | 4.1 | 4.3 | 13.9 | 4.0 | 4.1 | 26.3 | 3.5 |
| SL15 | 3.2 | 3.8 | 4.8 | 63.2 | 4.2 | 3.9 | 102.6 | 3.6 |
Decoding performance is presented as median error in centimeters. Data sets are ranked by number of clusters. MUA, multiunit activity.
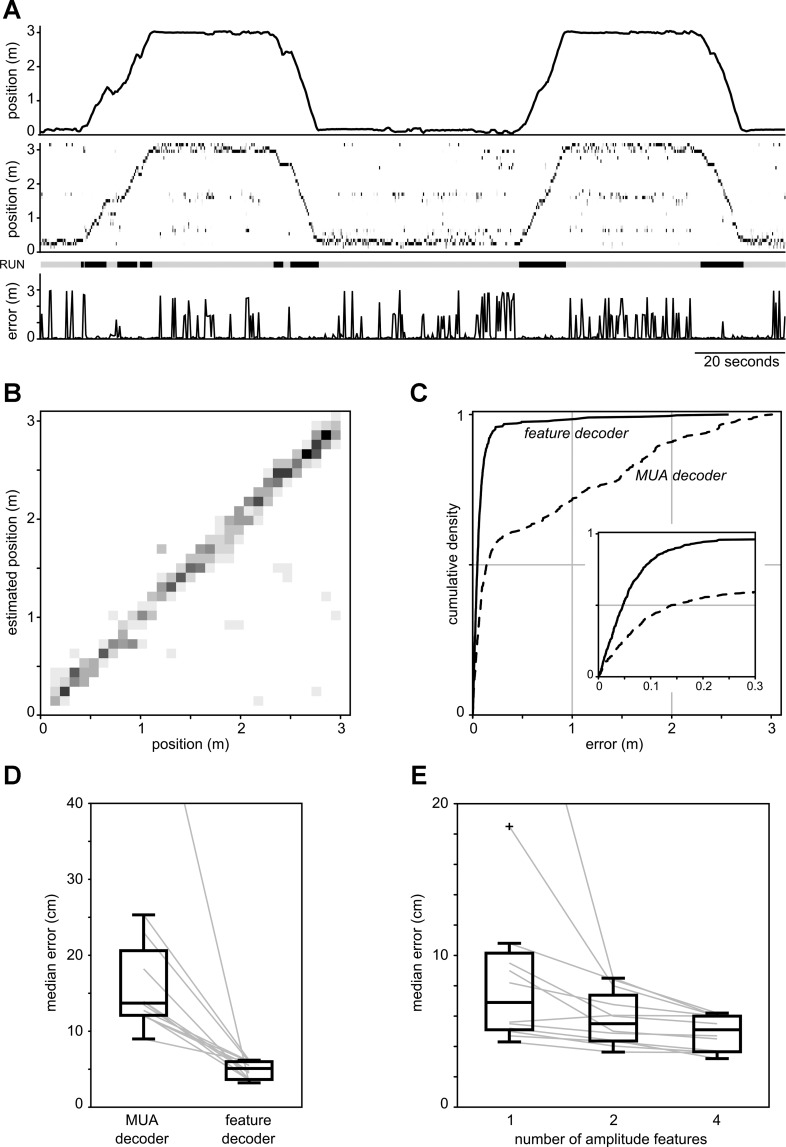
Figure 2, A–C, shows an example of decoding the position of a rat (data set SL14, see Table 1) using the peak amplitudes of recorded hippocampal spikes as feature vector. Qualitatively, our estimates of the rat's position on the track accurately follow its true position during periods of locomotion (Fig. 2A). To assess the decoding performance, we computed a confusion matrix (Fig. 2B) and the distribution of errors (Fig. 2C). In this example, the median decoding error is 3.6 cm and 90% of the errors fall within 10.3 cm. The confusion matrix shows a dominant diagonal structure, which indicates a high accuracy of decoding at most locations along the track. At the population level, the median decoding error across all data sets varied from 3.2 cm to 6.2 cm, with an average median error of 4.9 cm (see Table 2, column 2).
Fig. 2.
A: example of spike waveform feature decoding of a rat's position during 2 full laps on a 3-m-long track (data set SL14). Top: rat's true position. Middle: posterior probability distributions of the decoded position along the track computed in nonoverlapping 250-ms time bins. Darker shades of gray indicate higher probabilities. Bottom: decoding error. Note that decoding error is low during periods of active motion (“RUN,” indicated by thick bars above plot). B: confusion matrix showing the distribution of estimated positions for all positions on the track (data set SL14). Positions are binned in 10-cm bins for improved visualization. C: empirical cumulative distribution function (CDF) of decoding errors for data set SL14. Solid line, spike waveform feature decoder; dashed line, multiunit activity (MUA) decoder. Inset: detail of the CDFs. D: box plots comparing the distributions of median errors across all data sets for the spike waveform feature decoder and the MUA decoder. Gray lines represent data of individual data sets. E: box plots comparing decoding performance when 1, 2, or 4 amplitude features (from 1, 2, or 4 electrodes in tetrode) are used in the spike feature decoder. Outliers are indicated by “+” symbol.
In principle, the decoder has access to information conveyed by both the population firing rate and the specific spike amplitude vectors. To investigate the extent to which spike amplitude information aids in the decoding of position in the spike feature decoding approach, we compared the performance to a decoder based on spike timing alone using a single spatial tuning curve for each tetrode (MUA decoder; see Fraser et al. 2009). In all data sets the error distribution of the MUA decoder is significantly worse than the error distribution of the spike feature decoder (1-sided 2-sample Kolmogorov-Smirnov test, P < 10−5 for all data sets; see example in Fig. 2C). Overall, the median error was significantly reduced when spike amplitude information was included in our spike feature-based decoding approach (Fig. 2D; paired rank sum test, P = 4.9 × 10−4).
Next, we examined the dependence of decoding performance on the number of spike waveform features included in the analysis. Spike amplitudes on one or two electrodes of each tetrode were selected as features for decoding. Since the spike amplitude threshold was applied after electrode selection, fewer spikes were included in these data sets (see Table 4). For both the one-amplitude feature and two-amplitude feature case, decoding performance was higher than the corresponding MUA decoder (not shown). Increasing the number of amplitude features from one to two or from two to four significantly lowered the median decoding error (paired rank sum test: P = 9.8 × 10−4), with relative mean benefits of 26% and 14%, respectively.
Table 4.
Summary of experimental data for single-electrode and stereotrode simulation
| Single Electrode |
||||||
|---|---|---|---|---|---|---|
| Data Set | No. of Electrodes | Stereotrode, No. of Total Spikes | No. of total spikes | No. of sorted spikes | % Sorted spikes | No. of clusters |
| R1D2 | 14 | 55 K | 36 K | 3 K | 8 | 7 |
| R2D2 | 18 | 77 K | 53 K | 1 K | 3 | 4 |
| FK11 | 9 | 146 K | 104 K | 6 K | 6 | 1 |
| R1D1 | 14 | 132 K | 102 K | 9 K | 8 | 6 |
| R2D1 | 18 | 158 K | 121 K | 12 K | 10 | 6 |
| Esm02 | 17 | 237 K | 163 K | 25 K | 16 | 8 |
| Sat2 | 18 | 290 K | 218 K | 4 K | 2 | 2 |
| SL13 | 17 | 111 K | 68 K | 11 K | 16 | 6 |
| SL16 | 16 | 127 K | 75 K | 4 K | 6 | 3 |
| Esm01 | 18 | 170 K | 121 K | 4 K | 4 | 6 |
| SL14 | 14 | 180 K | 110 K | 10 K | 9 | 3 |
| SL15 | 17 | 280 K | 164 K | 11 K | 6 | 6 |
Data sets are ranked as in Table 1.
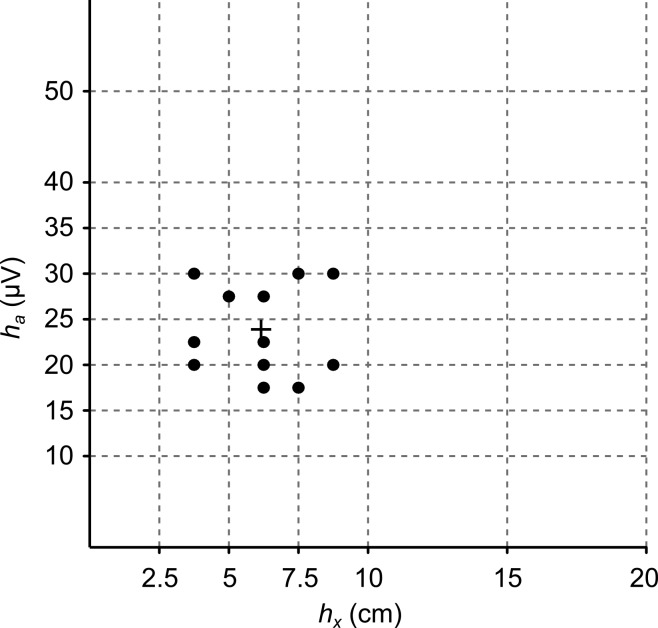
The decoding performance is critically dependent on the choice of kernel bandwidths ha and hx. Optimal bandwidths that minimized decoding error were selected separately for each decoder variant and each data set (Table 3; see methods). For the spike feature decoder, bandwidths clustered around hx = 6 cm and ha = 24 μV (Fig. 3). Decoding performance decreased gradually for increasing bandwidths. If the bandwidths were fixed at hx = 6 cm and ha = 24 μV for all data sets, the mean deviation from the median error obtained when optimized bandwidths were used was 5% (maximum: 17%). This suggests that even in cases where cross-validation is impractical, appropriate fixed bandwidth parameters selected a priori may still yield near-optimal decoding performance for distinct data sets.
Table 3.
Optimal bandwidths for decoder variants in Table 2
| Spike Feature Decoder |
Identity Decoder |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| All spikes |
Spikes in isolated clusters |
Spikes in hash |
MUA Decoder, All Spikes |
Isolated clusters + hash |
Isolated clusters |
Hash only |
All clusters |
||||
| Data Set | ha | hx | ha | hx | ha | hx | hx | hx | hx | hx | hx |
| R1D2 | 20 | 3.75 | 17.5 | 6.25 | 20 | 7.5 | 5 | 2.5 | 7.5 | 3.75 | 2.5 |
| R2D2 | 22.5 | 3.75 | 17.5 | 6.25 | 17.5 | 6.25 | 5 | 2.5 | 6.25 | 6.25 | 5 |
| FK11 | 20 | 6.25 | 25 | 7.5 | 25 | 8.75 | 17.5 | 5 | 6.25 | 11.25 | 6.25 |
| R1D1 | 17.5 | 6.25 | 20 | 7.5 | 20 | 6.25 | 2.5 | 5 | 3.75 | 2.5 | 3.75 |
| R2D1 | 27.5 | 5 | 20 | 7.5 | 17.5 | 6.25 | 2.5 | 2.5 | 3.75 | 2.5 | 5 |
| Esm02 | 22.5 | 6.25 | 32.5 | 8.75 | 25 | 5 | 5 | 3.75 | 5 | 5 | 5 |
| Sat2 | 20 | 8.75 | 20 | 7.5 | 30 | 7.5 | 8.75 | 8.75 | 6.25 | 7.5 | 8.75 |
| SL13 | 20 | 3.75 | 17.5 | 7.5 | 22.5 | 6.25 | 5 | 3.75 | 3.75 | 12.5 | 8.75 |
| SL16 | 30 | 8.75 | 30 | 7.5 | 20 | 6.25 | 12.5 | 7.5 | 7.5 | 10 | 6.25 |
| Esm01 | 30 | 7.5 | 45 | 10 | 27.5 | 7.5 | 5 | 7.5 | 6.25 | 6.25 | 7.5 |
| SL14 | 27.5 | 6.25 | 27.5 | 6.25 | 17.5 | 7.5 | 12.5 | 6.25 | 3.75 | 10 | 5 |
| SL15 | 17.5 | 7.5 | 27.5 | 5 | 17.5 | 7.5 | 17.5 | 2.5 | 3.75 | 11.25 | 3.75 |
Data sets are ranked by number of clusters. ha and hx, Bandwidths selected for feature dimensions (spike amplitude in microvolts) and stimulus dimensions (position in centimeters), respectively.
Fig. 3.
Optimal bandwidths hx and ha for spike feature decoder for all data sets. The mean optimal bandwidth is indicated by the “+” symbol. Dashed grid lines indicate bandwidth values used for cross-validation.
Comparison with Decoding Using Sorted Single Units
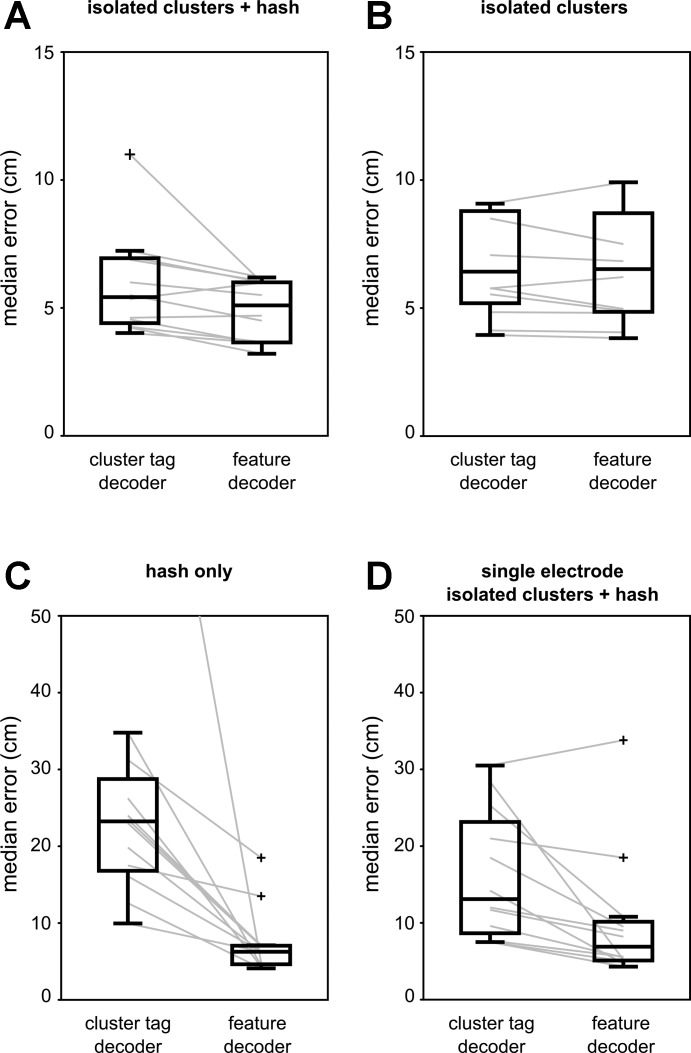
Next, we compared the spike feature decoding approach to the standard practice in which spikes recorded on tetrodes are first sorted into separate single units (“cluster tag decoder”; see Zhang et al. 1998). The automated sorting procedure produced a large number of putative clusters, of which 9–56 clusters across all data sets met the minimum quality criteria (Table 1), and the fraction of sorted spikes out of all putative pyramidal neuron spikes ranged from 6% to 21%. The remaining clusters on each tetrode were grouped into a separate hash cluster that was also included in the decoding. At the group level, the spike feature decoder resulted in a significantly lower median error than the cluster tag decoder (Fig. 4A; paired rank sum test, P = 0.0068), with an average improvement of 14%.
Fig. 4.
Comparison of the spike feature decoder to the standard single-unit decoder for 3 different decoding strategies. In all panels, box plots compare the median error distributions for the 2 decoders. Gray lines represent data of all individual data sets. Outliers are indicated by the “+” symbol. A: decoding using all spikes in isolated clusters and hash. B: decoding using only spikes in isolated clusters, excluding hash. C: decoding using only spikes in hash. D: comparison of spike feature decoder and cluster tag decoder using a reduced set of spike waveform features from a single electrode.
We next applied the spike feature decoding approach separately to spikes in the well-isolated clusters and spikes in the hash and compared the performance to the equivalent cluster tag decoder (Fig. 4B). For both decoders, the performance significantly decreased when only the well-isolated clusters or only the hash clusters were used (paired rank sum test; spike feature decoder: clusters + hash vs. clusters only P = 4.9 × 10−4, clusters + hash vs. hash only P = 4.9 × 10−4; cluster tag decoder: clusters + hash vs. clusters only P = 0.0024, clusters + hash vs. hash only P = 4.9 × 10−4). When only the well-isolated clusters were used for decoding, the spike feature decoder showed a tendency toward a lower median error (average benefit: 7%; paired rank sum test: P = 0.043). In contrast, the spike feature decoder performed significantly better than the cluster tag decoder when only the hash was used (average benefit: 64%; paired rank sum test: P = 4.9 × 10−4). This suggests that the improved performance of the spike feature decoder is mainly due to the extraction of more information from the hash.
For the purpose of decoding, clusters of sorted spikes do not need to correspond to single units (Ventura 2008). The cluster tag decoder may therefore benefit from using the original sorted spike clusters output by KlustaKwik, regardless of their quality and without grouping poorly isolated or multiunit clusters into a hash cluster. Indeed, in this case the cluster tag decoder and the spike feature decoder showed comparable performance (paired rank sum test: P = 0.8984).
Spike sorting quality depends on the number of (informative) spike waveform features. With fewer features, cluster isolation becomes poorer and hence decoding performance in the cluster tag decoder will decrease. The spike feature decoder taps into the same information in the spike waveform features as spike sorting, and we showed above that decoding performance decreases when fewer features are available (Fig. 2E). Here we investigated the relative performance of the spike feature decoder versus the cluster tag decoder when fewer features from a single electrode are available. Spike sorting proceeded as for tetrodes and resulted in fewer well-isolated clusters (Table 4). In this scenario, the spike feature decoder significantly outperformed the cluster tag decoder (Fig. 4D; paired rank sum test P = 0.0068), with an average improvement that is higher than the four-amplitude tetrode scenario (38% vs. 14%). This result suggests that the relative benefit of using the spike feature decoder may be higher when fewer spike features are available for spike sorting.
Online Encoding-Decoding Paradigm
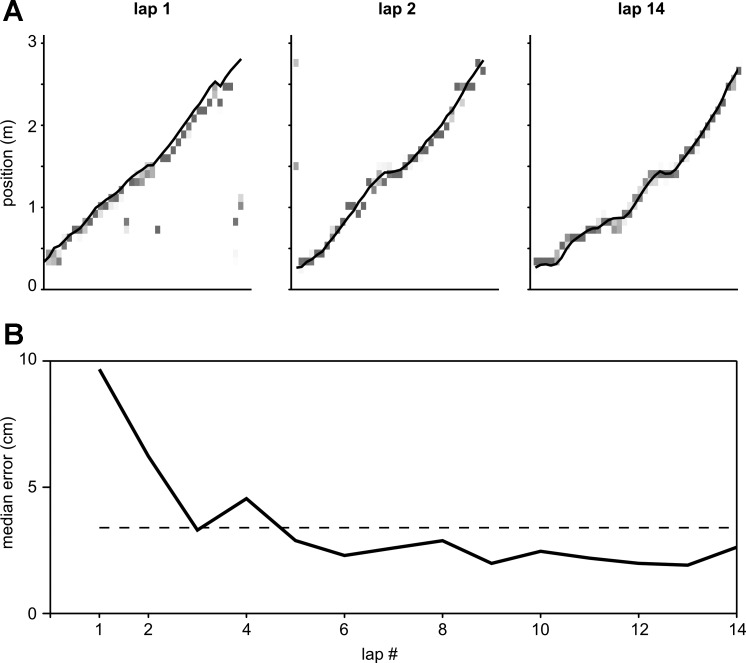
Both encoding and decoding phases of the spike feature decoding paradigm can be performed with minimal supervision, provided that suitable bandwidth parameters have been selected. As such, this new paradigm is well suited for online encoding and decoding of stimuli from ongoing neural activity as is required for brain-computer interfaces. As a demonstration we simulate an experiment in which the position of a rat is estimated as it explores a novel linear track for the first time. As soon as the rat enters the new environment neural data are acquired, and for every 250-ms time bin in which the rat is actively moving (speed > 10 cm/s) the recorded spikes and their waveform features are used to compute an estimate of position. Subsequently, the encoding model is updated with the newly acquired spikes, their waveform features, and the rat's physical location in the track. Thus at any given time decoding is performed with only the recorded spike data in the past. Figure 5A shows the position estimates for the first two laps and the last lap on the track for one data set (SL14). The estimates in the first lap of behavior are biased toward past positions as this is the only information available in the spiking. However, in subsequent laps the decoding error decreases and quickly reaches an asymptotic value (Fig. 5B). These results suggest that only a limited amount of spiking history may be necessary to achieve accurate decoding results when online real-time encoding and decoding is required.
Fig. 5.
Simulated minimally supervised online decoding. A: posterior probability distributions for 250-ms time bins in 3 epochs corresponding to the first 2 laps and the last lap on the track (data set SL14). Darker shades of gray indicate higher probabilities. Solid line is animal's true trajectory. B: median decoding error computed for each lap separately. Horizontal dashed line shows mean of the median decoding error statistics across all laps.
DISCUSSION
We have demonstrated a novel Bayesian neural decoding approach that implements a direct mapping between spike waveform features and a sensory stimulus or other covariates. This new approach has several important advantages over other methods. First, the spike feature decoder uses both the timing and waveform features of all available spike events to increase the amount of stimulus-related information that can be extracted from the neural signals. Second, there is no need for an intermediate spike sorting step that adds complexity and may lead to loss of information. Third, our approach defines a nonparametric encoding model to flexibly describe the relation between the stimulus and neural activity. And finally, the spike feature decoder allows both encoding and decoding stages to be performed with minimal supervision.
We tested the new decoding method on tetrode recordings from the hippocampus in freely behaving rats and showed that the animal's location on a track could be accurately estimated. In our tests we used spike peak amplitude as the feature for decoding; however, it is possible to use any other set of spike features as well, for example, waveform width, wave shape parameters, or PCs. Our approach takes advantage of the same spike waveform information as most spike sorting methods but does so without an explicit sorting step. Our results show a small benefit of the spike feature decoder compared with a standard cluster tag decoder, as the new approach extracts more information from the poorly separable hash spikes.
It is noteworthy to point out the key differences between our approach and other paradigms in which neural spiking data were used for decoding without a spike sorting step. Fraser et al. (2009) fitted MUA responses to movement with a spline function, and thus each electrode was treated as a single “virtual” unit. This approach may work well if only a few neurons are recorded on each electrode, or if all neurons contributing to the MUA have similar tuning properties. However, this method will likely perform poorly if the MUA contains spikes from many neurons with diverse responses, for example, in the hippocampus as we showed here.
Stark and Abeles (2007) presented another interesting approach in which MUA, defined as the root mean square of the 300 Hz to 6 kHz band of the local field potential, was used to predict arm movement in monkeys. In this method, spiking activity was not modeled explicitly and decoding was performed by a support vector machine classifier. Unlike our approach, the measure of MUA in Stark and Abeles (2007) does not separate the contributions of spike rate and amplitude, nor does it allow incorporation of other waveform features.
Ventura (2008) proposed a paradigm in which the identities of neurons are extracted implicitly from the spike train recorded on a single electrode by assuming a known parametric model for the neurons' tuning to the stimulus. This approach worked well in simulations but was not applied to a real data set, and it is not clear that the method will work equally well for populations of neurons with complex, nonparametric tuning functions. In addition, the decoding step assumes that the temporal evolution of the stimulus is smooth (Ventura 2008). This assumption is not necessarily true or known to be true, for example, if the goal is to decode a hidden stimulus with unknown temporal dynamics (Davidson et al. 2009; Kloosterman 2012).
More recently, Ventura proposed to exploit the information in covariates that modulate neuronal firing (“tuning information”) in addition to spike waveform information to improve spike sorting (Ventura 2009a) with application in decoding as well (Ventura 2009b). Ventura's approach uses a mapping between waveform features and covariates similar to what we have done in the spike feature decoder; however, parametric models are used rather than the nonparametric approach in our work.
Luczak and Narayanan (2005) avoided spike sorting by constructing a “spectral representation” of the neural data by considering a discrete space of spike waveform features (they used PCs) and time. The spectral representation of Luczak and Narayanan is similar to the spike feature-marked point process that is at the basis of our method. Using a combination of partial least squares to select stimulus-relevant features and linear discriminant analysis for classification, they demonstrated that the spectral representation can be used to successfully discriminate between two discrete auditory stimuli. In contrast, we used a naive Bayesian approach in which spiking statistics are explicitly modeled, and which we applied to the decoding of a continuously time-varying stimulus.
The main strength of the proposed spike feature decoding approach is that it provides straightforward stimulus estimation from information-rich spiking data in a minimally supervised manner. The method can be applied in online and real-time encoding/decoding scenarios, which makes it appealing for brain-machine interfaces and neural prosthetics that use chronic implants. By utilizing the information carried by all spikes, whether or not they can be uniquely assigned to a single neuron, the spike feature decoding approach is more robust to changes in signal quality as observed in chronic recording applications. In addition, nonstationarity of the neural signals, which is commonly encountered in long-term recordings, and slow changes in the encoding model can be easily handled by restricting the spike events that contribute to the encoding model to a finite temporal window.
The spike feature decoding approach is not fully unsupervised, as it requires the selection of appropriate kernel bandwidth parameters. In the present experimental data analysis, a cross-validation approach was employed to estimate the two bandwidth parameters for spike amplitude and position dimensions. We found that the decoding performance was relatively robust around suboptimal bandwidth parameters, suggesting that a priori selection of a fixed bandwidth may be appropriate.
For online applications it is important that the computations for encoding and decoding stages can be performed in real time. In our present implementation encoding is cheap (all recorded spikes are simply retained), whereas decoding is computationally intensive. The complexity of decoding scales with the total number of spike events incorporated in the encoding model. Several strategies can be used to decrease the computational burden. If precomputation of the rate function is practical, then this would avoid costly evaluation of the densities during decoding. If necessary, a trade-off can be made between decoding performance and computation speed by reducing the number of dimensions of the feature space. For example, using two amplitude features (from a stereotrode) instead of four features (from a tetrode) gives acceptable (albeit decreased) decoding performance. Alternatively, it is possible to reduce computational load of online density evaluation by excluding groups of low-information spike events based on their waveform properties. For example, in the hippocampus most stimulus-relevant information is carried by pyramidal neuron activity, and therefore in our tests we filtered out spikes from putative interneurons (peak-trough latency < 0.35 ms). This selection of spikes substantially reduced the computational burden without significantly affecting decoding performance. A further reduction can be achieved by increasing the spike amplitude threshold, excluding low-amplitude spikes that carry relatively little information. Another way to alleviate this problem is to find compact and efficient representations of the kernel density estimates of the rate functions (Girolami 2003; Huang and Chow 2006; Mitra et al. 2002; Zhou et al. 2003).
Our decoding algorithm is based on the statistical assumption that the spike events follow a spatio-temporal Poisson process (i.e., spike events are mutually independent in both spike feature space and time). This is equivalent to assuming an independent Poisson rate code for all neurons in the ensemble. In our example, the encoding model is constructed from multiple presentations of the same stimulus, and even if individual spike trains do not follow a Poisson distribution, the aggregate of many such spike trains will. Although the Poisson assumption is oversimplified for most experimental data, it provides us with a simple and tractable solution for decoding analysis. It is possible to relax the assumptions of Poisson statistics and independence—for example, to incorporate temporal dependence the spike history in each electrode can be represented as an augmented temporal feature. Alternatively, the spatial local dependence can be modeled by considering a Neyman-Scott process (Diggle 2003) or a Poisson cluster process (Bartlett 1964; Wolpert and Ickstadt 1998). These topics will be the subject of future investigations.
Our decoding paradigm can be extended in several ways. For instance, we can reformulate the Bayesian decoding problem within the state-space framework by inclusion of a smooth temporal prior (Brown et al. 1998). A temporal prior can be useful for reconstruction of stimuli or covariates that are known to change smoothly over time, such as the position of a rat or the movement of an arm. Spike feature decoding can also be used to extract information from ensemble spiking activity by estimating the mutual information and entropy of the stimulus and neural responses. Estimation of these information measures may provide a better understanding of the underlying principles of neural codes used in the brain (Jacobs et al. 2009; Quian Quiroga and Panzeri 2009).
GRANTS
This work was supported by National Institute of Mental Health Grant MH-061976 and Office of Naval Research MURI N00014-10-1-0936 Grant to M. A. Wilson. Z. Chen was supported by an Early Career Award from the Mathematical Biosciences Institute (MBI) and a National Science Foundation-CRCNS (Collaborative Research in Computational Neuroscience) grant.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: F.K. and M.A.W. conception and design of research; F.K. and S.P.L. performed experiments; F.K. and S.P.L. analyzed data; F.K., S.P.L., Z.C., and M.A.W. interpreted results of experiments; F.K. and S.P.L. prepared figures; F.K., S.P.L., and Z.C. drafted manuscript; F.K., S.P.L., Z.C., and M.A.W. edited and revised manuscript; F.K., S.P.L., Z.C., and M.A.W. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Greg Hale and Jun Yamamoto for contributing their data sets.
REFERENCES
- Bartlett MS. The spectral analysis of two-dimensional point processes. Biometrika 51: 299–311, 1964 [Google Scholar]
- Boloori AR, Jenks RA, Desbordes G, Stanley GB. Encoding and decoding cortical representations of tactile features in the vibrissa system. J Neurosci 30: 9990–10005, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botev ZI, Grotowski JF, Kroese DP. Kernel density estimation via diffusion. Ann Stat 38: 2916–2957, 2010 [Google Scholar]
- Brown EN, Frank LM, Tang D, Quirk MC, Wilson MA. A statistical paradigm for neural spike train decoding applied to position prediction from ensemble firing patterns of rat hippocampal place cells. J Neurosci 18: 7411–7425, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapin JK. Using multi-neuron population recordings for neural prosthetics. Nat Neurosci 7: 452–455, 2004 [DOI] [PubMed] [Google Scholar]
- Davidson TJ, Kloosterman F, Wilson MA. Hippocampal replay of extended experience. Neuron 63: 497–507, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle PJ. Statistical Analysis of Spatial Point Patterns (2nd ed.). Oxford, UK: Oxford Univ. Press, 2003 [Google Scholar]
- Fraser GW, Chase SM, Whitford A, Schwartz AB. Control of a brain-computer interface without spike sorting. J Neural Eng 6: 055004, 2009 [DOI] [PubMed] [Google Scholar]
- Georgopoulos AP, Schwartz AB, Kettner RE. Neuronal population coding of movement direction. Science 233: 1416–1419, 1986 [DOI] [PubMed] [Google Scholar]
- Girolami M. Probability density estimation from optimally condensed data samples. IEEE Trans Pattern Anal Mach Intell 25: 1253–1264, 2003 [Google Scholar]
- Goodman IN, Johnson DH. Information theoretic bounds on neural prosthesis effectiveness: the importance of spike sorting. In: Proc IEEE Int Conf Acoust Speech Signal Process 2008: 5204–5207, 2008 [Google Scholar]
- Gupta AS, Van der Meer MA, Touretzky DS, Redish AD. Hippocampal replay is not a simple function of experience. Neuron 65: 695–705, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KD, Henze DA, Csicsvari J, Hirase H, Buzsáki G. Accuracy of tetrode spike separation as determined by simultaneous intracellular and extracellular measurements. J Neurophysiol 84: 401–414, 2000 [DOI] [PubMed] [Google Scholar]
- Hochberg LR, Bacher D, Jarosiewicz B, Masse NY, Simeral JD, Vogel J, Haddadin S, Liu J, Cash SS, Van der Smagt P, Donoghue JP. Reach and grasp by people with tetraplegia using a neurally controlled robotic arm. Nature 485: 372–375, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Chow TW. Enhancing density-based data reduction using entropy. Neural Comput 18: 470–495, 2006 [DOI] [PubMed] [Google Scholar]
- Huys QJ, Zemel RS, Natarajan R, Dayan P. Fast population coding. Neural Comput 19: 404–441, 2007 [DOI] [PubMed] [Google Scholar]
- Jacobs AL, Fridman G, Douglas RM, Alam NM, Latham PE, Prusky GT, Nirenberg S. Ruling out and ruling in neural codes. Proc Natl Acad Sci USA 106: 5936–5941, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman F. Analysis of hippocampal memory replay using neural population decoding. Neuromethods 67: 259–282, 2012 [Google Scholar]
- Kloosterman F, Davidson TJ, Gomperts SN, Layton SP, Hale G, Nguyen DP, Wilson MA. Micro-drive array for chronic in vivo recording: drive fabrication. J Vis Exp 26: 1094, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luczak A, Narayanan NS. Spectral representation—analyzing single-unit activity in extracellularly recorded neuronal data without spike sorting. J Neurosci Methods 144: 53–61, 2005 [DOI] [PubMed] [Google Scholar]
- Mitra P, Murthy C, Pal S. Density-based multiscale data condensation. Pattern Anal Mach 24: 734–747, 2002 [Google Scholar]
- Nguyen DP, Layton SP, Hale G, Gomperts SN, Davidson TJ, Kloosterman F, Wilson MA. Micro-drive array for chronic in vivo recording: tetrode assembly. J Vis Exp 26: 1098, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paninski L, Pillow J, Lewi J. Statistical models for neural encoding, decoding, and optimal stimulus design. Prog Brain Res 165: 493–507, 2007 [DOI] [PubMed] [Google Scholar]
- Quian Quiroga R, Panzeri S. Extracting information from neuronal populations: information theory and decoding approaches. Nat Rev Neurosci 10: 173–185, 2009 [DOI] [PubMed] [Google Scholar]
- Sanger T. Neural population codes. Curr Opin Neurobiol 13: 238–249, 2003 [DOI] [PubMed] [Google Scholar]
- Schmitzer-Torbert N, Jackson J, Henze D, Harris K, Redish AD. Quantitative measures of cluster quality for use in extracellular recordings. Neuroscience 131: 1–11, 2005 [DOI] [PubMed] [Google Scholar]
- Schwartz AB, Cui XT, Weber DJ, Moran DW. Brain-controlled interfaces: movement restoration with neural prosthetics. Neuron 52: 205–220, 2006 [DOI] [PubMed] [Google Scholar]
- Scott DW. Multivariate Density Estimation: Theory, Practice, and Visualization. New York: Wiley, 1992 [Google Scholar]
- Stark E, Abeles M. Predicting movement from multiunit activity. J Neurosci 27: 8387–8394, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truccolo W, Eden UT, Fellows MR, Donoghue JP, Brown EN. A point process framework for relating neural spiking activity to spiking history, neural ensemble, and extrinsic covariate effects. J Neurophysiol 93: 1074–1089, 2005 [DOI] [PubMed] [Google Scholar]
- Ventura V. Spike train decoding without spike sorting. Neural Comput 20: 923–963, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura V. Automatic spike sorting using tuning information. Neural Comput 21: 2466–2501, 2009a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura V. Traditional waveform based spike sorting yields biased rate code estimates. Proc Natl Acad Sci USA 106: 6921–6926, 2009b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wand MP, Jones MC. Kernel Smoothing. London: Chapman and Hall, 1995 [Google Scholar]
- Wolpert R, Ickstadt K. Poisson/gamma random field models for spatial statistics. Biometrika 85: 251–267, 1998 [Google Scholar]
- Wood F, Black MJ. A nonparametric Bayesian alternative to spike sorting. J Neurosci Methods 173: 1–12, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Ginzburg I, McNaughton BL, Sejnowski TJ. Interpreting neuronal population activity by reconstruction: unified framework with application to hippocampal place cells. J Neurophysiol 79: 1017–1044, 1998 [DOI] [PubMed] [Google Scholar]
- Zhang X, King ML, Hyndman RJ. A Bayesian approach to bandwidth selection for multivariate density estimation. J Comput Stat Data Anal 50: 3009–3031, 2006 [Google Scholar]
- Zhou A, Cai Z, Wei L, Qian W. M-kernel merging: towards density estimation over data streams. Proc IEEE Int Conf Database Syst Adv Appl 2003: 285–292, 2003 [Google Scholar]
- Zhuang J, Truccolo W, Vargas-Irwin C, Donoghue JP. Decoding 3-D reach and grasp kinematics from high-frequency local field potentials in primate primary motor cortex. IEEE Trans Biomed Eng 57: 1774–1784, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]