Abstract
Expression of appropriate ion channels is essential to allow developing neurons to form functional networks. Our previous studies have identified LIM-homeodomain (HD) transcription factors (TFs), expressed by developing neurons, that are specifically able to regulate ion channel gene expression. In this study, we use the technique of DNA adenine methyltransferase identification (DamID) to identify putative gene targets of four such TFs that are differentially expressed in Drosophila motoneurons. Analysis of targets for Islet (Isl), Lim3, Hb9, and Even-skipped (Eve) identifies both ion channel genes and genes predicted to regulate aspects of dendritic and axonal morphology. Significantly, some ion channel genes are bound by more than one TF, consistent with the possibility of combinatorial regulation. One such gene is Shaker (Sh), which encodes a voltage-dependent fast K+ channel (Kv1.1). DamID reveals that Sh is bound by both Isl and Lim3. We used body wall muscle as a test tissue because in conditions of low Ca2+, the fast K+ current is carried solely by Sh channels (unlike neurons in which a second fast K+ current, Shal, also contributes). Ectopic expression of isl, but not Lim3, is sufficient to reduce both Sh transcript and Sh current level. By contrast, coexpression of both TFs is additive, resulting in a significantly greater reduction in both Sh transcript and current compared with isl expression alone. These observations provide evidence for combinatorial activity of Isl and Lim3 in regulating ion channel gene expression.
Keywords: aCC, central nervous system, Drosophila, muscle, RP3, Shaker
Introduction
The development of embryonic neurons is regulated by factors that are intrinsic or extrinsic to individual cells. It is, however, in the early stages of development, before axonal growth and circuit formation, that intrinsic factors predominate (Spana et al., 1995; Grosskortenhaus et al., 2005; Grosskortenhaus et al., 2006). These factors, which are likely determined by clonal lineage, control aspects of both morphological and functional (electrical) development. Studies on motoneuron specification, from flies to mammals, have shown that early developmental decisions, such as subclass identity, are dictated, at least in part, by a combinatorial code of LIM-HD transcription factors (TFs) (Thor and Thomas, 1997; Landgraf et al., 1999; Dasen et al., 2005; Landgraf and Thor, 2006; Dasen et al., 2008; De Marco Garcia and Jessell, 2008). However, whether early neuron-type specific ion channel gene expression is similarly influenced through combinatorial activity of these same TFs remains to be demonstrated.
Embryonic Drosophila motoneurons express a stereotypic mix of identified TFs, which are evolutionary conserved with mammals (Thor and Thomas, 1997; Thaler et al., 1999; Moran-Rivard et al., 2001; Esmaeili et al., 2002; Thaler et al., 2002). For example, the RP subgroup of motoneurons (RPs 1, 3–5), which innervate ventral and lateral muscles, express the TFs Isl (also known as Tail-up), Lim3, and Hb9 (also known as Extra-extra). Motoneurons (e.g., aCC) that project dorsally express Eve (Thor and Thomas, 1997; Landgraf et al., 1999; Landgraf and Thor, 2006). The presence or absence of individual TFs, particularly Isl and Eve, is a known determinant for both axonal projection, neurotransmitter phenotype, and neuron type-specific expression of ion channels (Thor and Thomas, 1997; Landgraf et al., 1999; Pym et al., 2006; Wolfram et al., 2012).
Our previous studies used DNA adenine methyltransferase identification (DamID) (van Steensel and Henikoff, 2000) to identify ion channel genes, in particular slowpoke (slo) and Sh, as targets of Eve and Islet, respectively (Pym et al., 2006; Wolfram et al., 2012). Here we now report the complete identified DamID-derived binding sites for Hb9 and Lim3, together with a reassessment of DamID data previously obtained for Eve and Isl, using Flybase release 5.47. Our analysis identifies ion channel genes as targets of all four TFs, and shows that some ion channel genes (e.g., Sh) are bound by multiple TFs (in this instance, Isl and Lim3). We further show that the combined action of Isl and Lim3, in regulating Sh expression, is additive. As such, these findings provide first direct experimental evidence to support combinatorial regulation of a specific ion channel gene.
Materials and Methods
Fly stocks.
Flies were maintained under standard conditions. For larval collections, flies were allowed to lay eggs onto grape juice agar plates. GAL424B (homozygous viable on the second chromosome) was used to express isl (2× UAS-isl, third chromosome) or Lim3 (1× UAS-Lim3, second chromosome) in body wall muscle (both kindly supplied by Dr. Stefan Thor). Coexpression was achieved using a genetic cross containing both transgenes (2× UAS-isl and 1× UAS-Lim3). Embryos of either sex were kept at 18°C to avoid embryonic lethality, which occurs with expression of either transgene in muscle at 25°C.
DamID analysis.
The construction of DamID constructs and transgenic Drosophila lines has been previously described (Pym et al., 2006; Wolfram et al., 2012). Briefly, the full-length TF-coding sequences were PCR-amplified from an embryonic cDNA library and cloned into pUASTattB-NDam. Preparation of Dam-methylated DNA from stage 17 embryos was performed as previously described (Pym et al., 2006) and gene-targets identified (Wolfram et al., 2012) using Flybase release 5.47 and a stringent false discovery rate (FDR) of 0.1%.
Electrophysiology.
Hatched larvae (1–4 h old) were dissected and the CNS removed (Wolfram et al., 2012). Muscles were treated with 1 mg/ml collagenase (Sigma) for 0.5 to 1 min before whole-cell patch recording. Larvae were visualized using a water-immersion lens (total magnification, 600×) combined with DIC optics (BX51W1 microscope; Olympus Optical). Recordings were made from muscle 6 in segments A3–4 using a Multiclamp 700B amplifier controlled by pClamp 10.2 (Molecular Devices). Recordings were sampled at 20 kHz and filtered at 2 kHz. The voltage protocol used a maintained holding potential of −60 mV and a −90 mV prepulse for 200 ms before a 50 ms step to 40 mV. Leak currents were subtracted online (P/4). Recordings were done in at least 4 animals, and at least 8 muscles were recorded from in total for each manipulation. Cell capacitance was determined by integrating the area under the capacity transients evoked by stepping from −60 to −90 mV (checked before and after recordings). External saline (Stewart et al., 1994) consisted of (in mm) as follows: 70 NaCl, 5 KCl, 0.1 CaCl2, 20 MgCl2·6H2O, 10 NaHCO3, 5 HEPES, 115 sucrose, 5 trehalose, pH 7.2. The calcium concentration was kept low to prevent activation of Ca2+-dependent K+ currents. Internal patch solution consisted of (in mm) as follows: 140 K+gluconate, 2 MgCl2·6H2O, 2 EGTA, 5 KCl, and 20 HEPES, pH 7.4.
Under conditions of low external Ca2+, recorded traces contained two types of K+ conductances: Kfast (Kf) inactivating type (carried by Sh) and a Kslow (Ks) noninactivating type. These can be separated based on holding potential: Kf is inactivated by sustained depolarization (Wu and Haugland, 1985). We find, however, that inactivation is often incomplete resulting in variability of separation. Thus, we adopted a computational approach. To determine magnitude of Kf, we fitted data to standard Hodgkin–Huxley ion channel models of the form: I = ḡm4h(V − E). We first fitted the kinetic parameters of both Kf and Ks simultaneously. The reversal potential was found as E = −67 mV. Channel activation (m) and inactivation (h) gates were calculated with dm/dt = (m∞ − m)/τm. The steady state for activation was defined as m∞ = 1/(1 + exp((V − V1/2)/k)), and we found Kf to have V1/2 = −25.1 mV and k = −12.0 mV, and Ks to have V1/2 = 5.4 mV and k = −24.7mV. The time constant was defined by τm = a + b/(1 + exp((V + c)/d)), and (a, b, c, d) was found for Kf as (3.15, 104.0, 66.2, 25.6) and for Ks as (2.7, 1.4, −78.0, 0.13). For inactivation of Kf, we found V1/2 = −60 mV and k = 6 mV, and (1.53, 7.62, 27.63, 26.12) for the time constant. Once this fit was achieved, it was used as a template. To measure change in magnitude of Kf, we allowed only the parameters of maximal conductance, ḡ, for each channel and time constant offset, a, for each gate, to vary. We used a nonlinear least-squares fitter (lsqcurvefit function) in MATLAB (MathWorks) that uses the large-scale trust-region reflective Newton method. The 95% confidence intervals of individual fits were observed and bad fits rejected. Once parameters for Kf were fitted for a given experiment, we calculated a “peak conductance” in nS by calculating the value of g = ḡm4h at time of the expected peak, tpcak = τm log , where p = 4 is the power of the activation gate, m.
qRT-PCR.
Muscle tissue was collected from newly hatched larvae. After RNA extraction (QIAGEN RNaesy Micro kit), cDNA was synthesized using the Fermentas Reverse Aid H minus First strand cDNA synthesis kit. RNA concentration was matched for control and experimental samples before cDNA synthesis. qRT-PCR was performed on a Roche LightCycler480 II (Roche) in dual-color mode using the LightCycler TaqMan Master reaction mix (Roche; 04535286001). The thermal profile used was 10 min at 95°C followed by 45 cycles of 10 s at 95°C, followed by 20 s at 60°C, and finally 10 s at 72°C. Results were analyzed by the Δ/Δ Ct method and are expressed as fold difference relative to control. Ct values used were the means of duplicates. TaqMan primers and probes (forward and reverse primers in 5′ to 3′ orientation) were as follows: rp49 primers CCAGTCGGATCGATCGATATGCTA and ACGTTGTGCACCAGGAA and probe GATGCCCAACATCGGTTACGG conjugated with Cy5; Sh primers TAGGTGATCAAGCAATTAATAAATTCAG and TAACAAATACACTAATTATGGCTACAACT and probe AGACCATTACCGGATAATGAGAAACAGAG conjugated with FAM. Levels of expressed transcription factor in muscle were also monitored by TaqMan qPCR.
Statistics.
Statistical significance was calculated using a one-way ANOVA with Tukey's post hoc test using confidence intervals of p ≤ 0.05 and p ≤ 0.01. All quantitative data shown are mean ± SEM.
Accession codes.
The DamID-chip data have been deposited under NCBI GEO accession GSE53446.
Results
Target genes include both ion channel and morphology-associated genes
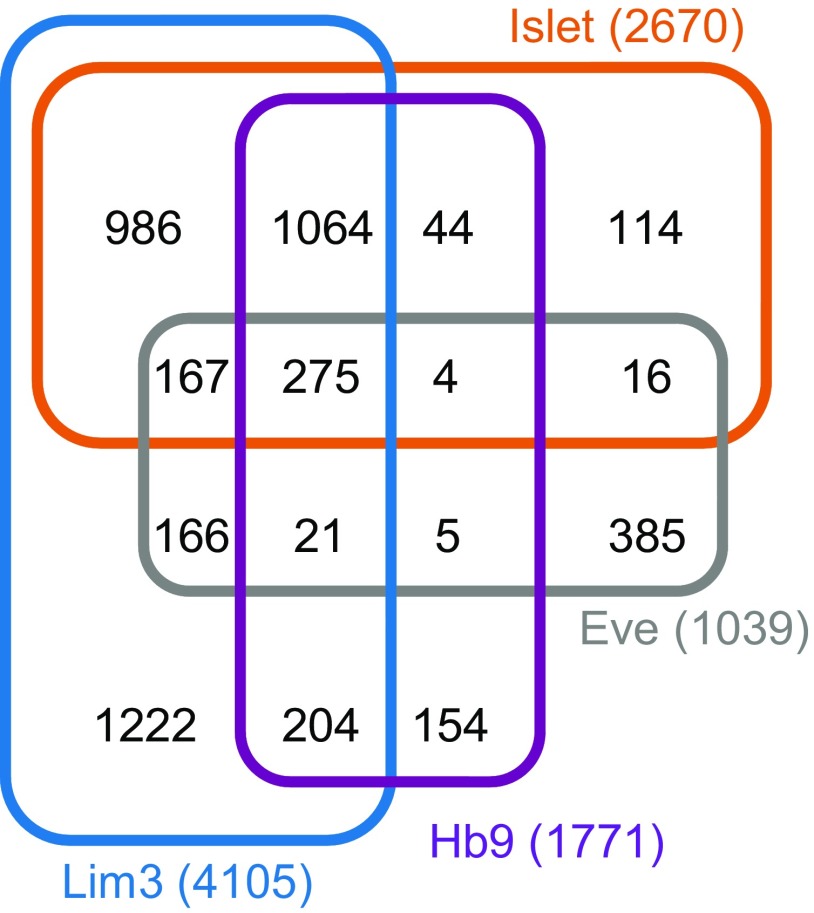
The LIM-HD transcription factors Isl and Eve control both axon guidance and ion-channel expression in developing neurons (Thor and Thomas, 1997; Landgraf et al., 1999; Pym et al., 2006; Wolfram et al., 2012). Together with other motoneuron-expressed TFs, notably Lim3 and Hb9, these TFs have been postulated to act combinatorially to set both structural and functional properties of embryonic neurons (Landgraf and Thor, 2006; Wolfram and Baines, 2013). However, to date, the gene targets of these TFs remain largely unknown and combinatorial activity remains to be definitively shown. We used DamID to describe binding sites for each of these TFs in Drosophila. Using an FDR of ≤0.1%, we identify 2670 genes (exhibiting one or more peaks of binding within 5 kb of the transcriptional unit) as targets of Isl; 4105 genes for Lim3, 1771 genes for Hb9, and 1039 genes for Eve. Notably, Islet, Lim3, and Hb9 bind a comparatively larger number of gene targets than does Eve. Analysis of targets indicates that these three TFs share a large number of common putative targets. In contrast, overlap with targets of Eve is relatively much smaller. This is predictable and validates our analysis because it reflects the observed overlap in neuron expression of these transcription factors. Lim3, Isl, and Hb9 are found colocalized in many motoneurons, whereas Eve is expressed in relatively few (Landgraf and Thor, 2006).
Consistent with the possibility of combinatorial regulation, many of target genes are bound by more than one TF (Fig. 1; Table 1). Gene ontology analysis reveals that targets include ion channels and also genes associated with both morphology (axonal and dendritic) and synapse formation (Table 1). This latter subset includes beat-Ic, Dscam, fra, Fas2, robo, and Sema1a, which have been studied in some detail (Baines et al., 2002; Corty et al., 2009; Mauss et al., 2009; Timofeev et al., 2012). One gene in particular, beat-Ic, has been shown to be downstream of the LIM-HD combinatorial code: regulation in this instance dictating motoneuron synaptic connectivity (Certel and Thor, 2004).
Figure 1.
Overlap of putative gene targets of Isl, Lim3, Hb9, and Eve determined by DamID. A four-way Venn diagram showing numbers of gene targets identified for each TF and overlap.
Table 1.
Selected genes identified by DamID as putative targets of Islet, Lim3, Hb9, and Eve
| Gene | Islet | Lim3 | Hb9 | Eve |
|---|---|---|---|---|
| Sh | ✓ | ✓ | ||
| Shab | ✓ | ✓ | ✓ | |
| Shawl | ✓ | |||
| Shal | ✓ | ✓ | ✓ | |
| slo | ✓ | ✓ | ||
| SK | ✓ | ✓ | ✓ | |
| KCNQ | ✓ | ✓ | ||
| Ih | ✓ | ✓ | ||
| Ca-alpha1T | ✓ | ✓ | ✓ | |
| Ca-alpha1D | ✓ | ✓ | ||
| Ca-beta | ✓ | ✓ | ||
| nAcRα-7E | ✓ | ✓ | ✓ | ✓ |
| nAcRα-30D | ✓ | ✓ | ||
| nAcRα-34E | ✓ | ✓ | ✓ | ✓ |
| nAcRα-96Aa | ✓ | ✓ | ||
| nAcRβ-96A | ✓ | |||
| beat-Ia | ✓ | ✓ | ||
| beat-Ib | ✓ | ✓ | ✓ | |
| beat-Ic | ✓ | |||
| beat-IIa | ✓ | ✓ | ||
| beat-IIb | ✓ | ✓ | ||
| beat-IIIa | ✓ | |||
| beat-IIIc | ✓ | ✓ | ✓ | |
| beat-Vb | ✓ | |||
| beat-Vc | ✓ | ✓ | ||
| beat-VI | ✓ | ✓ | ||
| beat-VII | ✓ | |||
| Sema-1a | ✓ | ✓ | ✓ | ✓ |
| Sema-1b | ✓ | ✓ | ✓ | |
| Sema-2a | ✓ | ✓ | ✓ | |
| Sema-2b | ✓ | ✓ | ✓ | |
| Sema-5c | ✓ | ✓ | ✓ | ✓ |
| NetA | ✓ | ✓ | ✓ | |
| NetB | ✓ | ✓ | ✓ | |
| fra | ✓ | ✓ | ✓ | |
| unc-5 | ✓ | ✓ | ✓ | |
| robo | ✓ | ✓ | ✓ | |
| robo3 | ✓ | |||
| Dscam | ✓ | ✓ | ✓ | |
| Fas1 | ✓ | ✓ | ✓ | |
| Fas2 | ✓ | ✓ | ✓ | |
| Fas3 | ✓ | ✓ | ✓ | ✓ |
| comm | ✓ | ✓ | ||
| Lar | ✓ | ✓ | ✓ | |
| shot | ✓ | ✓ | ✓ | ✓ |
Of the ion channel gene targets identified, there is considerable evidence to show that the voltage-gated K+ channel genes, Sh, Shal, Shab, the Ca2+-dependent K+ channel gene slo, and the Ca2+-channel gene Ca-α1D are expressed in motoneurons (Byerly and Leung, 1988; Leung and Byerly, 1991; Martínez-Padrón and Ferrús, 1997; Baines and Bate, 1998; Engel and Wu, 1998; Worrell and Levine, 2008). Synaptic excitation of motoneurons is also cholinergic (Baines et al., 1999); and in this regard, it is significant that our analysis identifies 5 nicotinic receptor-subunits (of a total of 10) (Sattelle et al., 2002). These genes are nAcRα-30D, nAcRα-7E, nAcRα-34E, nAcRα-96Aa, and nAcRβ-96A. We have previously validated three of the identified ion channel genes, Sh and slo/nAcRα-96Aα, as targets of Isl and Eve, respectively (Pym et al., 2006; Wolfram et al., 2012). It should be noted, however, that slo is now only identified (using stricter reassessment criteria) when we relax the FDR to ≤0.5%, indicative that our current target list (derived from an FDR of ≤0.1%) is restricted.
Combinatorial transcription factors share the same targets
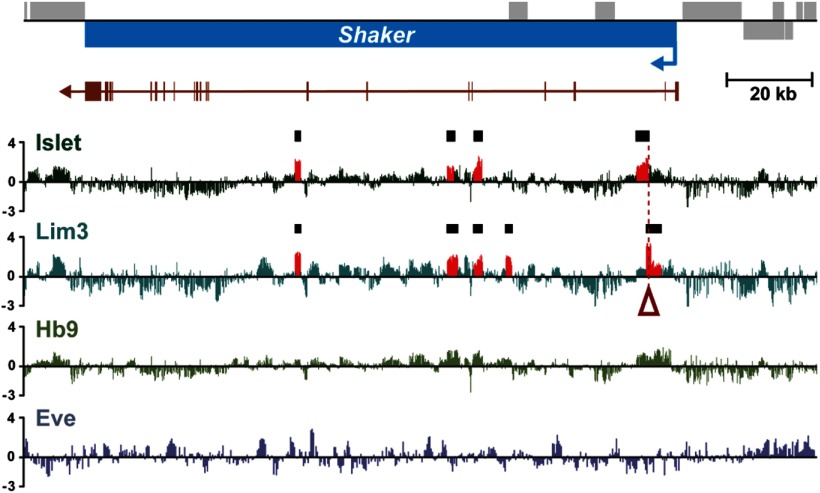
Our DamID shows that some genes are potentially bound by more than one TF. For example, Sh is a validated target of Isl (Wolfram et al., 2012) but also a putative target of Lim3 (Fig. 2; Table 1). Significantly, a mammalian Isl1:LhX3 (Isl/Lim3 homologs) ATTAGTTAATT “dimer” motif (Lee et al., 2008) underlies both Isl and Lim3 binding sites at this locus (Fig. 2). This overlap is consistent with the hypothesis of combinatorial regulation. We tested for this experimentally.
Figure 2.
DamID demonstrates direct binding of Isl and Lim3 to the Sh locus. The transcription unit of Sh is shown in blue; arrow indicates direction of transcription. Sh exons are shown immediately below in brown. Putative binding of each TF to this locus is indicted by the average of normalized log2-transformed ratios from multiple independent DamID experiments. Areas in red (and marked by the black boxes) represent a significant binding peak within the dataset (FDR < 0.1%) for both Isl and Lim3. No binding peaks were detected for Hb9 and Eve. Open red arrow indicates the location of a mammalian Isl1:LhX3 (Isl/Lim3 homologs) ATTAGTTAATT “dimer” motif (Lee et al., 2008).
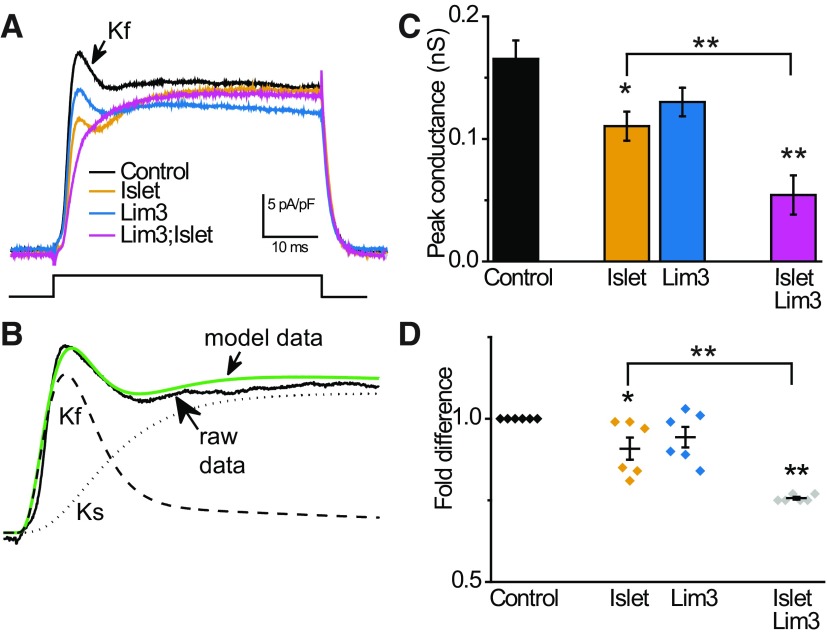
To determine how Lim3 influences expression of Sh and whether its effect is additive to that of Isl, we used body wall muscle in which Kf is carried entirely by Sh channels (in low external Ca2+) (Singh and Wu, 1990). Such analysis would be complicated in neurons because of the expression of an additional Kf encoded by Shal (Tsunoda and Salkoff, 1995) and homeostatic mechanisms (Baines et al., 2001), which maintain consistency in action potential firing through adjustment of ionic conductances. By contrast, muscle does not exhibit this type of homeostasis. To determine the amplitude of Kf in muscle whole-cell recordings, we modeled Kf and Ks, which allowed us to computationally determine peak conductances for both (see Materials and Methods). As previously shown, expression of isl in muscle is sufficient to reduce both the magnitude of Kf (Sh-dependent) (Fig. 3A–C) and the abundance of Sh transcript (Fig. 3D) consistent with transcriptional regulation (Wolfram et al., 2012). Expression of isl led to a significant reduction of Kf (0.17 ± 0.02 vs 0.11 ± 0.01 nS, p = 0.01, n ≥ 8, mean ± SE) and transcript (0.91 ± 0.03-fold difference, p = 0.02, n = 6). By contrast, expression of Lim3 did not statistically affect either Kf (0.13 ± 0.01 nS, p = 0.08, n = 10) or Sh transcript level (0.94 ± 0.03-fold reduction, p = 0.1; Fig. 2), even though the Lim3 transgene was expressed at increased levels relative to isl (∼30-fold compared with an ∼15-fold increase relative to control, no transgenic expression). Coexpression of both isl and Lim3 was, however, sufficient to reduce both Kf (0.05 ± 0.02 nS, p = 0.001, n = 9) and Sh transcript (0.76 ± 0.004-fold reduction, p = 6 × 10−14, n = 6) by an amount significantly greater than observed with isl alone (p = 0.015 for Kf and p = 0.001 for transcript; Fig. 3). In these coexpression experiments, qRT-PCR shows that both Lim3 and isl are up-regulated by ∼8- and ∼12-fold, relative to control. It should be noted that DamID shows that Lim3 is bound by Islet, which likely explains the significantly lower expression level compared with when Lim3 was overexpressed alone.
Figure 3.
Islet and Lim3 act combinatorially to regulate Sh expression. A, The fast K+ current (Kf) in body wall muscle is carried entirely by the Sh channel (in conditions of low Ca2+). Traces represent membrane current produced under voltage clamp for a step from −90 to 40 mV for 50 ms in control muscle and when Isl, Lim3, or both are expressed. Recordings are normalized to cell capacitance. B, Modeling of the outward K+ conductance allows Kf (carried by Sh) and Kslow (Ks) to be differentiated. Green line indicates model fit to capacitance-adjusted data (control muscle); model-derived Kf and Ks are shown. C, Model-derived peak conductances for Kf show that expression of Isl is sufficient to suppress the Sh-dependent Kf current, whereas Lim3 expression has no statistically significant effect. Coexpression of both Isl and Lim3 further reduces Kf to a level that is significantly different from Isl. D, qPCR shows that coexpression of Isl and Lim3 reduces Sh transcript to a level significantly different from Isl expression alone. In isolation, only Isl expression significantly reduces Sh transcript levels, mirroring the effect on Sh current. Values are mean ± SEM. *p ≤ 0.05. **p ≤ 0.01.
Together, our results are consistent with the previous demonstration of a direct gene–dosage relationship between Sh transcript and Sh-dependent Kf expressed in body wall muscle (Haugland and Wu, 1990). Attempts to verify this through higher TF transgene expression, often achieved by raising the temperature to 25°C, was not possible in our experiments because of lethality at this temperature. Thus, we conclude that repression of Sh expression by coexpressing both isl and Lim3 is additive, which is both predictive and supportive of combinatorial regulation.
Discussion
In contrast to specification of neuron identity, little is known concerning how embryonic neurons regulate ion channel gene expression before formation of neural networks. Recent studies, in a range of animals, suggest that members of a defined set of LIM-HD TFs regulate this early phase of expression. Collectively, these studies have led to the proposition that early neuron-type specific properties, including axonal and dendritic morphology, choice of synaptic target and expression of ion channels, are set by a combinatorial activity of TFs (Landgraf and Thor, 2006; Pym et al., 2006; Wolfram et al., 2012). For example, the presence of Lim3 is seemingly sufficient to subdivide Drosophila Isl-positive motoneurons into the ISNb and ISNd classes (Thor et al., 1999). Although there is evidence to show that ion channels are also regulated by LIM-HD TFs (Wolfram and Baines, 2013), definitive evidence to show combinatorial regulation is lacking. In this study, we show that the activity of two early expressed combinatorial TFs, Isl and Lim3, is additive such that their combined effect to suppress expression of the Sh K+ channel is significantly greater than either alone.
To show that Sh expression is regulated by both Isl and Lim3, we exploited muscle as a model tissue. There are several advantages of using muscle over neurons. First, Kf in muscle (in conditions of low external Ca2+) is carried solely by Sh channels (Singh and Wu, 1990). Second, muscle is isopotential (Jan and Jan, 1976), which significantly minimizes the impact of space clamp problems, which, in neurons, can be particularly problematic for fine-scale analysis of ionic currents. Third, studies in neurons can be complicated by homeostatic mechanisms, which maintain consistency in action potential firing. These mechanisms, not present in muscle, can modify expression of other channels to compensate for any enforced perturbation to membrane excitability that would be caused, in this instance, by manipulation of Sh expression (Baines et al., 2001; Wolfram et al., 2012). Validation of the effects we report here in muscle will, of course, require analysis of the effect of TF expression in central neurons. An attractive possibility, if endogenous levels of individual TFs can be accurately determined, is to use our understanding of Isl and Lim3 to predict levels of Sh-dependent K+ current in individual motoneurons, which can be subsequently confirmed by electrophysiology.
An important question to be addressed is how coexpression of Isl and Lim3 differs from expression of Isl alone in being able to repress Sh expression. The mammalian homologs, Isl1 and LhX3, have been shown to form a hexameric complex, together with the self-dimerizing cofactor NL1, to promote motoneuron fate (Lee et al., 2012). Although the LhX3/Isl1 complex and Isl1 or LhX3 response elements share A/T-rich sequences, each site is unique (Lee et al., 2008). Isl1 binds a DNA motif containing a core TAAT sequence (Yaden et al., 2005; Lee et al., 2008; Mazzoni et al., 2013). TAAT motif sites occur at a very high frequency in the genome; therefore, examining this site alone is not particularly informative. The first report to describe an Isl1 binding site (Boam and Docherty, 1989) identified a CTAATG, which is present at one of the sites of Isl binding at the Sh locus. Lee et al. (2008) reported a predicted site for LhX3 binding (AATTAATTA) and a hybrid motif (ATTAGCNTAATT), which is bound by Isl1:LhX3 dimers. We do not observe the Lhx3 (i.e., Lim3) motif but do find a highly similar ATTAGTTAATT “dimer” motif at a site where both Isl and Lim3 bind the Sh locus (coordinates 17950735–17950745; Fig. 2). Our identification of multiple genes for Isl and Lim3 in Drosophila affords significant opportunities to refine the identification of response elements in this model genome. This may allow identification of not only additional gene targets but will also have significant comparative value for identification of mammalian response elements.
In conclusion, sufficient studies have been reported to show that the TFs required for specification of neuron identity, such as Isl, Lim3, Hb9, and Eve, are present in all nervous systems and, moreover, that the developmental mechanisms they regulate are conserved. Our present DamID results, together with previous studies (Pym et al., 2006; Wolfram et al., 2012), provide experimental evidence that these regulatory factors orchestrate significant aspects of both the morphological and electrical development of embryonic neurons. For at least one gene, but likely more, this regulation is likely combinatorial. The use of Drosophila to describe the gene targets and underlying mechanisms through which these TFs operate will make a significant advance to understanding of neural circuit development.
Footnotes
This work was supported by The Wellcome Trust (090798 to R.A.B.; 068055/092545 to A.H.B.) and a Herchel Smith Postdoctoral Research Fellowship to T.D.S., C.G. was supported by the Epilepsy Foundation of America. A.H.B. was supported by the Gurdon Institute from The Wellcome Trust (092096) and CRUK (C6946/A14492). This project benefited from the Manchester Fly Facility, established by the University of Manchester and The Wellcome Trust (087742). We thank members of the R.A.B. and A.A.P. groups for advice.
The authors declare no competing financial interests.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/3.0), which permits unrestricted use, distribution and reproduction in any medium provided that the original work is properly attributed.
References
- Baines RA, Bate M. Electrophysiological development of central neurons in the Drosophila embryo. J Neurosci. 1998;18:4673–4683. doi: 10.1523/JNEUROSCI.18-12-04673.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines RA, Robinson SG, Fujioka M, Jaynes JB, Bate M. Postsynaptic expression of tetanus toxin light chain blocks synaptogenesis in Drosophila. Curr Biol. 1999;9:1267–1270. doi: 10.1016/S0960-9822(99)80510-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines RA, Uhler JP, Thompson A, Sweeney ST, Bate M. Altered electrical properties in Drosophila neurons developing without synaptic transmission. J Neurosci. 2001;21:1523–1531. doi: 10.1523/JNEUROSCI.21-05-01523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines RA, Seugnet L, Thompson A, Salvaterra PM, Bate M. Regulation of synaptic connectivity: levels of Fasciclin II influence synaptic growth in the Drosophila CNS. J Neurosci. 2002;22:6587–6595. doi: 10.1523/JNEUROSCI.22-15-06587.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boam DS, Docherty K. A tissue-specific nuclear factor binds to multiple sites in the human insulin-gene enhancer. Biochem J. 1989;264:233–239. doi: 10.1042/bj2640233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byerly L, Leung HT. Ionic currents of Drosophila neurons in embryonic cultures. J Neurosci. 1988;8:4379–4393. doi: 10.1523/JNEUROSCI.08-11-04379.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certel SJ, Thor S. Specification of Drosophila motoneuron identity by the combinatorial action of POU and LIM-HD factors. Development. 2004;131:5429–5439. doi: 10.1242/dev.01418. [DOI] [PubMed] [Google Scholar]
- Corty MM, Matthews BJ, Grueber WB. Molecules and mechanisms of dendrite development in Drosophila. Development. 2009;136:1049–1061. doi: 10.1242/dev.014423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasen JS, Tice BC, Brenner-Morton S, Jessell TM. A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell. 2005;123:477–491. doi: 10.1016/j.cell.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Dasen JS, De Camilli A, Wang B, Tucker PW, Jessell TM. Hox repertoires for motor neuron diversity and connectivity gated by a single accessory factor, FoxP1. Cell. 2008;134:304–316. doi: 10.1016/j.cell.2008.06.019. [DOI] [PubMed] [Google Scholar]
- De Marco Garcia NV, Jessell TM. Early motor neuron pool identity and muscle nerve trajectory defined by postmitotic restrictions in Nkx6.1 activity. Neuron. 2008;57:217–231. doi: 10.1016/j.neuron.2007.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel JE, Wu CF. Genetic dissection of functional contributions of specific potassium channel subunits in habituation of an escape circuit in Drosophila. J Neurosci. 1998;18:2254–2267. doi: 10.1523/JNEUROSCI.18-06-02254.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmaeili B, Ross JM, Neades C, Miller DM, 3rd, Ahringer J. The C. elegans even-skipped homologue, vab-7, specifies DB motoneurone identity and axon trajectory. Development. 2002;129:853–862. doi: 10.1242/dev.129.4.853. [DOI] [PubMed] [Google Scholar]
- Grosskortenhaus R, Pearson BJ, Marusich A, Doe CQ. Regulation of temporal identity transitions in Drosophila neuroblasts. Dev Cell. 2005;8:193–202. doi: 10.1016/j.devcel.2004.11.019. [DOI] [PubMed] [Google Scholar]
- Grosskortenhaus R, Robinson KJ, Doe CQ. Pdm and Castor specify late-born motor neuron identity in the NB7-1 lineage. Genes Dev. 2006;20:2618–2627. doi: 10.1101/gad.1445306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugland FN, Wu CF. A voltage-clamp analysis of gene-dosage effects of the Shaker locus on larval muscle potassium currents in Drosophila. J Neurosci. 1990;10:1357–1371. doi: 10.1523/JNEUROSCI.10-04-01357.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan LY, Jan YN. Properties of the larval neuromuscular junction in Drosophila melanogaster. J Physiol. 1976;262:189–214. doi: 10.1113/jphysiol.1976.sp011592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf M, Thor S. Development of Drosophila motoneurons: specification and morphology. Semin Cell Dev Biol. 2006;17:3–11. doi: 10.1016/j.semcdb.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Landgraf M, Roy S, Prokop A, VijayRaghavan K, Bate M. Even-skipped determines the dorsal growth of motor axons in Drosophila. Neuron. 1999;22:43–52. doi: 10.1016/S0896-6273(00)80677-7. [DOI] [PubMed] [Google Scholar]
- Lee S, Lee B, Joshi K, Pfaff SL, Lee JW, Lee SK. A regulatory network to segregate the identity of neuronal subtypes. Dev Cell. 2008;14:877–889. doi: 10.1016/j.devcel.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Cuvillier JM, Lee B, Shen R, Lee JW, Lee SK. Fusion protein Isl1-Lhx3 specifies motor neuron fate by inducing motor neuron genes and concomitantly suppressing the interneuron programs. Proc Natl Acad Sci U S A. 2012;109:3383–3388. doi: 10.1073/pnas.1114515109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung HT, Byerly L. Characterization of single calcium channels in Drosophila embryonic nerve and muscle cells. J Neurosci. 1991;11:3047–3059. doi: 10.1523/JNEUROSCI.11-10-03047.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Padrón M, Ferrús A. Presynaptic recordings from Drosophila: correlation of macroscopic and single-channel K+ currents. J Neurosci. 1997;17:3412–3424. doi: 10.1523/JNEUROSCI.17-10-03412.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauss A, Tripodi M, Evers JF, Landgraf M. Midline signalling systems direct the formation of a neural map by dendritic targeting in the Drosophila motor system. PLoS Biol. 2009;7:e1000200. doi: 10.1371/journal.pbio.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni EO, Mahony S, Closser M, Morrison CA, Nedelec S, Williams DJ, An D, Gifford DK, Wichterle H. Synergistic binding of transcription factors to cell-specific enhancers programs motor neuron identity. Nat Neurosci. 2013;16:1219–1227. doi: 10.1038/nn.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Rivard L, Kagawa T, Saueressig H, Gross MK, Burrill J, Goulding M. Evx1 is a postmitotic determinant of v0 interneuron identity in the spinal cord. Neuron. 2001;29:385–399. doi: 10.1016/S0896-6273(01)00213-6. [DOI] [PubMed] [Google Scholar]
- Pym EC, Southall TD, Mee CJ, Brand AH, Baines RA. The homeobox transcription factor Even-skipped regulates acquisition of electrical properties in Drosophila neurons. Neural Dev. 2006;1:3. doi: 10.1186/1749-8104-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattelle DB, Culetto E, Grauso M, Raymond V, Franks CJ, Towers P. Functional genomics of ionotropic acetylcholine receptors in Caenorhabditis elegans and Drosophila melanogaster. Novartis Found Symp. 2002;245:240–257. discussion 257–260. [PubMed] [Google Scholar]
- Singh S, Wu CF. Properties of potassium currents and their role in membrane excitability in Drosophila larval muscle fibers. J Exp Biol. 1990;152:59–76. doi: 10.1242/jeb.152.1.59. [DOI] [PubMed] [Google Scholar]
- Spana EP, Kopczynski C, Goodman CS, Doe CQ. Asymmetric localization of numb autonomously determines sibling neuron identity in the Drosophila CNS. Development. 1995;121:3489–3494. doi: 10.1242/dev.121.11.3489. [DOI] [PubMed] [Google Scholar]
- Stewart BA, Atwood HL, Renger JJ, Wang J, Wu CF. Improved stability of Drosophila larval neuromuscular preparations in haemolymph-like physiological solutions. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1994;175:179–191. doi: 10.1007/BF00215114. [DOI] [PubMed] [Google Scholar]
- Thaler JP, Lee SK, Jurata LW, Gill GN, Pfaff SL. LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell. 2002;110:237–249. doi: 10.1016/S0092-8674(02)00823-1. [DOI] [PubMed] [Google Scholar]
- Thaler J, Harrison K, Sharma K, Lettieri K, Kehrl J, Pfaff SL. Active suppression of interneuron programs within developing motor neurons revealed by analysis of homeodomain factor HB9. Neuron. 1999;23:675–687. doi: 10.1016/S0896-6273(01)80027-1. [DOI] [PubMed] [Google Scholar]
- Thor S, Thomas JB. The Drosophila islet gene governs axon pathfinding and neurotransmitter identity. Neuron. 1997;18:397–409. doi: 10.1016/S0896-6273(00)81241-6. [DOI] [PubMed] [Google Scholar]
- Thor S, Andersson SG, Tomlinson A, Thomas JB. A LIM-homeodomain combinatorial code for motor-neuron pathway selection. Nature. 1999;397:76–80. doi: 10.1038/16275. [DOI] [PubMed] [Google Scholar]
- Timofeev K, Joly W, Hadjieconomou D, Salecker I. Localized netrins act as positional cues to control layer-specific targeting of photoreceptor axons in Drosophila. Neuron. 2012;75:80–93. doi: 10.1016/j.neuron.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoda S, Salkoff L. Genetic analysis of Drosophila neurons: Shal, Shaw, and Shab encode most embryonic potassium currents. J Neurosci. 1995;15:1741–1754. doi: 10.1523/JNEUROSCI.15-03-01741.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotechnol. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- Wolfram V, Baines RA. Blurring the boundaries: developmental and activity-dependent determinants of neural circuits. Trends Neurosci. 2013;36:610–619. doi: 10.1016/j.tins.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfram V, Southall TD, Brand AH, Baines RA. The LIM-homeodomain protein islet dictates motor neuron electrical properties by regulating K(+) channel expression. Neuron. 2012;75:663–674. doi: 10.1016/j.neuron.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrell JW, Levine RB. Characterization of voltage-dependent Ca2+ currents in identified Drosophila motoneurons in situ. J Neurophysiol. 2008;100:868–878. doi: 10.1152/jn.90464.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CF, Haugland FN. Voltage-clamp analysis of membrane currents in larval muscle fibers of Drosophila: alteration of potassium currents in Shaker mutants. J Neurosci. 1985;5:2626–2640. doi: 10.1523/JNEUROSCI.05-10-02626.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaden BC, Savage JJ, Hunter CS, Rhodes SJ. DNA recognition properties of the LHX3b LIM homeodomain transcription factor. Mol Biol Rep. 2005;32:1–6. doi: 10.1007/s11033-004-4069-z. [DOI] [PubMed] [Google Scholar]