Abstract
The regenerative failure of mammalian optic axons is partly mediated by Socs3-dependent inhibition of Jak/Stat signaling (Smith et al., 2009, 2011). Whether Jak/Stat signaling is part of the normal regenerative response observed in animals that exhibit an intrinsic capacity for optic nerve regeneration, such as zebrafish, remains unknown. Nor is it known whether the repression of regenerative inhibitors, such as Socs3, contributes to the robust regenerative response of zebrafish to optic nerve damage. Here we report that Jak/Stat signaling stimulates optic nerve regeneration in zebrafish. We found that IL-6 family cytokines, acting via Gp130-coupled receptors, stimulate Jak/Stat3 signaling in retinal ganglion cells after optic nerve injury. Among these cytokines, we found that CNTF, IL-11, and Clcf1/Crlf1a can stimulate optic axon regrowth. Surprisingly, optic nerve injury stimulated the expression of Socs3 and Sfpq (splicing factor, proline/glutamine rich) that attenuate optic nerve regeneration. These proteins were induced in a Jak/Stat-dependent manner, stimulated each other's expression and suppressed the expression of regeneration-associated genes. In vivo, the injury-dependent induction of Socs3 and Sfpq inhibits optic nerve regeneration but does not block it. We identified a robust induction of multiple cytokine genes in zebrafish retinal ganglion cells that may contribute to their ability to overcome these inhibitory factors. These studies not only identified mechanisms underlying optic nerve regeneration in fish but also suggest new molecular targets for enhancing optic nerve regeneration in mammals.
Keywords: Jak, optic nerve, regeneration, retina, retinal ganglion cell, Stat
Introduction
The optic nerve is an accessible portion of the CNS that has served as a convenient system for studying CNS regeneration. Both positive and negative factors affect optic nerve regeneration in mammals (Fischer and Leibinger, 2012). Notable among these factors is cytokines, such as CNTF and LIF, which appear to enhance retinal ganglion cell (RGC) survival and axonal regeneration via activation of a Jak/Stat signaling cascade (Leibinger et al., 2009; Pernet et al., 2013b). Indeed, RGCs with constitutively active Stat3 exhibit robust axonal regeneration (Pernet et al., 2013a). Furthermore, SOCS3 inhibits Jak/Stat signaling, and its deletion enhances axonal regeneration (Smith et al., 2009). Thus, proteins, such as SOCS3, may contribute to regenerative failure in mammals. Interestingly, PTEN and KLF4 are also endogenous inhibitors of RGC axonal regeneration (Moore et al., 2009; Sun et al., 2011). Like that reported for KLF4 (Moore et al., 2009), the increased expression/activity of these inhibitors during development may contribute to the transition of RGCs from a growth-permissive state to one that is growth restrictive (Moore et al., 2009). Based on this idea, we wondered whether animals that retain a robust regenerative capacity into adulthood had reduced expression of regenerative inhibitors after nerve injury.
One animal that retains a robust regenerative response into adulthood is zebrafish. These fish can regenerate a damaged optic nerve that results in restoration of lost sight (Bernhardt, 1999; McDowell et al., 2004). This regenerative response is driven by the activation of positive effectors of axonal growth, including purpurin, Klf6, Klf7, Tuba1a, Igf1, pAkt, transglutaminase, and nitric oxide signaling (Matsukawa et al., 2004; Sugitani et al., 2006, 2012; Koriyama et al., 2007, 2009; Veldman et al., 2007, 2010). Although Jak/Stat signaling has been shown to stimulate optic nerve regeneration in mammals, there are no reports of it contributing to optic nerve regeneration in an animal, such as zebrafish, that naturally regenerate a damaged optic nerve. Based on studies in mammals (Smith et al., 2009; Pernet et al., 2013a, 2013b), we suspected that Jak/Stat signaling would stimulate optic nerve regeneration in zebrafish and that endogenous inhibitors of optic nerve regeneration would be repressed after nerve injury.
Here we report the surprising finding that zebrafish respond to optic nerve lesion by inducing the expression of Sfpq and Socs3a, two proteins that attenuate optic nerve regeneration. We found that these proteins are regulated by a Jak/Stat signaling cascade and that their knockdown enhances optic nerve regeneration. Our data suggest that fish overcome the actions of Sfpq and Socs3a by inducing a variety of IL-6-like cytokines in RGCs that act in an autocrine/paracrine fashion to stimulate Jak/Stat signaling. The identification of ligands, signaling cascades, and feedback inhibitory mechanisms operating on Jak/Stat signaling during zebrafish optic nerve regeneration shows a remarkable similarity to mechanisms controlling optic nerve regeneration in mammals. Importantly, our data suggest that, even when factors that decelerate axonal regrowth are highly induced, regeneration and restoration of visual function can ensue.
Materials and Methods
Animals and optic nerve lesion.
Zebrafish were kept at 26°C-28°C on a 14/10 h light/dark cycle. Fish of either sex were used in all experiments. All surgeries were performed on fish anesthetized in 0.02% tricaine methane sulfonate. Optic nerve surgeries and retinal explants were performed as previously described (Veldman et al., 2007). Briefly, fish were placed under a dissecting microscope for visualization, and the right optic nerve was exposed by gently pulling the eye out of the orbit and cutting the dorsal connective tissue. The optic nerve was cut using an iridectomy scissor, using care not to cut the ophthalmic artery. The eye was gently replaced in the orbit, and fish were placed in their home tank to recover. The left optic nerve remained intact and served as a control, except when bilateral optic nerve lesion was performed for analyzing restoration of visual function using an optokinetic response or phantom predator. In this case, sham-treated fish were used as controls.
Mice were anesthetized with isoflurane, and the right eye was visualized under a dissecting microscope. The conjunctiva around the globe was cut with scissors and retracted to expose the globe. The globe was rotated nasally to expose the optic nerve. The nerve was crushed with a forceps for 10 s taking special care not to interfere with the blood supply. The left eye served as an uninjured control.
Morpholino (MO) treatments.
MO-mediated gene knockdown in adult RGCs after optic nerve lesion was as previously described (Becker et al., 2004; Veldman et al., 2007). Briefly, 1 μl of a 1 mm MO (Gene Tools) solution was applied to a small piece of Gelfoam (Pharmacia) and placed at the cut site on the lesioned optic nerve. The following day, the fish was anesthetized and the Gelfoam removed. The following MOs were used: sfpq-MO1, 5′-ATCGGTCACGAGACATTATCACACA-3′; sfpq-MO2, 5′-CCATGCCACCGCGCATCCCCATTCC-3′ (Lowery et al., 2007); socs3a-ATG MO, 5′-TATCCAACTTGCTGTGGGTTATCAT-3′ (Veldman et al., 2007); socs3a-Sp MO, 5′-AAAAGAGCCTGGAAAAAAGAGAAAG-3′; socs3b-MO, 5′-TGTCAAGCCTACTATGCGTTACCAT-3′ (Veldman et al., 2007); gp130-MO, 5′-ACAGCCAATGATGTGAAGTGTCCAT-3′; clcf1-MO, 5′-CCTGACCAACTTTCCAGGGACACAT-3′; and crlf1a-MO, 5′-CAATAAGCAGATCATCTTACGAGGA-3′. The standard control MO from Gene Tools was used as a control for nonspecific effects. To verify the efficacy of the sfpq-, clcf1-, and crlf1a-targeting MOs, pTAL-gfp-reporter plasmids were created, which harbor the MO target sequence appended to the 5′ end of the gfp transcript. The plasmid (30 ng/ml) was injected into single-cell zebrafish embryos with lissamine tagged control MO or experimental MO (0.2 mm), or untagged MOs that were mixed with 3000 MW rhodamine dextran (Invitrogen) to help identify injected embryos. At 24 h after fertilization, embryos were assayed for GFP and rhodamine fluorescence. We also used TUNEL assay and BrdU incorporation to investigate whether MOs stimulated RGC death or proliferation, respectively, in the adult retina. This analysis showed that MOs had no effect on RGC death or proliferation.
Retinal explant assay.
Retinal explants were performed as described previously (Landreth and Agranoff, 1979; Veldman et al., 2007). Briefly, 0 or 4 d after the initial nerve transection, fish were dark adapted, killed by an overdose of tricaine, and retinas isolated. Retinas were cut into 0.5 mm squares with a razor blade and then digested with hyaluronidase (1 mg/ml) in L15 media (Invitrogen) for 15 min at room temperature. Explants were then rinsed 3 times with culture media and plated: one retina per 35 mm plate precoated with poly-l-lysine (100 μg/ml) and laminin (10 μg/ml). Explants were maintained at 28°C for 4 d in 0.5 ml of L15, 8% FCS, 3% zebrafish embryo extract, and 1× antibiotic/antimycotic in a humidified ambient air incubator. Adherent explants were quantified for neurite length and density as previously described (Veldman et al., 2007) with the following modifications: Axon density and length were measured using ImageJ software. Average density and lengths were calculated for each condition. In this study, 15–20 explants were generated from each retina and quantified. Only axons that could be clearly distinguished and traced for its full length were quantified (never <10/explant). Experiments were repeated 3 times using retinas from three different fish for each condition.
Inhibitors and recombinant proteins.
Jak inhibitors, P6 and JSI-124, were dissolved in DMSO. P6 (also known as Pyridone 6, EMD Chemicals) was used at 1 and 10 μm, and JSI-124 (also known as Cucurbitacin I, Indofine Chemical) was used at 10 nm and 1 μm. Control for inhibitors was DMSO (0.1% final). Recombinant rat CNTF and mouse IL-11 (R&D Systems) were reconstituted in PBS with 0.1% BSA/PBS and used at 50 pg/ml; 0.1% BSA/PBS was used as a control. Inhibitors or recombinant proteins were added to culture media when explants were plated.
RNA isolation, PCR, and in situ hybridization.
Total RNA was isolated from retinas using TRIzol (Invitrogen). Random primers and Superscript II reverse transcriptase (Invitrogen) were used to generate cDNA. qPCR was performed in triplicate (each time point or condition was determined on a single retina and averaged using 3 retinas from 3 different fish) with Absolute SYBR Green Fluorescein Master Mix (Thermo Scientific) on an iCycler real-time PCR detection system (Bio-Rad). The ΔΔCt method was used to determine relative expression of mRNAs in control and injured retinas and normalized to gapdh mRNA levels for fish and γ-Actin mRNA levels for mouse. Primers used in this study are listed in Table 1.
Table 1.
Primers used in this study
| Gene | Sequence 5′-3′ |
|---|---|
| atf3 | Forward: CCGTCAGAGATCAGTGCGTCAGCTTTG |
| Reverse: GTTCTGAGCGCGGACGATGCAGGTGG | |
| β-thymosin | Forward: CCTGCTGCTCCATCCACACAAGCACA |
| Reverse: GCTTGAGTTCGTCTGAAGTCTCTGG | |
| c-jun | Forward: TGGATACAACCACCAGGCTCT |
| Reverse: GTCACGTTCTTGGGACACAG | |
| clcf1 | Forward: GAAAGTTGGTCAGGTTGCTGTGC |
| Reverse: CATAAGTCCACACGTGTTGCTGC | |
| crlf1a | Forward: GGGATTCTGGGATCTAGGAAAGC |
| Reverse: TCCTTGAAGAACCTGGTTGCG | |
| cntfr | Forward: ACACCATCACCGACGCCTATGC |
| Reverse: GAAGCTCACACATCACATGATGG | |
| gfp | Forward: GCAAGCTGACCCTGAAGTTC |
| Reverse: AACACCGCCTAGAACTTCA | |
| gp130 | Forward: AATGAAGTTCGCCGATGGAGAGG |
| Reverse: CGTCTTCCTTGGGCATTTCGG | |
| gapdh | Forward: ATGACCCCTCCAGCATGA |
| Reverse: GGCGGTGTAGGCATGAAC | |
| Mouse γ-actin | Forward: ACCCAGGCATTGCTGACAGGATGC |
| Reverse: CCATCTAGAAGCATTTGCGGTGGACG | |
| il-11a | Forward: CTCCTCATCGCTGCTTCTCTCG |
| Reverse: TTGCGAAGTCACTGGCTCTGC | |
| il-11ra | Forward: GTTGGACTGTTGGTTTTGTTGG |
| Reverse: TGGATTGTGGGTAATGAAGGC | |
| klf6a | Forward: ATGGATGTTCTACCAATGTGC |
| Reverse: TGCTCCAAAGATCCTCCTGGCCCTC | |
| klf7a | Forward: ATGGACGTGTTGGCGAATTAC |
| Reverse: GGGGCTAGGCTCGTCGGATA | |
| lifrb | Forward: TCACAGTTGACCAGATGCTTGC |
| Reverse: ATGTGGGTTTTCTGAGGTGGG | |
| m17 | Forward: CTTGATTGCCGTTCAGTTAGTGC |
| Reverse: TTAAGATGCGCTCCGATTCAGT | |
| sfpq | Forward: TGCTCCCTTGAAGCAAGA |
| Reverse: AGACGACAACGTTGGGTG | |
| Mouse Sfpq | Forward: GCCGCTCCCGCCTCCAC |
| Reverse: CAGGCCTCCGCAAGAGAGAC | |
| socs-3a | Forward: CACTAACTTCTCTAAAGCAGGG |
| Reverse: GGTCTTGAAGTGGTAAAACG | |
| socs3b | Forward: GAAAACTCCCAAGATTGAGTCG |
| Reverse: TACTATGCGTTACCATGGCG | |
| tub1a | Forward: CGAGCTGTAAGATCTCTCCGCCAAG |
| Reverse: TCCTGTCCAGCACCAGGTCAATGAT | |
| tub1b | Forward: GCCATTCATCGAAAATACACATTTG |
| Reverse: CCCTATCTAAAACAAGGTCAATGAG | |
| tub1c | Forward: TTTTCCTCCTGCCTCCTGACATCC |
| Reverse: CAGCATCTTCCTTTCCTGTTATGAGCT |
In situ hybridizations were performed with antisense digoxigenin (DIG)-labeled RNA probes as described previously (Barthel and Raymond, 2000). The sfpq cDNA was kindly provided by Hazel Sive (Lowery et al., 2007). The gp130, clcf1, crlf1a, il-11a, cntfr, and il11-ra cDNAs were cloned by PCR into pCS2 and BSSK vectors. Antisense full-length probes were created from linearized plasmids using T3, T7, and Sp6 RNA polymerases (Promega). Sense control probes were also generated and showed no signal above background.
BrdU labeling, retina fixation, cryosectioning, and immunostaining.
BrdU labeling was accomplished by injecting 20 μl of BrdU (20 mm) intraperitoneally 3 h before death. Fish were overdosed with tricaine methane sulfonate. Eyes were isolated, the lens removed, and fixed in fresh 4% PFA or overnight at 4°C. After fixation, tissues were cryoprotected in phosphate-buffered 20% sucrose overnight and then embedded in Tissue-Tek OCT compound (Sakura Finetek). Embedded samples were stored frozen at −80°C until sectioned to 12 μm on a CM3050S cryostat (Leica). Sections were collected on Superfrost /Plus slides (Fisher Scientific), dried, and stored at −80°C.
Immunofluorescence protocols were previously described (Ramachandran et al., 2011). p-Stat3 immunofluorescence was performed using mouse antibody generated against a zebrafish phospho(Tyr708)-Stat3 protein (MBL) at 1:100 dilution. For BrdU and p-Stat3 immunostaining, we used an epitope retrieval protocol by placing slides in 100°C 10 mm citrate buffer, pH 6, for 40 min. Secondary antibody was conjugated to AlexaFluor-488 and used at a 1:500 dilution.
TUNEL asssy.
We used an in situ Cell Death Detection Kit fluorescein (Roche Applied Science) to detect apoptotic cells in control, Sfpq-, Socs3a-, and Gp130 MO-treated retinas. Twenty-four hours after optic nerve lesion and morpholino application, fish were killed with an overdose of tricaine and eyes collected, fixed, and cryosectioned as described above. TUNEL assay was performed following the manufacturer's protocol. Oubain-treated eyes served as a positive control.
RGC back-labeling.
RGC back-labeling for assaying optic axon regeneration was performed as previously described (Munderloh et al., 2009). Briefly, optic nerves were cut, and 7 d later regenerating nerves were lesioned ∼3 mm distal to the original cut site. Gelfoam impregnated with 1 μl (1 mg/ml) of Rhodamine dextran (MW 3000, Invitrogen) or Fluorescein dextran (MW 10,000, Invitrogen) was applied to the newly lesioned regenerating nerve. Fish were dark-adapted overnight, killed in tricaine, and retinas isolated. Retinas were then fixed in 4% PFA at 4°C, washed 3× with ice-cold PBS, placed on a superfrost/Plus slides (Fisher Scientific), and laid flat by making four slits with a scissors that were evenly spaced around the retinas' circumference. Whole-mount retinas were mounted in DABCO (1,4-diazabicyclo[2.2.2]octane) and visualized under a fluorescent microscope. Three fish were used for each condition per experiment, and the total number of fluorescent RGCs in each retinal quadrant was quantified. No significant differences were noted between retinal quadrants. Data are reported for the total retina.
Anterograde labeling of optic axons.
Fish were anesthetized in tricaine, and an incision was made near the cornea–sclera junction through which a small piece of Gelfoam, soaked in 1 μl (1 mg/ml) of Rhodamine dextran, was placed behind the lens. Six hours later, fish were killed by an overdose of tricaine, and the brain and eyes were carefully dissected, making sure the eyes with attached optic nerve, chiasm, and brain were all intact and connected. Tissue was fixed in 4% PFA and observed under a Leica MZ dissecting fluorescence microscope. Optic axon regeneration into the brain was assayed by quantifying the distance the fluorescent optic nerve extended beyond the optic chiasm using ImageJ software.
Optokinetic response.
Adult zebrafish were acclimated to the apparatus before behavior assays were performed. Lightly anesthetized fish was immobilized by gently clamping the fish between two pieces of sponge that was stabilized by two halves of a plastic pipe. The restrained fish was immersed in fish water in a custom-made Petri dish with a holder to keep restrained fish in place. The Petri dish was placed inside a custom-made circular drum, with vertical stripes (black and white, 8 mm wide) covering the inside of the drum wall. A light source was placed above the apparatus during video recording. Each fish was recorded for 2 min without drum rotation, followed by 2 min with drum rotation. Drum rotations were at 4 rpm and alternated between clockwise and counterclockwise rotations every 4 s. Movies were captured to a computer and replayed for quantification of spontaneous eye movements without rotating visual stimulus and eye movements tracking the rotating stripes. Background eye movements were subtracted from those obtained after a visual stimulus. A minimum of four fish were used for each experiment.
Phantom predator, escape response.
Zebrafish initiate an escape response when threatened by a potential predator. This innate behavior can be used to assess visual performance (Li and Dowling, 1997). The apparatus used consisted of an immobilized Petri dish (10 cm diameter) surrounded by a rotating drum (11.5 × 6.5 cm) covered with white paper. A black segment (6 × 6 cm) was marked on the paper as a threatening object. A post (3 cm diameter) that was placed in the center of the container prevented the fish from swimming directly from one side of the container to the other. The drum was illuminated from above with a white light source and rotated at 8 rpm for 2 min. A video camera mounted above the tank was used to record fish behavior before (2 min) and during (2 min) drum rotation. The video was captured to a computer and replayed for analysis. A minimum of four fish were used per condition for each experiment.
Statistical analyses.
ANOVA with Fisher's PLSD post hoc analysis was used for multiple parameter comparison; two-tailed Student's t test was used for single parameter comparison (see Figs. 2C,D,G,H, 4D, and 7F).
Figure 2.
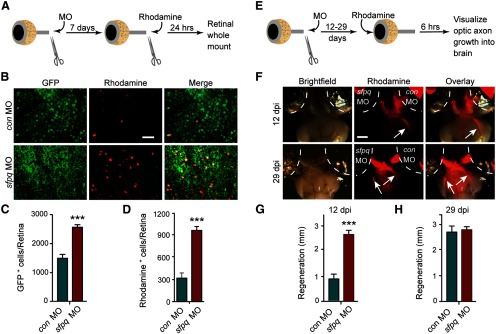
Sfpq inhibits optic nerve regeneration in vivo. A, Schematic of RGC back-labeling experiment for assaying optic nerve regeneration in vivo (see Materials and Methods). B, Representative pictures of retinal whole mounts from tuba1a:gfp transgenic fish shows Sfpq knockdown after optic nerve injury stimulates transgene expression and increases the number of rhodamine back-labeled RGCs. Scale bar, 50 μm. C, Quantification of GFP+ RGCs shown in B. ***p < 0.001, compared with control MO. n = 3. Error bars indicate SD. D, Quantification of rhodamine+ RGCs shown in B. ***p < 0.001, compared with control MO. n = 3. Error bars indicate SD. E, Schematic of anterograde labeling protocol (see Materials and Methods). F, Brightfield and fluorescent photomicrographs showing fluorescent optic axons regenerating to the optic tectum (arrow). Increased regeneration of optic axons in Sfpq knockdown RGCs at 12 dpi is indicated by rhodamine fluorescence in the brain. Brains are ventral side up. Scale bar, 500 μm. G, H, Quantification of the distance fluorescent optic nerves have regenerated past the optic chiasm at the indicated times. ***p < 0.001, compared with control MO. n = 4. Error bars indicate SD.
Figure 4.
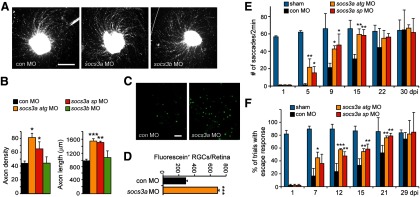
Socs3a knockdown enhances optic nerve regeneration and vision recovery. A, Axon outgrowth in a retinal explant that was prepared from a retina whose optic nerve was treated, in vivo, with a control, socs3a, or socs3b targeting MO. Scale bar, 500 μm. B, Quantification of axon density and lengths for retinal explants from control, socs3a, and socs3b targeting MO-treated optic nerves. *p < 0.05, compared with control MO. **p < 0.01, compared with control MO. ***p < 0.001, compared with control MO. n = 4. Error bars indicate SD. C, Socs3a knockdown enhances optic nerve regeneration in vivo. Back-labeling experiments were performed as schematized in Figure 2A. There is an increased number of back-labeled cells after Socs3a knockdown. Scale bar, 50 μm. D, Quantification of fluorescein+ RGCs shown in C. ***p < 0.001, compared with control MO. n = 3. Error bars indicate SD. E, Quantification of eye saccades tracking visual stimulus. *p < 0.05, compared with control MO. **p < 0.01, compared with control MO. n = 4. Error bars indicate SD. F, Quantification of escape response. *p < 0.05, compared with control MO. **p < 0.01, compared with control MO. ***p < 0.001, compared with control MO. n = 4. Error bars indicate SD.
Figure 7.
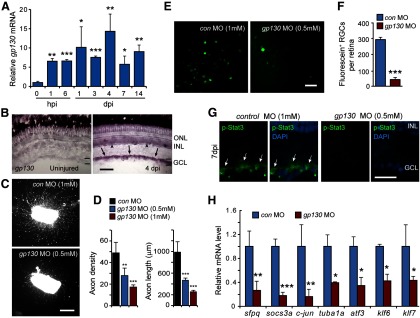
Gp130 stimulates Jak/Stat signaling during optic nerve regeneration. A, qPCR quantification of gp130 mRNA levels during optic nerve regeneration. mRNA levels are normalized to uninjured control. *p < 0.05, compared with uninjured control. **p < 0.01, compared with uninjured control. ***p < 0.001, compared with uninjured control. n = 3. Error bars indicate SD. B, In situ hybridization shows induction of gp130 in the GCL (arrows) and INL (arrowheads) after optic nerve injury. Scale bar, 50 μm. C, Retinal explants show that GP130 knockdown inhibits optic axon regeneration. Scale bar, 500 μm. D, Quantification of axon density and length for control and Gp130 knockdown retinal explants. **p < 0.01, compared with control MO. ***p < 0.001, compared with control MO. n = 4. Error bars indicate SD. E, In vivo back-labeling experiments, schematized in Figure 2A, indicate that Gp130 knockdown suppresses optic nerve regeneration. Scale bar, 50 μm. F, Quantification of the number of back-labeled RGCs shown in E. ***p < 0.001, compared with control MO. n = 3. Error bars indicate SD. G, p-Stat3 immunofluorescence (arrows) shows that Gp130 knockdown suppresses p-Stat3 expression at 7 d after optic nerve lesion. Nuclei are stained with DAPI (blue). Scale bar, 20 μm. H, qPCR shows that knockdown of Gp130 suppresses the induction of regeneration-associated genes. *p < 0.05, compared with control MO. **p < 0.01, compared with control MO. ***p < 0.001, compared with control MO. n = 3. Error bars indicate SD. ONL, Outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer.
Results
Injury-dependent Sfpq induction attenuates optic axon regeneration
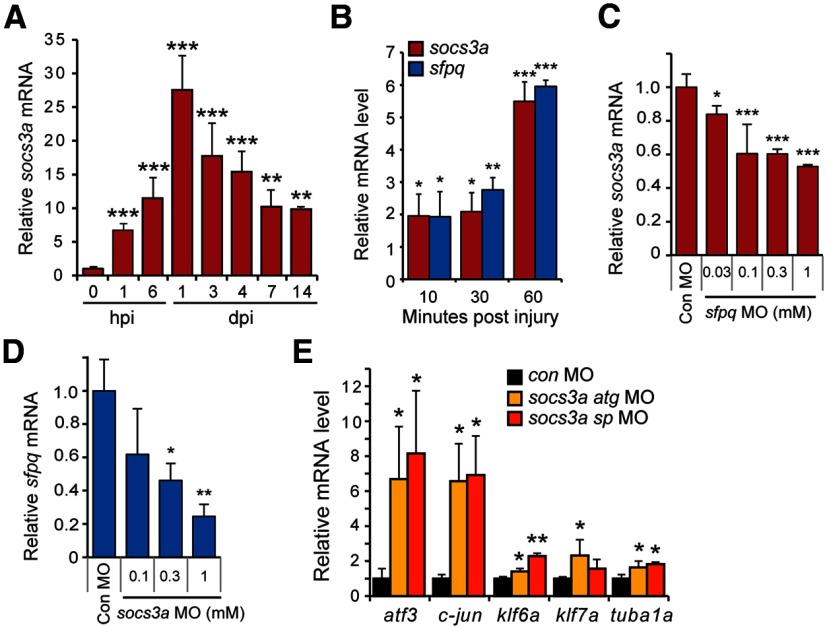
A microarray-based transcriptome analysis of gene expression during zebrafish optic nerve regeneration identified a number of candidate genes whose expression increased in RGCs and may encode proteins that regulate optic nerve regeneration in zebrafish (Veldman et al., 2007). qPCR was used to analyze the temporal induction of some of the most highly induced genes and identified sfpq as a transcript that was induced within 1 h post injury (hpi) (Fig. 1A). Sfpq is a multifunctional protein that is associated with RNA splicing, DNA synthesis, gene expression, DNA repair, and cell survival (Shav-Tal and Zipori, 2002). The rapid induction of sfpq mRNA was before the initiation of axonal growth that is noted ∼2 dpi (Bernhardt, 1989; Bernhardt et al., 1996; Becker et al., 2000; McCurley and Callard, 2010) and suggests it may participate in reprogramming RGCs to a growth-permissive state. Therefore, we decided to characterize its expression and function in more detail.
Figure 1.
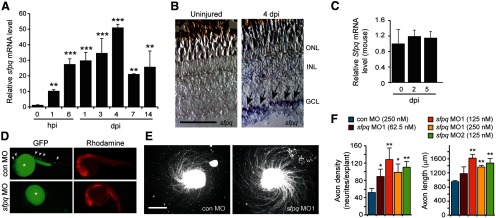
Sfpq inhibits RGC axon outgrowth in retinal explants. A, qPCR analysis of zebrafish sfpq mRNA induction in the retina after optic nerve injury. **p < 0.01, relative to uninjured control. ***p < 0.001, relative to uninjured control. n = 3. Error bars indicate SD. B, In situ hybridization shows injury-dependent sfpq mRNA induction in retinal ganglion cells (arrows). Scale bar, 50 μm. C, qPCR shows that Sfpq mRNA expression in the mouse retina is not regulated by optic nerve injury. D, A chimeric sCMV:sfpq-gfp transgene was injected into single-cell zebrafish embryos with rhodamine dextran (tracer) and a control or experimental MO, and transgene GFP expression assayed by fluorescence microscopy 24 h later. Shown are representative embyros. Arrowheads point to GFP expression (for quantification, see Results). Asterisk indicates autofluorescence in yolk. E, Axon outgrowth in a retinal explant that was prepared from a retina whose optic nerve was treated, in vivo, with a control or sfpq-targeting MO. Scale bar, 500 μm. F, Quantification of axon density and lengths for retinal explants from control and sfpq-targeting MO-treated optic nerves. *p < 0.05, compared with control MO. **p < 0.01, compared with control MO. n = 4. Error bars indicate SD. ONL, Outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer.
We used in situ hybridization assays to examine the spatial expression of sfpq in the uninjured and optic nerve-lesioned retina. sfpq RNA was undetectable in the uninjured retina and specifically induced in RGCs after optic nerve lesion (Fig. 1B). We wondered whether the rapid injury-dependent induction of sfpq gene expression may simply reflect a response to optic nerve injury and not necessarily be related to optic nerve regeneration. Therefore, we used qPCR to investigate whether this gene was induced in the mouse retina, where injured optic nerves do not regenerate. Although we detected basal expression of Sfpq mRNA, we found no evidence for injury-dependent induction of Sfpq gene expression in mice (Fig. 1C).
To investigate whether Sfpq expression had a consequence on optic axon regeneration, we knocked down its expression in RGCs using morpholino-modified antisense oligonucleotides (MOs) applied to the lesioned optic nerve stump as previously described (Veldman et al., 2007, 2010). One of the sfpq-targeting MOs was previously validated in zebrafish (Lowery et al., 2007); we tested the efficacy of the other using a chimeric sfpq-gfp transgene expressed in zebrafish embryos coinjected with either a control or sfpq-targeting MO (Fig. 1D). This analysis showed that almost all of the control MO-injected embryos expressed GFP (72 of 75), whereas none of the sfpq-targeting MO-injected embryos expressed GFP (0 of 80). Delivery of MOs to adult RGCs did not stimulate cell death (TUNEL assay) or proliferation (BrdU incorporation); however, after explanting retinas to tissue culture dishes, we noted that the sfpq-targeting MO increased the density and length of axons growing from them (Fig. 1E,F). Similar results were obtained with a second sfpq-targeting MO, suggesting the effect on axon regrowth is specific to Sfpq knockdown. These data suggested that Sfpq may attenuate optic axon regeneration.
We were surprised to find an attenuator of axonal regeneration induced in zebrafish RGCs after optic nerve injury and worried that this effect was specific to retinal explants grown in tissue culture. Therefore, we investigated whether Sfpq also attenuated optic axon regeneration in vivo. For these experiments, we took advantage of tuba1a:gfp transgenic fish that specifically express GFP in RGCs that are regrowing their damaged axons (Goldman and Ding, 2000; Goldman et al., 2001; Senut et al., 2004; Veldman et al., 2010). We lesioned the optic nerve of tuba1a:gfp fish, just behind the orbit, and applied either a control or sfpq-targeting MO to the optic nerve stump (Fig. 2A). Seven days later, the optic nerve was relesioned ∼3 mm distal to its original cut site and RGCs whose axons had regenerated this distance were identified by applying rhodamine-dextran–impregnated Gelfoam to the newly lesioned optic nerve stump (Fig. 2A). Whole-mount retinas were examined under a fluorescent microscope to visualize and quantify the number of GFP+ and rhodamine+ RGCs (Fig. 2B). Sfpq knockdown increased the number of RGCs with detectable GFP (Fig. 2B,C). Because induction of the tuba1a:gfp transgene is highly correlated with regenerating axons (Goldman and Ding, 2000; Goldman et al., 2001; Senut et al., 2004; Veldman et al., 2010), this increased GFP expression suggested the induction of a regeneration-associated gene expression program. Sfpq knockdown also increased the number of RGC cell bodies back-labeled with rhodamine+ (Fig. 2B,D), which suggested an increased rate of growth for regenerating axons.
The retinal explant and back-labeling experiments described above only measured regeneration over relatively short distances. To investigate whether the consequences of Sfpq knockdown persisted and affected optic axon regeneration into the brain, we used an anterograde labeling protocol (Fig. 2E). For these experiments, optic nerves were bilaterally lesioned with one optic nerve stump receiving a control MO and the other optic nerve stump receiving a sfpq-targeting MO. At 12 and 29 dpi, rhodamine dextran-saturated Gelfoam was inserted behind the lens into the vitreous, and 6 h later fish were killed, and eyes, with attached optic nerve and brain, were dissected and visualized under a fluorescent microscope (Fig. 2F). At 12 dpi, optic axons from Sfpq MO-treated RGCs, but not control MO-treated RGCs, were readily observed past the optic chiasm and heading to the tectum (Fig. 2F). By 29 dpi, we were unable to discern a significant difference in growth of optic axons from control and Sfpq MO-treated RGCs (Fig. 2F). The extent of regeneration in controls is consistent with previously published data using anterograde HRP tracing methods (Kaneda et al., 2008). Quantification revealed that, at 12 dpi, axons extended more than twofold further beyond the optic chiasm than control MO-treated RGC axons (Fig. 2G). This difference in growth just beyond the optic chiasm was no longer detectable by 29 dpi (Fig. 2F,H). These data suggest that Sfpq inhibits optic axon regeneration in vivo.
Sfpq knockdown enhances visual recovery and regeneration-associated gene expression after optic nerve lesion
The above data suggested that Sfpq knockdown enhanced optic nerve regeneration by increasing the number of RGCs activating regeneration-associated genes, like the tuba1a:gfp transgene, and also stimulated axonal growth as indicated by RGC back-labeling and anterograde labeling protocols (Fig. 2). We were curious whether this enhanced regeneration resulted in a more rapid restoration of vision. To investigate visual recovery, we monitored the optokinetic response of a restrained fish as it tracked alternating black and white stripes on a rotating wall (Fig. 3A). In addition, we monitored the escape response of a free swimming fish when presented with a phantom predator (large black bar on a rotating wall) (Fig. 3C). For these experiments, we compared sham operated animals with those whose optic nerves were bilaterally lesioned and treated with either control or sfpq-targeting MO. At 1 dpi, fish were essentially blind because no axons have regrown to tectal targets; therefore, little optokinetic response was detected (Fig. 3B). However, within 5 dpi, we were able to detect a remarkable enhancement of visual recovery after Sfpq knockdown (Fig. 3B). A similar analysis was done quantifying the escape response to a phantom predator, which also indicated that Sfpq knockdown enhanced visual recovery after optic nerve lesion (Fig. 3D). Thus, the enhanced axonal regeneration noted after Sfpq knockdown is reflected by a more rapid recovery of vision.
Figure 3.
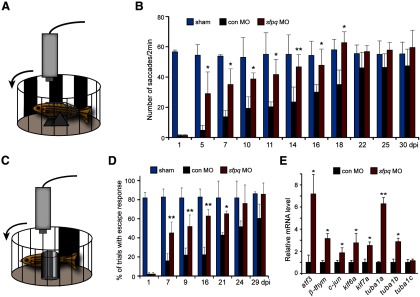
Sfpq knockdown enhances visual recovery and regeneration-associated gene expression after optic nerve lesion. A, Diagram of optokinetic apparatus. Fish received bilateral optic nerve lesions and control or sfpq MO was applied to both the left and right optic nerve stumps overnight. B, Quantification of eye saccades tracking visual stimulus in sham, control, or sfpq MO-treated fish. *p < 0.05, compared with control MO. **p < 0.01, compared with control MO. n = 6. Error bars indicate SD. C, Diagram of the apparatus used to measure the escape response to a phantom prey. D, Quantification of escape response. *p < 0.05, compared with control MO. **p < 0.01, compared with control MO. n = 4. Error bars indicate SD. E, qPCR analysis of the effects of control or sfpq MO on expression of genes associated with regeneration. *p < 0.05, compared with control MO. **p < 0.01, compared with control MO. n = 3. Error bars indicate SD.
The increased expression of the tuba1a:gfp transgene and the enhanced regrowth of optic axons after Sfpq knockdown and optic nerve injury suggested that Sfpq may suppress gene expression programs that regulate axonal regrowth in adult RGCs. To investigate this possibility, we lesioned the optic nerve and applied either a control or sfpq-targeting MO to the optic nerve stump overnight. At 3 dpi, RNA was purified from retinas and the expression of regeneration-associated genes (Goldman et al., 2001; Senut et al., 2004; Veldman et al., 2007, 2010; Saul et al., 2010) was assayed by qPCR. This analysis showed increased expression of regeneration-associated genes after Sfpq knockdown (Fig. 3E). Control RNAs, such as tuba1c and gapdh, are not regulated by optic nerve injury and remained unaffected by Sfpq knockdown. The promiscuous induction of regeneration-associated genes after Sfpq knockdown suggested that it may impinge on one or more signaling cascades that play a major role in reprogramming RGCs to a growth-permissive state after optic nerve injury.
Socs3a knockdown enhances optic nerve regeneration and recovery of vision
Our finding that Sfpq attenuates optic nerve regeneration in zebrafish was reminiscent of previous reports indicating that SOCS3 inhibits RGC axon regrowth after optic nerve injury in mammals (Park et al., 2009; Smith et al., 2009). There are two socs3 genes in zebrafish, socs3a and socs3b, both of which are induced after optic nerve injury (Veldman et al., 2007). We previously reported that knockdown of these genes had little effect on RGC axonal regeneration in retinal explants (Veldman et al., 2007). The different outcomes of Socs3 knockdown in zebrafish and mammals were surprising and prompted us to reinvestigate Socs3 knockdown in zebrafish. We realized that our earlier analysis of Socs3 knockdown was designed to detect growth inhibition and not growth enhancement because the control axon lengths were in the top bin for axon growth (Veldman et al., 2007, 2010). Therefore, we repeated these studies and quantified axon lengths without binning. This analysis revealed that in vivo knockdown of Socs3a enhanced RGC axonal regeneration in retinal explants, whereas knockdown of Socs3b had little effect (Fig. 4A,B). Socs3 MOs did not stimulate RGC death (TUNEL assay) or proliferation (BrdU incorporation).
Encouraged by the above result, we next investigated the consequence of Socs3a knockdown on optic nerve regeneration in vivo using the back-labeling protocol illustrated in Figure 2A, except that fluorescein dextran was used to back-label cells instead of rhodamine dextran. Whole-mount retinas were examined under a fluorescent microscope to visualize and quantify the number of back-labeled fluorescein+ RGCs. Similar to Sfpq knockdown (Fig. 2B,D), and consistent with our retinal explant studies (Fig. 4A,B), we found that MO-mediated Socs3a knockdown increased the number of fluorescein+ RGCs (Fig. 4C,D), suggesting that Soc3a knockdown stimulates optic nerve regeneration in zebrafish. TUNEL and BrdU labeling indicated no effect of Socs3 MOs on RGC death or proliferation.
We used the fish's optokinetic and escape response to investigate whether this enhanced optic nerve regeneration after Socs3a knockdown correlated with recovery of vision. These studies were performed as previously described for Sfpq knockdown (Fig. 3A,C). At 1 dpi, when fish are blind, we observed essentially no response to the stimulus; however, at later times, we found that the optokinetic and escape responses returned much faster in fish with Socs3a knockdown (Fig. 4E,F). The observation that two different previously validated socs3a-targeting MOs have similar effects on axon regrowth and visual recovery suggests these effects are specific. Together, our data indicate that Socs3a and Sfpq inhibit optic nerve regeneration and that this inhibition has a consequence on the rate of vision recovery.
Knockdown of Socs3a or Sfpq impacts the other's expression
Because our studies suggested that both Sfpq and Socs3a attenuate optic nerve regeneration, we wondered whether they acted in a common pathway. Consistent with this idea, qPCR suggested that socs3a gene expression was rapidly induced after optic nerve injury (Fig. 5A) and this induction was similar to that of sfpq (Figs. 1A and 5B). To investigate whether Socs3a and Sfpq exhibited a hierarchical relationship, we lesioned the optic nerve and applied either a socs3a- or sfpq-targeting MO to the optic nerve stump and assayed sfpq and socs3a injury-dependent induction, respectively, at 3 dpi. Interestingly, Sfpq knockdown partially suppressed injury-dependent socs3a mRNA induction (Fig. 5C), whereas Socs3a knockdown more severely inhibited injury-dependent sfpq mRNA induction (Fig. 5D). Although the mechanism underlying this Sfpq/Socs3a crosstalk remains unknown, we found that, like Sfpq (Fig. 3E), Socs3a also suppressed the expression of regeneration-associated genes (Fig. 5E). These studies indicate that Socs3a and Sfpq are immediate early genes, coordinately induced by optic nerve injury and capable of regulating each other's expression. Whether this regulation is direct or indirect remains to be determined. Nonetheless, their rapid induction and ability to inhibit gene expression programs driving optic nerve regeneration likely account for their effect on optic nerve regeneration.
Figure 5.
socs3a and sfpq are coordinately induced and regulate each other's expression. A, qPCR analysis of socs3a mRNA induction after optic nerve lesion. RNA levels are normalized to that found in uninjured control. **p < 0.01, compared with uninjured control. ***p < 0.001, compared with uninjured control. n = 4. Error bars indicate SD. B, qPCR analysis of socs3a and sfpq mRNA induction shortly after optic nerve lesion. *p < 0.05, compared with uninjured control. **p < 0.01, compared with uninjured control. ***p < 0.001, compared with uninjured control. n = 4. Error bars indicate SD. C, qPCR shows Sfpq knockdown suppresses socs3a mRNA expression at 3 dpi. *p < 0.05, compared with control MO. ***p < 0.001, compared with control MO. n = 3. Error bars indicate SD. D, qPCR shows Socs3a knockdown suppresses sfpq mRNA expression at 3 dpi. *p < 0.05, compared with control MO. **p < 0.01, compared with control MO. n = 3. Error bars indicate SD. E, qPCR shows that Socs3a knockdown stimulates expression of regeneration-associated genes at 3 dpi. mRNA levels are normalized to those obtained with control MO-treated optic nerves. *p < 0.05, compared with control MO. **p < 0.01, compared with control MO. n = 3. Error bars indicate SD.
Jak/Stat signaling is necessary for optic nerve regeneration
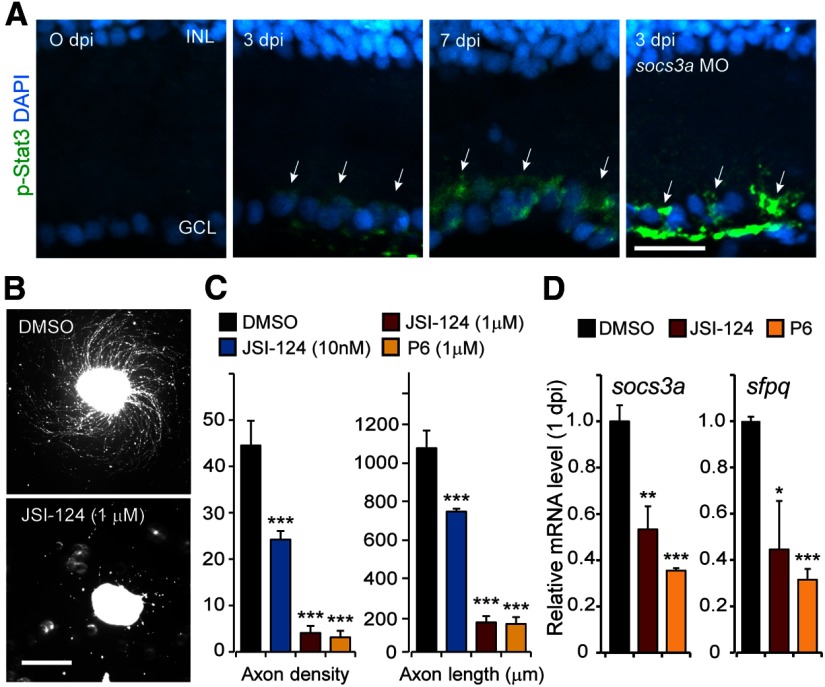
Jak/Stat signaling stimulates optic axon regeneration in mammals (Park et al., 2009; Smith et al., 2009; Sun et al., 2011; Pernet et al., 2013a), and socs3 gene expression often reflects Jak/Stat signaling activity (Babon and Nicola, 2012). Therefore, we investigated whether this signaling pathway was activated after optic nerve injury. We used an anti-phospho-Stat3 (p-Stat3) antibody that detects zebrafish Stat3 phosphorylated on Tyr708 (Tyr705 in mammals) to report Jak/Stat signaling (Yamashita et al., 2002). Immunofluorescence on retinal sections from control and optic nerve-lesioned fish indicated that p-Stat3 is first detectable in RGCs ∼3 dpi (Fig. 6A). We suspect that Stat3 activation may occur at earlier times (in part, because socs3a is induced within 1 hpi; Fig. 5A,B), but p-Stat3 levels may be below the limits of detection. Furthermore, because Socs3a is an inhibitor of Jak/Stat signaling, we suspected that injury-dependent induction of Socs3a may be contributing to the delayed detection of p-Stat3. To test this idea, we used socs3a-targeting MOs to knock down Socs3a protein expression after optic nerve lesion and assayed p-Stat3 at 3 dpi. Indeed, Socs3a knockdown dramatically enhanced p-Stat3 expression (Fig. 6A). Although we detected low levels of p-Stat3 in the nucleus where it can act on gene expression, p-Stat3 was most readily detected in the cytoplasm of RGCs after optic nerve lesion, perhaps suggesting roles for this protein beyond transcriptional regulation.
Figure 6.
Optic axon regeneration is mediated by a Jak/Stat3 signaling cascade. A, Immunofluorescence detection of p-Stat3 (green) in RGCs (arrows) during optic nerve regeneration is enhanced by Socs3a knockdown. Nuclei are stained with DAPI (blue). Scale bar, 20 μm. B, Axon outgrowth from retinal explants is inhibited by the Jak/Stat signaling inhibitor, JSI 124. Scale bar, 500 μm. C, Quantification of axon density and length for control, JSI-124-, and P6-treated retinal explants. ***p < 0.001, compared with the DMSO-treated controls. n = 4. Error bars indicate SD. D, Jak/Stat inhibitors JSI-124 and P6 inhibit injury-dependent induction of socs3a and sfpq mRNAs. *p < 0.05, compared with DMSO controls. **p < 0.01, compared with DMSO controls. ***p < 0.001, compared with DMSO controls. n = 4. Error bars indicate SD. INL, Inner nuclear layer; GCL, ganglion cell layer.
The above studies suggested that optic nerve lesion stimulated Jak/Stat signaling in RGCs but did not reveal whether injury-dependent induction of Jak/Stat signaling was necessary for optic axon regeneration. Therefore, we investigated whether the Jak inhibitors, JSI-124 and P6, affected RGC axon outgrowth in retinal explants. JSI-124 preferentially inhibits Jak2/Stat3 signaling, whereas P6 is a more general Jak inhibitor (Blaskovich et al., 2003; Pedranzini et al., 2006). Consistent with the idea that Jak/Stat signaling is necessary for optic axon regeneration, JSI-124 and P6 inhibited RGC axon outgrowth from retinal explants (Fig. 6B,C).
Because sfpq and socs3a gene expression are among the first genes induced after optic nerve injury, we were curious whether they were regulated in a Jak/Stat-dependent manner. We reasoned that the expression of these genes may be a more sensitive readout of Jak/Stat signaling than p-Stat3 expression. Therefore, we intravitreally injected eyes with either JSI-124 or P6 just before optic nerve lesion and 24 h later assayed sfpq and socs3a gene expression. This analysis showed that Jak/Stat signaling was active within 1 dpi and that it stimulated the expression of injury-responsive genes (Fig. 6D). Together, these data suggest an important role for Jak/Stat signaling in stimulating optic nerve regeneration and also activating gene expression programs that attenuate this regeneration.
Gp130-coupled receptors are necessary for enhanced Jak/Stat signaling during optic nerve regeneration
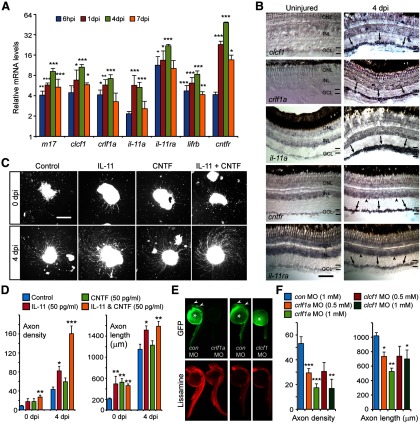
The above studies suggested that injury-dependent activation of Jak/Stat signaling was a key element underlying optic nerve regeneration in zebrafish. We suspected that Gp130-coupled receptors may mediate injury-dependent Jak/Stat activation because IL-6-like cytokines, like CNTF, that act through Gp130-coupled receptors and can stimulate RGC axon regrowth in mammals (Leaver et al., 2006; Pernet et al., 2013b). Therefore, we investigated whether we could detect gp130 gene expression in RGCs during optic nerve regeneration. qPCR and in situ hybridization assays showed that gp130 mRNA was very low in uninjured RGCs and rapidly induced after optic nerve injury (Fig. 7A,B).
We next investigated whether signaling via Gp130 was necessary for optic axon regeneration. For these studies, we assayed axon growth and density in retinal explants whose optic nerves were treated in vivo with a control or a gp130-targeting MO. Gp130 knockdown suppressed the total number of regenerating axons and reduced their length in a concentration-dependent manner (Fig. 7C,D). To confirm that signaling via Gp130-coupled receptors regulated optic nerve regeneration in vivo, we investigated the effect Gp130 knockdown would have on optic nerve regeneration using our back-labeling protocol described earlier (Fig. 2A). This analysis showed that Gp130 knockdown dramatically reduced the number of back-labeled RGCs (Fig. 7E,F) and suggests that Gp130 knockdown inhibits optic nerve regeneration.
Gp130-coupled receptors link extracellular cytokines to Jak/Stat signaling and gene activation. To determine whether Gp130 participates in Stat3 activation after optic nerve injury, we assayed p-Stat3 immunofluorescence in retinas from control and Gp130 MO-treated optic nerves. We observed a dramatic reduction in p-Stat3 expression after Gp130 knockdown (Fig. 7G). Furthermore, this Gp130-dependent regulation of Stat3 activity suppressed the expression of regeneration-associated genes (Fig. 7H). Therefore, Gp130-coupled cytokine receptors appear to be largely responsible for activating Jak/Stat3 signaling in RGCs after optic nerve injury, and this activation is necessary for the activation of gene expression programs driving optic nerve regeneration.
Cytokines acting via Gp130-coupled receptors regulate optic axon regeneration
The IL-6 family of cytokines signal through Gp130-coupled receptors (Hirano et al., 2000) and the IL-6 family member CNTF has been shown to stimulate RGC axon regrowth in mammals (Müller et al., 2007; Pernet et al., 2013b). Therefore, we suspected that IL-6 family members would contribute to optic nerve regeneration in zebrafish. Although a cntf gene has not been identified in zebrafish, we found that a variety of IL-6-like family member genes were induced in RGCs shortly after optic nerve lesion (Fig. 8A,B). These included genes encoding the alternative Cntf receptor composite ligand, Clcf1/Crlf1a (Elson et al., 2000), as well as those encoding M17 (Lif) and IL-11a. Furthermore, genes encoding the receptors for these ligands (cntfr, lifrb, and il-11ra) were also induced after optic nerve injury (Fig. 8A,B). The injury-dependent induction of IL-6 family members and their receptors in RGCs suggests an autocrine/paracrine mechanism of action.
Figure 8.
IL-6 family cytokines are induced in RGCs and stimulate optic axon regeneration. A, qPCR identifies candidate IL-6 family cytokines and receptors that are induced after optic nerve lesion. *p < 0.05, compared with uninjured control. **p < 0.01, compared with uninjured control. ***p < 0.001, compared with uninjured control. n = 3. Error bars indicate SD. B, In situ hybridization assays shows the spatial expression of clcf1, crlf1a, il-11a, cntfr, and il-11ra in uninjured retina and at 4 dpi (arrows point to positive signal in GCL and arrowheads point to positive signal in INL). Scale bar, 50 μm. C, Retinal explants were prepared from fish with (4 dpi) and without (0 dpi) a preconditioning optic nerve lesion. Recombinant IL-11 and/or CNTF were added as soon as retinas were placed in culture. Scale bar, 500 μm. D, Quantification of axon density and length for explants shown in C. *p < 0.05, compared with control. **p < 0.01, compared with control. n = 4. Error bars indicate SD. E, A chimeric sCMV:crlf1a-gfp transgene or a chimeric sCMV:clcf1-gfp transgene was injected into single-cell zebrafish embryos with a lissamine-labeled (red) control or experimental MO, and transgene GFP expression assayed by fluorescence microscopy 24 h later. Shown are representative embyros. Arrowheads point to GFP expression (for quantification, see Results). Asterisk indicates autofluorescence in yolk. F, Quantification of axon density and length for explants whose lesioned optic nerve was treated with control, Crlf1a, or Clcf1 MOs in vivo. *p < 0.05, compared with control. **p < 0.01, compared with control. ***p < 0.001, compared with control. n = 4. Error bars indicate SD. ONL, Outer nuclear layer; INL, inner nuclear layer; GCL ganglion cell layer.
To test whether IL-6 family members could stimulate optic axon regeneration, we treated retinal explants with recombinant mammalian IL-11 or CNTF. Explants were prepared either with or without a preconditioning optic nerve injury. Retinal explants prepared without a preconditioning optic nerve injury normally exhibit a meager regenerative response (Fig. 8C,D) (Landreth and Agranoff, 1979). However, regardless of how explants were prepared, treatment with IL-11 alone or in combination with CNTF stimulated axonal regrowth (Fig. 8C,D). Even preconditioned explants, which normally exhibit robust optic axon outgrowth, extended more and longer axons after treatment with IL-11 alone or IL-11 and CNTF in combination (Fig. 8C,D). Interestingly, CNTF had a more robust effect on axonal growth from explants prepared without a preconditioning nerve lesion than it did on explants with a preconditioned nerve injury (Fig. 8D). We also found that IL-11 and CNTF had an additive effect on axon density, but not axon length. Although the reason for this is not known, it may reflect the genes upon which they act.
Although zebrafish do not appear to harbor a Cntf encoding gene, we were intrigued by the observation that CNTF could stimulate optic axon regeneration in retinal explants and that the zebrafish cntfr gene is induced in RGCs after optic nerve injury. These data suggested to us that the alternative Cntfr composite ligand, Clcf1/Crlf1a, may contribute to optic axon regeneration in fish. Indeed, clcf1 and crlf1a gene expression is highly induced in RGCs after optic nerve injury (Fig. 8A,B). To test whether Clcf1/Crlf1a regulated optic axon regeneration, we applied control, clcf1-, or crlf1a-targeting MOs to lesioned optic nerves in vivo and assayed optic axon regrowth in retinal explants. We confirmed the efficacy of these MOs using a chimeric clcf1-gfp or crlf1a-gfp transgene expressed in zebrafish embryos coinjected with either control or experimental MO (Fig. 8E). This analysis showed that almost all of the control MO-injected embryos expressed GFP (65 of 67 for Crlf1a MO and 79 of 81 for Clcf1 MO), whereas none of the experimental MO-injected embryos expressed GFP (0 of 63 for Crlf1a MO and 0 of 90 for Clcf1 MO). We also tested whether MOs affected RGC survival or proliferation in adult retinas using TUNEL assay and BrdU immunofluorescence. No cell death or proliferation was detected in control Clcf1 or Crlf1a knockdown retinas. Consistent with the idea that the composite Cntf receptor ligand Clcf1/Crlf1a contributes to optic nerve regeneration in zebrafish, we found that knockdown of either Clcf1 or Crlf1a suppressed optic axon regrowth in retinal explants (Fig. 8F). Notable is the preferential effect Clcf1/Crlf1a knockdown had on axon density (Fig. 8F), which was also reflected in CNTF-treated explants (Fig. 8D). Because both these ligands act via the Cntf receptor, its activation may be regulating the number of responding RGCs by increasing their reprogramming to a growth-permissive state. Together, the above studies show that optic nerve injury stimulates the expression of IL-6 family cytokines and receptors in RGCs and identify IL-6 family members that are sufficient to stimulate optic axon regeneration in retinal explants. These studies suggest that IL-6 family members act in an autocrine/paracrine fashion to stimulate optic nerve regeneration in vivo.
Discussion
The studies reported here identify a Jak/Stat signaling cascade that regulates optic nerve regeneration in zebrafish (Fig. 9). We show that retinal injury stimulates the activation of this signaling cascade by inducing RGC expression of IL-6 family cytokines, such as IL-11 and Clcf1/Crlf1a, which act via Gp130-coupled receptors. This signaling cascade stimulates optic nerve regeneration by inducing expression of regeneration-associated genes, such as klf6, klf7, and tuba1a. However, it also activates the expression of genes encoding Sfpq and Socs3a that attenuate optic nerve regeneration. Thus, unlike mammals where basal Socs3 expression is sufficient to inhibit optic nerve regeneration, zebrafish activate an inhibitory pathway during optic nerve regeneration and still retain a robust regenerative response.
Figure 9.
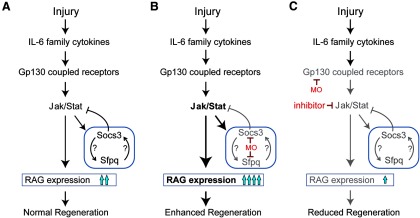
Model showing regulation of optic nerve regeneration by a cytokine/Jak/Stat signaling cascade in zebrafish. A, Cytokines normally act via Jak/Stat signaling to stimulate optic nerve regeneration by activating expression of regeneration-associated genes (RAG) and also by stimulating inhibitors of regeneration, such as Socs3a and Sfpq, that feedback to attenuate Jak/Stat signaling. B, Effect of MOs targeting Socs3a or Sfpq. Bigger arrows and bold lettering indicate superactivation. C, Effect of MOs targeting Gp130 or inhibitors of Jak/Stat signaling. Question marks indicate that the mechanism by which Socs3a and Sfpq regulate each other's expression is not understood and may be through direct or indirect pathways. Gray arrows and lettering indicate reduced activation of these components compared with the normal situation.
Jak/Stat signaling appears to be a major determinant of optic nerve regeneration in fish, and its lack of activation in mammals contributes to regenerative failure. Although this pathway and optic nerve regeneration can be activated in mammals with CNTF or constitutively active Stat3, the noted increase in regeneration is still incomplete and misguided (Leaver et al., 2006; Leibinger et al., 2013; Pernet et al., 2013a, 2013b). It is interesting that, during zebrafish optic nerve regeneration, a variety of IL-6 family cytokines are induced in RGCs, and it is tempting to speculate that this variety imparts not only robust Jak/Stat signaling but also unique characteristics on RGCs so a more robust and accurate regenerative response ensues. Thus, by activating multiple receptors, one might couple cytokine signaling to additional signal transduction cascades and/or recruit a variety of Stat proteins that target different gene expression programs. Importantly, IL-11 and Clcf1/Crlf1a cytokines, which are highly induced in fish RGCs and stimulate optic nerve regeneration, have never been tested in mammals. Their action alone or in combination with other cytokines may further enhance mammalian optic nerve regeneration.
The activation of gene expression programs underlying optic nerve regeneration appears to be mediated by Jak/Stat signaling in zebrafish RGCs. We previously reported that injury-dependent induction of klf6, klf7, and tuba1a gene expression is necessary for zebrafish optic nerve regeneration (Veldman et al., 2007, 2010), and here we report that these genes are regulated in a Jak/Stat-dependent manner. Interestingly, Klf6 and Klf7 gene expression in mammals also stimulates optic axon regeneration, but these genes are suppressed as RGCs mature and other genes, such as Klf4, Socs3, and Pten that inhibit optic nerve regeneration, are expressed (Moore et al., 2009; Smith et al., 2009; Sun et al., 2011), thus developing RGCs transition from a growth-permissive stage to one that is growth-restrictive, and this is reflected in the underlying genetic programs regulating axonal growth (Chen et al., 1995; Moore et al., 2009; Smith et al., 2009; Sun et al., 2011). This has led to the idea that optic nerve regeneration will require the activation of growth-promoting and the suppression of growth-inhibiting gene expression programs in adult RGCs, and recent studies support this contention (Kurimoto et al., 2010; de Lima et al., 2012). However, our studies in zebrafish indicate that, even in the face of a highly induced growth inhibitory gene expression program, regeneration can ensue. Although the details of how this occurs remain incompletely understood, robust activation of Jak/Stat signaling by multiple cytokines appears to play an important role.
We used p-Stat3 immunofluorescence to report Jak/Stat signaling in the injured retina. We think this immunofluorescence accurately reflects Jak/Stat signaling because knockdown of regulators of Jak/Stat signaling, such as Socs3 and Gp130, predictably affected p-Stat3 expression. We were surprised to find relatively low levels of p-Stat3 accumulation in RGC nuclei compared with the RGC cytoplasm. Although this low-level nuclear accumulation may be sufficient for gene activation, the cytoplasmic accumulation is intriguing. Cytoplasmic Stat3 has been reported to inhibit autophagy in mammalian cells (Shen et al., 2012), and enhanced autophagy may underlie RGC death after optic nerve injury (Russo et al., 2013). However, we were unable to detect increased RGC death after Gp130 knockdown, which reduced p-Stat3 expression. Thus, the functional significance of cytoplasmic p-Stat3 remains unknown.
We were quite surprised to find that Jak/Stat signaling not only stimulated optic nerve regeneration in fish but that it also stimulated the expression of decelerators of regeneration, such as Socs3a and Sfpq. We anticipated that in an animal, such as zebrafish, where regeneration was robust, these regenerative decelerators would be repressed. Sfpq is a multifunctional protein whose activity contributes to a diverse array of processes, such as RNA splicing, DNA synthesis, gene expression, DNA repair, and cell survival (Shav-Tal and Zipori, 2002). Sfpq is enriched in paraspeckles, which are RNA-protein structures found in the interchromatin space in the nucleus and may contribute to the regulation of gene expression (Fox and Lamond, 2010). During zebrafish development, Sfpq is necessary for neuronal survival and differentiation (Lowery et al., 2007). Our results suggest that, in the adult zebrafish retina, Sfpq inhibits optic nerve regeneration. This is a new function for Sfpq and was reminiscent of that reported for Socs3 deletion in mice (Smith et al., 2009). We also found Sfpq mRNA expressed in the mouse retina and suspect that it may collaborate with Socs3 in inhibiting optic nerve regeneration in mammals. Interestingly, we found that Socs3a and Sfpq regulate each other's expression, which may explain their shared ability to inhibit optic nerve regeneration in fish. Although these proteins regulate each other's expression, the mechanism underlying this effect remains unknown. There are no reports of Socs3 and Sfpq direct interactions or effects on each other's genes. However, both Sfpq and Socs3 have been reported to interact with the paraspeckle protein, NonO, which can regulate RNA expression (Song et al., 2008; Fox and Lamond, 2010). Furthermore, Sfpq has been shown to stimulate Jak/Stat signaling in Drosophila (Müller et al., 2005), suggesting a mechanism of action in zebrafish.
The finding that zebrafish induce the expression of inhibitors of optic nerve regeneration, like Sfpq and Socs3a, is intriguing and may reflect a mechanism by which Jak/Stat signaling is attenuated during optic nerve regeneration. Even though we could not detect p-Stat before 3 dpi, we suspect that Jak/Stat signaling is activated at a low level because inhibitors of this pathway suppressed injury-dependent induction of sfpq and socs3a at 1 dpi. This attenuated Jak/Stat signaling at early times after injury appears to be controlled by Sfpq/Soc3a signaling and may restrict gene activation programs to those that are most sensitive to this signaling pathway and needed at the earliest stages of RGC reprogramming. At later times when Jak/Stat signaling overcomes Sfpq/Socs3a attenuation, additional gene expression programs may be activated that impact later events. This timing may be ideal for matching gene expression with the changing cellular demands and environmental cues that occur over the time course of regeneration. Although we found an increased rate of optic nerve regeneration when this inhibitory system was knocked down, we do not know whether this was accompanied by an increase in errors of pathfinding or retinal-tectal mapping, which the visually mediated behaviors assayed in this study may accommodate.
In conclusion, the work reported here reveals a remarkable similarity in the signaling mechanisms underlying optic nerve regeneration in fish and mammals and suggests that zebrafish are a suitable model for uncovering mechanisms for enhancing regeneration in mammals. Our studies suggest that robust activation of Jak/Stat signaling is necessary for optic nerve regeneration. We have identified IL-11 and Clcf1/Crlf1a cytokines that regulate optic nerve regeneration in fish and may also enhance optic nerve regeneration in mammals. Furthermore, we discovered a new inhibitor of optic nerve regeneration, Sfpq, whose neutralization in mammals may facilitate regeneration.
Footnotes
This work was supported by Grant G2010013 from the American Health Assistance Foundation-National Glaucoma Research (BrightFocus Foundation) and NEI Grant RO1 EY018132. We thank Hazel Sive (Massachusetts Institute of Technology) for providing one of the sfpq MOs; members of the D.G. laboratory for comments and advice on these studies; and Randall Karr for fish care.
The authors declare no competing financial interests.
References
- Babon JJ, Nicola NA. The biology and mechanism of action of suppressor of cytokine signaling 3. Growth Factors. 2012;30:207–219. doi: 10.3109/08977194.2012.687375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel LK, Raymond PA. In situ hybridization studies of retinal neurons. Methods Enzymol. 2000;316:579–590. doi: 10.1016/S0076-6879(00)16751-5. [DOI] [PubMed] [Google Scholar]
- Becker CG, Meyer RL, Becker T. Gradients of ephrin-A2 and ephrin-A5b mRNA during retinotopic regeneration of the optic projection in adult zebrafish. J Comp Neurol. 2000;427:469–483. doi: 10.1002/1096-9861(20001120)427:3<469::AID-CNE12>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Becker CG, Lieberoth BC, Morellini F, Feldner J, Becker T, Schachner M. L1.1 is involved in spinal cord regeneration in adult zebrafish. J Neurosci. 2004;24:7837–7842. doi: 10.1523/JNEUROSCI.2420-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt R. Axonal pathfinding during the regeneration of the goldfish optic pathway. J Comp Neurol. 1989;284:119–134. doi: 10.1002/cne.902840109. [DOI] [PubMed] [Google Scholar]
- Bernhardt RR. Cellular and molecular bases of axonal regeneration in the fish central nervous system. Exp Neurol. 1999;157:223–240. doi: 10.1006/exnr.1999.7059. [DOI] [PubMed] [Google Scholar]
- Bernhardt RR, Tongiorgi E, Anzini P, Schachner M. Increased expression of specific recognition molecules by retinal ganglion cells and by optic pathway glia accompanies the successful regeneration of retinal axons in adult zebrafish. J Comp Neurol. 1996;376:253–264. doi: 10.1002/(SICI)1096-9861(19961209)376:2<253::AID-CNE7>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Blaskovich MA, Sun J, Cantor A, Turkson J, Jove R, Sebti SM. Discovery of JSI-124 (cucurbitacin I), a selective Janus kinase/signal transducer and activator of transcription 3 signaling pathway inhibitor with potent antitumor activity against human and murine cancer cells in mice. Cancer Res. 2003;63:1270–1279. [PubMed] [Google Scholar]
- Chen DF, Jhaveri S, Schneider GE. Intrinsic changes in developing retinal neurons result in regenerative failure of their axons. Proc Natl Acad Sci U S A. 1995;92:7287–7291. doi: 10.1073/pnas.92.16.7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lima S, Koriyama Y, Kurimoto T, Oliveira JT, Yin Y, Li Y, Gilbert HY, Fagiolini M, Martinez AM, Benowitz L. Full-length axon regeneration in the adult mouse optic nerve and partial recovery of simple visual behaviors. Proc Natl Acad Sci U S A. 2012;109:9149–9154. doi: 10.1073/pnas.1119449109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson GC, Lelièvre E, Guillet C, Chevalier S, Plun-Favreau H, Froger J, Suard I, de Coignac AB, Delneste Y, Bonnefoy JY, Gauchat JF, Gascan H. CLF associates with CLC to form a functional heteromeric ligand for the CNTF receptor complex. Nat Neurosci. 2000;3:867–872. doi: 10.1038/78765. [DOI] [PubMed] [Google Scholar]
- Fischer D, Leibinger M. Promoting optic nerve regeneration. Prog Retin Eye Res. 2012;31:688–701. doi: 10.1016/j.preteyeres.2012.06.005. [DOI] [PubMed] [Google Scholar]
- Fox AH, Lamond AI. Paraspeckles. Cold Spring Harb Perspect Biol. 2010;2:a000687. doi: 10.1101/cshperspect.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D, Ding J. Different regulatory elements are necessary for alpha1 tubulin induction during CNS development and regeneration. Neuroreport. 2000;11:3859–3863. doi: 10.1097/00001756-200011270-00051. [DOI] [PubMed] [Google Scholar]
- Goldman D, Hankin M, Li Z, Dai X, Ding J. Transgenic zebrafish for studying nervous system development and regeneration. Transgenic Res. 2001;10:21–33. doi: 10.1023/A:1008998832552. [DOI] [PubMed] [Google Scholar]
- Hirano T, Ishihara K, Hibi M. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene. 2000;19:2548–2556. doi: 10.1038/sj.onc.1203551. [DOI] [PubMed] [Google Scholar]
- Kaneda M, Nagashima M, Nunome T, Muramatsu T, Yamada Y, Kubo M, Muramoto K, Matsukawa T, Koriyama Y, Sugitani K, Vachkov IH, Mawatari K, Kato S. Changes of phospho-growth-associated protein 43 (phospho-GAP43) in the zebrafish retina after optic nerve injury: a long-term observation. Neurosci Res. 2008;61:281–288. doi: 10.1016/j.neures.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Koriyama Y, Homma K, Sugitani K, Higuchi Y, Matsukawa T, Murayama D, Kato S. Upregulation of IGF-I in the goldfish retinal ganglion cells during the early stage of optic nerve regeneration. Neurochem Int. 2007;50:749–756. doi: 10.1016/j.neuint.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Koriyama Y, Yasuda R, Homma K, Mawatari K, Nagashima M, Sugitani K, Matsukawa T, Kato S. Nitric oxide-cGMP signaling regulates axonal elongation during optic nerve regeneration in the goldfish in vitro and in vivo. J Neurochem. 2009;110:890–901. doi: 10.1111/j.1471-4159.2009.06182.x. [DOI] [PubMed] [Google Scholar]
- Kurimoto T, Yin Y, Omura K, Gilbert HY, Kim D, Cen LP, Moko L, Kügler S, Benowitz LI. Long-distance axon regeneration in the mature optic nerve: contributions of oncomodulin, cAMP, and pten gene deletion. J Neurosci. 2010;30:15654–15663. doi: 10.1523/JNEUROSCI.4340-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landreth GE, Agranoff BW. Explant culture of adult goldfish retina: a model for the study of CNS regeneration. Brain Res. 1979;161:39–55. doi: 10.1016/0006-8993(79)90194-X. [DOI] [PubMed] [Google Scholar]
- Leaver SG, Cui Q, Plant GW, Arulpragasam A, Hisheh S, Verhaagen J, Harvey AR. AAV-mediated expression of CNTF promotes long-term survival and regeneration of adult rat retinal ganglion cells. Gene Ther. 2006;13:1328–1341. doi: 10.1038/sj.gt.3302791. [DOI] [PubMed] [Google Scholar]
- Leibinger M, Müller A, Andreadaki A, Hauk TG, Kirsch M, Fischer D. Neuroprotective and axon growth-promoting effects following inflammatory stimulation on mature retinal ganglion cells in mice depend on ciliary neurotrophic factor and leukemia inhibitory factor. J Neurosci. 2009;29:14334–14341. doi: 10.1523/JNEUROSCI.2770-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibinger M, Müller A, Gobrecht P, Diekmann H, Andreadaki A, Fischer D. Interleukin-6 contributes to CNS axon regeneration upon inflammatory stimulation. Cell Death Dis. 2013;4:e609. doi: 10.1038/cddis.2013.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Dowling JE. A dominant form of inherited retinal degeneration caused by a non-photoreceptor cell-specific mutation. Proc Natl Acad Sci U S A. 1997;94:11645–11650. doi: 10.1073/pnas.94.21.11645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery LA, Rubin J, Sive H. Whitesnake/sfpq is required for cell survival and neuronal development in the zebrafish. Dev Dyn. 2007;236:1347–1357. doi: 10.1002/dvdy.21132. [DOI] [PubMed] [Google Scholar]
- Matsukawa T, Sugitani K, Mawatari K, Koriyama Y, Liu Z, Tanaka M, Kato S. Role of purpurin as a retinol-binding protein in goldfish retina during the early stage of optic nerve regeneration: its priming action on neurite outgrowth. J Neurosci. 2004;24:8346–8353. doi: 10.1523/JNEUROSCI.1809-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCurley AT, Callard GV. Time course analysis of gene expression patterns in zebrafish eye during optic nerve regeneration. J Exp Neurosci. 2010;2010:17–33. [PMC free article] [PubMed] [Google Scholar]
- McDowell AL, Dixon LJ, Houchins JD, Bilotta J. Visual processing of the zebrafish optic tectum before and after optic nerve damage. Vis Neurosci. 2004;21:97–106. doi: 10.1017/S0952523804043019. [DOI] [PubMed] [Google Scholar]
- Moore DL, Blackmore MG, Hu Y, Kaestner KH, Bixby JL, Lemmon VP, Goldberg JL. KLF family members regulate intrinsic axon regeneration ability. Science. 2009;326:298–301. doi: 10.1126/science.1175737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A, Hauk TG, Fischer D. Astrocyte-derived CNTF switches mature RGCs to a regenerative state following inflammatory stimulation. Brain. 2007;130:3308–3320. doi: 10.1093/brain/awm257. [DOI] [PubMed] [Google Scholar]
- Müller P, Kuttenkeuler D, Gesellchen V, Zeidler MP, Boutros M. Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature. 2005;436:871–875. doi: 10.1038/nature03869. [DOI] [PubMed] [Google Scholar]
- Munderloh C, Solis GP, Bodrikov V, Jaeger FA, Wiechers M, Málaga-Trillo E, Stuermer CA. Reggies/flotillins regulate retinal axon regeneration in the zebrafish optic nerve and differentiation of hippocampal and N2a neurons. J Neurosci. 2009;29:6607–6615. doi: 10.1523/JNEUROSCI.0870-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KK, Hu Y, Muhling J, Pollett MA, Dallimore EJ, Turnley AM, Cui Q, Harvey AR. Cytokine-induced SOCS expression is inhibited by cAMP analogue: impact on regeneration in injured retina. Mol Cell Neurosci. 2009;41:313–324. doi: 10.1016/j.mcn.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Pedranzini L, Dechow T, Berishaj M, Comenzo R, Zhou P, Azare J, Bornmann W, Bromberg J. Pyridone 6, a pan-Janus-activated kinase inhibitor, induces growth inhibition of multiple myeloma cells. Cancer Res. 2006;66:9714–9721. doi: 10.1158/0008-5472.CAN-05-4280. [DOI] [PubMed] [Google Scholar]
- Pernet V, Joly S, Jordi N, Dalkara D, Guzik-Kornacka A, Flannery JG, Schwab ME. Misguidance and modulation of axonal regeneration by Stat3 and Rho/ROCK signaling in the transparent optic nerve. Cell Death Dis. 2013a;4:e734. doi: 10.1038/cddis.2013.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernet V, Joly S, Dalkara D, Jordi N, Schwarz O, Christ F, Schaffer DV, Flannery JG, Schwab ME. Long-distance axonal regeneration induced by CNTF gene transfer is impaired by axonal misguidance in the injured adult optic nerve. Neurobiol Dis. 2013b;51:202–213. doi: 10.1016/j.nbd.2012.11.011. [DOI] [PubMed] [Google Scholar]
- Ramachandran R, Zhao XF, Goldman D. Ascl1a/Dkk/{beta}-catenin signaling pathway is necessary and glycogen synthase kinase-3{beta} inhibition is sufficient for zebrafish retina regeneration. Proc Natl Acad Sci U S A. 2011;108:15858–15863. doi: 10.1073/pnas.1107220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo R, Berliocchi L, Adornetto A, Amantea D, Nucci C, Tassorelli C, Morrone LA, Bagetta G, Corasaniti MT. In search of new targets for retinal neuroprotection: is there a role for autophagy? Curr Opin Pharmacol. 2013;13:72–77. doi: 10.1016/j.coph.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Saul KE, Koke JR, García DM. Activating transcription factor 3 (ATF3) expression in the neural retina and optic nerve of zebrafish during optic nerve regeneration. Comp Biochem Physiol A Mol Integr Physiol. 2010;155:172–182. doi: 10.1016/j.cbpa.2009.10.042. [DOI] [PubMed] [Google Scholar]
- Senut MC, Gulati-Leekha A, Goldman D. An element in the alpha1-tubulin promoter is necessary for retinal expression during optic nerve regeneration but not after eye injury in the adult zebrafish. J Neurosci. 2004;24:7663–7673. doi: 10.1523/JNEUROSCI.2281-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y, Zipori D. PSF and p54(nrb)/NonO-multi-functional nuclear proteins. FEBS Lett. 2002;531:109–114. doi: 10.1016/S0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- Shen S, Niso-Santano M, Adjemian S, Takehara T, Malik SA, Minoux H, Souquere S, Mariño G, Lachkar S, Senovilla L, Galluzzi L, Kepp O, Pierron G, Maiuri MC, Hikita H, Kroemer R, Kroemer G. Cytoplasmic STAT3 represses autophagy by inhibiting PKR activity. Mol Cell. 2012;48:667–680. doi: 10.1016/j.molcel.2012.09.013. [DOI] [PubMed] [Google Scholar]
- Smith PD, Sun F, Park KK, Cai B, Wang C, Kuwako K, Martinez-Carrasco I, Connolly L, He Z. SOCS3 deletion promotes optic nerve regeneration in vivo. Neuron. 2009;64:617–623. doi: 10.1016/j.neuron.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song KS, Kim K, Chung KC, Seol JH, Yoon JH. Interaction of SOCS3 with NonO attenuates IL-1beta-dependent MUC8 gene expression. Biochem Biophys Res Commun. 2008;377:946–951. doi: 10.1016/j.bbrc.2008.10.084. [DOI] [PubMed] [Google Scholar]
- Sugitani K, Matsukawa T, Koriyama Y, Shintani T, Nakamura T, Noda M, Kato S. Upregulation of retinal transglutaminase during the axonal elongation stage of goldfish optic nerve regeneration. Neuroscience. 2006;142:1081–1092. doi: 10.1016/j.neuroscience.2006.07.042. [DOI] [PubMed] [Google Scholar]
- Sugitani K, Ogai K, Hitomi K, Nakamura-Yonehara K, Shintani T, Noda M, Koriyama Y, Tanii H, Matsukawa T, Kato S. A distinct effect of transient and sustained upregulation of cellular factor XIII in the goldfish retina and optic nerve on optic nerve regeneration. Neurochem Int. 2012;61:423–432. doi: 10.1016/j.neuint.2012.06.004. [DOI] [PubMed] [Google Scholar]
- Sun F, Park KK, Belin S, Wang D, Lu T, Chen G, Zhang K, Yeung C, Feng G, Yankner BA, He Z. Sustained axon regeneration induced by co-deletion of PTEN and SOCS3. Nature. 2011;480:372–375. doi: 10.1038/nature10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman MB, Bemben MA, Thompson RC, Goldman D. Gene expression analysis of zebrafish retinal ganglion cells during optic nerve regeneration identifies KLF6a and KLF7a as important regulators of axon regeneration. Dev Biol. 2007;312:596–612. doi: 10.1016/j.ydbio.2007.09.019. [DOI] [PubMed] [Google Scholar]
- Veldman MB, Bemben MA, Goldman D. Tuba1a gene expression is regulated by KLF6/7 and is necessary for CNS development and regeneration in zebrafish. Mol Cell Neurosci. 2010;43:370–383. doi: 10.1016/j.mcn.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita S, Miyagi C, Carmany-Rampey A, Shimizu T, Fujii R, Schier AF, Hirano T. Stat3 controls cell movements during zebrafish gastrulation. Dev Cell. 2002;2:363–375. doi: 10.1016/S1534-5807(02)00126-0. [DOI] [PubMed] [Google Scholar]